Abstract
MicroRNAs (miRNAs) have been proposed as potential prognostic and diagnostic biomarkers in numerous types of cancer, including osteosarcoma (OS), which is the most common bone malignancy. The present study revealed that the expression of miR-185 was downregulated in OS tissues and cells. Overexpression of miR-185 significantly suppressed the proliferation and migration of OS cells. To further investigate the functional roles of miR-185 in OS, the downstream targets of miR-185 were predicted using the microRNA.org database. The results revealed that in cancer cells, hexokinase 2 (HK2), the rate-limiting enzyme of glycolysis, was a potential target of miR-185. Molecular analysis indicated that miR-185 binds to the 3′-untranslated region of HK2 mRNA. Overexpressed miR-185 downregulated the mRNA and protein levels of HK2 in OS cells. In addition, an inverse correlation between the expression of miR-185 and HK2 was reported in OS. Consistent with the downregulation of HK2 induced by miR-185, overexpression of HK2 in OS cells significantly attenuated the inhibitory effects of miR-185 on glucose consumption and lactate production, while depletion of miR-185 promoted the glycolysis of OS cells. Additionally, restoration of HK2 abolished the inhibitory effects of miR-185 on the proliferation of OS cells. In summary, these results revealed that miR-185 suppressed the glucose metabolism of OS cells; thus, miR-185 may be considered as a promising therapeutic target for the treatment of OS.
Keywords: microRNA-185, osteosarcoma, hexokinase 2, glycolysis
Introduction
Osteosarcoma (OS) is the most common form of bone malignancy in children and adults, and is characterized by high mortality and a poor curative rate (1–3). Neoadjuvant chemotherapy, surgical resection and limb preservation surgery are the leading therapeutic strategies employed for the treatment of OS (4). For high-grade OS, the 5-year survival rate of OS patients without cancer metastasis has been ≤60-70% due to effective treatments (5,6). The survival rate of OS patients with low-grade lesions is notably higher (7,8). Unfortunately, the majority of patients with OS are diagnosed at late stages and the prognosis of these patients remains poor (7,8). Molecular targeted therapy has been considered as a potential strategy for the treatment of OS (9). Therefore, it is important to identify the novel targets and the underlying molecular mechanisms that drive the pathogenesis of OS.
MicroRNAs (miRNAs/miRs) are short noncoding RNAs that regulate gene expression by binding with the 3′-untranslation region (3′-UTR) of the targeted mRNAs, consequently inhibiting the translation or inducing the degradation of mRNAs (10). Increasing evidence has demonstrated the regulatory function of miRNAs in the initiation and progression of OS (11,12). For instance, miR-449a is downregulated in OS, which promotes the apoptosis of OS cells via targeting B-cell lymphoma 2 (13). In addition, miR-20a is overexpressed in OS, which induces the proliferation of OS cells (14). Previously, miR-185 was reported to suppress the growth of human none small cell lung cancer by inducing cell cycle arrest (15). Additionally, miR-185 inhibits the growth of hepatocellular carcinoma cells through the DNA (cytosine-5)-methyltransferase 1 (DNMT1)/phosphatase and tensin homolog/protein kinase B signaling pathway (16). It has been demonstrated that miR-185 is a potential prognostic biomarker for early stage hepatocellular carcinoma (17). Interestingly, previous studies have revealed that miR-185 regulated the invasion, metastasis and radioresistance of nasopharyngeal carcinoma by targeting Wnt family member 2B in vitro (18,19). These studies indicate the important roles of miR-185 in numerous types of cancer; however, the function of miR-185 in OS remains unclear.
Aerobic glycolysis is considered as the hallmark of cancer cells, which regulates the metabolism of glucose into lactate as opposed to the mitochondrial oxidative phosphorylation pathway (20). The glucose transporters are responsible for the translocation of glucose across the plasma membrane. The metabolism of glucose in cancer cells is catalyzed by a variety of enzymes (21,22). During glycolysis, hexokinase 2 (HK2) is the first rate-limiting enzyme, which regulates the conversion of glucose into glucose-6-phosphate (23–28). These findings indicate that the expression of HK2 is associated with the glucose metabolism of cancer cells. Previous studies have demonstrated upregulation of HK2 in various cancers, which promotes the glucose consumption of cancer cells (29–32). Interestingly, HK2 has been reported to be the target of miRNAs in cancers (33–36). For instance, miR-181b downregulates HK2, consequently suppressing the glycolysis and proliferation of gastric cancer cells (36). miR-143 was reported to be a tumor suppressor in prostate cancer via targeting HK2 (35). In OS, miR-125b inhibits the aerobic glycolysis of OS cells by downregulating the expression of HK2 (37). These findings indicate that inhibition of HK2 may be a promising strategy for the treatment of cancer.
In the present study, the expression of miR-185 in OS tissues and cells was evaluated. Further molecular study revealed that miR-185 inhibited the expression of HK2 in OS cells. These findings suggested the potential regulatory function of miR-185 in OS.
Materials and methods
Cell culture
The normal human osteoplastic cell line (NHOst) and OS cell lines HOS, U2OS, Saos-2 and MG-63 were purchased from the American Type Culture Collection. Cells were cultured in Dulbecco's Modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum, 100 U/ml penicillin (Invitrogen; Thermo Fisher Scientific, Inc.) and 100 µg/ml of streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator containing 5% CO2.
Clinical samples
A total of 30 paired OS tissues and adjacent normal tissues (0.5–1 cm in diameter) were collected from the metaphyseal regions of long bones of the OS patients (age, 8–45 years old; female:male=1.35:1) at The Central Hospital of Chao Zhou between April 2015 and October 2016. The adjacent normal tissues were ≥5 cm away from the edge of the OS tissues. The exclusion criteria of the patients included: i) Received chemo- or radiotherapy prior to surgery; ii) patients were unsuitable for surgery; iii) Informed consent was not obtained; and iv) serious infection. The basic clinical characteristics of these patients were provided in Table I. The samples were snap-frozen in liquid nitrogen and stored at −80°C. Written informed consent was obtained from all the patients. Tissues were staged according to the Vanderbilt system (38). The procedures for tissue collection and the following experiments were approved by the Ethics Committee of The Third Affiliated Hospital of Southern Medical University.
Table I.
Clinical parameters of the osteosarcoma patients involved in this study.
| Clinical characteristics | Number |
|---|---|
| Age (years) | |
| ≤50 | 10 |
| >50 | 20 |
| Sex | |
| Male | 17 |
| Female | 13 |
| Tumor size (cm) | |
| ≤5 | 11 |
| >5 | 19 |
| Differentiation | |
| Moderate | 15 |
| Poor | 15 |
| TNM stage | |
| I–II | 18 |
| III–IV | 12 |
| Lymph node metastasis | |
| N0 | 16 |
| N1-3 | 14 |
TNM, tumor, node and metastasis.
miRNA transfection
miR-185 mimics (5′-UGGAGAGAAAGGCAGUUCCUGA), control miRNA (5′-GGUUCGUACGUACACUGUUCA-3′), miR-185 antagomir (5′-UCAGGAACUGCCUUUCUCUCCA-3′) and negative control miRNA (5′-CGGUACGAUCGCGGCGGGAUAUC-3′) were synthesized by Sangon Biotech Co., Ltd. For miRNA transfection, both U2OS and Saos-2 cells (10,000 cells per well) were seeded on 6-well plates with DMEM and 20 nm miRNAs were transfected into the cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. At 36 h post-transfection, the expression levels of miR-185 were determined by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The HA-HK2 transfection was also performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols.
RNA extraction and RT-qPCR
Total RNA was extracted from the tissues or cells using an RNeasy Mini kit (cat. no. 74104; Qiagen GmbH) according to the manufacturer's protocols. The concentration of RNA was determined using a NanoDrop 2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc). Extracted RNA (1 µg) was reverse transcribed using the Prime-Script RT kit (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocols. qPCR was performed using the miScript PCR system (Qiagen GmbH) according to the manufacturer's instructions. U6 RNA and GAPDH were used to normalize the expression of miR-185 and HK2, respectively. The PCR conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 58°C for 60 sec. The primer sequences used in this study were synthesized as: MiR-185 forward, 5′-CAATGGAGAGAAAGGCAGTTCC-3′ and reverse, 5′-AATCCATGAGAGATCCCTACCG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′; HK2 forward, 5′-ATGATCGCCTGCTTATTCACG-3′ and reverse, 5′-CGCCTAGAAATCTCCAGAAGGG-3′; actin forward, 5′-GTGACGTTGACATCCGTAAAGA-3′ and reverse, 5′-GCCGGACTCATCGTACTCC-3′. The experiments were performed with three independent repeats.
Cell proliferation assay
The proliferation of OS cells transfected with miR-185 mimics or control miRNA was determined using Cell Titer 96® Aqueous Cell Proliferation Assay kit (Promega Corporation) according to the manufacturer's protocols. Briefly, transfected OS cells (1,000 cells per well) were seeded in 96-well plates with DMEM. Cell proliferation was evaluated by adding MTT reagent to the medium. Following the incubation at 37°C for 3 h, the absorbance at 480 nm at day 1, 2, 3, 4 and 5 was determined using a microplate reader.
Luciferase reporter assay
Wild-type (WT) or mutant 3′-UTR of HK2 containing the putative miR-185 binding sites were amplified and inserted into pMIR-Luciferase-Reporter vectors (Promega Corporation) according to the manufacturer's protocols. The plasmids were transfected into OS cells in the presence of miR-185 mimics or control miRNA using Lipofectamine 3000. At 48 h post-transfection, luciferase activity was determined using Dual-GLO Luciferase Assay System (Promega Corporation) according to the manufacturer's protocols. The activity of Renilla was detected as the normalization. The experiments were performed in triplicate.
Western blotting
Following transfection for 48 h, total proteins of cells were extracted using radioimmunoprecipitation lysis buffer (Beyotime Institute of Biotechnology) and the protein concentration was determined using a BCA Assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of protein (20 µg) were separated using 15% SDS-PAGE and then transferred onto polyvinylidene difluoride membranes (EMD Millipore). The membrane was blocked with 5% non-fat milk for 1 h at room temperature and incubated with antibodies against HK2 (1:2,000; cat. no. GTX124375; GeneTex) at room temperature for 2 h. The goat anti rabbit secondary antibody conjugated with horseradish peroxidase (1:5,000; cat. no. 305005; Bio-Rad Laboratories, Inc.) was then applied for 1 h at room temperature. The bands were visualized using an enhanced chemiluminescence protein detection kit (EMD Millipore) and exposed using X-ray film (Kodak, Inc.). β-actin was used as the loading control (1:3,000; cat. no. AC004; ABclonal Biotech Co., Ltd.).
Determination of glucose consumption and lactate production
To evaluate glucose uptake and lactate production, OS cells (10,000 cells) were seeded in the 6-well plates with DMEM and transfected with miRNAs. At 48 h post-transfection, the culture medium was collected. The glucose consumption and lactate production were determined using an Amplex® Red Glucose/Glucose oxidase Assay kit (Invitrogen; Thermo Fisher Scientific, Inc.) and lactate assay kit (Sigma-Aldrich; Merck KGaA) according to the manufacturer's protocols.
Colony formation
OS cells were transfected with miR-185 mimics or control miRNA and seed in the 6-well plates with the density of 1,000 cells per well. Cells were maintained in DMEM containing 10% FBS. Following culture for 10 days, colonies were washed with PBS and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology) for 15 min at room temperature. The number of colonies was counted with the light microscopy (magnification, ×100; Olympus Corporation).
Wound-healing assay
OS cells transfected with miR-185 mimics or control miRNA were seeded in a 6-well plate (~10,000 cells per well). Following culture for 24 h, cells were wounded with a 200 µl pipette tip and the cell debris was removed by washing with PBS. Cells were cultured with fresh DMEM for 24 h at 37°C. Subsequently, the migration of the cells was observed using a light microscope (Olympus Corporation; magnification, ×40).
Statistical analysis
The data were presented as the mean + standard deviation from three independent repeats and analyzed with the GraphPad Prism software (Version 5.0; GraphPad Software, Inc.). Statistical comparisons between two groups were performed using a Student's t-test. Comparisons between multiple groups were performed using a one-way analysis of variance followed by a Newman-Keuls post-hoc comparison. The correlation between the expression of miR-185 and HK2 was analyzed with the Spearman's correlation analysis P<0.05 was considered to indicate a statistically significant difference.
Results
miR-185 is downregulated in OS tissues and cells
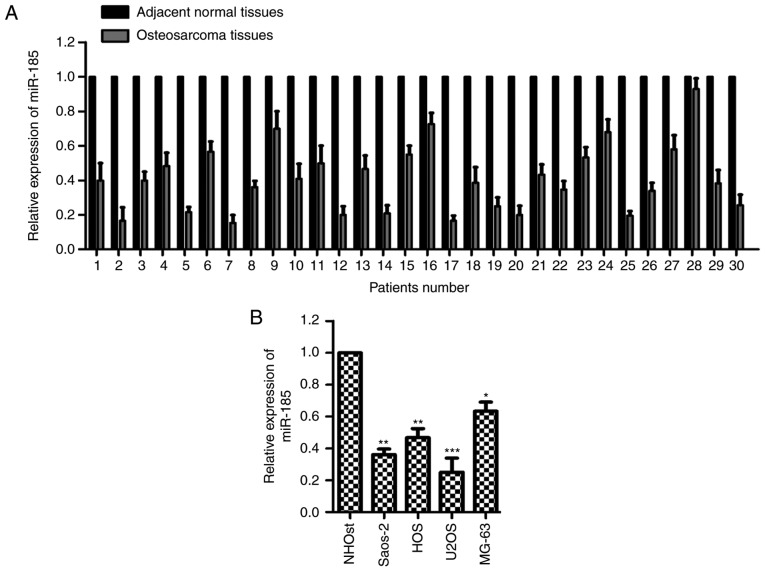
To characterize the potential function of miR-185 in OS, the expression of miR-185 was determined in paired OS and adjacent normal tissues. The expression levels of miR-185 were notably reduced in OS tissues compared with the adjacent noncancerous tissues (Fig. 1A). Additionally, the expression of miR-185 was significantly downregulated in OS cells compared with normal cells (P<0.05; Fig. 1B). These data revealed the downregulated expression of miR-185 in OS. The relative expression levels of miR-185 in U2OS and Saos-2 cells were the lowest; thus, these two cell lines were selected for further cytological study to overexpress miR-185.
Figure 1.
miR-185 is downregulated in OS tissues and cells. (A) Expression levels of miR-185 were determined using RT-qPCR in paired OS and adjacent normal tissues. (B) RT-qPCR was performed to compare the expression of miR-185 in normal and OS cells. *P<0.05, **P<0.01, ***P<0.001 vs. NHOst. miR, microRNA; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative chain reaction.
miR-185 suppresses the proliferation of OS cells
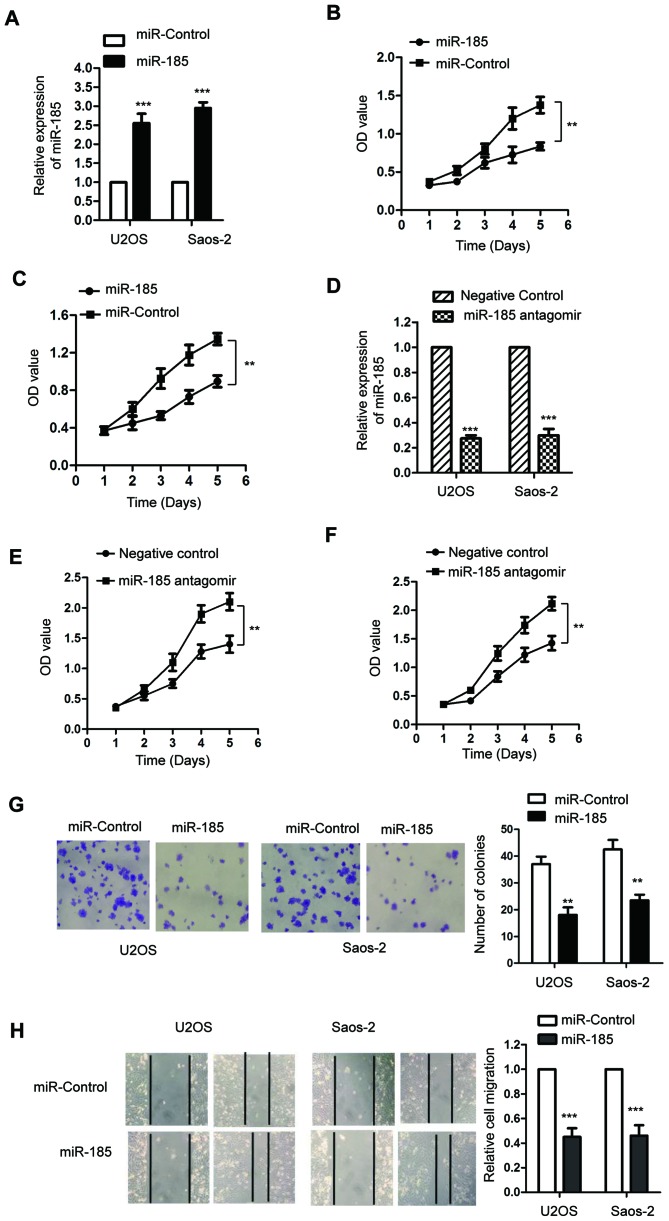
To investigate the function of miR-185 in the progression of OS, both U2OS and Saos-2 cells were transfected with miR-185 mimics or the control miRNA. The expression levels of miR-185 were significantly upregulated following the transfection of miR-185 in U2OS and Saos-2 cells compared with the control (P<0.05; Fig. 2A). To determine the effect of miR-185 on the proliferation of OS cells, an MTT assay was performed and the results revealed that miR-185 overexpression significantly inhibited the growth of OS cells compared with the control (P<0.05; Fig. 2B and C). To further investigate the regulatory function of miR-185 on the growth of OS cells, U2OS and Saos-2 cells were transfected with miR-185 antagomir or negative control. Transfection of miR-185 antagomir significantly downregulated the expression of miR-185 (P<0.05; Fig. 2D). Furthermore, the results of MTT assay revealed that inhibition of miR-185 significantly promoted the proliferation of U2OS and Saos-2 cells compared with the control (Fig. 2E and F). Consistent with these results, the results of colony formation assay also indicated that miR-185 upregulation inhibited the colony formation ability of U2OS and Saos-2 cells compared with the control (Fig. 2G). To further investigate the effect of miR-185 on the migration of OS cells, a wound-healing assay was performed using U2OS and Saos-2 cells transfected with miR-185 mimics or control miRNA. Overexpression of miR-185 in U2OS and Saos-2 cells significantly inhibited the migration ability compared with the control group (P<0.05; Fig. 2H). These findings indicated that miR-185 could inhibit the growth of OS cells.
Figure 2.
Overexpression of miR-185 suppresses the proliferation and migration of OS cells. (A) Overexpression of miR-185 in both U2OS and Saos-2 cells was confirmed by RT-qPCR. (B and C) Transfection of miR-185 significantly suppressed the proliferation of U2OS and Saos-2 cells. (D) U2OS and Saos-2 cells were transfected using negative control miRNA or miR-185 antagomir; downregulation of miR-185 was confirmed using RT-qPCR. (E and F) The proliferation of OS cells transfected with miR-185 antagomir was evaluated using MTT assay. (G) Transfection with miR-185 mimics significantly inhibited the colony formation ability of OS cells compared with the control. Magnification, ×10. (H) Overexpression of miR-185 inhibited the migration of U2OS and Saos-2 cells. **P<0.01, ***P<0.001 vs. miR-control. miR, microRNA; OD, optical density; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative chain reaction.
miR-185 downregulates the expression of HK2 by binding to the 3′-UTR of HK2
To further investigate the regulatory function of miR-185 in OS, the targets of miR-185 were identified using bioinformatic analysis (http://34.236.212.39/microrna/getMirnaForm.do). HK2 was predicted to be a potential target of miR-185. The putative binding sequence of miR-185 at the 3′-UTR of HK2 was indicated in Fig. 3A. To confirm this potential binding interaction, a luciferase reporter assay was performed using a luciferase vector containing the WT or mutant 3′-UTR of HK2 in the presence of miR-185 mimics or control miRNA. The results revealed that miR-185 significantly inhibited the luciferase activity of WT but not the mutant 3′-UTR of HK2 (P<0.05; Fig. 3B and C). Furthermore, the mRNA and protein levels of HK2 were also determined in U2OS and Saos-2 cells transfected with miR-185 mimics or control miRNA. Ectopic expression of miR-185 significantly downregulated the mRNA levels of HK2 in U2OS and Saos-2 cells compared with the control (P<0.05; Fig. 3D). Consistent with these results, the protein levels of HK2 were also reduced in OS cells overexpressing miR-185 (Fig. 3E). These results demonstrated that miR-185 inhibited the expression of HK2 in OS cells. To confirm the association between the expression of miR-185 and HK2, the mRNA levels of HK2 in OS tissues were determined using RT-qPCR compared with adjacent normal tissues. The data revealed that the expression of HK2 was significantly upregulated in OS tissues compared with the control (P<0.05; Fig. 3F). The results of Spearman's correlation analysis indicated that the expression levels of miR-185 and HK2 in OS tissues were inversely correlated (Fig. 3G). These results demonstrated that HK2 could be a potential downstream target of miR-185 in OS cells.
Figure 3.
HK2 is a potential downstream target of miR-185. (A) Predicted target sequence of miR-185 in the 3′-UTR of HK2. (B and C) OS cells were transfected with wild type or mutant 3′-UTR of HK2 in the presence of miR-185. Luciferase assay was performed to confirm the binding between miR-185 and the 3′-UTR of HK2. **P<0.01, ***P<0.001 vs. miR-control. (D) U2OS and Saos-2 cells were transfected with miR-185 mimics or control miRNA, and the mRNA levels of HK2 were determined using RT-qPCR. **P<0.01 vs. miR-control. (E) Overexpression of miR-185 in OS cells suppressed the protein expression of HK2. (F) RT-qPCR was performed to evaluate the expression of miR-185 in paired OS and adjacent normal tissues. ***P<0.001. (G) Spearman's correlation test revealed the inverse correlation between the expression of miR-185 and HK2 in OS tissues. r=−0.6192, P<0.001. HK2, hexokinase 2; miR, microRNA; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative chain reaction; UTR, untranslated region.
miR-185 inhibits the glucose metabolism of OS cells
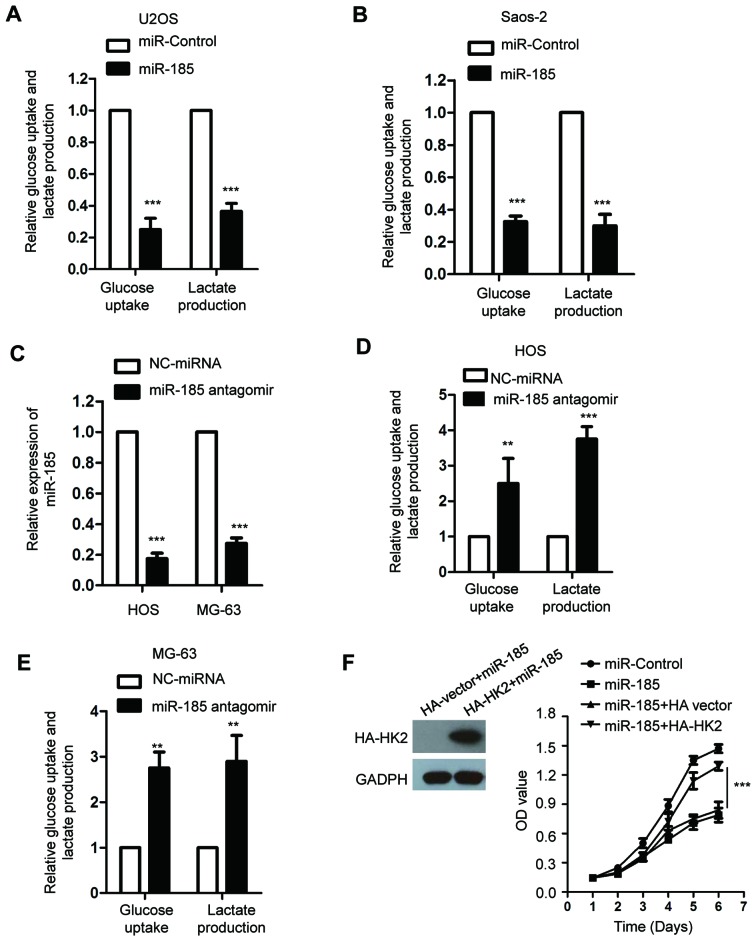
HK2 serves important roles in the glycolysis of cancer cells (39). As miR-185 downregulated the expression of HK2, we hypothesized that miR-185 could regulate the glycolysis of OS cells. To confirm this, the glucose consumption and lactate production of U2OS and Saos-2 cells transfected with miR-185 or control miRNA were determined. The data revealed that overexpression of miR-185 significantly suppressed glucose uptake and lactate production of OS cells compared with the control (P<0.05; Fig. 4A and B). To further investigate the inhibitory effect of miR-185 on the glycolysis of OS cells, the endogenous expression of miR-185 was abolished by transfecting miR-185 antagomir into HOS and MG-63 cells, which express miR-185 at a relatively higher level among the OS cell lines we used. The results revealed that miR-185 was significantly downregulated in OS cells transfected with miR-185 antagomir (P<0.05; Fig. 4C). In addition, inhibition of miR-185 significantly promoted the glycolysis of HOS and MG-63 cells compared with the control (Fig. 4D and E). To determine whether miR-185 inhibited the growth of OS cells through downregulating HK2, the expression of HK2 was restored by transfecting U2OS cells with HA-HK2. The ectopic expression of HK2 suppressed the inhibitory effect of miR-185 on the growth of OS cells (Fig. 4F).
Figure 4.
miR-185 inhibits the glucose consumption and lactate production of OS cells. (A and B) U2OS and Saos-2 cells were transfected with miR-185 or control miRNA, and the glucose consumption and lactate production of the cells were determined. ***P<0.001 vs. miR-control. (C) The endogenous expression of miR-185 in OS cells was suppressed by transfecting with miR-185 antagomir. The knockdown efficiency was confirmed using reverse transcription-quantitative chain reaction. ***P<0.001 vs. NC-miRNA. (D and E) HOS and MG-63 cells were transfected with miR-185 antagomir; the glucose consumption and lactate production of the cells were determined. **P<0.01, ***P<0.001 vs. NC-miRNA. (F) U2OS cells were transfected with the indicated expression vectors, and the cell proliferation was evaluated with an MTT assay. ***P<0.001; miR-185+HA-HK2 vs. miR-185+HA vector. miR/miRNA, microRNA; NC, negative control; OD, optical density; OS, osteosarcoma; ns, no significance.
Discussion
The regulatory functions of miRNAs in the progression of cancers have been highlighted in previous studies (40–42). miR-185 was reported to function as a tumor suppressor in numerous types of cancer, including hepatocellular carcinoma and non-small cell lung cancer (15,16). In the present study, the expression levels of miR-185 in OS were evaluated, and the downstream targets of miR-185 were identified. The results indicated that miR-185 was significantly downregulated in OS tissues and cells. Ectopic overexpression of miR-185 suppressed the growth and migration of OS cells. These findings suggested the potential tumor suppressive role of miR-185 in OS.
To identify the underlying molecular mechanisms of miR-185-mediated inhibition in the proliferation of OS cells, the potential targets of miR-185 were predicted. A previous study identified AKT1 as one of the targets of miR-185 in non-small cell lung cancer (43). The potential tumor suppressive function of miR-185 through the E2F1 and DNMT1 pathways was also revealed in triple-negative breast cancer (44). Furthermore, recent studies reported that miR-185 targeted WNT2B to regulate the invasion and metastasis of nasopharyngeal carcinoma (18,19). Additionally, miR-185 suppressed the proliferation of pancreatic cells by targeting a transcriptional co-activator with a PDZ-binding motif (45). miR-185 was also reported to inhibit the Wnt/β-catenin pathway in colorectal cancer (46).
In the present study, HK2 was revealed to be a direct target of miR-185 in OS cells. Overexpressed miR-185 suppressed the mRNA and protein levels of HK2. In addition, an inverse correlation between the expression of miR-185 and HK2 was also observed in OS tissues. HK2 controls the first step of glycolysis, consequently inducing the ATP-dependent phosphorylation of glucose into glucose-6-phasphatase (47). Upregulated HK2 has been detected in cancer cells, and was associated with poor prognosis of patients (48,49). HK2 was determined to be involved in the initiation and development of Kras-driven lung cancer and ErbB2-driven breast cancer in HK2 conditional knockout mice (50). In addition, previous studies suggested that HK2 was a target of numerous miRNAs in cancers, which could be associated with cancer progression (35,36). Furthermore, recent findings indicated that miR-155 inhibited HK2-mediated glycolysis and sensitized lung cancer cells to irradiation (51). HK2 was also revealed as a target of miR-143 in prostate cancer (35). These results indicated that HK2 was regulated by miRNAs in numerous types of cancer and consequently regulated the progression of tumor. In the present study, miR-185 overexpression suppressed the glucose consumption and lactate production of OS cells, while restoration of HK2 rescued the growth of OS cells. As HK2 is involved in aerobic glycolysis, these results suggested that miR-185 could regulate the proliferation of OS cells in a HK2-dependent manner.
Due to the tumor suppressive role of miR-185 in OS, it might be interesting to identify the upstream regulators of miR-185. Accumulating evidence suggested that some small molecules or natural compounds, such as docosahexaenoic acid, resveratrol, curcumin, regulated the expression of miRNAs (52). Further studies could be conducted to identify the molecules that regulate the expression of miR-185 in OS. Additionally, to further characterize the potential tumor suppressive function of miR-185 in OS, in vivo experiments may also be performed. Additionally, as numerous downstream targets of miR-185 have been identified in a variety of cancers (43,44,53–55), the regulatory function of miR-185 on its potential targets in OS could be explored.
In conclusion, the results of the present study demonstrated that miR-185 inhibited the proliferation and glycolysis of OS cells through targeting HK2. The suppressive function of miR-185 on the growth of OS cells indicated that miR-185 may be a novel therapeutic candidate for the treatment of OS in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
CL and HL designed the study. CL performed the experiments. LC performed the luciferase assay. HL wrote the manuscript.
Ethics approval and consent to participate
Written informed consent was provided by all the patients enrolled in this study. The procedures for tissue collection and the following experiments were approved by the Ethics Committee of The Third Affiliated Hospital of Southern Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Friebele JC, Peck J, Pan X, Abdel-Rasoul M, Mayerson JL. Osteosarcoma: A Meta-analysis and review of the literature. Am J Orthop (Belle Mead NJ) 2015;44:547–553. [PubMed] [Google Scholar]
- 2.Abarrategi A, Tornin J, Martinez-Cruzado L, Hamilton A, Martinez-Campos E, Rodrigo JP, González MV, Baldini N, Garcia-Castro J, Rodriguez R. Osteosarcoma: Cells-of-Origin, cancer stem cells, and targeted therapies. Stem Cells Int. 2016;2016:3631764. doi: 10.1155/2016/3631764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189–195. doi: 10.1016/j.canlet.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Moore DD, Luu HH. Osteosarcoma. Cancer Treat Res. 2014;162:65–92. doi: 10.1007/978-3-319-07323-1_4. [DOI] [PubMed] [Google Scholar]
- 5.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 6.Bernthal NM, Federman N, Eilber FR, Nelson SD, Eckardt JJ, Eilber FC, Tap WD. Long-term results (>25 years) of a randomized, prospective clinical trial evaluating chemotherapy in patients with high-grade, operable osteosarcoma. Cancer. 2012;118:5888–5893. doi: 10.1002/cncr.27651. [DOI] [PubMed] [Google Scholar]
- 7.Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment-where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Geller DS, Gorlick R. Osteosarcoma: A review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8:705–718. [PubMed] [Google Scholar]
- 9.Zhou W, Hao M, Du X, Chen K, Wang G, Yang J. Advances in targeted therapy for osteosarcoma. Discov Med. 2014;17:301–307. [PubMed] [Google Scholar]
- 10.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7:147–154. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang L, Shrestha S, LaChaud G, Scott MA, James AW. Review of microRNA in osteosarcoma and chondrosarcoma. Med Oncol. 2015;32:613. doi: 10.1007/s12032-015-0613-z. [DOI] [PubMed] [Google Scholar]
- 12.Sampson VB, Yoo S, Kumar A, Vetter NS, Kolb EA. MicroRNAs and Potential Targets in Osteosarcoma: Review. Front Pediatr. 2015;3:69. doi: 10.3389/fped.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Zhou J, Chen X, Yang B, Wang D, Yang P, He X, Li H. miRNA-449a is downregulated in osteosarcoma and promotes cell apoptosis by targeting BCL2. Tumour Biol. 2015;36:8221–8229. doi: 10.1007/s13277-015-3568-y. [DOI] [PubMed] [Google Scholar]
- 14.Yuan G, Zhao Y, Wu D, Gao C, Jiao Z. miRNA-20a upregulates TAK1 and increases proliferation in osteosarcoma cells. Future Oncol. 2018;14:461–469. doi: 10.2217/fon-2017-0490. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi Y, Forrest AR, Maeno E, Hashimoto T, Daub CO, Yasuda J. MiR-107 and MiR-185 can induce cell cycle arrest in human non small cell lung cancer cell lines. PLoS One. 2009;4:e6677. doi: 10.1371/journal.pone.0006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qadir XV, Han C, Lu D, Zhang J, Wu T. miR-185 inhibits hepatocellular carcinoma growth by targeting the DNMT1/PTEN/Akt pathway. Am J Pathol. 2014;184:2355–2364. doi: 10.1016/j.ajpath.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhi Q, Zhu J, Guo X, He S, Xue X, Zhou J, Hu B, Li H, Chen S, Zhao H, Kuang Y. Metastasis-related miR-185 is a potential prognostic biomarker for hepatocellular carcinoma in early stage. Biomed Pharmacother. 2013;67:393–398. doi: 10.1016/j.biopha.2013.03.022. [DOI] [PubMed] [Google Scholar]
- 18.Liu C, Li G, Ren S, Su Z, Wang Y, Tian Y, Liu Y, Qiu Y. miR-185-3p regulates the invasion and metastasis of nasopharyngeal carcinoma by targeting WNT2B in vitro. Oncol Lett. 2017;13:2631–2636. doi: 10.3892/ol.2017.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li G, Wang Y, Liu Y, Su Z, Liu C, Ren S, Deng T, Huang D, Tian Y, Qiu Y. miR-185-3p regulates nasopharyngeal carcinoma radioresistance by targeting WNT2B in vitro. Cancer Sci. 2014;105:1560–1568. doi: 10.1111/cas.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akram M. Mini-review on glycolysis and cancer. J Cancer Educ. 2013;28:454–457. doi: 10.1007/s13187-013-0486-9. [DOI] [PubMed] [Google Scholar]
- 21.Li XB, Gu JD, Zhou QH. Review of aerobic glycolysis and its key enzymes-new targets for lung cancer therapy. Thorac Cancer. 2015;6:17–24. doi: 10.1111/1759-7714.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review) Oncol Lett. 2012;4:1151–1157. doi: 10.3892/ol.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coelho RG, Calaca IC, Celestrini DM, Correia-Carneiro AH, Costa MM, Zancan P, Sola-Penna M. Hexokinase and phosphofructokinase activity and intracellular distribution correlate with aggressiveness and invasiveness of human breast carcinoma. Oncotarget. 2015;6:29375–29387. doi: 10.18632/oncotarget.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dai W, Wang F, Lu J, Xia Y, He L, Chen K, Li J, Li S, Liu T, Zheng Y, et al. By reducing hexokinase 2, resveratrol induces apoptosis in HCC cells addicted to aerobic glycolysis and inhibits tumor growth in mice. Oncotarget. 2015;6:13703–13717. doi: 10.18632/oncotarget.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng Y, Lu J. Targeting hexokinase 2 in castration-resistant prostate cancer. Mol Cell Oncol. 2015;2:e974465. doi: 10.4161/23723556.2014.974465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu CL, Qin L, Liu HC, Candas D, Fan M, Li JJ. Tumor cells switch to mitochondrial oxidative phosphorylation under radiation via mTOR-mediated hexokinase II inhibition-a Warburg-reversing effect. PLoS One. 2015;10:e0121046. doi: 10.1371/journal.pone.0121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viticchie G, Agostini M, Lena AM, Mancini M, Zhou H, Zolla L, Dinsdale D, Saintigny G, Melino G, Candi E. p63 supports aerobic respiration through hexokinase II. Proc Natl Acad Sci USA. 2015;112:11577–11582. doi: 10.1073/pnas.1508871112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Y, Lu N, Qiao C, Ni T, Li Z, Yu B, Guo Q, Wei L. FV-429 induces apoptosis and inhibits glycolysis by inhibiting Akt-mediated phosphorylation of hexokinase II in MDA-MB-231 cells. Mol Carcinog. 2016;55:1317–1328. doi: 10.1002/mc.22374. [DOI] [PubMed] [Google Scholar]
- 29.Katabi MM, Chan HL, Karp SE, Batist G. Hexokinase type II: A novel tumor-specific promoter for gene-targeted therapy differentially expressed and regulated in human cancer cells. Hum Gene Ther. 1999;10:155–164. doi: 10.1089/10430349950018959. [DOI] [PubMed] [Google Scholar]
- 30.Brown RS, Goodman TM, Zasadny KR, Greenson JK, Wahl RL. Expression of hexokinase II and Glut-1 in untreated human breast cancer. Nucl Med Biol. 2002;29:443–453. doi: 10.1016/S0969-8051(02)00288-3. [DOI] [PubMed] [Google Scholar]
- 31.Goel A, Mathupala SP, Pedersen PL. Glucose metabolism in cancer. Evidence that demethylation events play a role in activating type II hexokinase gene expression. J Biol Chem. 2003;278:15333–15340. doi: 10.1074/jbc.M300608200. [DOI] [PubMed] [Google Scholar]
- 32.Jin Z, Gu J, Xin X, Li Y, Wang H. Expression of hexokinase 2 in epithelial ovarian tumors and its clinical significance in serous ovarian cancer. Eur J Gynaecol Onco. 2014;35:519–524. [PubMed] [Google Scholar]
- 33.Peschiaroli A, Giacobbe A, Formosa A, Markert EK, Bongiorno-Borbone L, Levine AJ, Candi E, D'Alessandro A, Zolla L, Finazzi Agrò A, Melino G. miR-143 regulates hexokinase 2 expression in cancer cells. Oncogene. 2013;32:797–802. doi: 10.1038/onc.2012.100. [DOI] [PubMed] [Google Scholar]
- 34.Guo W, Qiu Z, Wang Z, Wang Q, Tan N, Chen T, Chen Z, Huang S, Gu J, Li J, et al. MiR-199a-5p is negatively associated with malignancies and regulates glycolysis and lactate production by targeting hexokinase 2 in liver cancer. Hepatology. 2015;62:1132–1144. doi: 10.1002/hep.27929. [DOI] [PubMed] [Google Scholar]
- 35.Zhou P, Chen WG, Li XW. MicroRNA-143 acts as a tumor suppressor by targeting hexokinase 2 in human prostate cancer. Am J Cancer Res. 2015;5:2056–2063. [PMC free article] [PubMed] [Google Scholar]
- 36.Li LQ, Yang Y, Chen H, Zhang L, Pan D, Xie WJ. MicroRNA-181b inhibits glycolysis in gastric cancer cells via targeting hexokinase 2 gene. Cancer Biomark. 2016;17:75–81. doi: 10.3233/CBM-160619. [DOI] [PubMed] [Google Scholar]
- 37.Wu Y, He H, Wu B, Wen J, Guo Z, Luo Y, Cao G. miR-125b suppresses the aerobic glycolysis of osteosarcoma HOS cells by downregulating the expression of hexokinase-2. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2017;33:1365–1370. (In Chinese) [PubMed] [Google Scholar]
- 38.Cates JMM. Simple staging system for osteosarcoma performs equivalently to the AJCC and MSTS systems. J Orthop Res. 2018;36:2802–2808. doi: 10.1002/jor.24032. [DOI] [PubMed] [Google Scholar]
- 39.Garcia SN, Guedes RC, Marques MM. Unlocking the potential of HK2 in cancer metabolism and therapeutics. Curr Med Chem. 2018 Dec 12; doi: 10.2174/0929867326666181213092652. doi: 10.2174/0929867326666181213092652 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 40.Gentilin E, Degli Uberti E, Zatelli MC. Strategies to use microRNAs as therapeutic targets. Best Pract Res Clin Endocrinol Metab. 2016;30:629–639. doi: 10.1016/j.beem.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 41.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101:2309–2315. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, Ma Y, Hou X, Liu Y, Li K, Xu S, Wang J. MiR-185 acts as a tumor suppressor by targeting AKT1 in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8:11854–11862. [PMC free article] [PubMed] [Google Scholar]
- 44.Tang H, Liu P, Yang L, Xie X, Ye F, Wu M, Liu X, Chen B, Zhang L, Xie X. miR-185 suppresses tumor proliferation by directly targeting E2F6 and DNMT1 and indirectly upregulating BRCA1 in triple-negative breast cancer. Mol Cancer Ther. 2014;13:3185–3197. doi: 10.1158/1535-7163.MCT-14-0243. [DOI] [PubMed] [Google Scholar]
- 45.Xia D, Li X, Niu Q, Liu X, Xu W, Ma C, Gu H, Liu Z, Shi L, Tian X, et al. MicroRNA-185 suppresses pancreatic cell proliferation by targeting transcriptional coactivator with PDZ-binding motif in pancreatic cancer. Exp Ther Med. 2018;15:657–666. doi: 10.3892/etm.2017.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong-Xu W, Jia L, Su-Juan Z. MicroRNA-185 is a novel tumor suppressor by negatively modulating the Wnt/β-catenin pathway in human colorectal cancer. Indian J Cancer. 2015;52(Suppl 3):E182–E185. doi: 10.4103/0019-509X.186576. [DOI] [PubMed] [Google Scholar]
- 47.Wilson JE. Isozymes of mammalian hexokinase: Structure, subcellular localization and metabolic function. J Exp Biol. 2003;206:2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- 48.Anderson M, Marayati R, Moffitt R, Yeh JJ. Hexokinase 2 promotes tumor growth and metastasis by regulating lactate production in pancreatic cancer. Oncotarget. 2016;8:56081–56094. doi: 10.18632/oncotarget.9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katagiri M, Karasawa H, Takagi K, Nakayama S, Yabuuchi S, Fujishima F, Naitoh T, Watanabe M, Suzuki T, Unno M, Sasano H. Hexokinase 2 in colorectal cancer: A potent prognostic factor associated with glycolysis, proliferation and migration. Histol Histopathol. 2017;32:351–360. doi: 10.14670/HH-11-799. [DOI] [PubMed] [Google Scholar]
- 50.Patra KC, Wang Q, Bhaskar PT, Miller L, Wang Z, Wheaton W, Chandel N, Laakso M, Muller WJ, Allen EL, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24:213–228. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lv X, Yao L, Zhang J, Han P, Li C. Inhibition of microRNA-155 sensitizes lung cancer cells to irradiation via suppression of HK2-modulated glucose metabolism. Mol Med Rep. 2016;14:1332–1338. doi: 10.3892/mmr.2016.5394. [DOI] [PubMed] [Google Scholar]
- 52.Lin Q, Ma L, Liu Z, Yang Z, Wang J, Liu J, Jiang G. Targeting microRNAs: A new action mechanism of natural compounds. Oncotarget. 2017;8:15961–15970. doi: 10.18632/oncotarget.14392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao L, Zhang Y, Liu J, Yin W, Jin D, Wang D, Zhang W. MiR-185 inhibits cell proliferation and invasion of non-small cell lung cancer by targeting KLF7. Oncol Res. 2018 May 1; doi: 10.3727/096504018X15247341491655. 10.3727/096504018X15247341491655 (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afshar S, Najafi R, Sedighi Pashaki A, Sharifi M, Nikzad S, Gholami MH, Khoshghadam A, Amini R, Karimi J, Saidijam M. MiR-185 enhances radiosensitivity of colorectal cancer cells by targeting IGF1R and IGF2. Biomed Pharmacother. 2018;106:763–769. doi: 10.1016/j.biopha.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 55.Jiang CY, Ruan Y, Wang XH, Zhao W, Jiang Q, Jing YF, Han BM, Xia SJ, Zhao FJ. MiR-185 attenuates androgen receptor function in prostate cancer indirectly by targeting bromodomain containing 8 isoform 2, an androgen receptor co-activator. Mol Cell Endocrinol. 2016;427:13–20. doi: 10.1016/j.mce.2016.02.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.