Abstract
Mycobacterium bovis is responsible for bovine tuberculosis in both animals and humans. Despite being one of the most important global zoonotic disease, data related to the ecology and pathogenicity of bovine tuberculosis is scarce, especially in developing countries. In this report, we examined the dynamics of M. bovis transmission among dairy cattle in the Nile Delta of Egypt. Animals belonging to 27 herds from 7 governorates were tested by the Single Intradermal Comparative Skin Tuberculin (SICST), as a preliminary screen for the presence of bovine tuberculosis. Positive SICST reactors were identified in 3% of the animals spread among 40% of the examined herds. Post-mortem examination of slaughtered reactors confirmed the presence of both pulmonary and/or digestive forms of tuberculosis in > 50% of the examined animals. Targeted and whole-genome analysis of M. bovis isolates indicated the emergences of a predominant spoligotype (SB0268) between 2013–2015, suggesting a recent clonal spread of this isolate within the Nile Delta. Surprisingly, 2 isolates belonged to M. bovis BCG group, which are not allowed for animal vaccination in Egypt, while the rest of isolates belonged to the virulent M. bovis clonal complex European 2 present in Latin America and several European countries. Analysis of strain virulence in the murine model of tuberculosis indicated the emergence of a more virulent strain (MBE4) with a specific genotype. More analysis is needed to understand the molecular basis for successful spread of virulent isolates of bovine tuberculosis among animals and to establish genotype/phenotype association.
Subject terms: Bacterial genetics, Pathogens, Infectious diseases
Introduction
Mycobacterium bovis is the most common causative agent of bovine tuberculosis (bTB), an important infectious disease of cattle all over the world. The Office International des Epizooties (OIE) identified bTB as a list B transmissible disease of public health importance and of high impact on the international trade of animals and animal products1. Although national campaigns of “test and slaughter strategy” have reduced the incidence of the infection worldwide, bTB remains an important public health concern because of its zoonotic potential and re-emergence in animals and humans2. Bovine tuberculosis is a chronic, debilitating infection that infects a wide range of hosts including domesticated and wild animals3. Unlike M. tuberculosis, M. bovis has an unusually extensive host range including humans as recognized by the World Health Organization4 with a greater zoonotic potential in developing countries. M. bovis is also the progenitor of the Bacillus Calmette–Guérin (BCG), the only licensed tuberculosis vaccine and the gold standard for protection against childhood disseminated tuberculosis5. Despite its worldwide use for humans5, BCG is not approved for use in cattle vaccination in most of the world. In Egypt and other African countries, bTB represents a significant health and economic problem6,7 that requires further analysis on both genetic and genomic levels of M. bovis. For example, in Egypt the animal-level prevalence of bTB in cattle and buffaloes during the 1980s ranged between 6.9% and 26.2% then was reduced to 2.6% during the 1990s6. However, recent reports on the ecology, genotypes and virulence of M. bovis isolates circulating in Egypt are lacking and hence, the focus of this report.
M. bovis is a highly clonal pathogen8, where clonal complexes are defined based on chromosomal deletion, spoligotyping and MIRU-VNTR to define dynamics of disease transmission9,10. Recently, Whole-Genome Sequencing (WGS) provided new insights into host-pathogen interactions and the dynamics of disease transmission11–13. For example, sequencing the genomes of clinical isolates of M. bovis provided an accurate estimates of strain geographical distribution and evolution14–18. Such approaches are not common on isolates from Africa and the Middle East, contratry to the analysis of isolates from Euroupe8. In this report, we analyzed the genomic diversity and virulence of M. bovis isolates from selected dairy herds from the Nile Delta of Egypt. Among examined herds, bTB reached up to 41% on the herd-level but only 3.4% on the animal-level. Both genotyping and WGS indicate the genomic diversity of Egyptian isolates with predominance of isolates being closely related to clonal complex “European 2”. Interestingly, 2 isolates of M. bovis were found to be BCG isolates. Further molecular dating analysis allowed us to understand the spatial dynamics and phylogeography of the bTB infection in the Nile Delta, which was further verified by historical events. In vitro drug sensitivity testing and mice virulence assays further confirmed strain diversity despite being associated with a specific clonal complex. Overall, we utilized WGS to gain novel insights onto polymorphism and virulence of M. bovis isolates circulating in the Nile Delta.
Results
Prevalence of bovine tuberculosis in the Nile Delta
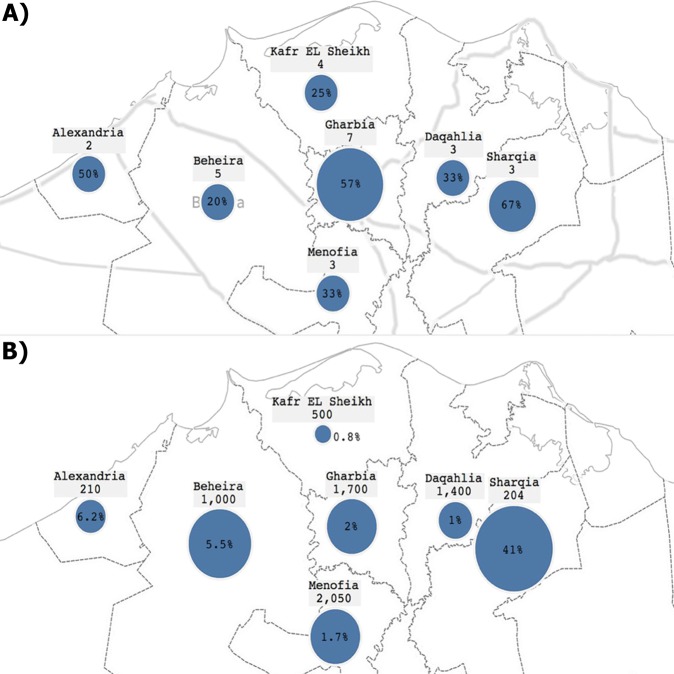
Our study design is based on the examination of dairy herds with previous history of bTB followed by the slaughtering of the positive reactors for lesion collection, when possible. The single intradermal comparative skin test (SICST) screening of dairy herds included herds from 7 governorates located in the Nile Delta (Fig. 1). A total of 7,064 animals belonging to 27 herds were tested, and only 242 (3.43%) were considered reactors by the applied test standards. However, significant variations in disease prevalence were present within governorates surveyed in this study. For example, the prevelance of bTB was 41% at Sharqia but only 0.8% at Kafr EL Sheikh, at the animal-level. At the herd-level (presence of at least one positive reactor in the herd), Gharbia had the highest number of positive herds (4 out of 7 herds). Only 2 herds were examined from Alexandria and only one herd was positive. The overall prevalence of ≥1 positive reactor in herds was 11 among all 27 examined herds (40.74%) (Table 1). Finally, the age distribution of the tested animals per governorate showed most to be 3–5 years old, the most productive time in a cow life cycle.
Figure 1.
Geographical map of the Nile Delta displaying the prevalence of bovine tuberculosis within several governorates. (A) Map showing prevalence of bTB at the herd-level per governorate, (B) Map showing prevalence of bTB at the animal-level per governorate. Each governorate shows a number of the total cattle tested and the circle size represents the magnitude of positive results.
Table 1.
The number of herds and animals per governorate with age distribution (parenthesis refer to percentages).
| Governorate | Herd-level | Animal level | |||||
|---|---|---|---|---|---|---|---|
| Number tested animals | Age distribution | Number positive animals | |||||
| Number tested herds | Number positive herds | “<3” | “3–5” | “>5” | |||
| Gharbia | 7 | 4 (57.14) | 1700 | 170 (10.00) | 1100 (64.71) | 430 (25.29) | 35 (2.06) |
| Daqahlia | 3 | 1 (33.33) | 1400 | 70 (5.00) | 930 (66.43) | 400 (28.57) | 16 (1.14) |
| Menofia | 3 | 1 (33.33) | 2050 | 200 (9.76) | 1350 (65.85) | 500 (24.39) | 35 (1.71) |
| Kafr EL Sheikh | 4 | 1 (25.00) | 500 | 70 (14.00) | 230 (46.00) | 200 (40.00) | 4 (0.80) |
| Sharqia | 3 | 2 (66.67) | 204 | 21 (10.29) | 151 (74.02) | 32 (15.69) | 84 (41.18) |
| Alexandria | 2 | 1 (50.00) | 210 | 11 (5.24) | 176 (83.81) | 23 (10.95) | 13 (6.19) |
| Beheira | 5 | 1 (20.00) | 1000 | 179 (17.9) | 654 (65.4) | 167 (16.70) | 55 (5.50) |
| Total | 27 | 11 (40.74) | 7064 | 721 (10.21) | 4591 (64.99) | 1752 (24.80) | 242 (3.43) |
Nature of lesions associated with bovine tuberculosis
To confirm the initial diagnosis with SICST and to culture isolates from positive reactors, 70 SICST-positive animals were slaughtered and examined by local health authorities. Necropsy lesions from animals are summarized in Table 2. Overall, the highest percentage of the examined tissues showed mixed visible granulomatous lesions (28.57%) in 2 or more systems (e.g. pulmonary and digestive) with the presence of the characteristic lymphocytic infiltration when histology was performed (Fig. 2). Animals with only pulmonary or only digestive lesions also occupied significant proportions of reactors with 25% and 14%, respectively. On the other hand, generalized infections (involvement of multiple systems) were less prevalent (4.29%). Interestingly, M. bovis was cultured from 44.29% of the slaughtered animals, even when lesions were not visible (Table 2). As expected, primary isolates of M. bovis were cultured mainly from mixed and pulmonary lesions, while most of the mycobacterium-negative cultures (N = 15) were from cases with non-visible lesions (NVL, N = 19). Colony morphology, biochemical testing, acid-fast staining and PCR genotyping were used to distinguish M. bovis primary isolates (N = 31) from other unclassified slow-growing mycobacteria. Finally, only 11 M. bovis isolates were sub-cultured successfully after original isolation (designated MBE for M. bovis from Egypt) and were subjected for drug susceptibility testing (DST). Interestingly, 2 isolates (MBE4 and MBE12) were resistant to Isoniazid (INH) while the rest of isolates were fully sensitive to all tested drugs Isoniazid (INH), Rifampicin (RIF), Ethambutol (EMB) and Streptomycin (SM).
Table 2.
Types and numbers of lesions and culturing results of animals underwent post-mortem examination in slaughterhouses (parenthesis refers to percentages).
| Site of infection | Types of Isolated Mycobacteria | Total | ||
|---|---|---|---|---|
| M. bovis | Unidentified Slow Grower AFB* | Negative culture | ||
| Pulmonary | 10 (55.56) | 0 (0.00) | 8 (44.44) | 18 (25.71) |
| Digestive | 4 (40.00) | 1 (10.00) | 5 (50.00) | 10 (14.29) |
| Mixed | 12 (60.00) | 0 (0.00) | 8 (40.00) | 20 (28.57) |
| Generalized | 3 (100) | 0 (0.00) | 0 (0.00) | 3 (4.29) |
| NVL* | 2 (10.53) | 2 (10.53) | 15 (78.95) | 19 (27.14) |
| Total | 31 (44.29) | 3 (4.29) | 36 (51.43) | 70 |
*Non-visible lesions indicative of bovine tuberculosis, *Acid-Fast Bacilli.
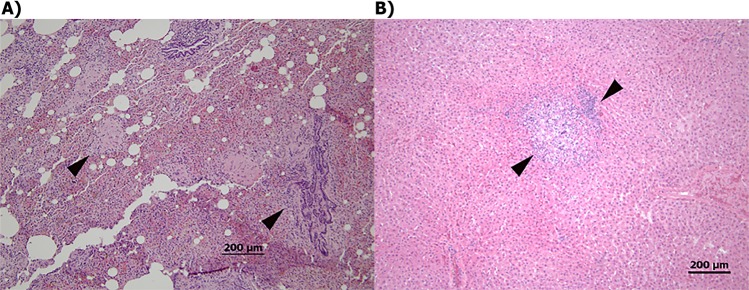
Figure 2.
Histology of bovine lungs following post-mortem examination. Tissue sections were taken from bTB positive cases at a participating slaughterhouse. (A) Lung with lymphohistiocytic inflammation showing aggregates of macrophages and lymphocytes (arrow heads) and (B) Liver with granulomatous inflammatory responses (arrow heads). All sections were stained with H&E and scale bar representing 200 um.
Molecular characteristics of M. bovis isolates
To gain more insights onto the epizootiology of bTB in the Nile Delta, DNA was extracted from all MBE isolates and subjected to various genotyping protocols. First, restriction fragment length polymorphism (RFLP) based on the gyrB gene confirmed that all isolates belong M. tuberculosis complex and could be either M. bovis or M. bovis–BCG. Subsequently, we used PCR to detect the presence of the region of deletion-1 (RD1), identified previously in the genome of M. tuberculosis and M. bovis19,20. Interestingly, all the MBE isolates were virulent M. bovis strains except MBE9 and MBE10 which belong to the M. bovis BCG species with the characteristic 196 bp band21.
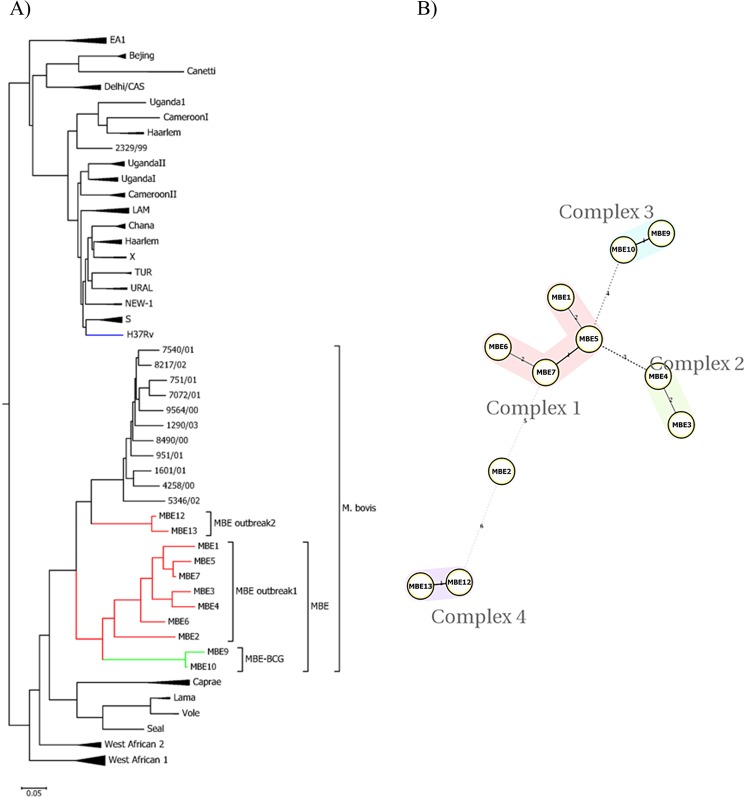
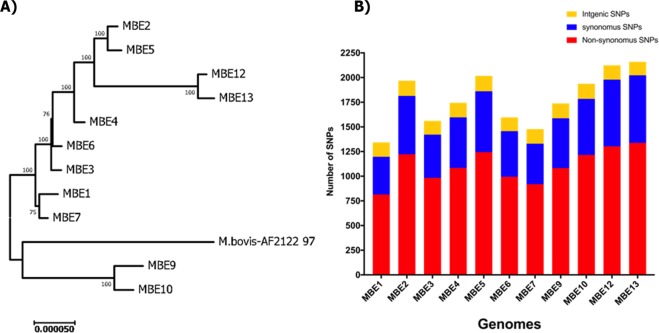
Twelve-locus Mycobacterial Interspersed Repetitive Units-Variable Number of Tandem Repeats (MIRU-VNTR) analysis22 was perfomed on all MBE isolates to further delineate relationships between isolates from the Nile Delta and those circulating worldwide. As expected, all MBE isolates were clustered with the virulent M. bovis except MBE 9 and 10 (BCG cluster) which were closely related but not in the same cluster (Fig. 3A). Interestingly, two isolates (MBE12 and MBE13) which were collected in a more recent outbreak (2 years after the first outbreak) in the Nile Delta showed a distinct but close cluster to the main cluster of MBE Outbreak 1, a clear indication of continous evolution and clonal exapansion of M. bovis within the Nile Delta. Further analysis of MIRU-VNTR profiles with minimum spaning tree (MST) algorithm for network analysis23, suggested that MBE isolates could be classified into 4 main complexes (Fig. 3B); with pairs of MBE12/MBE13 and MBE3/MBE4 occupying their own complex with a significant genetic distance from the rest of other complexes. Only the MBE2 isolate did not belong to any of the other complexes when MST was used.
Figure 3.
Phylogenetic tree of the clinical M. bovis isolates based on the MIRU-VNTR. (A) A dendogram was generated using the neighbor-joining algorithm using tools available from the MIRU-VNTRplus identification database, (B) Minimum spanning tree of 11 M. bovis isolates based on the MIRU-VNTR spoligotyping results, categorized based on the identified complexes. Each complex is defined by its number and color code.
Diversity of M. bovis isolates
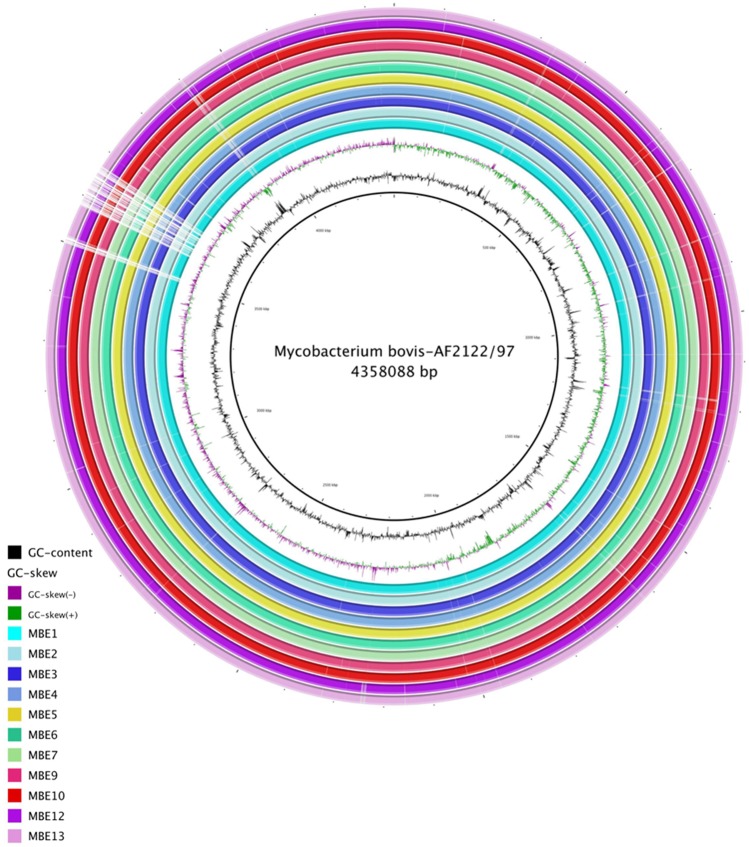
To better characterize variations among MBE isolates on the single nucleotide level, we used Next-generation sequencing technology (Illumina, MiSeq 2000) to generate draft sequences of the MBE isolates. An average of 7,043,390 reads per genome were mapped against the reference genome M. bovis AF2122/97 with almost 97.5% reads mapped to the reference genome. Overall sequence analysis indicated that all MBE isolates had genomes which were sequenced on an average of 269 times and an average G + C content of 65.52% representing 97.1% of a genome size estimated to be around 4,231,045 bp (Fig. 4). The calculated average nucleotide identity (ANI) of all the MBE isolates to the reference genome were >99.95 where most of genomes were compiled into ~ 40 contigs with DeNovo assembly. More comparative statistical features of the MBE genomes relative to the reference genome M. bovis AF2122/97 are detailed in (Table 3). The number of predicted single nucleotide polymorphism (SNPs) for the MBE consensus sequences is shown in (Fig. 5). Interestingly, the majority of SNPs (>50%) are non-synonymous (nSNP). Specifically, when 137 virulence related genes were selected for dN/dS analysis, all ratios were >1.0 (Table 3), an indication of a positive selective pressure for virulence genes encoded within M. bovis isolates analyzed in this study. The number of predicted SNPs for the MBE2, MBE5, MBE12 and MBE13 consensus sequences was higher than that predicted for the other MBE sequences when aligned to the standard reference genome M. bovis AF2122/97. On the other hand, phylogenetic analysis of SNPs from all the isolates confirmed the divergence of MBE10 and MBE9 strains (BCG-like) compared to virulent isolates of M. bovis including the distinctive sub-cluster of MBE12 and MBE13 (Fig. 5). However, whole genome alignments with progressiveMUAVE24 did not identify major genomic rearrangements among MBE isolates, as expected from members of the M. tuberculosis complex due to their clonal nature. Approximately 2–4 locally collinear blocks (LCBs) were found among all MBE genomes relative to the homologous reference genome (Supplemental Fig. 2). Overall, all MBE field isolates clustered with M. bovis AN5 originally a Brazilian strain used to produce PPD worldwide25,26 while the BCG-Like isolates clustered with Russian BCG strain which included the deletion of the RD1 region19 (Supplemental Fig. 1).
Figure 4.
Genomic organization of M. bovis from isolates from the Nile Delta. Circular representation showing (from inner to outer), % G + C, GC skew and the homology based on BLASTn+ analysis of the 11 MBE genomes sequentially aligned to the M. bovis reference genome strain AF2122/97 (check color legend). The figure was generated using BRIG 0.95. Gene mapping was done using BLASTn with an E-value cut-off 1e-5.
Table 3.
Summary of next generation sequencing results.
| Parameters | MBE1 | MBE2 | MBE3 | MBE4 | MBE5 | MBE6 | MBE7 | MBE9 | MBE10 | MBE12 | MBE13 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Year of isolation | 2014 | 2014 | 2014 | 2014 | 2014 | 2014 | 2014 | 2014 | 2014 | 2015 | 2015 |
| Governorate | Gharbia | Gharbia | Beheira | Menofia | Gharbia | Beheira | Gharbia | Alexandria | Alexandria | Menofia | Menofia |
| Tissue of isolation | Lung LN | Lung Tissue | Pre-scapular LN | Head LN | Generalized | Retropharyngeal LN | Mesenteric LN | Retro-pharyngeal LN | Mesenteric LN | Mesenteric LN | Mesenteric LN |
| Breed | Cross breed | Cross breed | Holstein | Holstein | Holstein | Holstein | Holstein | Native | Holstein | Native | Native |
| Age in years | 3 | 5 | 3–5 | 5 | 3–5 | >5 | 5 | >5 | 1–3 | >5 | >5 |
| Consensus length (Mbp) | 4.22 | 4.24 | 4.23 | 4.23 | 4.25 | 4.23 | 4.23 | 4.23 | 4.23 | 4.22 | 4.22 |
| % of bases | 97.10 | 97.37 | 97.02 | 97.55 | 97.51 | 97.31 | 97.39 | 96.99 | 97.31 | 96.59 | 96.01 |
| % G + C | 65.48 | 65.55 | 65.52 | 65.52 | 65.55 | 65.51 | 65.49 | 65.51 | 65.53 | 65.55 | 65.55 |
| # of reads | 2652522 | 9724874 | 2190220 | 2546898 | 7073852 | 2381816 | 2412476 | 3250964 | 3342700 | 22446678 | 19454286 |
| Average Coverage | 140.00 | 514.35 | 115.36 | 135.38 | 374.50 | 126.49 | 127.89 | 170.80 | 175.53 | 584.96 | 498.29 |
| % of mapped reads | 95.91 | 96.75 | 96.14 | 97.16 | 97.08 | 96.53 | 96.69 | 95.77 | 96.72 | 96.41 | 95.8 |
| Mean mapped read length | 241.59 | 240.7 | 240.41 | 240.56 | 239.94 | 241.3 | 240.27 | 241.43 | 238.4 | 120.67 | 119.91 |
| Mean paired read distance | 439.76 | 418.69 | 443.67 | 429.86 | 451.43 | 445.05 | 453.91 | 407.74 | 417.31 | 172.10 | 173.46 |
| # of contigs | 90 | 58 | 83 | 73 | 61 | 75 | 83 | 81 | 71 | 85 | 83 |
| N50 | 104531 | 150169 | 119829 | 119870 | 155216 | 117225 | 117190 | 119771 | 118077 | 107878 | 115326 |
| ANI to M. bovis AF2122/97 | 99.96 | 99.95 | 99.96 | 99.96 | 99.96 | 99.97 | 99.95 | 99.95 | 99.96 | 99.95 | 99.95 |
| # of InDels against M. bovis Af2122/97 | 53 | 61 | 53 | 44 | 68 | 51 | 54 | 30 | 27 | 286 | 343 |
| # of SNPs against M. bovis Af2122/97 | 1343 | 1968 | 1561 | 1744 | 2017 | 1597 | 1477 | 1738 | 1937 | 2124 | 2160 |
| Non-synonymous SNP | 815 | 1222 | 984 | 1084 | 1244 | 995 | 919 | 1082 | 1216 | 1303 | 1338 |
| Synonymous SNP | 382 | 592 | 438 | 513 | 617 | 462 | 411 | 505 | 568 | 676 | 685 |
| Intergenic SNP | 146 | 154 | 139 | 147 | 156 | 140 | 147 | 151 | 153 | 145 | 137 |
| dN/dS* | 1.8059 | 1.7800 | 1.7220 | 1.8060 | 1.7798 | 1.7363 | 1.7086 | 1.9140 | 1.9483 | 1.6203 | 1.8364 |
| In-silico Spoligotyping | SB0268 | SB0268 | SB0268 | SB0268 | SB0268 | SB0268 | SB0268 | SB0120 | SB0120 | unreported | unreported |
*Calculations of dN/dS ratio is based on selected 137 virulence related genes listed in Supplemental Table 1.
Figure 5.
Whole genome sequence analysis of SNPs of M. bovis isolates. (A) A phylogenetic tree inferred using the neighbor-joining method based on the predicted SNPs of each isolate compared with the M. bovis AF2122/97 reference genome. Numbers at each branch represent bootstrap values and branches correspond to partitions with >50% bootstrap replicates. The tree is drawn to a scale with evolutionary distances computed using the maximum composite likelihood method in MEGA7. (B) Histogram of the number of SNPs (synonymous, non-synonymous and intergenic) for each examined M. bovis isolate compared to the reference genome.
To better trace the origin of MBE isolates within the Nile Delta, we used the SpoTyping tool that utilizes WGS to identify the characteristic spoligotype for each isolate27. The majority of MBE isolates fell within the SB0268 (MBE1-MBE7) which is the spoligotype of the Brazilian strain M. bovis AN5 and also found in united Kingdom and Mexico28. The MBE9 and MBE10 belonged to the SB0120 spoligotype, which is the spoligotype of BCG-like human M. bovis isolates previously found in Tunisia and Italy29,30. Finally, the MBE12 and MBE13 spoligotypes are of unknown SB number, suggesting a new spoligotype. Interestingly, the in silico analysis of the clonal complexes identified that all virulent MBE isolates were missing the spacer 21 plus the presence of the guaA SNP, a characteristic for the Clonal Complex European 231. This clonal complex is known to be prevalent in Brazil8,31, Portugal, Spain, and at a low frequency in both France and Italy31.
Dynamics of bovine tuberculosis transmission in the Nile Delta
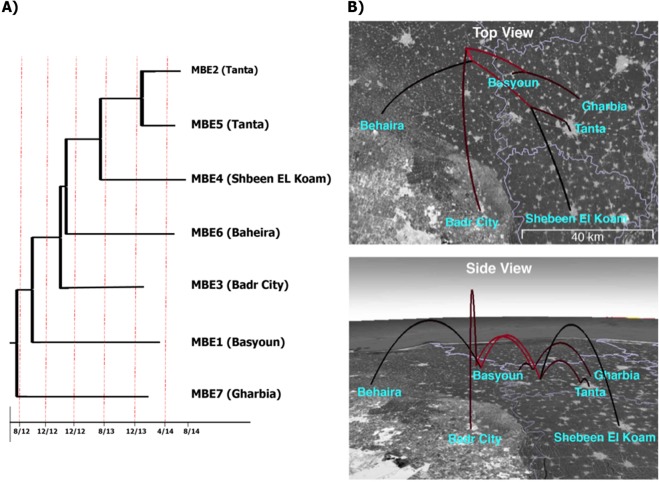
To better evaluate the dynamics of bTB transmission within the Nile Delta, we traced both herd historical records and molecular dating records of virulent isolates of M. bovis from Egypt. Our initial analysis incorporating all MBE isolates confirmed the monophyly of the non-BCG isolates from Egypt (Supplemental Fig. 3). Molecular timing analysis estimated that those isolates have originated about 3–4 years prior to the most recent sampled isolate. Subsequent phylogeographic analysis indicated a single independent transmission into each of the localities from which isolates were derived (Fig. 6). For example, the two isolates from Tanta are each other’s closest relatives, supporting a scenario where bTB was spread among individuals within the same region. While we are unable to identify the original source of the infection in the region, the phylogeographic model suggests transmission among these localities all within a matter of months during this particular outbreak. Dates of first case diagnosis and the start of the local bTB outbreak confirmed the scenario proposed by molecular dating analysis.
Figure 6.
Divergence times and phylogeographic model of bovine tuberculosis in the Nile Delta. The examined outbreak originated from a single common ancestor in early 2012, and rapidly spread among the sampled localities. (A) Divergence time estimates. Tip names indicate the isolate name and locality. (B) Phylogeographic model of the studied outbreak of bovine tuberculosis. The phylogeny from A is projected onto a map of the Nile Delta region of Egypt. Red colored branches represent more ancestral branches of the phylogeny. Maps were created by SPREAD V.1.0.780.
Virulence of M. bovis isolates
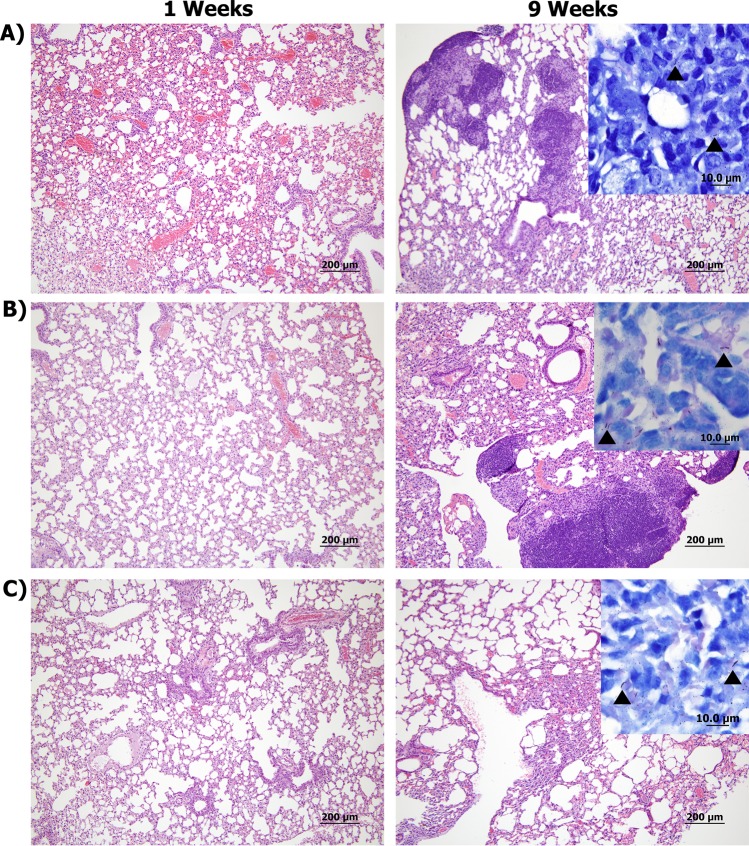
To evaluate potential differences in virulence among examined MBE isolates, mycobacterial colonization levels and histological analysis of murine tissues were analyzed. The selected isolates (MBE4, MBE5, and MBE7) were representing different geographical locations with disinctive genotypes (based on phylogenetic and molecular timing analysis). Mouse groups were sampled at 1, 3, and 9 weeks post-infection to represent early, middle, and progressive stages of infection following aerosol infection. As expected, low bacterial burden was found in all examined tissues at 1-week post-infection (WPI) with MBE5 and MBE7 (Fig. 7), suggesting decreased ability to initiate infection compared to MBE4. This low-colonization profile for MBE5 and MBE7 continued at 3 and 9 WPI. For the standard M. bovis isolate (AF2122/97), colonization levels increased at 3 WPI but peaked by 9 WPI, as expected. Histologically, mild inflammatory responses in murine lungs, spleen and liver were noticeable (score 1–2) by 3 WPI with the reference M. bovis strain AF2122/97. However, as infection progressed, the inflammatory responses were intensified in lungs by 9 WPI (score 3) with the reference strain (Fig. 8). Interestingly, lesions from mice infected with MBE4 were first observed in spleen by 3 WPI (score 2) but then more noticeable in the lungs by 9 WPI (score 2–3). Such unique histological profile could suggest a tendency of MBE4 to establish systemic infection by early infection of spleen, similar to the standard virulent strain. Unlike MBE4, all other tested clinical isolates (MBE5, MBE7) did not induce measurable levels of inflammation throughout the 9 WPI (score 0–1), consistent with their low levels of bacterial colonization.
Figure 7.
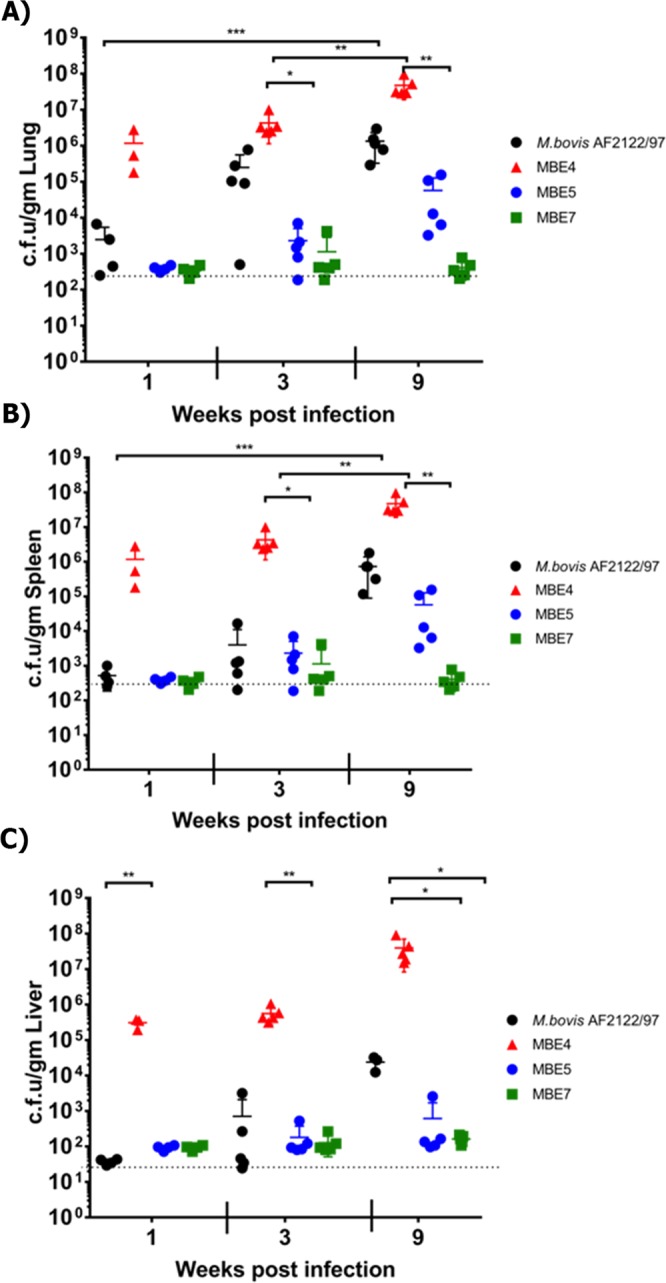
Tissue colonization of MBE isolates. Groups of BALB/c mice were infected by aerosol route with either MBE4, MBE5 and MBE7 in comparison with the reference M. bovis strain AF2122/97. Lungs, spleens and livers from infected animals were harvested and cultured at different weeks post infection. Each circle represents the colonization level for each organ from one animal. Asterisks (* for p < 0.05 and ** for p < 0.005) indicate statistical significant difference in colonization level between each clinical isolate compared to the reference strain.
Figure 8.
Histopathology of mice infected with clinical and reference isolates of M. bovis. Tissues sections stained with H&E collected from mice lungs infected with M. bovis AF2122/97 (A), MBE4 (B), and MBE7 (C) at 1 (left) and 9 (right) weeks post infection are shown at 40 × magnification (scale bar = 200 μm). Insets showing Ziehl-Neelsen-stained lung sections are also included, with arrowheads indicating acid-fast bacilli at 1000× magnification (scale bar = 10 μm). No bTB-associated granuloma infiltrates or acid-fast bacilli were found in any tissues in the naive group. Only tissue sections from mice infected with MBE7 were shown because of histopathology similarity between both MBE5 and MBE7 infections.
To further analyze the difference among MBE isolates; a detailed analysis of SNPs and insertions/deletions regions (InDels) was done to profile potential correlates with increased virulence. Several SNPs were identified for MBE4 including the absence of specific SNPs from MBE4 while those SNPs were present in the rest of the isolates. Interestingly, 5 SNPs were present only in the PE_PGRS14 of MBE5 and MBE7 isolates, all were confirmed by Sanger sequencing. The PE_PGRS14 gene is a member of the M. tuberculosis complex PE family, PGRS subfamily of glycine-rich proteins involved in virulence32, as shown from its upregulation during acute phase of macrophage infection33,34. In addition, MBE4 had the least number of InDels (n = 34) from the reference virulent strain when compared to MBE5 (n = 53) and MBE7 (n = 37). Among the InDels of MBE4, a fragment of 106 bp length (Supplemental Table 4) was deleted only from PGRS14 genes but present in the rest of isolates. It is noteworthy to mention here that several SNPs and InDels (Supplemental Tables 3 and 4) were present throughout the genomes of MBE4, MBE5 and MBE7 but not associated with PE-PGRS14 with potential impact on strain virulence. For example, a unique insertion was found in MBE4 associated with moeB1 (BQ2027_MB3231C), a probable molybdenum cofactor biosynthesis protein with potential impact on virulence35.
Discussion
In Africa and other developing countries, M. bovis accounts for approximately 10–20% of cases of human tuberculosis36. The global prevalence of human M. bovis infection is higher among patients with extra-pulmonary tuberculosis, since the pathogen is frequently acquired via oral ingestion and gastrointestinal disease is an important clinical manifestation37. Understanding disease transmission dynamics of M. bovis in cattle could eliminate such infections in humans. In Egypt, a previous investigation of health centers indicated presence of M. bovis in 0.4% to 6.4% of sputum-positive samples38. In this report, we examined the status of bTB infection in the Nile Delta of Egypt on both ecological and genomic levels. Our investigation traced cases of bTB in dairy herds of 7 governorates with a previous history of M. bovis infection. Historically, the General Organization of Veterinary Services (GOVS) indicated that bTB in slaughtered cattle in the Egyptian abattoirs was 0.05% in 1989. In subsequent years, the percentage of positive reactors in some governorates reached 11–23%39. In the present study, the SICST test indicated that 40.74% of the herds were tested positive for M. bovis-infection. At the animal-level, only 3.25% of animals were SICST-positive, similar to earlier reports6,7. Bovine tuberculosis may be kept under control by a national campaign of continuous testing and slaughtering of reactors, but the impact of emerging new virulent isolates remains to be assessed. As expected, animals >3 years-old were more often SICST-positive compared to younger animals. This may be due to the long incubation period and stress of high milk production in this age class40. Interestingly, 27% of SICST reactors had no visible bTB lesions, which was significantly higher than earlier studies41,42. This high level of potential false-positive SICST reactors could be attributed to infection with mycobacteria other than M. bovis or poor sensitivity of the postmortem examination43. More importantly, pulmonary and mixed type lesions were the most often observed lesions, suggesting the respiratory route to be the most important route of infection in the studied herds.
A surprising finding in our study was the isolation of BCG isolates from suspected cases of bTB in the Nile Delta. To our knowledge, previous isolation of BCG-like isolates from animals accounted for 26% of isolates in one study based on spoligotyping without any further confirmation to these isolates identity44, however, isolation of M. bovis BCG itself from animals is expected to be rare. In fact, BCG isolates were noticed in human infections with M. bovis29, mainly with the same spoligotype of the BCG isolates described in this study (SB0120). Other than illegal vaccination of animals with M. bovis BCG, this finding might imply a potential transmission from immune-compromised individual vaccinated earlier with BCG. More focused analysis of the virulent MBE isolates indicated their association with SB0268 spoligotype, suggesting limited genetic variations among isolates of M. bovis circulating in Egypt. Nevertheless, MIRU-VNTR indicated the presence of 4 complexes with more diversity than discovered by spoligotyping. Such analysis provided enough justification for WGS to uncover further information that could not be detected by analysis of limited sequence targets.
As expected, WGS showed some subtle differences amongst the 11 MBE genomes. Comparative genome analysis of the MBE genomes against a standard M. bovis reference (AF2122/97) identified additional genome-wide features that could not be deduced from specific gene analyses, as it provided higher levels of analysis at a single nucleotide level (SNPs). Further analysis of other SNPs indicated that recent isolates (MBE12 and MBE13) had a higher number of SNPs and constitute a separate clade of isolates from those belong to the first outbreak (MBE1-MBE10), another indication of the emergence of new foci of infection with their own potential virulence characteristics. Interestingly, one of the isolates (MBE4) has a unique genotype compared to other isolates (e.g. MBE5, and MBE7) from the Nile Delta including 5 SNPs and a 106 bp deletion in the PE_RGRS14 gene alone. PE_PGRS14 (Rv0834c) is thought to be upregulated in human lung granulomas compared to in-vitro grown bacteria34. Members of the PGRS subfamily of PE proteins are thought to be associated with cell wall or surface exposed45 with a potential role in mycobacterial pathogenesis46 and the evasion of host defenses47. However, given the wide polymorphism within MBE4 genotype (additional 22 SNPs outside the PGRS14 gene), it is difficult to establish virulence association without further studies. The fact that spleens from MBE4-infected mice showed inflammatory responses during the first 3 weeks of infection could suggest a highly successful isolate for establishing systemic infection during early stages of the disease. Earlier reports suggested that early dissemination of M. tuberculosis to the spleen, is usually associated with increased immune responses to the infection48, as evident from our histopathology analysis. Analysis of further time points with targeted gene deletions could reveal the potential virulence for MBE5 and MBE7 isolates, a logical extension of this project.
Overall, we were able to use all SNPs identified in all isolates to predict the dynamics of M. bovis isolates within the Nile Delta using a molecular timing algorithm. This approach is very helpful to show how M. bovis was able to transfer among dairy herds within a relatively short period of time (3–4 years), another reason to establish better control programs for bovine tuberculosis. Moreover, identified SNPs indicated the potential origin of recent outbreak of bTB in the Nile Delta from isolates present in Latin America and several European countries. The apparent lack of isolate genotypes from African countries with documented animal trade with Egypt (e.g. Ethiopia) and a known history of enzootic bTB49,50 was to some extent, surprising. It is possible that isolates from Africa are not well-represented in the M. bovis databases or historical trades with European countries allowed for successful spread within the Nile Delta. Analysis of more isolates from African countries, including Egypt, could further improve our understanding of the nature of M. bovis transmission and virulence.
Materials and Methods
Identifying naturally infected animals with bovine tuberculosis
The present study was based on the screening of herds with known history of bTB between 2013 to 2015, conducted by the Veterinary Serum and Vaccine Research Institute (VSVRI) and General Organization of Veterinary Services (GOVS) in Egypt. A total of 27 dairy herds from 7 governorates (Fig. 1) were included in this study based on previous reporting of suspected cases of bTB. The initial herd testing included whole-herd screening using Single Intradermal Comparative Skin Tuberculin (SICST) test administered in one side of the neck according to the OIE manual for diagnostic tests and vaccines for terrestrial animals51. Positive reactors (≥4 mm skin induration in bovine tuberculin injection site compared to avian tuberculin injection site) were subjected to culling then postmortem examination to inspect lungs, liver, kidneys, udder and regional lymph nodes. All tissue specimens were carefully examined for any gross lesions associated with bTB such as granuloma formation or caseation and calcification of parenchymal lymph nodes52,53.
Bacterial isolation and genomic DNA extraction
All organs and tissues showing gross lesions were collected. Samples were decontaminated by Hexadecyl-Pyridinium Chloride (HPC) as previously described54,55. Briefly, HPC was added to the homogenized tissues at room temperature for 15 min, tissue suspensions were centrifuged for 15 min at 1000Xg and the obtained sediments were inoculated into 2 glycerinated and 2 pyruvate Lowenstein-Jenson slants. All slants were incubated at 37 °C in inclined position for overnight, then vertically for at least 6–8 weeks. The obtained colonies were observed for morphological character and for pigment production56. A total of 31 M. bovis isolates were obtained from clinical samples taken from suspected tuberculous lesions from animals that scored positive in the intradermal tuberculin test, only 11 isolates were successfully sub-cultured. All the isolates (designated MBE for M. bovis Egypt) were grown in 10 ml of Middlebrook 7H9 liquid media supplemented with 0.36% sodium pyruvate and 10% ADC (Albumin-Dextrose-Catalase) for 4–5 weeks using BSL3 practices according to our approved biosafety protocol from the University of Wisconsin-Madison. High-quality genomic DNA (gDNA) samples were isolated as detailed before57 and its quality verified by both NanoDrop (Thermo Scientific, Wilmington, DE) and gel electrophoresis. In addition to clinical isolates, DNA samples from cultures of M. bovis AF2122/97, M. bovis-BCG (Pasteur) and M. tuberculosis H37Rv were isolated to serve as controls. All cultures of M. bovis isolates were handled under BSL3 environment at the University of Wisconsin-Madison.
Drug susceptibility testing
All clinical isolates were subjected to a standard drug susceptibility test against 4 first-line drugs isoniazid (INH), rifampicin (RIF), ethambutol (EMB) and streptomycin (SM) using the disk elusion assay (DEA)58. The following drug concentrations were used, INH, 0.2 μg/mL; RIF, 5.0 μg/mL; SM, 10.0 μg/mL; EMB, 5.0 μg/mL, according to Clinical and Laboratory Standards Institute (CLSI) guidelines M24-A258. Each strain was scored as resistant to a specific drug if its growth rate was >1% compared to the control isolate growth.
Molecular typing
The MBE isolates were genotyped using the PCR-RFLP of gyrB gene for general species identification among members of M. tuberculosis complex as described before59,60. In the first step, a 1020 bp fragment of the gyrB gene was amplified with the specific primers MTUB60. In the second step, amplicons were digested with restriction enzymes RsaI, TaqI, and SacII and the generated band pattern was examined to match members of the M. tuberculosis complex. To differentiate between members of the M. bovis cluster (virulent M. bovis and vaccine M. bovis-BCG), the RD1 typing were done utilizing RD1-F: AAGCGGTTGCCGCCGACCGACC, RD1-R: CTGGCTATATTCCTGGGCCCGG, and RD-1-R2: GAGGCGATCTGGCGGTTTGGGG primers61. Additionally, all MBE isolates were analyzed using the Mycobacterial Interspersed Repetitive Units-Variable Number of Tandem Repeats (MIRU-VNTR) method22,62–64. A panel of 12 MIRU loci was targeted for MIRU-VNTR genotyping62 and results were analyzed using a MIRU-VNTRplus database65.
Whole genome sequencing and comparative genomic analysis
Next-generation sequencing using the Illumina-MiSeq 2000 platform was performed at the University of Wisconsin-Madison Biotechnology Center as detailed before66. Raw sequence reads with average read length of 250 bp, were assembled against the sequence of the reference strain M. bovis Af2122/9767 using CLC-Bio Genomic Workbench version 8.0.1. Single nucleotide polymorphisms (SNPs) and insertion and deletion (InDels) polymorphisms were also identified using algorithms implemented by the CLC Genomics Workbench 8.0.1. However, repetitive genomic regions were not excluded from SNP predictions which could lead to discovery of false SNPs. For SNP predictions, parameters were set as follows: average base quality filter cutoff 15, central base quality filter cutoff 20, minimum sequence coverage 20, minimum variation frequency cutoff 50%, and maximum variation 2. All SNPs predicted from PE_PGRS14 that differed between genomes (N = 5) were further confirmed by Sanger sequencing68. For the In-Del analysis, parameters were set as follows: minimum sequence coverage was set to 10 and minimum variation frequency cutoff was 50%. The concatenated SNP files for each strain were aligned with MEGA V7.0.2669 and used to build a Neighbor-Joining phylogenetic tree (Fig. 5) with 1000 bootstrap replications and Jukes-Cantor substitution model70. The dN/dS ratio was calculated based on the codons of the 137 virulence related genes (Supplemental Table 1) using the Nei and Gojobori (Jukes-Cantor)71 and incorporating a statistic developed in Ota and Nei72 method using SNAP v2.1.1 implemented in HIV sequence database (https://www.hiv.lanl.gov/content/sequence/SNAP/SNAP.html).
To provide a robust measurement of genetic distance among bacterial genomes, the average nucleotide identity (ANI) was calculated through ANI calculator on EzGenome (http://www.ezbiocloud.net/tools/ani). Multiple whole-genomic alignments were performed using the Harvest suit73 allowing the identification of the gene or the intergenic region where differences are located. A phylogenomic tree (Supplemental Fig. 2) was constructed using a total of 67 M. bovis genomes retrieved from the NCBI public database for comparative purposes (Supplementary Table 2). The phylogenomic tree is based on the genomic polymorphism found at the core genome sequence shared with a similarity over a threshold between all genomes included in the analysis (Supplementary Fig. 1). The dendogram is visualized with CLC-Bio Genomic Workbench version 8.0.1. To analyze genome-wide rearrangements, sequences of all MBE genomes were aligned against the M. bovis AF2122/97 using MAUVE multiple alignment software with the progressive alignment option24,74,75 (Supplementary Fig. 2).
For the detection of in silico spoligotypes of MBE, we used SpoTyping-v2.127 by analyzing raw sequence reads obtained from Illumina MiSeq 2000 platform. SpoTyping is implemented with the Python language and BLAST algorithm. The binary and octal code representation of the spoligotypes were compared using the M. bovis Spoligotype Database (www.mbovis.org). For the identification of M. bovis clonal complexes76, In silico analysis of the 4 M. bovis clonal complexes were investigated among the MBE genomes consisted in the detection of RDAf1 and deletion of spacer 30 for African 1 using 3 previously described primers77. Detection of RDAf2 and deletion of spacers 3–7 for African 2 using another set of 3 previously described primers49. Detection of RDEu1 and deletion of spoligotype spacer 11 for European 1 using a pair of previously described primers78. Detection of guaA gene SNP at 3,765,573 position according to the reference genome (M. bovis AF2122/97) and deletion of spoligotype spacer 21 were used for the identification of European 2 clonal complex76.
Molecular timing
To analyze the patterns of bTB introduction and transmission throughout the Nile Delta, we employed an approach based on molecular clock and phylogeographic analyses. To establish an estimate of the age of the most recent common ancestor (MRCA) of the MBE isolates, we first analyzed the genomic SNP dataset that incorporated previously published bTB genomes from around the world, using BEAST 2.4.7. We used a GTR model of molecular evolution, which was identified as the best fitting model with jModelTest79. We placed a lognormal prior on the substitution rate (mean = 0.005 substitutions per site per year, SD = 0.3), and allowed these rates to vary among lineages, following previously estimated rates. We ran two chains of 100 million generations, assessing convergence using Tracer v1.6. We used the ages resulting from this first analysis as the basis for a phylogeographic analysis of a single outbreak of MBE isolates (isolates MBE1–7), which was estimated to be about 3.4 years prior to the most recently collected sample (September 2014). We therefore placed a prior under a normal distribution with an offset of 7 years on the MRCA of these isolates, for which we also indicated the tip dates. We placed a conservative exponential prior on rates of geographical spread and allowed these to vary among branches. Due to the smaller sampling size of this second analysis, we ran two chains of 20 million generations. We used Spread v1.0 to visualize the phylogeographic model80 available at https://github.com/phylogeography/SPREAD.
Virulence determination
Male and female BALB/c mice groups (n = 14) at 5–6 weeks age were infected using the M. bovis field isolates MBE4, MBE5, and MBE7 in comparison with the M. bovis AF2122/97 reference strain. Approximately 50–100 CFU were administered by aerosol using the Glas-Col inhalation system (Glas-Col, LLC, Terre Haute, IN). The infectious dose for each group was confirmed by plating lungs of an infected mouse at 1-day post-challenge. At 1, 3 and 9 weeks’ post-challenge 4–5 mice were sacrificed from each group for bacteriology and histopathology as detailed before81. Briefly lung, spleen, and liver samples were homogenized in PBS, brought up to a total volume of 2 ml before plating undiluted and 10-fold serial dilutions of samples onto 7H10 Middlebrook plates. For histopathology, sections were cut from embedded samples and stained with H&E or Ziehl–Neelsen for acid-fast bacilli. Histological lesions were scored by a trained pathologist blinded to the expperimental groups using a severity scale of 0 to 5 where 1, minimal; 2, mild; 3, moderate; 4, severe; and 5, massive. Images of lungs at 40× magnification were also analyzed in Photoshop CS2 (Adobe, San Jose, CA) to determine the percent of the inflamed area of the lung as compared to the total area of the lung for each animal post challenge. For statistical analysis, One-way ANOVA with Bonferroni’s post-test was used in Prism 5.01 (GraphPad Software). The post-test P values are as follows: *p < 0.05; **p < 0.01; ***p < 0.001.
Ethics statement
All procedures for animal testing, euthanasia, and sample collection were carried out at the governmental veterinary regional slaughterhouses, General Organization of Veterinary Services (GOVS), Nile Delta, Egypt. Euthanasia of animals was carried out in regional abattoirs in accordance with the recommendations by the Code of Practice for the Care and Handling of Dairy Cattle (http://www.nfacc.ca/codes-of-practice/dairy-cattle). All animal experiments were approved of by the Institutional Animal Care and Use Committee, University of Wisconsin-Madison. All laboratory procedures and techniques described in this report were conducted in accordance with the relevant guidelines and regulation of the University of Wisconsin-Madison.
Supplementary information
Acknowledgements
The authors would like to thank Dr. Michael T. Collins and Dr. Caitlin Pepperell for the constructive feedback on this manuscript. This work was partially supported by grants USAID (0210-22310-005-27G), Animal Formula Fund #WIS01814 USDA and JSOR project#2005 from the Academy of Scientific Research and Rechnology (ASRT), Egypt.
Author Contributions
All the authors are claiming authorship as they made substantial contributions to conception and design, acquisition of data, analysis and interpretation of data. Authors also gave final approval of the version to be submitted and any revised version. Conception or design of the work: H.A., E.N., A.M.T. Data collection: H.A., A.A., E.H., E.N., A.M.T. Data analysis and interpretation: H.A., D.S., H.S., A.M.T. Drafting the article: H.A., D.S., A.M.T. Critical revision of the article: H.A., D.S., A.M.T. Final approval of the version to be published: H.A., A.M.T.
Data Availability
The genome sequences and annotated sequence files for all MBE isolates reported here have been deposited in GenBank under the accession numbers (MBE1: QFZD00000000, MBE2: QFZC00000000, MBE3: QFZB00000000, MBE4: QFZA00000000, MBE5: QFYZ00000000, MBE6: QFYY00000000, MBE7: QFYX00000000, MBE9: QFYW00000000, MBE10: QFYV00000000, MBE12: QFYT00000000, MBE13: QFYU00000000).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48106-3.
References
- 1.Proceedings of the International Conference on Mycobacterium bovis. Dublin, Ireland, 20–21 August 1991. Vet Microbiol. 1994;40:1–205. doi: 10.1016/0378-1135(94)90041-8. [DOI] [PubMed] [Google Scholar]
- 2.Thoen CO, Lobue PA, Enarson DA, Kaneene JB, de Kantor IN. Tuberculosis: a re-emerging disease in animals and humans. Veterinaria italiana. 2009;45:135–181. [PubMed] [Google Scholar]
- 3.Hope JC, Villarreal-Ramos B. Bovine TB and the development of new vaccines. Comparative immunology, microbiology and infectious diseases. 2008;31:77–100. doi: 10.1016/j.cimid.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Grange JM, Collins CH. Bovine tubercle bacilli and disease in animals and man. Epidemiol Infect. 1987;99:221–234. doi: 10.1017/S0950268800067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pai M, et al. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 6.Cosivi O, Meslin FX, Daborn CJ, Grange JM. Epidemiology of Mycobacterium bovis infection in animals and humans, with particular reference to Africa. Rev.Sci.Tech. 1995;14:733–746. doi: 10.20506/rst.14.3.875. [DOI] [PubMed] [Google Scholar]
- 7.Ayele WY, Neill SD, Zinsstag J, Weiss MG, Pavlik I. Bovine tuberculosis: an old disease but a new threat to Africa. International Journal of Tuberculosis & Lung Disease. 2004;8:924–937. [PubMed] [Google Scholar]
- 8.Zimpel CK, et al. Complete Genome Sequencing of Mycobacterium bovis SP38 and Comparative Genomics of Mycobacterium bovis and M. tuberculosis Strains. Front Microbiol. 2017;8:2389. doi: 10.3389/fmicb.2017.02389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Campos S, et al. Limitations of spoligotyping and variable-number tandem-repeat typing for molecular tracing of Mycobacterium bovis in a high-diversity setting. J Clin Microbiol. 2011;49:3361–3364. doi: 10.1128/JCM.00301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roring S, et al. Development of variable-number tandem repeat typing of Mycobacterium bovis: comparison of results with those obtained by using existing exact tandem repeats and spoligotyping. J Clin Microbiol. 2002;40:2126–2133. doi: 10.1128/JCM.40.6.2126-2133.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biek R, et al. Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathog. 2012;8:e1003008. doi: 10.1371/journal.ppat.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruning-Fann CS, et al. Use of whole-genome sequencing and evaluation of the apparent sensitivity and specificity of antemortem tuberculosis tests in the investigation of an unusual outbreak of Mycobacterium bovis infection in a Michigan dairy herd. J Am Vet Med Assoc. 2017;251:206–216. doi: 10.2460/javma.251.2.206. [DOI] [PubMed] [Google Scholar]
- 13.Perea Razo CA, et al. Molecular epidemiology of cattle tuberculosis in Mexico through whole-genome sequencing and spoligotyping. PLoS One. 2018;13:e0201981. doi: 10.1371/journal.pone.0201981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lasserre M, et al. Whole genome sequencing of the monomorphic pathogen Mycobacterium bovis reveals local differentiation of cattle clinical isolates. BMC Genomics. 2018;19:2. doi: 10.1186/s12864-017-4249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price-Carter M, et al. Whole Genome Sequencing for Determining the Source of Mycobacterium bovis Infections in Livestock Herds and Wildlife in New Zealand. Front Vet Sci. 2018;5:272. doi: 10.3389/fvets.2018.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patane JS, et al. Patterns and processes of Mycobacterium bovis evolution revealed by phylogenomic analyses. Genome Biol Evol. 2017 doi: 10.1093/gbe/evx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orloski K, Robbe-Austerman S, Stuber T, Hench B, Schoenbaum M. Whole Genome Sequencing of Mycobacterium bovis Isolated From Livestock in the United States, 1989-2018. Front Vet Sci. 2018;5:253. doi: 10.3389/fvets.2018.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barbier, M. & Wirth, T. The Evolutionary History, Demography, and Spread of the Mycobacterium tuberculosis Complex. Microbiol Spectr4, 10.1128/microbiolspec.TBTB2-0008-2016 (2016). [DOI] [PubMed]
- 19.Mahairas GG, Sabo PJ, Hickey MJ, Singh DC, Stover CK. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M-bovis. Journal of Bacteriology. 1996;178:1274–1282. doi: 10.1128/jb.178.5.1274-1282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gordon SV, et al. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Molecular.Microbiology. 1999;32:643–655. doi: 10.1046/j.1365-2958.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 21.Warren RM, et al. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int J Tuberc Lung Dis. 2006;10:818–822. [PubMed] [Google Scholar]
- 22.Teeter LD, et al. Evaluation of 24-locus MIRU-VNTR genotyping in Mycobacterium tuberculosis cluster investigations in four jurisdictions in the United States, 2006–2010. Tuberculosis (Edinb) 2017;106:9–15. doi: 10.1016/j.tube.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allix-Beguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darling AE, Mau B, Perna N. T. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roperto S, et al. Proteomic analysis of protein purified derivative of Mycobacterium bovis. J Transl Med. 2017;15:68. doi: 10.1186/s12967-017-1172-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia Pelayo MC, et al. Gene expression profiling and antigen mining of the tuberculin production strain Mycobacterium bovis AN5. Vet Microbiol. 2009;133:272–277. doi: 10.1016/j.vetmic.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Xia E, Teo YY, Ong RT. SpoTyping: fast and accurate in silico Mycobacterium spoligotyping from sequence reads. Genome Med. 2016;8:19. doi: 10.1186/s13073-016-0270-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandoval-Azuara SE, et al. Whole genome sequencing of Mycobacterium bovis to obtain molecular fingerprints in human and cattle isolates from Baja California, Mexico. Int J Infect Dis. 2017;63:48–56. doi: 10.1016/j.ijid.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Lari N, Bimbi N, Rindi L, Tortoli E, Garzelli C. Genetic diversity of human isolates of Mycobacterium bovis assessed by spoligotyping and Variable Number Tandem Repeat genotyping. Infect Genet Evol. 2011;11:175–180. doi: 10.1016/j.meegid.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 30.Lamine-Khemiri H, et al. Genotypic characterization by spoligotyping and VNTR typing of Mycobacterium bovis and Mycobacterium caprae isolates from cattle of Tunisia. Trop Anim Health Prod. 2014;46:305–311. doi: 10.1007/s11250-013-0488-y. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez-Campos S, et al. European 2–a clonal complex of Mycobacterium bovis dominant in the Iberian Peninsula. Infect Genet Evol. 2012;12:866–872. doi: 10.1016/j.meegid.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Mohareer K, Tundup S, Hasnain SE. Transcriptional regulation of Mycobacterium tuberculosis PE/PPE genes: a molecular switch to virulence? J Mol Microbiol Biotechnol. 2011;21:97–109. doi: 10.1159/000329489. [DOI] [PubMed] [Google Scholar]
- 33.Dubnau E, Fontan P, Manganelli R, Soares-Appel S, Smith I. Mycobacterium tuberculosis genes induced during infection of human macrophages. Infect Immun. 2002;70:2787–2795. doi: 10.1128/IAI.70.6.2787-2795.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rachman H, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74:1233–1242. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacGurn, J. A. & Cox, J. S. A Genetic Screen for M. tuberculosis Mutants Defective for Phagosome Maturation Arrest Identifies Components of the ESX-1 Secretion System. Infection and Immunity, IAI (2007). [DOI] [PMC free article] [PubMed]
- 36.Muller B, et al. Zoonotic Mycobacterium bovis-induced tuberculosis in humans. Emerg Infect Dis. 2013;19:899–908. doi: 10.3201/eid1906.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Reilly LM, Daborn CJ. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber Lung Dis. 1995;76:1–46. doi: 10.1016/0962-8479(95)90591-X. [DOI] [PubMed] [Google Scholar]
- 38.Cosivi O, et al. Zoonotic tuberculosis due to Mycobacterium bovis in developing countries. Emerging infectious diseases. 1998;4:59–70. doi: 10.3201/eid0401.980108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GOVS. Annual Report of the General Organization of Veterinary Services (GOVS). Report about TB eradication, (1989).
- 40.Broughan JM, et al. A review of risk factors for bovine tuberculosis infection in cattle in the UK and Ireland. Epidemiol Infect. 2016;144:2899–2926. doi: 10.1017/S095026881600131X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Hagan MJ, et al. Risk factors for visible lesions or positive laboratory tests in bovine tuberculosis reactor cattle in Northern Ireland. Prev Vet Med. 2015;120:283–290. doi: 10.1016/j.prevetmed.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Menin A, et al. Asymptomatic cattle naturally infected with Mycobacterium bovis present exacerbated tissue pathology and bacterial dissemination. PLoS ONE. 2013;8:e53884. doi: 10.1371/journal.pone.0053884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corner LA. Post mortem diagnosis of Mycobacterium bovis infection in cattle. Vet Microbiol. 1994;40:53–63. doi: 10.1016/0378-1135(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 44.Haddad N, et al. Spoligotype diversity of Mycobacterium bovis strains isolated in France from 1979 to 2000. J Clin Microbiol. 2001;39:3623–3632. doi: 10.1128/JCM.39.10.3623-3632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brennan MJ, Delogu G. The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends in Microbiology. 2002;10:246–249. doi: 10.1016/s0966-842x(02)02335-1. [DOI] [PubMed] [Google Scholar]
- 46.Tian C, Jian-Ping X. Roles of PE_PGRS family in Mycobacterium tuberculosis pathogenesis and novel measures against tuberculosis. Microb Pathog. 2010;49:311–314. doi: 10.1016/j.micpath.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Brennan, M. J. The Enigmatic PE/PPE Multigene Family of Mycobacteria and Tuberculosis Vaccination. Infect Immun85, 10.1128/IAI.00969-16 (2017). [DOI] [PMC free article] [PubMed]
- 48.Chackerian AA, Alt JM, Perera TV, Dascher CC, Behar SM. Dissemination of Mycobacterium tuberculosis is influenced by host factors and precedes the initiation of T-cell immunity. Infect Immun. 2002;70:4501–4509. doi: 10.1128/IAI.70.8.4501-4509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berg S, et al. African 2, a clonal complex of Mycobacterium bovis epidemiologically important in East Africa. J Bacteriol. 2011;193:670–678. doi: 10.1128/JB.00750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ameni G, et al. Transmission of Mycobacterium tuberculosis between farmers and cattle in central Ethiopia. PLoS One. 2013;8:e76891. doi: 10.1371/journal.pone.0076891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de la Rua-Domenech R, et al. Ante mortem diagnosis of tuberculosis in cattle: a review of the tuberculin tests, gamma-interferon assay and other ancillary diagnostic techniques. Res Vet Sci. 2006;81:190–210. doi: 10.1016/j.rvsc.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Thoen C, LoBue P, de K. I. The importance of Mycobacterium bovis as a zoonosis. Vet.Microbiol. 2006;112:339–345. doi: 10.1016/j.vetmic.2005.11.047. [DOI] [PubMed] [Google Scholar]
- 53.Berg S, et al. The burden of mycobacterial disease in ethiopian cattle: implications for public health. PLoS ONE. 2009;4:e5068. doi: 10.1371/journal.pone.0005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambrosio SR, de Deus Oliveira EM, Rodriguez CA, Ferreira Neto JS, Amaku M. Comparison of three decontamination methods for Mycobacterium bovis isolation. Braz J Microbiol. 2008;39:241–244. doi: 10.1590/S1517-83822008000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medeiros L, Marassi CD, Duarte RS, da Silva MG, Lilenbaum W. Comparison of decontamination methods for primary isolation of Mycobacterium bovis in paucibacillary bovine tissues. Lett Appl Microbiol. 2012;54:182–186. doi: 10.1111/j.1472-765X.2011.03185.x. [DOI] [PubMed] [Google Scholar]
- 56.Niemann S, Richter E, Rüsch-Gerdes S. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis. J Clin Microbiol. 2000;38:152–157. doi: 10.1128/jcm.38.1.152-157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghosh P, et al. Genome-wide analysis of the emerging infection with Mycobacterium avium subspecies paratuberculosis in the Arabian camels (Camelus dromedarius) PLoS One. 2012;7:e31947. doi: 10.1371/journal.pone.0031947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.CLSI. (CLSI document M24-A2. Wayne: Clinical and Laboratory Standards Institute, 2011).
- 59.Chimara, E., Ferrazoli, L. & Leão, S. C. Mycobacterium tuberculosis complex differentiation using gyrB-restriction fragment length polymorphism analysis. Mem Inst Oswaldo Cruz99, 745–748, doi:/S0074-02762004000700014 (2004). [DOI] [PubMed]
- 60.Niemann S, Harmsen D, R++sch-Gerdes S, Richter E. Differentiation of Clinical Mycobacterium tuberculosis Complex Isolates by gyrB DNA Sequence Polymorphism Analysis. Journal of Clinical Microbiology. 2000;38:3231–3234. doi: 10.1128/jcm.38.9.3231-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y, et al. A simple and efficient multiplex PCR assay for the identification of Mycobacterium genus and Mycobacterium tuberculosis complex to the species level. Yonsei Med J. 2013;54:1220–1226. doi: 10.3349/ymj.2013.54.5.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frothingham R, Meeker-O’Connell WA. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144(Pt 5):1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 63.Mazars E, et al. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA. 2001;98:1901–1906. doi: 10.1073/pnas.98.4.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Supply P, et al. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- 65.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:W326–331. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu C-Y, Wu C-W, Talaat AM. Genome-wide sequence variations among Mycobacterium avium subspecies paratuberculosis isolates: A better understanding of Johne’s disease transmission dynamics. Front Microbiol. 2011;2:236. doi: 10.3389/fmicb.2011.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malone, K. M. et al. Updated Reference Genome Sequence and Annotation of Mycobacterium bovis AF2122/97. Genome Announc5, 10.1128/genomeA.00157-17 (2017). [DOI] [PMC free article] [PubMed]
- 68.Chen J, et al. Identification of novel mutations associated with cycloserine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother. 2017;72:3272–3276. doi: 10.1093/jac/dkx316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falconnet M. Phylogenetic distances for neighbour dependent substitution processes. Math Biosci. 2010;224:101–108. doi: 10.1016/j.mbs.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 71.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 72.Ota T, Nei M. Variance and covariances of the numbers of synonymous and nonsynonymous substitutions per site. Mol Biol Evol. 1994;11:613–619. doi: 10.1093/oxfordjournals.molbev.a040140. [DOI] [PubMed] [Google Scholar]
- 73.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/PREACCEPT-2573980311437212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakakibara Y, Osana Y, Popendorf K. [Development of a large-scale comparative genome system and its application to the analysis of mycobacteria genomes] Nihon Hansenbyo Gakkai Zasshi. 2007;76:251–256. doi: 10.5025/hansen.76.251. [DOI] [PubMed] [Google Scholar]
- 75.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith NH. The global distribution and phylogeography of Mycobacterium bovis clonal complexes. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2012;12:857–865. doi: 10.1016/j.meegid.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 77.Müller B, et al. African 1, an epidemiologically important clonal complex of Mycobacterium bovis dominant in Mali, Nigeria, Cameroon, and Chad. Journal of bacteriology. 2009;191:1951–1960. doi: 10.1128/JB.01590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith NH, et al. European 1: a globally important clonal complex of Mycobacterium bovis. Infect Genet Evol. 2011;11:1340–1351. doi: 10.1016/j.meegid.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 79.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 80.Bielejec F, Rambaut A, Suchard MA, Lemey P. SPREAD: spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics. 2011;27:2910–2912. doi: 10.1093/bioinformatics/btr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marcus SA, Sidiropoulos SW, Steinberg H, Talaat AM. CsoR Is Essential for Maintaining Copper Homeostasis in Mycobacterium tuberculosis. PLoS One. 2016;11:e0151816. doi: 10.1371/journal.pone.0151816. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences and annotated sequence files for all MBE isolates reported here have been deposited in GenBank under the accession numbers (MBE1: QFZD00000000, MBE2: QFZC00000000, MBE3: QFZB00000000, MBE4: QFZA00000000, MBE5: QFYZ00000000, MBE6: QFYY00000000, MBE7: QFYX00000000, MBE9: QFYW00000000, MBE10: QFYV00000000, MBE12: QFYT00000000, MBE13: QFYU00000000).