Abstract
MicroRNAs (miRNAs) are small, noncoding RNAs that are ~22 nucleotides in length. Accumulating evidence has revealed that miRNAs act as oncogenes or tumor suppressors in various human cancers. In order to investigate the role of miR-195- in squamous cell lung cancer (SQCLC) cells, and to determine the underlying mechanism, the present study utilized RT-qPCR, western blot analysis, luciferase assay, MTT assay, cell migration assay, and in vitro angiogenesis techniques. The results obtained revealed that miR-195-5p acted as a tumor suppressor in SQCLC cells. The expression levels of miR-195 were decreased in two SQCLC cell lines (H520 and SK-Mes-1) compared with a normal lung cell line, and miR-195 directly targeted the 3′-untranslated region of vascular endothelial growth factor (VEGF) in SQCLC cells. Additionally, miR-195 upregulation suppressed the viability and migration of SQCLC cells. Furthermore, miR-195 inhibited the growth and tube formation of endothelial vascular cells. Collectively, the findings indicated that miR-195 downregulated VEGF, and that targeting this miRNA may provide an effective approach to inhibit angiogenesis in tumors.
Keywords: microRNA-195, squamous cell lung cancer, vascular endothelial growth factor
Introduction
MicroRNAs (miRNAs/miRs) are small, noncoding RNAs that negatively regulate gene expression at the post-transcriptional level (1,2). As previous studies have reported, miRNAs bind to the 3-untranslated region (3′-UTR) of target mRNAs (3), resulting in degradation or inhibited translation of the target mRNA (4–6). miRNAs have been reported to serve important roles in tumorigenesis (7,8).
Lung cancer has been a major health problem in developed countries for several decades, and has emerged recently as the leading cause of cancer death in many developing countries (9). Squamous cell lung cancer (SQCLC) is a common type of lung cancer, accounting for ~40,000 deaths in the USA in 2013 (10). The 5-year survival of patients with SQCLC is only 16% (10). It has been reported that patients with SQCLC tend to be older, typically displaying advanced stages of the disease (11,12). SQCLC is closely associated with smoking, with the majority of patients exhibiting centrally located tumors (12) that are locally aggressive, and which frequently lack actionable genetic alterations. As a result, targeted agents developed for lung adenocarcinoma are largely ineffective against SQCLC. Despite the efforts that have been made in the study of lung cancer in recent decades, the molecular mechanisms underlying SQCLC are yet to be determined.
miR-195 is a member of the miR-15/16 family, which comprises a group of miRNAs: miR-195, miR-15a, miR-15b, miR-16-1 and miR-16-2 (13). The functions of miR-195 in non-small cell lung carcinoma (NSCLC) cells targeting insulin-like growth factor 1 receptor (14) and checkpoint kinase 1 (CHEK1) (15) have been previously reported; additionally, it has been revealed to be a tumor suppressor that inhibited tumor cell viability and migration (15). miR-195 has been demonstrated to suppress osteosarcoma cell metastasis by targeting cyclin D1 (CCND1) (16), the role of miR-195 in SQCLC remains unclear.
In the present study, miR-195 was revealed to serve an important role in tumorigenesis. First, it was observed that the levels of miR-195 were decreased in SQCLC cell lines, whereas it was highly expressed in a control cell line. Second, as it was previously reported that vascular endothelial growth factor (VEGF) was a direct target of miR-195 (17), the present study revealed that miR-195 regulated the expression of VEGF in SQCLC cells. Collectively, the results of the present study suggested that miR-195 functioned as a tumor suppressor in SQCLC.
Materials and methods
Cell lines
Two squamous carcinoma cell lines [H520 and SK-Mes-1 (Mes-1)] and a normal lung bronchus epithelial cell line (Beas-2B) were obtained from the American Type Culture Collection and cultured in RPMI 1640 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; PAN-Biotech GmbH), 100 U/ml penicillin, and 100 U/ml streptomycin at 37°C in an atmosphere of 5% CO2. The human umbilical vein endothelial cells (HUVECs) were isolated from umbilical cord vein by collagenase treatment. The HUVECs were cultured in EGM2 (Lonza Group, Ltd.). 293T cells obtained from the American Type Culture Collection were maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1% penicillin/streptomycin (complete media) and incubated at 37C and 5% CO2.
Plasmids
miR-195 mimic (forward: 5′UAGCAGCACAGAAAUAUUGGC3′, reverse: 3′AUCGUCGUGUCUUUAUAACCG5′) and inhibitor (forward: 5′GCCAAUAUUUCUGUGCUGCUA3′), and their respective negative controls (NCs) (micrON™ mimic Negative Control #22: 5′UUUGUACUACACAAAAGUACUG3′, 3′AAACAUGAUGUGUUUUCAUGAC5′ and micrOFF™ inhibitor Negative Control #22: 5′CAGUACUUUUGUGUAGUACAAA3′) were obtained from Guangzhou RiboBio Co., Ltd. The wild-type (WT) and mutant (MUT) 3′-UTR of VEGF were inserted into the GV272 (http://www.genechem.com.cn/service/index.php?ac=gene&at=vector_search&keyword=gv272) firefly luciferase plasmid by Shanghai GeneChem Co., Ltd.
Luciferase assay
Targets can version 7.2 (http://www.targetscan.org/) and miRanda (http://www.microrna.org/) were used for miRNA target prediction and analysis. In order to confirm the association between miR-195 and VEGF, the following experiment was designed: The full-length WT VEGF 3′-UTR was cloned (Shanghai GeneChem Co., Ltd.) and inserted into a luciferase reporter plasmid (GV272), downstream from the firefly luciferase gene. CV045 Renilla was co-transfected as internal reference. A mutated VEGF 3′-UTR (Shanghai GeneChem Co., Ltd.), cloned as control, was inserted into the same plasmid backbone. Subsequently, miRNA mimic or inhibitor, or their respective NC RNAs, with CV045 (http://www.genechem.com.cn/service/index.php?ac=gene&at=vector_search&keyword=CV045) were co-transfected with the constructed plasmids into 293T cells. The 293T cells were cultured for 24 h, and subsequently harvested for Renilla and firefly luciferase activity assays using the Dual-Luciferase Report Assay system (cat. no. E1910; Promega Corporation). The firefly luciferase activity to Renilla luciferase activity.
miRNA transfection
miR-195 NC, miR-195 mimic, miR-195 inhibitor NC or miR-195 inhibitor were purchased from Guangzhou RiboBio Co., Ltd. Mes1 cells or HUVECs were seeded in 12-well plates (8×104 cells/well) for 12 h. For transfection, cells were transfected with 100 nM miR-195 NC, miR-195 mimic, miR-195 inhibitor NC or miR-195 inhibitor using Lipofectamine® 2000 (2 µl; 11668-019, Invitrogen; Thermo Fisher Scientific, Inc.). After 24 h Samples were collected for quantification of miRNA or protein expression.
Construction of Mes-1-miR-control (Mes1-control), Mes-1-miR-195 mimic (Mes1-195) and Mes-1-miR-195-inhibitor (Mes1-195-inhibitor) cell lines
LV3(H1/GFP&Puro) expressing plasmids encoding miR-195 mimic, miR-195 inhibitor and miR-control, and polybrene were purchased from Shanghai GenePharma Co., Ltd. 293T cells were seeded in 10-cm plates (1×106 cells per well) and incubated at 37°C overnight, and subsequently the cells were transfected with the packaging plasmid (containing the vsv-g, rev, and rre genes) and the target plasmids. The packaging plasmid and the target plasmids were purchased from Shanghai GenePharma Co., Ltd. Following culture for 72 h, the lentiviral supernatant was harvested. Mes-1 cells were seeded into 6-cm plates (2×105 cells per well), incubated overnight, and subsequently the cells were infected with lentiviral supernatant s(1×107 TU/ml) containing miR-195-mimic, miR-195-inhibitor or miR-control, and 5 µg/ml polybrene at 37°C in an atmosphere of 5% CO2 for 12 h. The culture medium was then removed and replaced with fresh medium containing 10% FBS; the incubation was continued for a further 48 h. The infected cells expressed GFP, and transformed cells were selected in the presence of puromycin (10 µg/ml). Therefore, three stable cell lines, Mes1-control, Mes1-195 and Mes1-195-inhibitor, were generated. The transduction efficiency was evaluated via reverse transcription-quantitative PCR (RT-qPCR) analysis to determine miR-195 expression.
RNA isolation and RT-qPCR
Total cells were collected and homogenized in TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), and subsequently total RNA was extracted from the cultured cells using TRIzol, according to the manufacturer's protocol. miRNAs was extracted with an miRNA isolation kit was (Qiagen GmbH). miRNAs and total RNA were reverse-transcribed into cDNA using a Reverse Transcription kit (Takara Biotechnology Co., Ltd.). The reverse transcription reaction was performed under the following conditions: 37°C for 30 min; then 85°C for 5 sec; and holding at 4°C. For qPCR amplification of the cDNA, the following thermocycling conditions were used: 30 sec at 95°C, followed by 40 cycles of 95°C for 10 sec and 60°C for 1 min. qPCR was performed using a SYBR Premix Ex Taq II kit (Takara Biotechnology Co., Ltd.) on a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories Inc.). Sense and antisense primer sequences for VEGF and β-actin were as follows: VEGF, forward 5′-ATCTTCAAGCCATCCTGTGTGC-3′, reverse 5′-GCTCACCGCCTCGGCTTGT-3′; and β-actin, forward 5′-CACATCGCTCAGACACCA-3′, reverse 5′-ATGGCAACAATATCCACTTT-3′. The miRNA primers (RT, and forward and reverse primers for miR-195- and U6) were purchased from Guangzhou RiboBio Co., Ltd. Relative expression of miR-195-5p was evaluated using the 2−∆∆Cq method (18) with U6 small nuclear RNA used for normalization; β-actin used for normalization of VEGF expression.
Western blot analysis
The cells were lysed in RIPA buffer (Beijing Solarbio Science & Technology Co., Ltd.) supplemented with protease inhibitors (cat. no. 4693116001; Roche Diagnostics). Following cell lysis, the lysates were centrifuged at 12,000 × g at 4°C for 20 min. The protein concentration was measured using the BCA method. The proteins (40 µg per lane) were separated via 12% SDS-PAGE and subsequently transferred on to PVDF membranes (Roche Diagnostics). Membranes were blocked with 5% non-fat milk powder in TBS-T buffer (20 mM Tris-HCl, pH 7.4, 137 mM NaCl, and 0.1% Tween) for 1 h at room temperature. The samples were incubated with primary antibodies at 4°C overnight, followed by subsequent incubation with horseradish peroxidase-conjugated secondary antibodies. The films were developed using an enhanced chemiluminescence system (EMD Millipore). ImageJ version 1.48 (National Institutes of Health) was used to quantify protein expression. Polyclonal anti-VEGF (1:1,000; cat. no. sc-152; Santa Cruz Biotechnology, Inc.) and anti-β-actin (1:1,000; cat. no. 8457; Cell Signaling Technology, Inc.) primary antibodies, and mouse anti-rabbit secondary antibodies (1:10,000; cat. no. sc-2357; Santa Cruz Biotechnology, Inc.) were used during the study.
Cell migration assays
Cell migration assays were performed using Transwell chambers. The cells were suspended in serum-free F12 medium (Gibco; Thermo Fisher Scientific, Inc.) and seeded in the upper chambers at a total density of 0.8×105 cells/well; 500 µl F12 medium with 10% FBS was added to the lower chambers, and the Transwell plates were incubated at 37°C for 12 h. Cells that remained on the upper surfaces of the membranes were removed using cotton swabs, and the migrated cells on the lower surface were fixed with 4% paraformaldehyde for 10 min, followed by staining with 0.1% crystal violet (Beyotime Institute of Biotechnology) for 15 min at room temperature. Images of migrated cells were captured using a light microscope (BX61; Olympus Corporation). Digital images (magnification, ×10) of the underside of the inserts were acquired with the microscope. A total of 5 random fields were captured from each membrane.
Cell viability
The MTT assay was used to examine the viability of Mes-1 cells that were stably transfected as aforementioned. A total of 2,000 cells/well were plated in 96-well plates and incubated at 37°C. Cell viability was measured by MTT reagent, Cells was measured after cultured 1, 2, 3 and 4 days. They were gently washed with PBS, and 20 µl MTT (5 mg/ml) were added in the cell culture. After 4 h incubation, the media were discarded, and 150 µl DMSO was added in each well to dissolve the precipitates and the absorbance at 560 nm was measured using a microplate reader (Promega Corporation).
In vitro angiogenesis assay
Human umbilical vein endothelial cells (HUVECs) were plated in 24-well plates (1×105 cells/well), and cultured with EGM2 (Lonza Group Ltd.) overnight at 37°C in an atmosphere containing 5% CO2. Cells was transfected with miR-195 NC, miR-195 mimic, miR-195 inhibitor NC or miR-195 inhibitor. Following incubation for 6 h, the HUVECs were digested and seeded in 48-well plates(0.3×105 cells/well) containing 50 µl solidified Matrigel™, and incubated at 37°C for 9 h. The cells were stained with 3 µM calcein-acetoxymethyl (Invitrogen; Thermo Fisher Scientific, Inc.) for 30 min at 37°C in the presence of 5% CO2. Formation of capillary-tubule structures was observed, and digital images were captured under a light microscope (magnification, ×100). The branch points of the formed tubes were quantified using Image-Pro Plus 6.0 software (Media Cybernetics, Inc.) from at least five fields.
Statistical analysis
At least three repeats were conducted for each experiment. Data were analyzed with GraphPad Prism 7.0 software (GraphPad Software, Inc.). Data were analyzed using Student's t-test or one-way analysis of variance followed by Dunnett's post hoc test for multiple comparisons. Data are presented as the mean ± standard deviation of three independent experiments. P≤0.05 was considered to indicate a statistically significant difference.
Results
VEGF is upregulated in SQCLCs
RT-qPCR analysis revealed that VEGF was upregulated in the SQCLC cell lines, H520 and Mes-1, compared with in the normal lung bronchus epithelial cell line, Beas-2B (Fig. 1A). Furthermore, the expression of miR-195 in the two SQCLC cell lines was significantly downregulated (P<0.001; Fig. 1B).
Figure 1.


Expression of VEGF and miR-195 in SQCLC. Expression of (A) VEGF and (B) miR-195-5p in SQCLC and control cell lines, as determined via reverse transcription-quantitative PCR analysis. Data are presented as the mean ± standard deviation of three independent experiments. ***P<0.001. VEGF, vascular endothelial growth factor; SQCLC, squamous cell lung cancer; miR-195-5p, microRNA-195-5p; Mes-1, SK-Mes-1.
VEGF is a target of miR-195
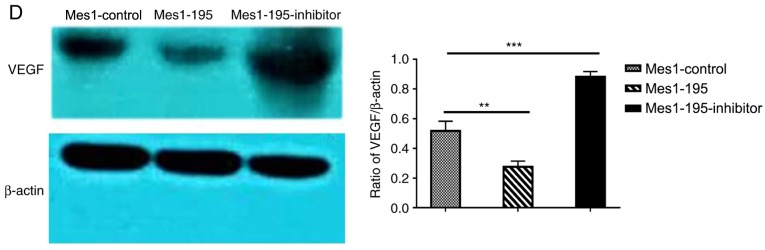
The position of the miR-195 target site in VEGF, and the mutated site that was designed in the clone, are presented in Fig. 2A. 293T cells were transfected with miR-195 mimic, miR-195 NC, miR-195 inhibitor or miR-195 inhibitor NC; the transfection efficiency was presented in Fig. 2B. The results of the luciferase activity experiments revealed that the levels of luciferase activity were significantly reduced in the miR-195 mimic group cells compared with the miR-195 NC group (Fig. 2B). Conversely, the levels of luciferase activity in the miR-195 inhibitor group cell were not significantly different to those in the miR-195 inhibitor NC group (Fig. 2B). Of note, the inhibitory effects of miR-195 mimic, as determined from the luciferase assay, were lost when the predicted binding site was mutated (Fig. 2B). Subsequently, the Mes-1 cell line was used to generate stable cells with altered miR-195 expression, Mes1-195, Mes1-195-inhibitor and Mes1-control, via lentiviral transduction. The expression levels of miR-195 and VEGF were evaluated in these cell lines. The results demonstrated that, compared with Mes1-control, miR-195 was significantly upregulated in the Mes1-195 cell line, and downregulated in the Mes1-195-inhibitor cell line (Fig. 2C). Subsequently, western blot analysis was conducted to investigate the protein expression levels of VEGF for the different experimental groups. VEGF was significantly downregulated in the Mes1-195 cell line, whereas it was upregulated in the Mes1-195-inhibitor cell line compared with the Mes1-control cell line (Fig. 2D). Collectively, these data indicated that miR-195 directly interacted with the VEGF 3′-UTR, inhibiting VEGF expression.
Figure 2.


VEGF is a target of miR-195 in SQCLC cells. (A) Position of the miR-195 binding site in VEGF, and the Mut sequence. (B) 293T cells were co-transfected with a firefly luciferase reporter containing the WT or Mut VEGF 3′-UTR, and miR-195 mimic, miR-195 inhibitor or the corresponding NCs. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01, ***P<0.001, ****P<0.0001. (C) Relative levels of miR-195 in Mes-1 cells infected with lentivirus encoding miR-control, miR-195 mimic or miR-195 inhibitor plasmids, determined by reverse transcription-quantitative PCR. (D) Western blot analysis of changes in VEGF protein expression levels in Mes-1 cells infected with the aforementioned lentiviruses. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01; ***P<0.001. miR-, microRNA; VEGF, vascular endothelial growth factor; SQCLC, squamous cell lung cancer; Mes-1/Mes1, SK-Mes-1; WT, wild-type; Mut, mutant; NC, negative control; Mes1-control, Mes-1 cells transfected with miR-control; Mes1-195, Mes-1 cells transfected with miR-195 mimic; Mes1-195-inhibitor, Mes-1 cells transfected with miR-195 inhibitor.
miR-195 inhibits cell viability
Following generation of the Mes1-control, Mes1-195 and Mes1-195-inhibitor cell lines, they were subsequently used to measure cell viability, which was determined using an MTT assay. The results revealed that the number of viable cells was significantly reduced in the Mes1-195 cell line compared with the Mes1-control cell line (Fig. 3A). Conversely, the number of viable cells in the Mes1-195-inhibitor group was significantly increased compared with the Mes1-control group (Fig. 3B).
Figure 3.

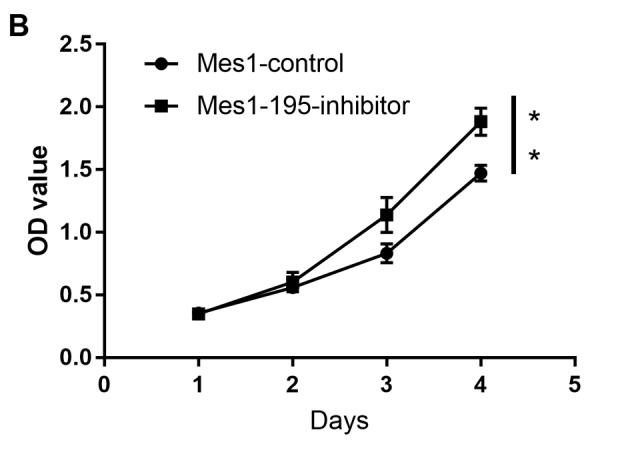
Effects of miR-195 on Mes-1 cell viability. Viability of (A) Mes1-195 and (B) Mes1-195-inhibitor cells compared with Mes1-control cells, as determined via an MTT assay. Data are presented as the mean ± standard deviation of three independent experiments. **P<0.01 vs. Mes1-control. miR-, microRNA; Mes1-control, Mes-1 cells transfected with miR-control; Mes1-195, Mes-1 cells transfected with miR-195 mimic; Mes1-195-inhibitor, Mes-1 cells transfected with miR-195 inhibitor; OD, optical density.
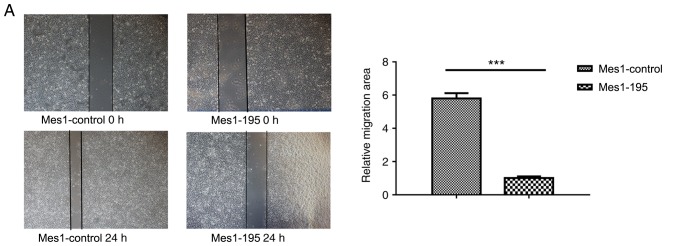
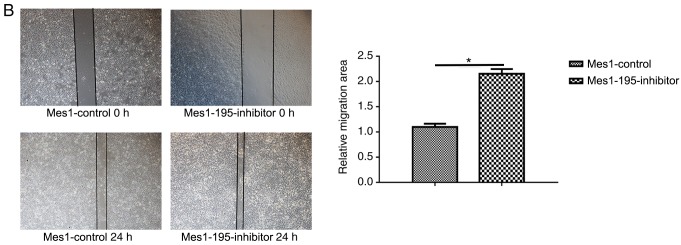
Expression of miR-195 affects cell migration Subsequently, wound healing and Transwell assays were performed using the cell lines
A wound healing assay revealed that miR-195 overexpression significantly inhibited cell migration by ~6-fold in the Mes-1 cell line (Fig. 4A). Conversely, when miR-195 expression was inhibited, cell migration was significantly promoted by ~2-fold compared with Mes1-control cells (Fig. 4B).
Figure 4.
Effects of miR-195 on Mes-1 cell viability migration. Wound healing assays were performed to measure the migration of Mes-1 cells transfected with (A) miR-195 mimic and (B) miR-195 inhibitor compared with the control. Quantitative analysis of cell migration is presented on the right. (C) Transwell assays were performed with the aforementioned cell lines; quantitative analysis of the number of invasive cells is presented on the right. Data are presented as the mean ± standard deviation of three independent experiments. *P<0.05, ***P<0.001 vs. Mes1-control. miR-, microRNA; Mes-1/Mes1, SK-Mes-1; Mes1-control, Mes-1 cells transfected with miR-control; Mes1-195, Mes-1 cells transfected with miR-195 mimic; Mes1-195-inhibitor, Mes-1 cells transfected with miR-195 inhibitor.
In the Transwell assay, it was observed that an increase in miR-195 led to a significant reduction in cell invasion, whereas a decrease in miR-195 promoted cell invasion (Fig. 4C).
miR-195 inhibits angiogenesis in vitro
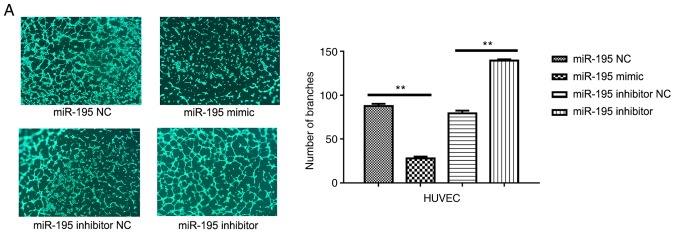
To investigate the effects of miR-195 on angiogenesis, HUVECs were transfected with miR-195 mimic (Fig. 5). It was revealed that miR-195 overexpression significantly inhibited the ability of the HUVECs to form capillary-like tubules on a Matrigel coating (Fig. 5A). Conversely, when miR-195 inhibitor was overexpressed in HUVECs, it was revealed that the cells exhibited a significantly enhanced capacity for tubule formation (Fig. 5A). The protein expression levels of VEGF in the transfected HUVECs were also detected via western blot analysis (Fig. 5B). It was observed that the presence of the miR-195 mimic led to a significant reduction in the expression levels of VEGF compared with the control, whereas the presence of the miR-195 inhibitor led to an increase in the VEGF levels compared with the control. The transfection efficiency of the transfected HUVECs was presented in Fig. 5C.
Figure 5.
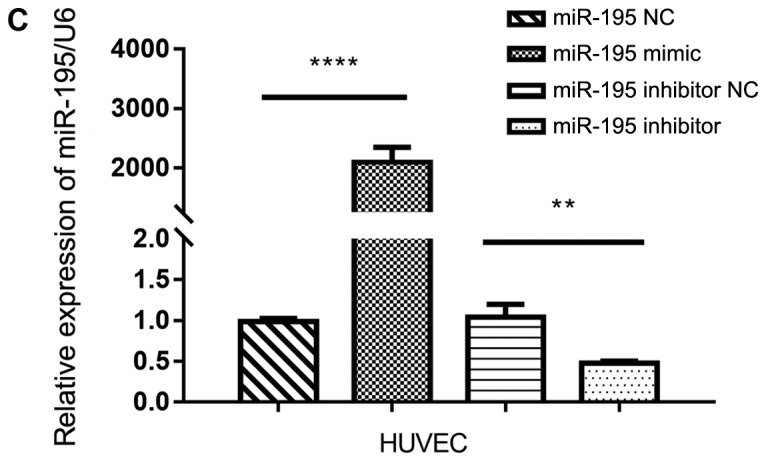
miR-195 suppresses angiogenesis in the tumor microenvironment. (A) Representative images of HUVECs in Matrigel. Quantitative analysis of the number of branches is presented on the right. Data are presented as the mean ± standard deviation. of three independent experiments. **P<0.01. (B) Western blot analysis of the protein expression levels of VEGF in transfected cells; quantitative analysis of expression is presented on the right. Data are presented as the mean ± standard deviation of three independent experiments. ***P<0.001. VEGF, vascular endothelial growth factor. (C) Relative levels of miR-195 in HUVECs transfected with miR-195 NC, miR-195 mimic, miR-195 inhibitor, or miR-195 inhibitor NC, as determined via reverse transcription-quantitative PCR analysis. **P<0.01, ****P<0.0001. Data are presented as the mean ± standard deviation of three independent experiments. HUVECs, human umbilical vein endothelial cells; miR-195, microRNA-195; NC, negative control; VEGF, vascular endothelial growth factor.
Discussion
At present, there are no effective targeted drug therapies for SQCLC. Angiogenesis is critical for tumor progression, whereas metastasis is the major cause of tumor recurrence and patient mortality (17). Therefore, miRNAs that induce anti-angiogenic or anti-metastatic effects may provide novel targets for anticancer therapies. The function of miR-195 has been reported in prostate cancer (19), hepatocellular carcinoma (HCC) (20), osteosarcoma (21) and breast cancer (22). Accumulating evidence has indicated an important role for miRNAs in the progression of NSCLC (23,24); however, the role of miR-195 has not been determined in SQCLC. A role for miR-195 was first identified in HCC by Wang et al (17). The effects of miR-195 on the expression of CHEK1 (15), CCND1 (16), ribosomal protein S6 kinase b1 (19) and PHD finger protein 19 (20) have also previously been reported.
The present study was a preliminary investigation into the function of miR-195 in SQCLC. In this study, the tumor-suppressive function of miR-195 in SQCLC development and progression in vitro was revealed. Downregulated expression of miR-195 was observed in SQCLC cell lines compared with the control. Upon overexpression of miR-195, suppression of Mes-1 cell viability, migration, invasion and angiogenesis in HUVECs was reported. Conversely, when the expression of miR-195 was downregulated, opposing effects were observed. In addition, VEGF was identified as a target of miR-195 in SQCLC cells. Collectively, these data suggested that miR-195 is a potential therapeutic target for SQCLC.
In conclusion, the results of the present study have revealed that miR-195 was significantly decreased in SQCLC cell lines, and that miR-195 overexpression suppressed the viability, migration and invasion of Mes-1 cells, and angiogenesis of HUVECs, potentially by targeting VEGF. miR-195 has been reported to promote apoptosis or inhibit proliferation in various types of cancer (15–17,19). The present study suggested that miR-195 may be potentially involved in the pathophysiology of SQCLC.
Acknowledgements
Not applicable.
Funding
This study was supported by a Cancer Translational Medicine Seed Fund (grant no. 1606), and the Science & Technology Development Fund of the Tianjin Education commission for Higher Education (grant no. 2017ZD11).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
HLL conducted the experimental work and designed the study, reviewed the literature and drafted the manuscript. YLC, YL and CGL contributed to the design and coordination of experimental work, and acquisition of data. TTQ, MB, ZFZ and RJ participated in the study design, data collection, analysis of data and preparation of the manuscript. HLL, CLW and YJS analyzed the data and drafted the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gandellini P, Doldi V, Zaffaroni N. microRNAs as players and signals in the metastatic cascade: Implications for the development of novel anti-metastatic therapies. Semin Cancer Biol. 2017;44:132–140. doi: 10.1016/j.semcancer.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou W, Wang S, Ying Y, Zhou R, Mao P. miR-196b/miR-1290 participate in the antitumor effect of resveratrol via regulation of IGFBP3 expression in acute lymphoblastic leukemia. Oncol Rep. 2017;37:1075–1083. doi: 10.3892/or.2016.5321. [DOI] [PubMed] [Google Scholar]
- 4.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Semin Cancer Biol. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Kai K, Dittmar RL, Sen S. Secretory microRNAs as biomarkers of cancer. Semin Cell Dev Biol. 2018;78:22–36. doi: 10.1016/j.semcdb.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Nambara S, Mimori K. MicroRNA in various aspects of cancer development. Gan To Kagaku Ryoho. 2017;44:362–366. (In Japanese) [PubMed] [Google Scholar]
- 7.Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene. 2014;33:135–147. doi: 10.1038/onc.2013.54. [DOI] [PubMed] [Google Scholar]
- 8.Su SF, Chang YW, Andreu-Vieyra C, Fang JY, Yang Z, Han B, Lee AS, Liang G. miR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene. 2013;32:4694–4701. doi: 10.1038/onc.2012.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Zhu J, Zhang YH, Chen YS, Ding LL, Kensler TW, Chen JG. Lung cancer in a rural area of China: Rapid rise in incidence and poor improvement in survival. Asian Pac J Cancer Prev. 2015;16:7295–7302. doi: 10.7314/APJCP.2015.16.16.7295. [DOI] [PubMed] [Google Scholar]
- 10.Yamano S, Gi M, Tago Y, Doi K, Okada S, Hirayama Y, Tachibana H, Ishii N, Fujioka M, Tatsumi K, Wanibuchi H. Role of deltaNp63posCD44vpos cells in the development of N-nitroso-tris-chloroethylurea-induced peripheral-type mouse lung squamous cell carcinomas. Cancer Sci. 2016;107:123–132. doi: 10.1111/cas.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang YC, Zhou Q, Wu YL. Emerging challenges of advanced squamous cell lung cancer. ESMO Open. 2016;1:e000129. doi: 10.1136/esmoopen-2016-000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Funai K, Yokose T, Ishii G, Araki K, Yoshida J, Nishimura M, Nagai K, Nishiwaki Y, Ochiai A. Clinicopathologic characteristics of peripheral squamous cell carcinoma of the lung. Am J Surg Pathol. 2003;27:978–984. doi: 10.1097/00000478-200307000-00013. [DOI] [PubMed] [Google Scholar]
- 13.Griffiths-Jones S, Saini HK, Van Dongen S, Enright AJ. Enrigh: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Wang Y, Lan H, Li J. MiR-195 inhibits the growth and metastasis of NSCLC cells by targeting IGF1R. Tumour Biol. 2014;35:8765–8770. doi: 10.1007/s13277-014-2140-5. [DOI] [PubMed] [Google Scholar]
- 15.Liu B, Qu J, Xu F, Guo Y, Wang Y, Yu H, Qian B. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6:9445–9456. doi: 10.18632/oncotarget.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han K, Chen X, Bian N, Ma B, Yang T, Cai C, Fan Q, Zhou Y, Zhao TB. MicroRNA profiling identifies MiR-195 suppresses osteosarcoma cell metastasis by targeting CCND1. Oncotarget. 2015;6:8875–8889. doi: 10.18632/oncotarget.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R, Zhao N, Li S, Fang JH, Chen MX, Yang J, Jia WH, Yuan Y, Zhuang SM. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology. 2013;58:642–653. doi: 10.1002/hep.26373. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Cai C, Chen QB, Han ZD, Zhang YQ, He HC, Chen JH, Chen YR, Yang SB, Wu YD, Zeng YR, et al. miR-195 inhibits tumor progression by targeting RPS6KB1 in human prostate cancer. Clin Cancer Res. 2015;21:4922–4934. doi: 10.1158/1078-0432.CCR-15-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu H, Hu YW, Zhao JY, Hu XM, Li SF, Wang YC, Gao JJ, Sha YH, Kang CM, Lin L, et al. MicroRNA-195-5p acts as an anti-oncogene by targeting PHF19 in hepatocellular carcinoma. Oncol Rep. 2015;34:175–182. doi: 10.3892/or.2015.3957. [DOI] [PubMed] [Google Scholar]
- 21.Cai H, Zhao H, Tang J, Wu H. Serum miR-195 is a diagnostic and prognostic marker for osteosarcoma. J Surg Res. 2015;194:505–510. doi: 10.1016/j.jss.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 22.Igglezou M, Vareli K, Georgiou GK, Sainis I, Briasoulis E. Kinetics of circulating levels of miR-195, miR-155 and miR-21 in patients with breast cancer undergoing mastectomy. Anticancer Res. 2014;34:7443–7447. [PubMed] [Google Scholar]
- 23.Lin L, Huang Y, Gong W. Inhibition of miR-92b suppresses nonsmall cell lung cancer cells growth and motility by targeting RECK. Mol Cell Biochem. 2014;387:171–176. doi: 10.1007/s11010-013-1882-5. [DOI] [PubMed] [Google Scholar]
- 24.Nishikawa R, Goto Y, Kojima S, Enokida H, Chiyomaru T, Kinoshita T, Sakamoto S, Fuse M, Nakagawa M, Naya Y, et al. Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int J Oncol. 2014;45:401–410. doi: 10.3892/ijo.2014.2437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.