Abstract
Filamentous fungi play an important role in human health and industrial/agricultural production. With the increasing number of full genomes available for fungal species, the study of filamentous fungi has brought about a wider range of genetic manipulation opportunities. However, the utilization of traditional methods to study fungi is time consuming and laborious. Recent rapid progress and wide application of a versatile genome editing technology, i.e., the CRISPR (clustered regularly interspaced short palindromic repeat)–Cas9 (CRISPR-related nuclease 9) system, has revolutionized biological research and has many innovative applications in a wide range of fields showing great promise in research and application of filamentous fungi. In this review, we introduce the CRISPR/Cas9 genome editing technology focusing on its application in research of filamentous fungi and we discuss the general considerations of genome editing using CRISPR/Cas9 system illustrating vector construction, multiple editing strategies, technical consideration of different sizes of homology arms on genome editing efficiency, off-target effects, and different transformation methodologies. In addition, we discuss the challenges encountered using CRISPR/Cas9 technology and give the perspectives of future applications of CRISPR/Cas9 technology for basic research and practical application of filamentous fungi.
Keywords: Filamentous fungi, CRISPR/Cas9, Genome editing, Off-target
Introduction
Filamentous fungi have widely been used for expression of heterologous proteins, and production of antibiotics and organic acid (Jiang et al. 2013; Osiewacz 2002). At present, the filamentous fungi used for production of enzymes and recombinases are mainly Aspergillus niger, Aspergillus oryzae, Rhopzus oryzae, and Aspergillus nidulans. Filamentous fungi produce abundant secondary metabolites, such as paclitaxel and swainsonine (SW), which have become important clinical therapeutics.
Filamentous fungi such as A. nidulans and Neurospora crassa are often used as model eukaryotic microorganisms and play a critical role in basic research. N. crassa is one of the earliest filamentous fungi used for genetic studies. Metarhizium anisopliae, Pyricularia oryzae, and Aspergillus fumigatus are important pathogens of animals, plants, and humans, respectively. As a multicellular eukaryotic microorganism, filamentous fungi have a genetic background that is more complex than those of bacteria and other prokaryotes, and their genetic manipulation is often difficult. This has led to a relatively slow progress in molecular biology and genetic research of filamentous fungi.
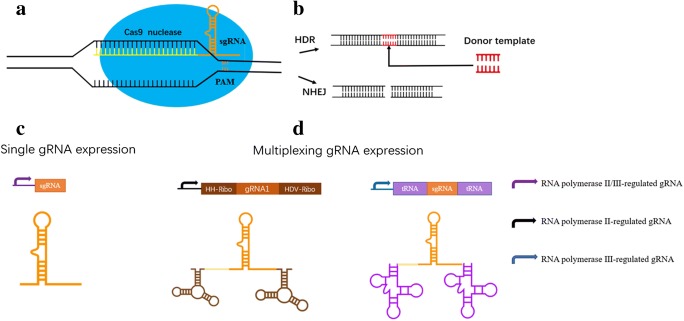
Interestingly, the breakthrough of the manipulation of eukaryotic organisms began with exploration of the much simpler bacterial genomes through rudimentary bioinformatic analysis. Mojica et al. (2005) found, based on the available DNA sequence data base, that clustered regularly interspaced short palindromic repeats (CRISPR) loci from bacterial DNA data base matched the sequences of bacterial phage and made an important conjecture that CRISPR must be part of the bacterial immune system used by the host bacteria to guard against repeated infection of the same phage (Lander 2016). With the continuous deepening of research, accumulated information on CRISPR/Cas9 system suggest there are two categories of CRISPR/Cas9 systems, which can be further divided into 6 types and 19 subtypes (Makarova et al. 2015; Shmakov et al. 2015; Shmakov et al. 2017). Current major research results and efforts gravitated toward type II CRISPR/Cas9 system which includes Cas9 (nuclease), mature crRNA (CRISPR associate RNA), tracrRNA (trans-activating crRNA), and RNaseIII. The type II system is simpler than other CRISPR systems and has been widely used. Jinek et al. (2012) further modified and optimized the bacterial type II CRISPR system by linking crRNA and tracrRNA into a single-guide RNA (sgRNA), which can efficiently direct the Cas9 protein to the target sequences to cut the DNA. After constructed exogenous CRISPR/Cas9 system is introduced into the recipient cells, the crRNA-tracrRNA (sgRNA) complex further interacts with Cas9 to form a crRNA-tracrRNA-Cas9 complex. The Cas9 protein contains two key domains, HNH and RuvC, which cleave the two strands of the DNA through binding to the target sequences and then cleaves the DNA duplex under the guidance of sgRNA. The HNH domain can cleave the DNA strand complementary to the crRNA, and the RuvC domain is responsible for the cleavage of the DNA that is not complementary to the crRNA (Jinek et al. 2014; Nishimasu et al. 2014; Sternberg et al. 2014). Therefore, sgRNA is responsible for identifying the 20 nucleotide sequences upstream of the protospacer adjacent motif (PAM) site, and the Cas9 protein is responsible for shearing 3–4 bp upstream of PAM to form blunt ends (Fig. 1a).
Fig. 1.
Schematic illustration of Cas9/gRNA genome editing. a sgRNA-mediated Cas9 protein can bind to target sequences of site and cut DNA double strands. b When the DNA double-strand breaks occur, the cell initiates the self-repair mechanism. The NHEJ-dominated repair pathway will cause the random loss, insertion, and replacement of bases at the breakage point, resulting in gene mutation. The HR pathway will accurately edit the target gene guided by the donor DNA fragment. c Single-promoter-driven gRNA expression cassette. d multiple gRNA expression cassettes can be constructed by concatenating 2 or more of gRNA linked together by linkers, which can then be enzymatically processed into multiple single gRNAs thus targeting multiple sites
When DNA double-strand breaks (DSBs) occur, the genomic DNA initiate self-repair mechanism resulting in genome mutation, non-homologous end-joining (NHEJ) as the dominant repair pathway which will cause random loss, insertion, and replacement of bases at the DNA breakage points, the homologous repairing (HR) pathway will precisely edit the gene of interest with the help of exogenous donor fragments (Sander and Joung 2014) (Fig. 1b). Currently, CRISPR/Cas9 technology has optimized and utilized for genome editing in numerous organisms (Cong et al. 2013; Mali et al. 2013a; DiCarlo et al. 2013), generating great impacts in humanity from industrial biotechnology (van Erp et al. 2015) to plant breeding (Bortesi and Fischer 2015; Schiml and Puchta 2016) and show great promise in treatment of human disease (Cai et al. 2016).
This review covers the latest developments in application of CRISPR/Cas9-mediated genome editing in filamentous fungi (Fig. 2), highlighting the general technical issues of using CRISPR/Cas9-based genome editing approaches in fungi (Grzybek et al. 2018; Wang et al. 2017; Shi et al. 2017). In addition, future development and challenges of CRISPR/Cas9 technology are discussed.
Fig. 2.
The history of the development and application of CRISPR/Cas9 technology in filamentous fungi; different colors represent different promoter-driven gRNA expression cassettes
CRISPR/Cas9 genome editing in filamentous fungi
In recent years, the birth of CRISPR/Cas9 genome editing technology has shown great promise to revolutionize the field of fungal research. Dicarlo et al. (2013) first introduced the CRISPR/Cas9 genome editing system into Saccharomyces cerevisiae. Subsequently, Liu et al. (2015) applied the CRISPR/Cas9 genome editing system to Trichoderma reesei; Matsu-ura et al. (2015) and Nodvig et al. (2015) applied this system to the model fungus N. crassa and A. nidulans, respectively. Since then, the CRISPR/Cas9 genome editing system has found wide applications genetically alteration of many filamentous fungi (Table 1).
Table 1.
The application of the CRISPR/Cas9 system in filamentous fungi
| Species | Cas9 expression (selection marker, promoter) | Delivery method | Editing method | Application and efficiency | Reference |
|---|---|---|---|---|---|
| T. reesei | Codon-optimized Cas9, ura5, pdc/cbh1 | PMT | NHEJ/HDR | Single/multiple-gene disruption ⩾ 93%/4.2–45% | Liu et al. 2015 |
| P. oryzae | Codon-optimized Cas9, Bar, tef1 | PMT | NHEJ/HDR | Single-gene disruption, 36.1–80.5% | Arazoe et al. 2015 |
| Aspergillus | pFC332, pyrG/argB/hph/ble, gpdA | PMT | NHEJ | Single-gene disruption | Nodvig et al. 2015 |
| N. crassa | Codon-optimized Cas9, Bar, Trpc | PMT | NHEJ/HDR | Single-gene disruption | Matsu-Ura et al. 2015 |
| A. fumigatus | Human codon-optimized Cas9, hph/ble, amy | PMT | NHEJ | Single-gene disruption, 25%-53% | Fuller et al. 2015 |
| P. sojae | Human-optimized codons Cas9, G418, Ham34 | PMT | NHEJ/HDR | Single-gene disruption | Fang and Tyler 2016 |
| U. maydis | Codon-optimized Cas9, ip, Otef (modified tef1)/hsp70, | PMT | NHEJ | Single/double-gene disruption, 50%-90% | Schuster et al. 2016 |
| A. fumigatus | Human codon-optimized Cas9, pyr4/hph, niiA/gpdA | PMT | HDR | Single/double-gene disruption, 95%-100% | Zhang et al. 2016 |
| A. oryzae | Codon-optimized Cas9, pyrG, amyB | PMT | NHEJ | Single-gene disruption, 10–20% | Katayama et al. 2016 |
| P. chrysogenum | Human codon-optimized Cas9, amdS, xlnA | PMT | NHEJ/HDR | Single-gene disruption, 100% | Pohl et al. 2016 |
| C. albicans | Codon-optimized Cas9, Nat, ENO1 | Lithium acetate | NHEJ/HDR | Single-gene disruption | Min et al. 2016 |
| A. fumigatus | pFC332, pyrG, TetON | PMT | NHEJ/HDR | Weber et al. 2017 | |
| A. niger | pFC332, pyrG/hph, tef1 | PMT | NHEJ/HDR | Single-gene disruption, 37.5–100% | Kuivanen et al. 2016 |
| U. maydis | Codon-optimized Cas9, ip, Otef (modified tef1)/hsp70 | PMT | NHEJ | Multiple-gene disruption, 50–90% | Schuster et al. 2018 |
|
M. thermophila M. heterothallica |
Codon-optimized Cas9, Bar, tef1 | PMT/AMT | NHEJ/HDR | Single/multiple-gene disruption, 15–95% | Liu et al. 2017 |
| T. atroroseus | pFC330, hph, tef1 | PMT | NHEI | Single-gene disruption | Nielsen et al. 2017 |
| A. carbonarius | pFC332, hph, tef1, | AMT | NHEJ/HDR | Single-gene disruption, 27% | Weyda et al. 2017 |
| A. niger | pFC332, hph, tef1 | PMT | NHEJ | Single-gene disruption | Kuivanen et al. 2017 |
| Ganoderma lucidum | Codon-optimized Cas9, ura3, gpdA | PMT | NHEI | Single-gene disruption, | Qin et al. 2017 |
| B. bassiana | Codon-optimized Cas9, gfp/ura5/bar, gpdA | PMT | NHEJ/HDR | Single/multiple-gene disruption, 5–50% | Chen et al. 2017 |
| A. alternata | pFC332, pyr4/hph, gpdA | PMT | NHEJ | Single-gene disruption | Wenderoth et al. 2017 |
| S. bambusicola | Codon-optimizedCas9, hph, TrpC | PMT | NHEJ | Single-gene disruption | Deng et al. 2017a |
| S. bambusicola | Codon-optimizedCas9, hph, TrpC | PMT | NHEJ/HDR | Single-gene disruption, 32% | Deng et al. 2017b |
| Nodulisporium sp. | Codon-optimized Cas9, Bar, TrpC | PMT | NHEJ/HDR | Single-gene disruption | Zheng et al. 2017 |
| Mucor circinelloides | SpCas9, pyr4 | PMT | NHEJ/HDR | Single/double-gene disruption, 100% | Nagy et al. 2017 |
| Fusarium graminearum | Codon-optimized Cas9, fludioxonil, gpdA | PMT | NHEJ/HDR | Single-gene disruption, 1–10% | Gardiner and Kazan 2018 |
| Leptosphaeria maculans | Human codon-optimized Cas9, Ip/G418/hph, act1 | AMT | NHEJ | Single-gene disruption | Idnurm et al. 2017 |
| A. niger | pFC332, hph, tef1 | PMT | NHEJ | Single-gene disruption | Kuivanen and Richard 2018 |
| C. albicans | SpCas9, Nat/Phloxine B | Electroporation | NHEJ | Double-gene disruption, 53–98% | Shapiro et al. 2018 |
| Aspergilli | pFC332, argB/pyrG, tef1 | AMT | HDR | Multiple-gene disruption, 15–90% | Nodvig et al. 2018 |
| B. dermatitidis | pFC332, hph, tef1 | AMT | NHEJ | Single/double-gene disruption, 22–73% | Kujoth et al. 2018 |
| A. niger | Codon-optimized Cas9, hph/amdS, glaA | PMT | NHEL/HDR | Single-gene disruption, 33.3–100% | Zheng et al. 2018 |
| C. albicans | Codon-optimized Cas9, Nat, ENO1 | Electroporation | HDR | Single/double-gene disruption, 25–100% | Vyas et al. 2018 |
| P. sojae | Human-optimized codons Cas9, G418, Ham34 | PMT | NHEJ | Double-gene disruption | Miao et al. 2018 |
| F. oxysporum | pFC332, hph | PMT | NHEJ/HDR | Single-gene disruption, 20–53.8% | Wang et al. 2018 |
| Ustilaginoidea virens | Codon-optimized Cas9, G418, pdc/cbh1 | AMT PMT | NHEJ | Multiple-gene disruption, 60%-90% | Liang et al. 2018 |
| Cordyceps militaris | Codon-optimized Cas9, 5-FOA/blpR, tef1 | AMT/PMT | NHEJ/HDR | Single-gene disruption, 87.2–84.3% | Chen et al. 2018 |
| Cryptococcus neoformans | Codon-optimized Cas9, Ntc, tef1 | Electroporation | HDR | Single-gene disruption, 96.5–100% | Wang 2018 |
| Sclerotinia sclerotiorum | Codon-optimized Cas9, hph, tef1 | PMT | NHEJ/HDR | Single-gene disruption, 38–100% | Li et al. 2018 |
| Aspergillus novofumigatus | pFC332, pyrG, tef1 | PMT | NHEJ | Single-gene disruption | Matsuda et al. 2018 |
| A. niger | pFC332, argB/pyrG, tef1 | PMT | HDR | Single-gene disruption, 100% | Leynaud-Kieffer et al. 2019 |
| A. oryzae | Codon-optimized Cas9, pyrG, amyB | PMT | HDR | Single/double-gene disruption, 50–100% | Katayama et al. 2019 |
| A. alternata | pFC332, hph, tef1 | PMT | NHEJ | Single-gene disruption | Igbalajobi et al. 2019 |
| A. niger | Codon-optimized Cas9, pyrG | PMT | NHEJ/HDR | Single/multiple-gene disruption 100% | Kuivanen et al. 2019 |
| M.r circinelloides | SpCas9, pyr4 | PMT | HDR | Multiple-gene disruption 100% | Nagy et al. 2019 |
| L. maculans | Human codon-optimized Cas9, hph, act1 | AMT | NHEJ | Double-gene disruption | Darma et al. 2019 |
| A. gossypii | Human-optimized codons Cas9, G418, tef1 | Electroporation | NHEJ | Single-gene disruption, 44–85% | Jiménez et al. 2019 |
| Duddingtonia flagrans | pFC332, hph, tef1 | PMT | HDR | Single-gene disruption | Loubna et al. 2019 |
Cas9 expression
The Cas9 protein is an important component of the CRISPR/Cas9 system that performs endonuclease function and has a total length of approximately 1400 amino acids. The CRISPR/Cas9 system was discovered in bacteria or Archaea. Therefore, when the CRISPR/Cas9 system is used in fungi, genomes encoding the Cas9 protein are usually fungal codon-optimized and a nuclear localization signal is added at both ends of the Cas9 gene. Therefore, amino acid sequence of Cas9 can be bacterial codon-optimized (Liu et al. 2015), human codon-optimized (DiCarlo et al. 2013; Fuller et al. 2015; Fang and Tyler 2016; Zhang et al. 2016; Pohl et al. 2016; Idnurm et al. 2017), or fungal codon-optimized (Nodvig et al. 2015; Nodvig et al. 2018; Generoso et al. 2016). The classical nuclear localization sequence (NLS) SV40 has been shown to function in many organisms (Nodvig et al. 2015). However, Wang et al. (2018) and Fang and Tyler (2016) demonstrated that this nuclear localization signal is not applicable to Fusarium oxysporum and Phytophthora sojae, for which they used an endogenous nuclear localization signal to fuse with Cas9 and successfully applied to F. oxysporum and P. sojae. In order to detect the expression and localization of optimized Cas9, most of the studies have fused the enhanced green fluorescent protein (eGFP) gene to the optimized Cas9. Thereafter, it was verified whether the Cas9 protein was expressed in the recipient fungus by detecting fluorescence (Fang and Tyler 2016; Wang et al. 2018; Chen et al. 2017; Chen et al. 2018).
The successful expression of the Cas9 gene depends on the types of promoter used. The strength of the promoter in driving transcription is an important factor affecting the expression efficiency of Cas9 exogenous genes, so the selection of a suitable promoter is of great significance for the function of the CRISPR/Cas9 system. Constitutive promoters are the most common and are not induced by external factors. It is a continuous expression system in all tissues and organs in an organism. The RNA transcription and protein expression levels of such promoter-initiated genes are relatively stable and have no spatial or temporal specificity. In general, most studies have used constitutive promoters to drive expression of Cas9 protein in filamentous fungi, such as the trpC promoter of the A. nidulans tryptophan synthesis gene trpC, the gpdA promoter of the A. nidulans glyceraldehyde-3-phosphate dehydrogenase gene gpdA, and TEF1 the promoter of the A. nidulans translation elongation factor 1α (Matsu-Ura et al. 2015; Nodvig et al. 2015; Arazoe et al. 2015; Sarkari et al. 2017). In addition to the three common promoters (trpC, gpdA and TEF1), xlnA, Ham34, amyB, niiA, Otef (modified tef1), and hsp70 have also been successfully used for Cas9 expression in filamentous fungi (Zhang et al. 2016; Pohl et al. 2016; Katayama et al. 2016; Schuster et al. 2018). If mutants generated using the CRISPR system are used in studies of pathogenesis and secondary metabolites, expression of the Cas9 nuclease itself cannot exert adverse effects on the recipient strain. However, studies have shown that the expression of Cas9 as a foreign gene in recipient organisms may have negative effects (Miyagishi and Taira 2002). Therefore, it is important to ensure that Cas9 protein has no adverse effect on filamentous fungi by testing the Cas9 expression strain’s growth, development, stress resistance, and virulence and other biological characteristics (Schuster et al. 2016; Deng et al. 2017a; Deng et al. 2017b).
Guide RNA expression
RNA polymerase III promoter–regulated gRNA
The gRNA in the CRISPR/Cas9 system does not contain a cap structure and a poly A tail, and requires a clear transcription starting point. In most cases, gRNA is driven by RNA polymerase type III U6 promoters that have been shown to have base preference and persistence in driving transcription (Gao and Zhao 2014). Bioinformatic analysis has shown that U6 snRNA gene sequences are highly conserved from yeast to mammals. Arazoe et al. (2015) first compared human U6 snRNA and analyzed two endogenous U6 promoters of P. oryzae and successfully knocked out P. oryzae’s SRS2 gene with an efficiency of 36.1–80.5%. CRISPR/Cas9 system with U6 promoter to transcribe gRNA has been used for genetic manipulation in most filamentous fungi (Fig. 1c). The gRNA in the CRISPR/Cas9 system requires a well-defined transcription start site, and the transcription of gRNA in higher organisms is usually driven by the Pol III U6 promoter. In addition to the U6 promoter, Schuster et al. (2018) optimized the CRISPR/Cas9 system and tested the expression efficiency of tRNA promoter–driven gRNA (ptRNALeu-TAA, ptRNAGlyGCC, ptRNATyrGTA, and ptRNAGlyTCC). The four tRNA promoters have higher driving efficiencies than the U6 promoter, indicating that tRNA promoters can be used for gRNA expression as well (Liang et al. 2018).
However, the genome editing efficiency of different tRNA promoters varies greatly in a strain-dependent manner. In some cases, tRNA-based genome editing efficiency remains controversial. In Yarrowia lipolytica, the use of tRNA as the sgRNA promoter brings about 30% gene fragmentation efficiency (Schwartz et al. 2016). Therefore, tRNA is preferably used to excise sgRNA from the primary transcript by its endogenous processing mechanism, rather than acting as a promoter. In order to find a more efficient promoter, Zheng et al. (2018) found that 5S rRNA is a basic cellular component that is highly conserved and abundant in cells. Therefore, they used the 5S rRNA gene of A. niger and its 338-bp upstream sequence as a promoter and fused with the sgRNA sequence to construct an sgRNA expression cassette. Meanwhile, in order to avoid the interference of 5S rRNA on the structure and function of sgRNA, an 88-bp HDV ribozyme gene from Trichoderma atroviride was added between the 5S rRNA gene and sgRNA. Experiments have shown that the CRISPR/Cas9 system based on the 5S rRNA promoter shows a gene disruption efficiency close to 96%, which is significantly higher than the 15% and 23% destruction efficiency based on the PhU6 or PanU6 promoter system. The selection of promoters for sgRNA expression represents an important technical limitation in the development of the CRISPR/Cas9 genome editing system.
RNA polymerase II–regulated gRNA
The gRNAs such as those produced from expression from the U3 and U6 promoter are transcribed using RNA polymerase III (pol III) in most organisms. However, the U6 or U3 promoter transcribes gRNA with many limitations. Firstly, U6 snRNA and U3 snRNA are housekeeping genes that are ubiquitously expressed. Therefore, they cannot be used to produce cell type or tissue-specific gRNA. Secondly, U6 and U3 promoters are not suitable for conventional in vitro transcription of gRNA because RNA polymerase III is not commercially available. In addition, the CRISPR target sequences recognized by the U6 and U3 promoter are constrained with certain sequence specificity and must be G(N)20GG and A(N)20GG (Gao and Zhao 2014; Nissim et al. 2014).
Gao and Zhao (2014) used the nuclease activity of ribozymes to design the artificial gene RGR (ribozyme-gRNA-ribozyme), a 5′-terminal hammerhead (HH) and a 3′-terminal hepatitis D virus (HDV), which flank sgRNA. This system was introduced into yeast under the control of the RNA polymerase II–transcribed alcohol dehydrogenase 1 (ADH1) promoter to achieve targeted DNA cleavage. It has been shown that in this system targeting sequence is no longer limited to G(N)20GG or A(N)20GG. Subsequently, Nodvig et al. (2018) first constructed ribozyme-gRNA-ribozyme using the A. nidulans strong constitutive gpdA promoter (PgpdA) and the trpC terminator (TtrpC), and successfully applied it to species in the filamentous fungus genus Aspergillus. Wenderoth et al. (2017) knocked out the key genes pksA and brm2 controlling the melanin biosynthetic pathway in Alternaria alternata using the same method and successfully obtained gene mutant strains. In order to optimize Cas9 and gRNA expression, Nodvig et al. (2018) introduced the strong constitutive promoter gpdA and tef1-controlled mRFP into the NHEJ deletion strain NID1; in comparison, it was found that the tef1 promoter is more efficient than gpdA promoter. Recently, Kujoth et al. (2018) adapted RNA Pol II system to achieve double gene knockout on Blastomyces dermatitidis by Agrobacterium-mediated transformation. It is further proven that the knock-out efficiency of the single-promoter-driven double gRNA is higher than that of dual-promoter.
However, the aforementioned methods are relatively complex and difficult to operate and may increase the burden on the carrier and may even bring additional foreign substances in the cells to cause toxic effects. Subsequently, Wang et al. (2015) used the Golden Gate method (another RNA Pol II–driven system) to design a miRsh-sgRNA cassette that is regulated by an RNA polymerase II promoter. The miRsh-sgRNA cassette has been experimentally proven to be effective in expressing gRNA. The gRNA cassette could generate gRNA targeting the p53 gene and achieve specific cleavage of the P53 target gene under Cas9 protein mediated. This miRsh-sgRNA structure regulated by the RNA polymerase class II promoter allows for more controlled and safer gene editing in the future using the CRISPR/Cas9 system. This effective method has not been used in the filamentous fungi, and its utility in the filamentous fungi warrants further investigation.
Other strategies for generating gRNA
In addition to the above methods, two methods can be used to generate functional gRNA. One strategy is by using the SNR52 promoter of S. cerevisiae, which was shown to transcribe the initial gRNA with a leader sequence that can be excised during processing to form a functional gRNA (Matsu-Ura et al. 2015; Fuller et al. 2015). In addition, the plasmid-free CRISPR-Cas9 system is also capable of genome editing of fungal cells.
The plasmid-free CRISPR-Cas9 system assembles Cas9 protein and sgRNA to form a stable RNP in vitro, and transfers the complex to fungal protoplasts by PEG- or electroporation-mediated methods for genome editing. There are several advantages to using this system for genome editing compared with plasmid transformation. Cas9 and sgRNA are capable of forming stable ribonucleoproteins in vitro, and thus, RNA degradation is less likely than plasmid transformation. In addition, the method is capable of assessing the target cleavage efficiency of different sgRNAs in vitro, so efficient sgRNA can be selected for research. This RNP-mediated genome editing technique has been successfully applied to some species (Min et al. 2016; Grahl et al. 2017; Wang 2018; Al Abdallah et al. 2017; Kuivanen and Richard 2018).
Multiplexing gRNA expression
The above reports all used CRISPR/Cas9 technology to knock out a single gene, but in practical applications, some fungal traits are usually controlled by multiple genes. There is a great need for simultaneous knockouts of multiple genes, and the traditional methods for obtaining multiple gene mutants are time consuming and laborious. The cumbersome ZFN and TALEN technologies are also limited in the ability to knock out multiple genes at the same time. In contrast, the CRISPR/Cas9 technology can be used to knock out multiple genes.
The core problem of knockout of multiple genes lies in multiple targeting sites. Therefore, this can be eliminated as long as the problem of multiple gRNA expression cassettes is solved. Currently, multi-gene simultaneous targeting mainly focuses on the following three methods: (1) Liu et al. (2015) introduced the vector containing the Cas9 expression cassette into the recipient fungal cells by the Agrobacterium-mediated method, and then introduced multiple mature gRNA synthesized in vitro into the Cas9-positive cells through protoplast-mediated methods. This method has been applied to A. fumigatus (Zhang et al. 2016), A. niger (Kuivanen et al. 2016), and Beauveria bassiana (Chen et al. 2017). (2) The tRNA-spacer system has been successfully used for multiple sgRNA expression in other organisms, including plants, yeast, human cells, and fruit flies (Xie et al. 2015; Schwartz et al. 2016; Port and Bullock 2016; Qi et al. 2016; Dong et al. 2017). Specifically, the gRNAs are concatenated, interspaced by 5′ and 3′ splice sites of a tRNA that are recognized by RNase P and RNase Z. The mature gRNAs are then released through the action of RNase P and RNase Z (Fig. 1d). Nodvig et al. (2015) used this mechanism to construct a multiple gRNAs expression vector and verified that the polymerase III promoter PAf-U3 was more efficient than PgpdA. (3) In addition, Kujoth et al. (2018) knocked out two genes, PRA1 and ZRT1, of B. dermatitidis using two gRNA expression cassettes driven by the RNA polymerase II promoter PgpdA (Fig. 1d).
Delivery of the Cas9 and sgRNA expression cassettes into fungal cells
CRISPR/Cas9 system targeting fungal genome must be delivered to fungal cells by vectors carrying Cas9 and sgRNA expression cassettes. Typically, the Cas9 protein expression cassette and the sgRNA expression cassette are carried by a single vector or a double vector. Zhang et al. (2016) validated the efficiency of two different methods of expression in the model fungus A. fumigatus. They show that the accuracy and efficiency of using a single-vector expression system is significantly higher than that of a dual-vector expression system. This may be due to the fact that the ratio of Cas9 and sgRNA expression cassettes transformed into fungal cells cannot be precisely controlled. Therefore, most studies first stably transduced Cas9 expression cassettes into recipient cells and screened for positive strains expressing Cas9 protein; then, the Cas9-positive cells are transfected with the in vitro transcribed gRNA for genome editing (Liu et al. 2015; Chen et al. 2017; Kuivanen et al. 2016; Kuivanen et al. 2017). The polyethylene glycol (PEG) transformation and Agrobacterium-mediated transformation (AMT) are the two most common methods for transforming CRISPR/Cas9 systems into fungal cells. Since PEG-mediated transformation methods are the easiest for many fungi, most studies use this method to transform Cas9 and gRNA expression cassettes into fungal cells (Li et al. 2018; Matsuda et al. 2018; Miao et al. 2018). Agrobacterium-mediated traditional genome editing is also a common method. However, this method is not common in CRISPR/Cas9 system–mediated fungal genome editing. Recently, Weyda et al. (2017) compared these two approaches and demonstrated that Agrobacterium-mediated CRISPR/Cas9 systems can also be used for efficient genome editing in fungal cells.
Regulation of NHEJ or HR pathways
DSBs generated by Cas9 can be directly subjected to indel mutagenesis by non-homologous end-joining repair (NHEJ); or homology repair (HR) can be achieved if a DNA repair template (donor DNA) is provided (Ma et al. 2016; Doudna and Charpentier 2014; Shalem et al. 2015). The NHEJ repair route is different from the HR repair pathway (Fig. 1b). The poor fidelity of the NHEJ repair pathway can occur in the G1 phase of the entire cell cycle, and HR repair is a precise repair pathway that occurs only during DNA replication, and the NHEJ repair pathway can be directly connected at the ends of the DNA double-strand breaks, in which the ku70 and ku80 proteins play a major role, and the HR repair pathway needs to provide target site for homologous donor DNA fragments (Gorbunova and Levy 1997; Branzei and Foiani 2008). In addition, the possibility of homologous recombination repair will increase by approximately 1000 times if there are nearby homologous DNA fragments during DNA damage repair (Rouet et al. 1994). Based on the above principles, the CRISPR/Cas9 system can use the HR pathway for accurate target editing, such as the introduction of specific point mutations at target sites, insertion of a desired sequence (Feng et al. 2014), or the exact replacement of a sequence with the desired sequence. Recently, Liu et al. (2015) first established a CRISPR/Cas9 system in T. reesei by adding homologous arms of different sizes to the left and right of the selectable marker, and demonstrated that when a ≥ 600-bp homology arm is added around the selectable marker, the frequency of homologous recombination is almost 100%, enabling CRISPR/Cas9-mediated gene knockout in T. reesei. Subsequently, Zhang et al. (2016) established the MMEJ-CRISPR system in the ku80 deletion strain A1160PgpdA-Cas9 (ZC03). The A1160PgpdA-Cas9 (ZC03) strain eliminates the effects of the NHEJ repair pathway and ensures Cas9 expression. They added short (approximately 35 bp) homology arms near the PAM site to the two sides of the selectable marker HPH and co-transformed with the linear PU6-3-pksP-sgRNA fragment with a mutation efficiency of about 67%. Compared with traditional homologous integration methods with homology arms of at least 500-bp or 1000-bp length (da Silva Ferreira et al. 2006), the data indicates that the CRISPR/Cas9 system with two very short homology arms on the flanks is sufficient to repair the resulting cleavage to achieve the efficient genome editing. Liu et al. (2017) targeted the carbon catabolic repression (CCR) transcription factor cre-1 in Myceliophthora thermophila and Myceliophthora heterothallica, demonstrating that, compared with the traditional HR (20%), frequency up to 95% can be achieved by the CRISPR/Cas9 system with repair HR after template co-transformation. The basic method of homologous repair template construction is to add 39–1000 bp upstream and downstream homologous segments of the target gene bracketing the selectable marker, so that the DSBs generated by the CRISPR/Cas9 system can be repaired through the HR pathway and ultimately increase the mutation efficiency of the target gene. Recent studies have shown that circular homologous repair templates are more efficient than linearized templates. This may be due to the fact that circular homologous repair templates cannot directly enter the NHEJ pathway because there are no free ends available for binding to the NHEJ protein and thus may be beneficial for HR repair. In addition, the lack of a free end in a circular gene-targeting substrate also protects the cyclic repair template from exonuclease action, therefore, extending the time that can be used as a repair template (Nodvig et al. 2018).
Interestingly, Shapiro et al. (2018) established a CRISPR-Cas9-based gene-driven array (GDA) platform at C. albicans, which inserts two different gRNA in the middle of homologous arms to carry out the genome editing on the adenine biosynthesis gene ADE2. When Cas9 cleaves the ORF, the gRNA module is flanked by regions that are homologous to the upstream and downstream sequences of the Cas9-targeting locus, and the entire ORF is deleted and replaced with the targeted gRNA, resulting in a “driven allele.” When the driver-containing haploid cells are mated with wild-type cells, the gRNA-modified locus will initiate another round of cleavage, which converts the incoming wild-type allele into a driver variant. Once the cells contain a functional driver, they can easily convert the heterozygous deletion into a homozygous deletion, facilitating the rapid generation of homozygous deletion mutants in the diploid pathogen. In addition to C. albicans, gene-driven platforms may be able to be adapted to other clinically relevant but often overlooked fungal pathogens such as C. auris in the future.
Features of CRISPR/Cas9-induced mutations
Most studies have shown that about half of the CRISPR/Cas9-induced mutations are single-base (mostly A and T) insertions; the rest are small deletions (1–50 bp), and base substitutions of two or more bases insertion are very rare (Feng et al. 2014; Ma et al. 2015). The CRISPR/Cas9 system–mediated single-gene knockouts in the filamentous fungi are mostly single-gene insertions or small fragment deletions (Kujoth et al. 2018). If two or more sites are targeted in a gene or a chromosomal region, a few hundred kilobase fragment deletions may occur between target sites (Zhao et al. 2016).
Off-target effects
The CRISPR/Cas9 system has the characteristics of high specificity, simplicity in operation, and high efficiency and has been widely used in the biological research and biomedical application. However, the off-target effects of the CRISPR/Cas9 system are a major concern. Previous studies have shown that there are up to five mismatch target genes for each gRNA (Cradick et al. 2013; Fu et al. 2013). Off-target effects are mainly divided into the following types: (1) in most cases, Cas9/gRNA does not recognize more than three mismatched DNA sites; (2) Cas9/gRNA does not recognize and edit DNA sites with any number of mismatches (within 10–12 bp) near the PAM; (3) the higher the Cas9/gRNA concentration, the greater the possibility of off-target effects (Hsu et al. 2013; Mali et al. 2013b; Pattanayak et al. 2013). In order to avoid off-target effects, Wang and colleagues designed plasmid DNA, which may remove the inserted Cas9 gene after genome editing (Wang et al. 2016). Recently, Nagy et al. (2017) developed a CRISPR-Cas9 system specifically for transient expression. This system assembles Cas9 protein and sgRNA to form a stable RNP in vitro, and transfers the complex to fungal protoplasts by PEG- or electroporation-mediated methods for genome editing. This method enables the in vitro evaluation of target cleavage efficiency of different sgRNAs, so efficient sgRNAs can be selected for research. Furthermore, the transformation of RNP mitigates the possibility of integration of genetic material into non-targeted regions of the genome. Methods based on next-generation sequencing technologies such as GUIDE-seq (Tsai et al. 2015), Digenome-seq (Kim et al. 2015), and ChIP-seq (Kuscu et al. 2014) can be also used to identify the off-target site. The above strategies can be used to provide guidance for increasing the specificity of the CRISPR/Cas9 system in filamentous fungi.
Application of CRISPR/Cas9 in industrial strains
Enzymes produced by filamentous fungi play an important role in industrial production, for example, in paper, food, feed, textiles, and detergents. Filamentous fungi are used in food fermentation and industrial production of recombinant proteins, and they also serve as hosts for industrially valuable secondary metabolites. Obviously, the production of various important enzymes by industrial strains has become the focus of industry and academia. However, wild-type strains cannot produce the desired enzymes at an industrial level. Therefore, genetic engineering is needed to enhance these fungi involved in industrial production but traditional continuous modification of multiple genes using a limited number of selection markers is laborious. To solve this problem, researchers have developed different methods to increase their enzyme yield but most methods are only suitable for a limited number of fungi. With the development of more and more fungal genome sequences and omics data, the study of industrial fungi has brought a wider range of genetic manipulation opportunities, and the use of CRISPR/Cas9 genome editing techniques in fungi producing industrial enzymes greatly improves the ability of these fungi to produce industrial enzymes.
For example, T. reesei is the main source of the most widely used commercial lignocellulolytic enzyme preparations, a potential cell factory for heterogeneous protein and secreting proteins, often used to produce a variety of enzymes or metabolites. Initially, in order to establish the CRISPR/Cas9 genome editing technology in T. reesei, Liu et al. (2015) optimized the Cas9 gene of S. pyogenes and linked it to the nuclear localization signal SV40. The lae1 and vib1 genes were successfully knocked out by in vitro synthesis of gRNA. Later, Leynaud-Kieffer et al. (2019) used the CRISPR/Cas9 genome editing technology to knock out the SxlR gene, a negative regulator of xylanase activity. The results indicate that deletion of the SxlR gene results in a significant increase in gene expression encoding the GH11 endoxylanase, whereas overexpression of this gene results in a decrease in xylanase activity but does not affect cellulase activity. Industrial fungi producing cellulase also include A. oryzae. In Japan, A. oryzae is an important industrial filamentous fungus for traditional fermentation, production of enzymes and heterologous proteins. In order to efficiently produce these enzymes and compounds, it is necessary to genetically engineer the A. oryzae strain but continuous (single) changes to multiple genes using conventional methods are time consuming. In 2016, Katayama et al. (2016) developed the CRISPR/Cas9 method for mutagenesis of A. oryzae but the efficiency of this method is very low. In order to improve the editing efficiency of the CRISPR/Cas9 genome editing technology in A. oryzae strains, Katayama et al. (2019) used a plasmid containing the AMA1 autonomously replicating sequence to increase the mutation efficiency of A. oryzae wild and industrial strains to 50–100%. The AMA1 plasmid was present in multiple copies in the A. oryzae strain, thereby increasing the expression of Cas9 and sgRNA, and ultimately increasing the mutation efficiency of the A. oryzae strain. In addition, AMA1-based plasmids are discarded when there is no resistance to stress so this approach avoids the impact of the CRISPR/Cas9 system on the strain. In theory, the AMA1-based CRISPR/Cas9 genome editing technology will allow for infinite rounds of genetic engineering. The AMA1 autonomously replicating sequence can be expressed in a variety of filamentous fungi, including not only Aspergillus species but also P. chrysogenum, Talaromyces atroroseus, and Gaemannomyces graminis.
In addition to its use in these enzyme-producing fungi, the CRISPR/Cas9 genome editing technology can also be used in other industrial fungi. As we all know, Ashbya gossypii is an industrial fungus used in the production of vitamin B2. To date, a large number of genetic engineering and biotechnology tools are available for A. gossypii. For example, Cre-loxP technology is applied to A. gossypii. However, a large number of modifications greatly reduced the ability of the strain to produce spores and a large number of loxP scars were produced after multiple rounds of operation (Ledesma-Amaro et al. 2015). In 2019, Jiménez et al. (2019) knocked out the ADE2 gene of A. gossypii using a one-vector CRISPR/Cas9 system controlled by the yeast promoter SNR52, and verified the presence of an 8-bp deletion in the ade2 mutant by DNA sequencing. The stability of this mutation was then confirmed because no phenotypic changes were observed after five-spore formation. Thereafter, in order to verify the stability of the system, Jiménez et al. (2019) transferred the CRISPR/Cas9 vector containing the sgA754-dA754 fragment into strain A754. PCR identification and DNA sequencing confirmed that the system can eliminate loxP scars in A. gossypii. These examples above show that the CRISPR/Cas9 system has been effectively applied to industrial fungi, and the use of the CRISPR/Cas9 genome editing system can further enhance the applicability of these industrial fungi.
Conclusion and perspectives
Compared with ZFN and TALEN systems, the CRISPR/Cas9 system has several advantages. (1) The CRISPR/Cas9 system is widely used: the requirement for the selection of target sites in this system is to have a PAM sequence, which is widely present in the genome and can find several target sites in any gene, so that almost all genes can be edited. (2) The CRISPR/Cas9 system consists of simple components: sgRNA and Cas9 proteins are the two major components of the system. They can be placed on a transformation vector as needed, or placed on different transformation vectors, or assembled directly in vitro. (3) The CRISPR system can reduce the selection of markup applications: For example, a limiting factor in mutagenesis in A. fumigatus stems from a limited number of dominant selection markers. When we need to mutate three or more genes in a row, the only selection markers available for A. fumigatus are hph, ble, and ptrA. The CRISPR/Cas9 system can edit multiple genes at the same time, and it is possible to obtain mutants with multiple site mutations in one transformation, which greatly improves the efficiency of genome editing.
However, at present, there are still deficiencies in this technology, such as off-target effect, needs to improve editing efficiency, and need to construct a carrier system that is applicable to various fungi. There is also a risk that the system will not stop the genome editing function after the error gene is repaired, but may continue to modify the normal gene to cause off-target effects. To address this problem, Nagy et al. (2017) developed the CRISPR-Cas9 system specifically for transient expression. This system not only evaluates the target cleavage efficiency of different sgRNAs in vitro but also reduces the possibility of integration of genetic material into non-targeted regions of the genome. These advantages greatly reduce off-target effects of the CRISPR system.
Cho et al. (2014) has proposed that Cas9 inhibitory protein can be used to disable the runaway editing function by inhibiting its activity when Cas9 completes genome editing, but it is in its early stage and further experiments are needed to verify and identify better techniques. Based on the principle of DNA double-strand break and chromosomal translocation, Yin et al. (2019) developed a new method PEM-seq with higher sensitivity and comprehensive and quantitative evaluation of gene editing based on high-throughput sequencing methods. Compared with previous methods based on second-generation sequencing to assess the off-target activity of Cas9, PEM-seq not only can sensitively identify the off-target sites of Cas9 but also can accurately quantify the cutting efficiency of CRISPR/Cas9 at the target site, thus finding more efficient Cas9 cutting site. Although these methods have not yet been established in fungi, with the continuous development and improvement of CRISPR/Cas9 technology, it is certain that the technology will play a greater role in the future research for genetic modification of filamentous fungi.
Acknowledgements
We are grateful to Prof. Zekun Guo for the help in the manuscript preparation.
Authors’ contributions
HL and QYG planned the review; RJS, LS, EXH, QZ, and BYZ analyzed the literature; RJS, YZ, EXH, and YLZ wrote the first version of the manuscript; RJS and YNT prepared the figures; HL, QYG, and RJS revised the final version of the manuscript. All authors accepted the final version of the manuscript.
Funding
This work was supported by the grants from the National Natural Science Foundation (No. 31201958) and the Key project of Natural Science Foundation of Shaanxi province (No. 2017JZ004) and the Special Fund for Agro-scientific Research in the Public Interest (No. 201203062).
Compliance with ethical standards
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Runjie Song and Qing Zhai contributed equally to this work.
Contributor Information
Qingyun Guo, Email: guoqingyunqh@163.com.
Hao Lu, Phone: +86-29-87092429, Email: luhao@nwsuaf.edu.cn.
References
- Al Abdallah Q, Ge W, Fortwendel JR. A simple and universal system for gene manipulation in Aspergillus fumigatus: in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere. 2017;2(6):e00446-17. doi: 10.1128/mSphere.00446-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arazoe T, Ogawa T, Miyoshi K, Yamato T, Ohsato S, Sakuma T, Yamamoto T, Arie T, Kuwata S. Tailor-made TALEN system for highly efficient targeted gene replacement in the rice blast fungus. Biotechnol Bioeng. 2015;112:1335–1342. doi: 10.1002/bit.25559. [DOI] [PubMed] [Google Scholar]
- Bortesi L, Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Cai L, Fisher AL, Huang H, Xie Z. CRISPR-mediated genome editing and human diseases. Genes Dis. 2016;3:244–251. doi: 10.1016/j.gendis.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lai Y, Wang L, Zhai S, Zou G, Zhou Z, Cui C, Wang S. CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus Beauveria bassiana. Sci Rep. 2017;8:45763. doi: 10.1038/srep45763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BX, Wei T, Ye ZW, Yun F, Kang LZ, Tang HB, Guo LQ, Lin JF. Efficient CRISPR-Cas9 gene disruption system in edible-medicinal mushroom Cordyceps militaris. Front Microbiol. 2018;9:1157. doi: 10.3389/fmicb.2018.01157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41:9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darma R, Lutz A, Elliott CE, Idnurm A. Identification of a gene cluster for the synthesis of the plant hormone abscisic acid in the plant pathogen Leptosphaeria maculans. Fungal Genet Biol. 2019;130:62–71. doi: 10.1016/j.fgb.2019.04.015. [DOI] [PubMed] [Google Scholar]
- Deng H, Gao R, Liao X, Cai Y. Characterization of a major facilitator superfamily transporter in Shiraia bambusicola. Res Microbiol. 2017;168:664–672. doi: 10.1016/j.resmic.2017.05.002. [DOI] [PubMed] [Google Scholar]
- Deng H, Gao R, Liao X, Cai Y. Genome editing in Shiraia bambusicola using CRISPR-Cas9 system. J Biotechnol. 2017;259:228–234. doi: 10.1016/j.jbiotec.2017.06.1204. [DOI] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Xie K, Chen Y, Yang Y, Mao Y. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem Biophys Res Commun. 2017;482:889–895. doi: 10.1016/j.bbrc.2016.11.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- van Erp PB, Bloomer G, Wilkinson R, Wiedenheft B. The history and market impact of CRISPR RNA-guided nucleases. Curr Opin Virol. 2015;12:85–90. doi: 10.1016/j.coviro.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Tyler BM. Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol Plant Pathol. 2016;17:127–139. doi: 10.1111/mpp.12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci U S A. 2014;111:4632–4637. doi: 10.1073/pnas.1400822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller KK, Chen S, Loros JJ, Dunlap JC. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot Cell. 2015;14:1073–1080. doi: 10.1128/EC.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56:343–349. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Kazan K. Selection is required for efficient Cas9-mediated genome editing in Fusarium graminearum. Fungal Biol. 2018;122:131–137. doi: 10.1016/j.funbio.2017.11.006. [DOI] [PubMed] [Google Scholar]
- Generoso WC, Gottardi M, Oreb M, Boles E. Simplified CRISPR-Cas genome editing for Saccharomyces cerevisiae. J Microbiol Methods. 2016;127:203–205. doi: 10.1016/j.mimet.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Gorbunova V, Levy AA. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 1997;25:4650–4657. doi: 10.1093/nar/25.22.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahl N, Demers EG, Crocker AW, Hogan DA. Use of RNA-protein complexes for genome editing in non-albicans Candida species. mSphere. 2017;2(3):e00218–e00217. doi: 10.1128/mSphere.00218-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzybek M, Golonko A, Górska A, Szczepaniak K, Strachecka A, Lass A, Lisowski P. The CRISPR/Cas9 system sheds new lights on the biology of protozoan parasites. Appl Microbiol Biotechnol. 2018;102(11):4629–4640. doi: 10.1007/s00253-018-8927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idnurm A, Urquhart AS, Vummadi DR, Chang S, Van de Wouw AP, Lopez-Ruiz FJ. Spontaneous and CRISPR/Cas9-induced mutation of the osmosensor histidine kinase of the canola pathogen Leptosphaeria maculans. Fungal Biol Biotechnol. 2017;4:12. doi: 10.1186/s40694-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbalajobi O, Yu Z, Fischer R. Red- and blue-light sensing in the plant pathogen Alternaria alternata depends on phytochrome and the white-collar protein LreA. MBio. 2019;10:e00371–e00319. doi: 10.1128/mBio.00371-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Zhu W, Wang Y, Sun C, Zhang KQ, Yang J. Molecular tools for functional genomics in filamentous fungi: recent advances and new strategies. Biotechnol Adv. 2013;31:1562–1574. doi: 10.1016/j.biotechadv.2013.08.005. [DOI] [PubMed] [Google Scholar]
- Jiménez A, Muñoz-Fernández G, Ledesma-Amaro R, Buey RM, Revuelta JL (2019) One-vector CRISPR/Cas9 genome engineering of the industrial fungus Ashbya gossypii. Microb Biotechnol 0(0):1-9. doi: 10.1111/1751-7915.13425 [DOI] [PMC free article] [PubMed]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Tanaka Y, Okabe T, Nakamura H, Fujii W, Kitamoto K, Maruyama J. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus oryzae. Biotechnol Lett. 2016;38:637–642. doi: 10.1007/s10529-015-2015-x. [DOI] [PubMed] [Google Scholar]
- Katayama T, Nakamura H, Zhang Y, Pascal A, Fujii W, Maruyama JI. Forced recycling of an AMA1-based genome-editing plasmid allows for efficient multiple gene deletion/integration in the industrial filamentous fungus Aspergillus oryzae. Appl Environ Microbiol. 2019;85(3):e01896–e01818. doi: 10.1128/AEM.01896-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(237-243):1–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- Kuivanen J, Richard P. NADPH-dependent 5-keto-D-gluconate reductase is a part of the fungal pathway for D-glucuronate catabolism. FEBS Lett. 2018;592:71–77. doi: 10.1002/1873-3468.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen J, Wang YJ, Richard P. Engineering Aspergillus niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb Cell Factories. 2016;15:210. doi: 10.1186/s12934-016-0613-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen J, Arvas M, Richard P. Clustered genes encoding 2-keto-l-gulonate reductase and l-idonate 5-dehydrogenase in the novel fungal d-glucuronic acid pathway. Front Microbiol. 2017;8:225. doi: 10.3389/fmicb.2017.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuivanen J, Korja V, Holmström S, Richard P. Development of microtiter plate scale CRISPR/Cas9 transformation method for Aspergillus niger based on in vitro assembled ribonucleoprotein complexes. Fungal Biol Biotechnol. 2019;6:3. doi: 10.1186/s40694-019-0066-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujoth GC, Sullivan TD, Merkhofer R, Lee TJ, Wang H, Brandhorst T, Wuthrich M, Klein BS. CRISPR/Cas9-mediated gene disruption reveals the importance of zinc metabolism for fitness of the dimorphic fungal pathogen Blastomyces dermatitidis. MBio. 2018;9(2):e00412–e00418. doi: 10.1128/mBio.00412-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Lander ES. The heroes of CRISPR. Cell. 2016;164:18–28. doi: 10.1016/j.cell.2015.12.041. [DOI] [PubMed] [Google Scholar]
- Ledesma-Amaro R, Lozano-Martínez JA, Revuelta JL. Engineering Ashbya gossypii for efficient biolipid production. Bioengineered. 2015;6(2):119–123. doi: 10.1080/21655979.2015.1011525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leynaud-Kieffer LMC, Curran SC, Kim I, Magnuson JK, Gladden JM, Baker SE, Simmons BA. A new approach to Cas9-based genome editing in Aspergillus niger that is precise, efficient and selectable. PLoS One. 2019;14(1):e0210243. doi: 10.1371/journal.pone.0210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhang Y, Zhang Y, Yu PL, Pan H, Rollins JA. Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen Sclerotinia sclerotiorum. MBio. 2018;9(3):e00567–e00518. doi: 10.1128/mBio.00567-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Han Y, Wang C, Jiang C, Xu JR. Targeted deletion of the USTA and UvSLT2 genes efficiently in Ustilaginoidea virens with the CRISPR-Cas9 system. Front Plant Sci. 2018;9:699. doi: 10.3389/fpls.2018.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Chen L, Jiang Y, Zhou Z, Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015;1:15007. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Gao R, Li J, Lin L, Zhao J, Sun W, Tian C. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol Biofuels. 2017;10:1. doi: 10.1186/s13068-016-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubna Y, Valentin W, Nicole H, Xi Y, Hildebrand HG, Birgit S, Marius K, Birgit HF, Andrew D, Reinhard F. Intercellular communication is required for trap formation in the nematode-trapping fungus Duddingtonia flagrans. PLoS Genet. 2019;15:e1008029. doi: 10.1371/journal.pgen.1008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhu Q, Chen Y, Liu YG. CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol Plant. 2016;9:961–974. doi: 10.1016/j.molp.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Bai T, Phippen C, Nodvig CS, Kjaerbolling I, Vesth TC, Andersen MR, Mortensen UH, Gotfredsen CH, Abe I, Larsen TO Novofumigatonin biosynthesis involves a non-heme iron-dependent endoperoxide isomerase for orthoester formation. Nat Commun. 2018;9:2587. doi: 10.1038/s41467-018-04983-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsu-Ura T, Baek M, Kwon J, Hong C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol Biotechnol. 2015;2:4. doi: 10.1186/s40694-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Li X, Lin D, Liu X, Tyler BM. Oxysterol-binding protein-related protein 2 is not essential for Phytophthora sojae based on CRISPR/Cas9 deletions. Environ Microbiol Rep. 2018;10:293–298. doi: 10.1111/1758-2229.12638. [DOI] [PubMed] [Google Scholar]
- Min K, Ichikawa Y, Woolford CA, Mitchell AP. Candida albicans Gene deletion with a transient CRISPR-Cas9 system. mSphere. 2016;1(3):e00130–e00116. doi: 10.1128/mSphere.00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Nagy G, Szebenyi C, Csernetics A, Vaz AG, Toth EJ, Vagvolgyi C, Papp T. Development of a plasmid free CRISPR-Cas9 system for the genetic modification of Mucor circinelloides. Sci Rep. 2017;7:16800. doi: 10.1038/s41598-017-17118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy G, Vaz AG, Szebenyi C, Takó M, Tóth EJ, Csernetics A, Bencsik O, Szekeres A, Homa M, Ayaydin F, Galgóczy L, Vágvölgyi C, Papp T. CRISPR-Cas9-mediated disruption of the HMG-CoA reductase genes of Mucor circinelloides and subcellular localization of the encoded enzymes. Fungal Genet Biol. 2019;129:30–39. doi: 10.1016/j.fgb.2019.04.008. [DOI] [PubMed] [Google Scholar]
- Nielsen ML, Isbrandt T, Rasmussen KB, Thrane U, Hoof JB, Larsen TO. Mortensen UH. Genes linked to production of secondary metabolites in Talaromyces atroroseus revealed using CRISPR-Cas9. PLoS One. 2017;12:e169712. doi: 10.1371/journal.pone.0169712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodvig CS, Nielsen JB, Kogle ME, Mortensen UH. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One. 2015;10:e133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodvig CS, Hoof JB, Kogle ME, Jarczynska ZD, Lehmbeck J, Klitgaard DK, Mortensen UH. Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet Biol. 2018;115:78–89. doi: 10.1016/j.fgb.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Osiewacz HD. Genes, mitochondria and aging in filamentous fungi. Ageing Res Rev. 2002;1(3):425–442. doi: 10.1016/s1568-1637(02)00010-7. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl C, Kiel JA, Driessen AJ, Bovenberg RA, Nygard Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth Biol. 2016;5:754–764. doi: 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- Port F, Bullock SL. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods. 2016;13:852–854. doi: 10.1038/nmeth.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Zhu T, Tian Z, Li C, Zhang W, Song R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016;16:58. doi: 10.1186/s12896-016-0289-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Xiao H, Zou G, Zhou Z, Zhong J. CRISPR-Cas9 assisted gene disruption in the higher fungus Ganoderma species. Process Biochem. 2017;56:57–61. [Google Scholar]
- Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkari P, Marx H, Blumhoff ML, Mattanovich D, Sauer M, Steiger MG. An efficient tool for metabolic pathway construction and gene integration for Aspergillus niger. Bioresour Technol. 2017;245:1327–1333. doi: 10.1016/j.biortech.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Schiml S, Puchta H. Revolutionizing plant biology: multiple ways of genome engineering by CRISPR/Cas. Plant Methods. 2016;12:8. doi: 10.1186/s13007-016-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Schweizer G, Reissmann S, Kahmann R. Genome editing in Ustilago maydis using the CRISPR-Cas system. Fungal Genet Biol. 2016;89:3–9. doi: 10.1016/j.fgb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- Schuster M, Schweizer G, Kahmann R. Comparative analyses of secreted proteins in plant pathogenic smut fungi and related basidiomycetes. Fungal Genet Biol. 2018;112:21–30. doi: 10.1016/j.fgb.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Schwartz CM, Hussain MS, Blenner M, Wheeldon I. Synthetic RNA polymerase III promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synth Biol. 2016;5:356–359. doi: 10.1021/acssynbio.5b00162. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Chavez A, Porter C, Hamblin M, Kaas CS, DiCarlo JE, Zeng G, Xu X, Revtovich AV, Kirienko NV, Wang Y, Church GM, Collins JJ. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat Microbiol. 2018;3:73–82. doi: 10.1038/s41564-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi TQ, Liu GN, Ji RY, Shi K, Song P, Ren LJ, Huang H, Ji XJ. CRISPR/Cas9-based genome editing of the filamentous fungi: the state of the art. Appl Microbiol Biotechnol. 2017;101(20):7435–7443. doi: 10.1007/s00253-017-8497-9. [DOI] [PubMed] [Google Scholar]
- Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Hartl A, Heinekamp T, Brakhage AA, Goldman GH. The akuB (KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell. 2006;5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, Aryee MJ, Joung JK. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas VK, Bushkin GG, Bernstein DA, Getz MA, Sewastianik M, Barrasa MI, Bartel DP, Fink GR (2018) New CRISPR Mutagenesis Strategies Reveal Variation in Repair Mechanisms among Fungi. mSphere 3(2): e00154-18. 10.1128/mSphere.00154-18 [DOI] [PMC free article] [PubMed]
- Wang P. Two distinct approaches for CRISPR-Cas9-mediated gene editing in Cryptococcus neoformans and related species. mSphere. 2018;3:e00208–e00218. doi: 10.1128/mSphereDirect.00208-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li X, Zhao Y, Li J, Zhou Q, Liu Z. Generation of cell-type-specific gene mutations by expressing the sgRNA of the CRISPR system from the RNA polymerase II promoters. Protein Cell. 2015;6:689–692. doi: 10.1007/s13238-015-0169-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wei D, Zhu X, Pan J, Zhang P, Huo L, Zhu X. A ‘suicide’ CRISPR-Cas9 system to promote gene deletion and restoration by electroporation in Cryptococcus neoformans. Sci Rep. 2016;6:31145. doi: 10.1038/srep31145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Chen H, Tang X, Zhang H, Chen W, Chen YQ. Molecular tools for gene manipulation in filamentous fungi. Appl Microbiol Biotechnol. 2017;101(22):8063–8075. doi: 10.1007/s00253-017-8486-z. [DOI] [PubMed] [Google Scholar]
- Wang Q, Cobine PA, Coleman JJ. Efficient genome editing in Fusarium oxysporum based on CRISPR/Cas9 ribonucleoprotein complexes. Fungal Genet Biol. 2018;117:21–29. doi: 10.1016/j.fgb.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J, Valiante V, Nodvig CS, Mattern DJ, Slotkowski RA, Mortensen UH, Brakhage AA. Functional reconstitution of a fungal natural product gene cluster by advanced genome editing. ACS Synth Biol. 2017;6:62–68. doi: 10.1021/acssynbio.6b00203. [DOI] [PubMed] [Google Scholar]
- Wenderoth M, Pinecker C, Voss B, Fischer R. Establishment of CRISPR/Cas9 in Alternaria alternata. Fungal Genet Biol. 2017;101:55–60. doi: 10.1016/j.fgb.2017.03.001. [DOI] [PubMed] [Google Scholar]
- Weyda I, Yang L, Vang J, Ahring BK, Lubeck M, Lubeck PS. A comparison of Agrobacterium-mediated transformation and protoplast-mediated transformation with CRISPR-Cas9 and bipartite gene targeting substrates, as effective gene targeting tools for Aspergillus carbonarius. J Microbiol Methods. 2017;135:26–34. doi: 10.1016/j.mimet.2017.01.015. [DOI] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JH, Liu MZ, Liu Y, Wu JC, Gan TT, Zhang WW, Li Y, Zhou YH, Hu JZ. Optimizing genome editing strategy by primer-extension-mediated sequencing. Cell Discov. 2019;5:18. doi: 10.1038/s41421-019-0088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Meng X, Wei X, Lu L. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet Biol. 2016;86:47–57. doi: 10.1016/j.fgb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Zhang C, Liu W, Gao W, Liu C, Song G, Li WX, Mao L, Chen B, Xu Y, Li X, Xie C. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci Rep. 2016;6:23890. doi: 10.1038/srep23890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YM, Lin FL, Gao H, Zou G, Zhang JW, Wang GQ, Chen GD, Zhou ZH, Yao XS, Hu D. Development of a versatile and conventional technique for gene disruption in filamentous fungi based on CRISPR-Cas9 technology. Sci Rep. 2017;7:9250. doi: 10.1038/s41598-017-10052-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zheng P, Zhang K, Cairns TC, Meyer V, Sun J, Ma Y (2018) 5S rRNA promoter for guide RNA expression enabled highly efficient CRISPR/Cas9 genome editing in Aspergillus niger. ACS Synth Biol. 10.1021/acssynbio.7b00456 [DOI] [PubMed]