Abstract
Complement factor H-related 3 (CFHR3) belongs to the human factor H protein family and is associated with various human diseases, including nephropathy, age-related macular degeneration and atypical hemolytic uremic syndrome. However, to the best of our knowledge, the role of CFHR3 in hepatocellular carcinoma (HCC) remains largely unknown. In the present study, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis were performed to determine mRNA and protein expression levels of CFHR3 in HCC and normal adjacent tissue. In addition, CFHR3 was overexpressed in Huh-7 cells and cell counting kit-8 assay was used to determine cell viability. Cell proliferation and apoptosis were assessed using flow cytometry, RT-qPCR and western blotting. The results demonstrated that mRNA (2−ΔΔCq) and protein expression levels of CFHR3 were significantly lower in tumor tissue compared with in adjacent tissue. Additionally, CFHR3 overexpression decreased cell viability, inhibited cell proliferation and significantly increased apoptosis. It was also identified that CFHR3 could downregulate the expression of Ki67. The results suggested that CFHR3 induced apoptosis by downregulating the expression of survivin and B cell lymphoma 2, upregulating the expression of Bcl-2-associated X and promoting caspase-3 activity. Western blotting revealed that CFHR3 significantly inhibited the protein expression levels of phosphorylated (p)-phosphoinositide 3-kinase (PI3K), p-protein kinase B (Akt) and p-mammalian target of rapamycin (mTOR). Overexpression of CFHR3 suppressed proliferation and promoted apoptosis of HCC cells by inhibiting the PI3K/Akt/mTOR signaling pathway.
Keywords: CFHR3, HCC, apoptosis, proliferation, PI3K/Akt/mTOR
Introduction
Liver cancer is one of the commonest types of cancer, with ~850,000 new cases being diagnosed annually worldwide (1). Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer and accounts for ~90% of all liver cancer cases (2). HCC is the second leading cause of cancer related mortality in China (3). The high rate of mortality could be explained by many risk factors, including hepatitis B and C virus infections as well as excessive alcohol intake and chronic liver injury, all of which can contribute to the development of HCC (4–6). A previous study demonstrated that complement factor H-related (CFHR) 3 is highly expressed in normal liver tissue, but not in other tissues (7). This tissue-specific expression led to the hypothesis that CFHR3 may serve a role in HCC.
CFHR3 belongs to the human factor H protein family, which contains complement regulators, including complement factor H (CFH) and CFH-like protein 1 as well as other CFHR proteins (8). CFH family proteins have individual folding domains that are called short consensus repeats. These are also reflected in the tandem arrangement of the CFH gene and the five CFHR genes. Notably, CFHR3 and CFHR4 are similar, and have overlapping functions (9–11). Previous studies have reported that CFH genes are associated with several human diseases, including age-related macular degeneration, atypical hemolytic uremic syndrome and membranoproliferative glomerulonephritis type II (12–15). These diseases are increasingly thought to be caused by CFH polymorphisms, homozygous or heterozygous mutations and CFHR1/3 deletion (16–18).
Using in-situ hybridization, it was revealed that many tumor cells, including bladder, cervical and renal cancer cell lines, can produce and secrete human CFHR proteins (19,20). However, normal epithelial keratinocytes and colon cancer cell lines do not express CFHR proteins (21). Several studies have suggested that tumor cells evade the immune system by using CFH and its associated proteins (22–24). Cui et al (25), suggested that abnormal expression of the CFHR1 and CFHR3 gene may be related to cisplatin resistance in U251/CP2 gliomas. However, to the best of the authors' knowledge, no studies have been conducted on the relationship between CFHR3 expression and HCC.
The phosphoinositide 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway is known to be regulated in HCC (26). Thus, in the present study, the aim was to determine whether CFHR3 could inhibit liver cancer cells via the PI3K/Akt/mTOR pathway.
Materials and methods
Patients and tissue
Between May 2005 and July 2017, 42 HCC and adjacent normal tissues samples were collected from patients with HCC (age: 55.1±9.2 years; 23 male, 19 female), undergoing surgical resection at the Yantai Infectious Disease Hospital (Yantai, China). Inclusion criteria were HCC diagnosed according to Clinical diagnosis and staging of Primary Hepatocellular carcinoma (27). Exclusion criteria were patients receiving hormones and/or antineoplastic drugs, radiotherapy or chemotherapy within 2 weeks prior to surgery. Tissues were confirmed as HCC following pathological examination. The normal liver tissues were taken at a distance of >2 cm from the edge of the tumor. All HCC specimens and corresponding adjacent normal tissues were collected within 0.5 h of surgical resection. One section of the paired tissues was used for pathological diagnosis and stored in 4% formaldehyde, while the section was stored in liquid nitrogen for later experimentation. This study was approved by the Ethics Committee of Yantai Infectious Disease Hospital (Yantai, China). Written informed consent was obtained from the patients.
The Cancer Genome Atlas (TCGA) database
Another cohort of patients with HCC was obtained from the TCGAdatabase (https://cancergenome.nih.gov/) via the Genomic Data Commons Data Portal. The expression values of genes from RNA-seq data were scaled with log2 (FPKM + 0.01). The CFHR3 expression in human tissues. Using NCBI database (https://www.ncbi.nlm.nih.gov/gene/10878), RNA-seq was performed on tissue samples from 95 human individuals representing 27 different tissues in order to determine tissue-specificity of all protein-coding genes.
Cell culture
The Huh-7 cell line was purchased from Procell Life Science & Technology Co., Ltd., (Wuhan, China). Cells were cultured in RPMI 1640 medium (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) containing 20% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C and 5% CO2. The cells were digested with 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.) and passaged when cells reached 80–90% confluence.
Cell transfection
CFHR3 overexpression and negative control (NC) plasmids were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). Cells were seeded in 6-well plates (3×105 cells/well) one day in RPMI 1640 medium before transfection. Transient transfection was carried out using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Cells were transfected with 20 µM CFHR3 overexpression or NC plasmids using 5 µl Lipofectamine® 3000, in serum-free medium at 25°C for 10 min. Cells treated with 0.1% PBS were used as the control. Following a 6 h incubation at 37°C, the cells were cultured in RPMI 1640 medium containing 10% FBS. Cells were used for subsequent experiments 3 day later.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to examine the gene expression levels of CFHR3, Ki67, survivin, caspase-3, Bcl-2-associated X (Bax), B cell lymphoma 2 (Bcl-2), PI3K, Akt and mTOR. Total RNA was extracted from tissues samples or cultured cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), according to manufacturer's instructions and reversed transcribed to cDNA using the QuantiTect Reverse Transcription kit (Qiagen GmbH, Hilden, Germany). The RT conditions were as follows: 37°C for 60 min and 95°C for 5 min. qPCR was carried out using the SYBR qPCR Mix (Toyobo Life Science, Osaka, Japan), with GAPDH as the internal reference. The thermocycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec, 60°C for 10 sec and 72°C for 10 sec. The primer sequences are listed in Table I. The relative mRNA expression levels were calculated using the 2−ΔΔCq method (28).
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer | Sequence |
|---|---|---|
| CFHR3 | Forward | 5′-TGGGCATTAGTCAAGAATACAGTAAAA-3′ |
| Reverse | 5′-ATTAATGCCGCTTCAATATGACTTT-3′ | |
| Ki67 | Forward | 5′-AAAGTGCCCAAGCCATAGA-3′ |
| Reverse | 5′-CACCATTTGCCAGTTCCTC-3′ | |
| Survivin | Forward | 5′-TCATCCTCACTGCGGCTGTC-3′ |
| Reverse | 5′-AGGTCATCTCGGCTGTTCCTG-3′ | |
| Caspase-3 | Forward | 5′-ACTGGACTGTGGCATTGAG-3′ |
| Reverse | 5′-GAAACAATACATGGAATCTG-3′ | |
| Bax | Forward | 5′-GCGAATTGGAGATGAACT-3′ |
| Reverse | 5′-GTGAGCGAGGCGGTGAGGAC-3′ | |
| Bcl-2 | Forward | 5′-GGTTGCCTTATGTATTTGTTTG-3′ |
| Reverse | 5′-CCTCCGCAATGCTGAAAG-3′ | |
| PI3K | Forward | 5′-ATGCCTGCTCTGTAGTGGTGG-3′ |
| Reverse | 5′-CATTGAGGGAGTCGTTGTGC-3′ | |
| Akt | Forward | 5′-TTGTCATGGAGTACGCCAACG-3′ |
| Reverse | 5′-ACAGCCCGAAGTCTGTGATCTT-3′ | |
| mTOR | Forward | 5′-CCAATCATTCGCATTCAGTCC-3′ |
| Reverse | 5′-AACAAACTCATGTCCGTTGCTG-3′ | |
| GAPDH | Forward | 5′-AATCTCATCACCATATTCCA-3′ |
| Reverse | 5′-CCTGCTTCACCACCTTGTTG-3′ |
Akt, protein kinase B; Bax, Bcl-2-associated X; Bcl-2, B cell lymphoma 2; CFHR3, complement factor H-related 3; mTOR, mammalian target of rapamycin; PI3K, phosphoinositide 3-kinase.
Western blotting
Total protein was extracted from tissues or cultured cells using lysis RIPA buffer (Thermo Fisher Scientific, Inc.), followed by centrifugation at 18,000 × g for 5 min at 4°C. Protein concentration was determined using an Enhanced BCA Protein Assay kit (Beyotime Institute of Biotechnology, Shanghai, China). Proteins (20 µg/lane) were separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes, which were blocked with 5% milk in Tris-buffered saline containing 0.2% Tween-20 (TBST) at room temperature for 2 h. Proteins were incubated with primary antibodies: Rabbit anti-CFHR3 (1:1,000; cat. no. 16583-1-AP; ProteinTech Group, Inc., Chicago, IL, USA), rabbit anti-Bcl-2 (1:1,000; cat. no. ab32124; Abcam, Cambridge, MA, USA), rabbit anti-Bax (1:1,000; cat. no. ab32503; Abcam), rabbit anti-Ki67 antibody (1:1,000; cat. no. ab16667; Abcam), rabbit anti-survivin (1:5,000; cat. no. ab76424; Abcam), rabbit anti-caspase-3 (1:500; cat. no. ab13847; Abcam), rabbit anti-PI3K antibody (1:500; cat. no. orb137259; Wuhan Booute Co. Ltd., Wuhan, China), rabbit anti-p-PI3K antibody (1:1,000; cat. no. 4228; Cell Signaling Technology, Inc., Danvers, MA, USA), rabbit anti-pan-Akt (1:500; cat. no. ab8805; Abcam), rabbit anti-p-Akt antibody (1:2,000; cat. no. 4060; Cell Signaling Technology, Inc.), rabbit anti-mTOR (1:1,000; cat. no. ab137341; Abcam), rabbit anti-p-mTOR antibody (1:1,000; cat. no. 5536; Cell Signaling Technology, Inc.) and rabbit anti-GAPDH (1:1,000; cat. no. ab9485; Abcam) at 4°C overnight. Subsequently, membranes were washed with TBST and incubated with goat anti-rabbit horseradish peroxidase-conjugated antibodies (1:2,000; SA00001-2, ProteinTech Group, Inc., Chicago, IL, USA) at room temperature for 90 min. The protein bands were visualized using an ECL system (Amersham; GE Healthcare, Chicago, IL, USA). Densitometric analysis of western blots was performed using Quantity One® software version 2.4 (Bio-Rad, Laboratories, Inc., Hercules, CA, USA).
Cell Counting Kit-8 (CCK-8) cell viability analysis
Cells were dissociated using 0.25% trypsin, 0, 12, 24 and 48 h after transfection. Then, cells were seeded in 96-well plates at a density of 1×104 cells/well and 10 µl CCK-8 solution was added to the cells for 2 h at 37°C. The optical density was measured at a wavelength of 450 nm using a microplate reader (Bio-Rad, Laboratories, Inc.).
Flow cytometry
Cells were harvested 48 h after transfection, and then digested by 0.25% trypsin without EDTA and collected in 1.5 ml Eppendorf tubes. Cells were washed twice using washing buffer, and then incubated with Annexin V-FITC and propidium iodide (cat. no. 40302ES20; Yeasen, Shanghai, China) in the dark at 25°C for 20 min. Binding buffer was added to each tube and apoptosis analyzed within 1 h. The apoptosis rate is derived from the addition of right upper quadrant and right lower quadrant together.
Cell proliferation was measured using carboxyfluorescein succinimidyl ester (CFSE) (29). Cells were labeled with CellTrace ™ CFSE kit (C34554; Invitrogen; Thermo Fisher Scientific, Inc.) for 10 min at 37°C and then washed twice with phosphate-buffered saline. The cells were incubated for at ≥10 min before analysis to allow the CellTrace™ reagent to undergo acetate hydrolysis. Fluorescence was measured using a flow cytometer and Flow Jo version 10.0 software (FlowJo LLC, Ashland, OR, USA).
Statistical analysis
GraphPad Prism software version 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used to perform statistical analyses. All data are presented as the mean ± standard deviation. One-way analysis of variance followed by Tukey's post hoc test was used to compare the means of multiple groups. Correlation between CFHR3 expression levels and tumor diameter was analyzed by Pearson's correlation coefficient. P<0.05 was considered to indicate a statistically significant difference.
Results
CFHR3 mRNA and protein expression levels in HCC and adjacent tissue
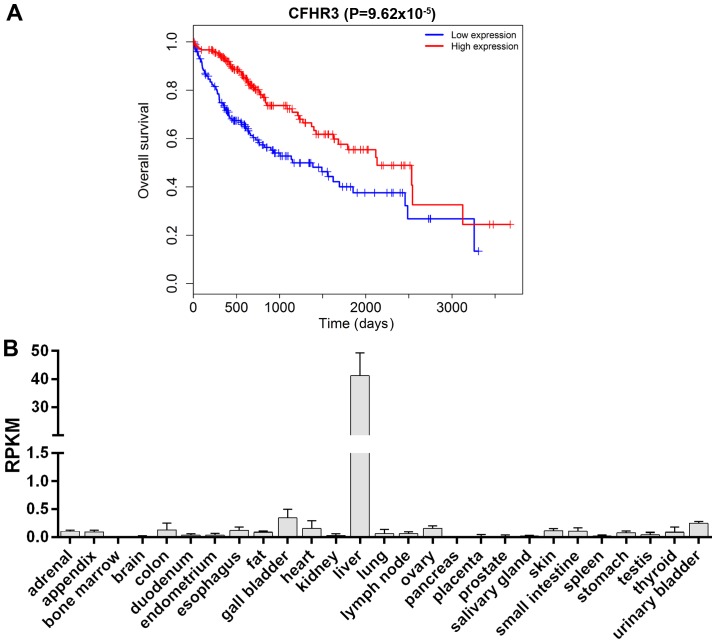
Survival analysis of patients with HCC from the TCGA database is shown in Fig. 1A. The overall survival of patients with HCC was significantly improved in the CFHR3 high expression group compared with the low expression group (P=9.62×10−5). CFHR3 expression levels in 27 different normal tissues were also compared (Fig. 1B) using the HPA RNA-seq normal tissues from NCBI (https://www.ncbi.nlm.nih.gov/gene/10878). The data demonstrated that CFHR3 was only highly expressed in liver tissues, suggesting that CFHR3 may be tissue-specific and related to the progression of liver cancer.
Figure 1.
(A) Survival analysis in patients with hepatocellular carcinoma with high or low CFHR3 expression. The data was obtained from The Cancer Genome Atlas database. (B) CFHR3 expression levels in 27 different normal tissues. The data was obtained from the National Center for Biotechnology Information gene database. CFHR3, complement factor H-related 3; RPKM, reads per kilobase million.
In the present study, 42 patients with HCC were recruited, and CFHR3 mRNA expression levels in HCC and adjacent tissues were detected by RT-qPCR. Western blot analysis of CFHR3 protein expression levels was performed on tissue samples from six patients. The results revealed that CFHR3 mRNA expression levels (2−ΔΔCT) were lower in cancer tissue compared with in adjacent normal liver tissue (P<0.05; Fig. 2A). This was consistent with the western blot results, which demonstrated that CFHR3 protein expression levels were significantly decreased in HCC tissue (P<0.01; Fig. 2B). However, the correlation between CFHR3 expression levels and tumor diameter in the 42 HCC cases was not significant (P>0.05; Fig. 2C).
Figure 2.
CFHR3 expression levels in HCC and adjacent tissue. (A) A total of 42 patients with HCC were recruited and relative CFHR3 mRNA expression levels were detected in HCC and adjacent tissue using reverse transcription-quantitative polymerase chain reaction. *P<0.05. (B) CFHR3 protein expression levels were evaluated in the HCC and adjacent tissue of six patients with HCC by western blot analysis. (C) Correlation between CFHR3 expression levels and tumor diameter in 42 HCC cases. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. Adjacent. A, adjacent; C, cancer; CFHR3; complement factor H-related 3; HCC, hepatocellular carcinoma.
CFHR3 expression in HCC cells
Huh-7 cells were transfected with the NC plasmid, and the RT-qPCR and western blot results demonstrated that transfection had no effect on CFHR3 expression levels compared with the control group. Cells transfected with the CFHR3 overexpression plasmid had significantly higher CFHR3 mRNA and protein expression levels compared with the control or NC group (P<0.01; Fig. 3A and B).
Figure 3.
Effect of CFHR3 on the cell viability of Huh-7 cells. Huh-7 cells were transfected with the CFHR3 overexpression plasmid and transfection efficiency was confirmed by (A) western blot analysis and (B) reverse transcription-quantitative polymerase chain reaction. (C) Cell viability was determined using the cell counting kit-8 assay. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01 vs. Con. CFHR3, complement factor H-related 3; Con, control; NC, negative control.
CFHR3 inhibits cell viability
Cell viability was detected using CCK-8 assay. The results revealed that at 24 and 48 h post-transfection cell viability was significantly decreased in the CFHR3 overexpression group compared with in the control or NC group (P<0.05; Fig. 3C). Therefore, the results suggested that cell viability can be inhibited by CFHR3.
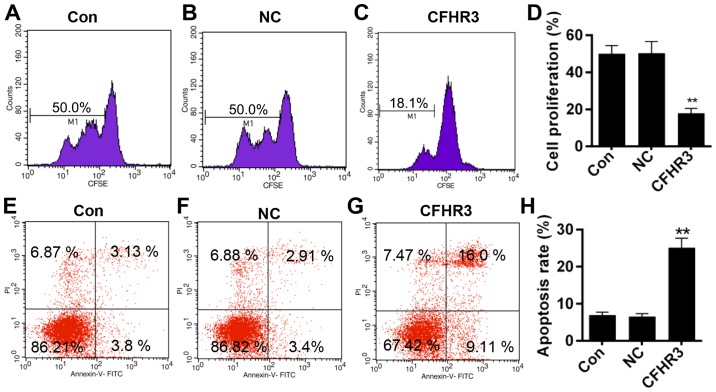
Effect of CFHR3 on cell proliferation and apoptosis
Cell proliferation and apoptosis were measured using flow cytometry. CFHR3 overexpression significantly decreased the cell fluorescence intensity (M1 value; P<0.01; Fig. 4A-D), indicating that CFHR3 inhibited cell proliferation. In addition, the results also demonstrated that CFHR3 overexpression significantly increased the apoptosis rate compared with the control or NC group (P<0.01; Fig. 4E-H).
Figure 4.
Effect of CFHR3 on Huh-7 cell proliferation and apoptosis. (A-C) Cell proliferation was measured by flow cytometric analysis of CFSE. (D) Quantification of the cell proliferation data. (E-G) Apoptosis was detected by flow cytometric analysis of Annexin V-FITC- and PI-stained cells. (H) Quantification of the apoptosis rate. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. Con. Con, control; NC, negative control; CFHR3, complement factor H-related 3; CFSE, carboxyfluorescein succinimidyl ester; FITC, fluorescein isothiocyanate; PI, propidium iodide.
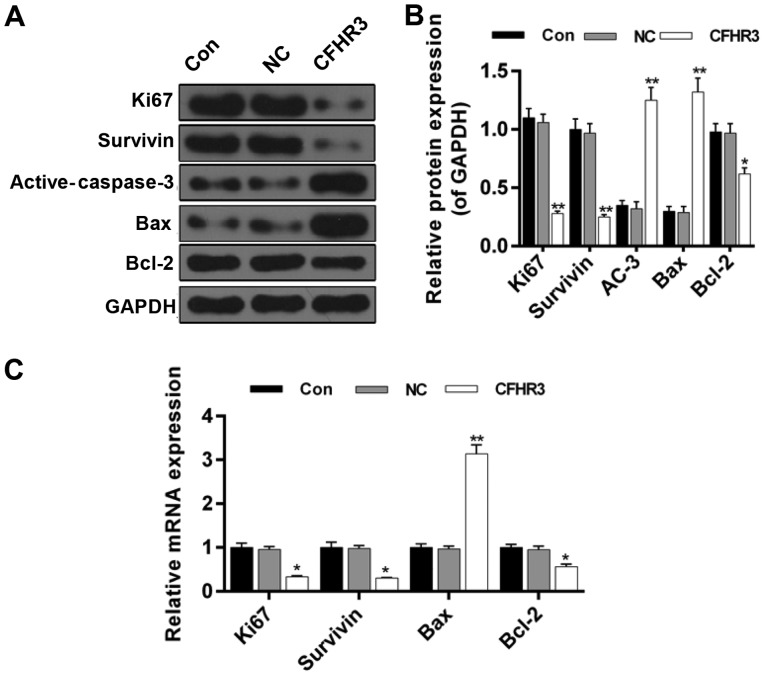
CFHR3 regulates the expression of proliferation- and apoptosis-related genes
In order to confirm the effects of CFHR3 on cell proliferation and apoptosis, protein and mRNA expression levels of proliferation- and apoptosis-related proteins and genes were detected using western blotting and RT-qPCR, respectively. The western blot results revealed that the protein expression levels of Ki67, survivin and Bcl-2 were significantly downregulated in the CFHR3 overexpression group (P<0.05; Fig. 5A and B). However, protein expression levels of active-caspase-3 and Bax were significantly increased in the CFHR3 group compared with in the control or NC group (P<0.01; Fig. 5A and B). A similar pattern was observed for mRNA expression levels. CFHR3 overexpression significantly decreased the mRNA expression levels of Ki67, survivin and Bcl-2 (P<0.05; Fig. 5C), whereas active-caspase-3 and Bax were significantly increased (P<0.05; Fig. 5C).
Figure 5.
Effect of CFHR3 on the expression of proliferation- and apoptosis-related genes and proteins in Huh-7 cells. (A) Western blotting was performed to determine the protein expression levels of Ki67, survivin, active-caspase-3, Bax and Bcl-2. (B) Summary of the relative protein expression levels. (C) Reverse transcription-quantitative polymerase chain reaction was performed to determine the mRNA expression levels of Ki67, survivin, Bax and Bcl-2. Expression levels were normalized to GAPDH. Data are presented as the mean ± standard deviation from three independent experiments. *P<0.05, **P<0.01 vs. Con. AC-3, active-caspase-3; Bax, Bcl-2-associated X; Bcl-2, B cell lymphoma 2; CFHR3, complement factor H-related 3; Con, control; NC, negative control.
CFHR3 inhibits PI3K/Akt/mTOR signaling
The protein expression levels of PI3K, Akt and mTOR were detected in order to determine the potential involvement of the PI3K/Akt/mTOR signaling pathway in HCC. The results demonstrated no noticeable differences in the protein expression levels of PI3K, Akt and mTOR among the control, NC and CFHR3 groups. However, CFHR3 significantly inhibited the phosphorylation of PI3K, Akt and mTOR (P<0.01; Fig. 6). Taken together, the results suggested that CFHR3 can inhibit liver cancer cell growth via the PI3K/Akt/mTOR pathway.
Figure 6.
Effect of CFHR3 on the PI3K/Akt/mTOR signaling pathway in Huh-7 cells. Western blotting was used to determine the protein expression levels of (A and B) p-PI3K, (C and D) p-Akt and (E and F) p-mTOR. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 vs. Con. Akt, protein kinase B; Con, control; mTOR, mammalian target of rapamycin; NC, negative control; p, phosphorylated; PI3K, phosphoinositide 3-kinase.
Discussion
CFHR3 is mainly studied in the context of IgA nephropathy, age-related macular degeneration and atypical hemolytic uremic syndrome (30–32). Previous studies have demonstrated that CFH family proteins may be important in cancer (19,20,33); however, little is known about the role of CFHR3 in cancer. Researchers have suggested that high expression levels of CFHR3 could increase the risk of neuroblastoma and glioma (19,34). CFHR3 is tissue-specific and is related to the progression of liver cancer. In the present study, the expression of CFHR3 in different normal human tissues was investigated and it was demonstrated that CFHR3 was highly expressed in normal liver tissues. In addition, high expression of CFHR3 in patients with HCC resulted in an improvement in overall survival. Therefore, it was hypothesized that CFHR3 may inhibit the occurrence and development of HCC. To further validate this, 42 patients with HCC were recruited, and expression of CFHR3 in HCC and adjacent normal tissues were detected. The results demonstrated that liver cancer tissue had a lower CFHR3 mRNA (2−ΔΔCq) and protein levels compared with normal tissue. However, no significant correlation between CFHR3 expression level and tumor diameter was identified in the 42 HCC cases. The sample size in this study was relatively small, which may affect the results obtained.
Liver cancer is one of the most common forms of cancer. Surgical resection and chemotherapy are the most common strategies to treat HCC (35–38). However, the prognosis for patients with liver cancer has not been improved significantly. Thus, it is necessary to develop new approaches to treat liver cancer. In the present study, CFHR3 was overexpressed in HCC cells. This resulted in a significant inhibition of cell viability and proliferation, whereas apoptosis was significantly upregulated. The protein and mRNA expression levels of proliferation- and apoptosis-related genes were also detected. The results suggested that CFHR3 inhibited proliferation and induced apoptosis by downregulating the expression of Ki67, Bcl-2 and survivin, and upregulating the expression of Bax and active-caspase-3. Ki67 is a cell proliferation marker (39). Survivin is an important member of the inhibitor of apoptosis protein family and serves a critical role in the inhibition of apoptosis (40,41). Caspase-3 is a key protein of the caspase family and has a major role in the execution-phase of apoptosis (42).
The occurrence and development of primary liver cancer is closely related to cell signaling pathways. The signal transduction pathways in HCC form a complex network, which involves multi-factor and multi-protein pathways. Cell growth is associated with several major molecular pathways, including Ras/mitogen-activated protein kinase kinase, PI3K/Akt/mTOR, Wnt/β-catenin, Janus kinase/signal transducer and activator of transcription (43–47). Among these four pathways, the PI3K/Akt/mTOR pathway is most closely related to HCC (48). PI3K is a heterodimer that is composed of a regulatory p85 subunit and a catalytic p110 subunit. PI3K is found in the cytoplasm, and when activated, aggregates to the cell membrane and interacts with the pleckstrin homology domain of Akt, leading to its activation. mTOR is a downstream molecule of the PI3K/Akt pathway, which controls cell growth and proliferation by regulating the cell cycle. Dysregulation of the cell cycle can lead to cell transformation and tumor deterioration (49,50). In the present study, the results revealed that CFHR3 significantly inhibited the phosphorylation and thereby inactivation of PI3K, Akt and mTOR.
However, only the effects of CFHR3 overexpression were demonstrated in the present study. Therefore, using a knock-down of CFHR3 could strengthen the results. In addition, the TCGA database contains an abundant of clinical information. It would be useful to conduct multivariate Cox regression analysis to identify the effect of other important prognostic factors, including serum α-fetoprotein, tumor stage, cirrhosis, vascular tumors and inflammation on overall survival rates, using blood samples. This analysis would help evaluate the independent prognostic value of CFHR3 mRNA expression. In addition, an association between CFHR3 expression and tumor diameter could further confirm the results obtained from the in vitro experiments. Future experiments in vivo will also aid in validating the results and investigations probing into the definite mechanism underlying the effect of CFHR3 will be of interest.
In conclusion, the results from the present study suggested that CFHR3 functions as a tumor suppressor. CFHR3 may suppress proliferation and promote apoptosis via the PI3K/Akt/mTOR signaling pathway.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
HL made substantial contributions to the conception and design of the present study. LZ and PW performed data acquisition, analysis and interpretation. PW and HL drafted the article and critically revised it for important intellectual content. All authors approved the final version of the published article. LZ agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of the study are appropriately investigated and resolved.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Yantai Infectious Disease Hospital (Yantai, China). Written informed consent was obtained from patients.
Patient consent for publication
Written informed consent was obtained from patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.European Association For The Study Of The Liver; European Organisation For Research and Treatment Of Cancer, corp-author. EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Chan SL, Wong AM, Lee K, Wong N, Chan AK. Personalized therapy for hepatocellular carcinoma: Where are we now? Cancer Treat Rev. 2016;45:77–86. doi: 10.1016/j.ctrv.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Fan D. Hepatitis B in China. Lancet. 2007;369:1582–1583. doi: 10.1016/S0140-6736(07)60723-5. [DOI] [PubMed] [Google Scholar]
- 5.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: New estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 6.Sartorius K, Sartorius B, Aldous C, Govender PS, Madiba TE. Global and country underestimation of hepatocellular carcinoma (HCC) in 2012 and its implications. Cancer Epidemiol. 2015;39:284–290. doi: 10.1016/j.canep.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zipfel PF, Skerka C, Hellwage J, Jokiranta ST, Meri S, Brade V, Kraiczy P, Noris M, Remuzzi G. Factor H family proteins: On complement, microbes and human diseases. Biochem Soc Trans. 2002;30:971–978. doi: 10.1042/bst030a097a. [DOI] [PubMed] [Google Scholar]
- 9.Hellwage J, Jokiranta TS, Koistinen V, Vaarala O, Meri S, Zipfel PF. Functional properties of complement factor H-related proteins FHR-3 and FHR-4: Binding to the C3d region of C3b and differential regulation by heparin. FEBS Lett. 1999;462:345–352. doi: 10.1016/S0014-5793(99)01554-9. [DOI] [PubMed] [Google Scholar]
- 10.McRae JL, Duthy TG, Griggs KM, Ormsby RJ, Cowan PJ, Cromer BA, McKinstry WJ, Parker MW, Murphy BF, Gordon DL. Human factor H-related protein 5 has cofactor activity, inhibits C3 convertase activity, binds heparin and C-reactive protein, and associates with lipoprotein. J Immunol. 2005;174:6250–6256. doi: 10.4049/jimmunol.174.10.6250. [DOI] [PubMed] [Google Scholar]
- 11.Kotarsky H, Hellwage J, Johnsson E, Skerka C, Svensson HG, Lindahl G, Sjobring U, Zipfel PF. Identification of a domain in human factor H and factor H-like protein-1 required for the interaction with streptococcal M proteins. J Immunol. 1998;160:3349–3354. [PubMed] [Google Scholar]
- 12.Appel GB, Cook HT, Hageman G, Jennette JC, Kashgarian M, Kirschfink M, Lambris JD, Lanning L, Lutz HU, Meri S, et al. Membranoproliferative glomerulonephritis type II (dense deposit disease): An update. J Am Soc Nephrol. 2005;16:1392–1403. doi: 10.1681/ASN.2005010078. [DOI] [PubMed] [Google Scholar]
- 13.Venables JP, Strain L, Routledge D, Bourn D, Powell HM, Warwicker P, Diaz-Torres ML, Sampson A, Mead P, Webb M, et al. Atypical haemolytic uraemic syndrome associated with a hybrid complement gene. PLoS Med. 2006;3:e431. doi: 10.1371/journal.pmed.0030431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zipfel PF, Edey M, Heinen S, Jozsi M, Richter H, Misselwitz J, Hoppe B, Routledge D, Strain L, Hughes AE, et al. Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet. 2007;3:e41. doi: 10.1371/journal.pgen.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monteferrante G, Brioschi S, Caprioli J, Pianetti G, Bettinaglio P, Bresin E, Remuzzi G, Noris M. Genetic analysis of the complement factor H related 5 gene in haemolytic uraemic syndrome. Mol Immunol. 2007;44:1704–1708. doi: 10.1016/j.molimm.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 17.Hageman GS, Hancox LS, Taiber AJ, Gehrs KM, Anderson DH, Johnson LV, Radeke MJ, Kavanagh D, Richards A, Atkinson J, et al. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: Characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38:592–604. doi: 10.1080/07853890601097030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kavanagh D, Richards A, Atkinson J. Complement regulatory genes and hemolytic uremic syndromes. Annu Rev Med. 2008;59:293–309. doi: 10.1146/annurev.med.59.060106.185110. [DOI] [PubMed] [Google Scholar]
- 19.Kinders R, Jones T, Root R, Bruce C, Murchison H, Corey M, Williams L, Enfield D, Hass GM. Complement factor H or a related protein is a marker for transitional cell cancer of the bladder. Clin Cancer Res. 1998;4:2511–2520. [PubMed] [Google Scholar]
- 20.Junnikkala S, Jokiranta TS, Friese MA, Jarva H, Zipfel PF, Meri S. Exceptional resistance of human H2 glioblastoma cells to complement-mediated killing by expression and utilization of factor H and factor H-like protein 1. J Immunol. 2000;164:6075–6081. doi: 10.4049/jimmunol.164.11.6075. [DOI] [PubMed] [Google Scholar]
- 21.Corey MJ, Kinders RJ, Poduje CM, Bruce CL, Rowley H, Brown LG, Hass GM, Vessella RL. Mechanistic studies of the effects of anti-factor H antibodies on complement-mediated lysis. J Biol Chem. 2000;275:12917–12925. doi: 10.1074/jbc.275.17.12917. [DOI] [PubMed] [Google Scholar]
- 22.Fedarko NS, Fohr B, Robey PG, Young MF, Fisher LW. Factor H binding to bone sialoprotein and osteopontin enables tumor cell evasion of complement-mediated attack. J Biol Chem. 2000;275:16666–16672. doi: 10.1074/jbc.M001123200. [DOI] [PubMed] [Google Scholar]
- 23.Jozsi M, Manuelian T, Heinen S, Oppermann M, Zipfel PF. Attachment of the soluble complement regulator factor H to cell and tissue surfaces: Relevance for pathology. Histol Histopathol. 2004;19:251–258. doi: 10.14670/HH-19.251. [DOI] [PubMed] [Google Scholar]
- 24.Fedarko NS, Jain A, Karadag A, Van Eman MR, Fisher LW. Elevated serum bone sialoprotein and osteopontin in colon, breast, prostate, and lung cancer. Clin Cancer Res. 2001;7:4060–4066. [PubMed] [Google Scholar]
- 25.Cui L, Fu J, Pang JC, Qiu ZK, Liu XM, Chen FR, Shi HL, Ng HK, Chen ZP. Overexpression of IL-7 enhances cisplatin resistance in glioma. Cancer Biol Ther. 2012;13:496–503. doi: 10.4161/cbt.19592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabatini DM. mTOR and cancer: Insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 27.Ayuso C, Rimola J, Vilana R, Burrel M, Darnell A, García-Criado Á, Bianchi L, Belmonte E, Caparroz C, Barrufet M, Bruix J, Brú C. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol. 2018;101:72–81. doi: 10.1016/j.ejrad.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Hoefel D, Grooby WL, Monis PT, Andrews S, Saint CP. A comparative study of carboxyfluorescein diacetate and carboxyfluorescein diacetate succinimidyl ester as indicators of bacterial activity. J Microbiol Methods. 2003;52:379–388. doi: 10.1016/S0167-7012(02)00207-5. [DOI] [PubMed] [Google Scholar]
- 30.Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J, Sanna-Cherchi S, Men CJ, Julian BA, Wyatt RJ, et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet. 2011;43:321–327. doi: 10.1038/ng.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munch J, Bachmann A, Grohmann M, Mayer C, Kirschfink M, Lindner TH, Bergmann C, Halbritter J. Effective immunosuppressive management with belatacept and eculizumab in post-transplant aHUS due to a homozygous deletion of CFHR1/CFHR3 and the presence of CFH antibodies. Clin Kidney J. 2017;10:742–746. doi: 10.1093/ckj/sfx053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakri NM, Ramachandran V, Kee HF, Subrayan V, Isa H, Ngah NF, Mohamad NA, Mooi CS, Mun CY, Ismail P, et al. Association of copy number variations in complement factor H-Related genes among age-related macular degenerative subjects. Kaohsiung J Med Sci. 2017;33:602–608. doi: 10.1016/j.kjms.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junnikkala S, Hakulinen J, Jarva H, Manuelian T, Bjorge L, Butzow R, Zipfel PF, Meri S. Secretion of soluble complement inhibitors factor H and factor H-like protein (FHL-1) by ovarian tumour cells. Br J Cancer. 2002;87:1119–1127. doi: 10.1038/sj.bjc.6600614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan FH, Pandian V, Ramraj S, Natarajan M, Aravindan S, Herman TS, Aravindan N. Acquired genetic alterations in tumor cells dictate the development of high-risk neuroblastoma and clinical outcomes. BMC Cancer. 2015;15:514. doi: 10.1186/s12885-015-1463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanazaki K, Kajikawa S, Shimozawa N, Mihara M, Shimada K, Hiraguri M, Koide N, Adachi W, Amano J. Survival and recurrence after hepatic resection of 386 consecutive patients with hepatocellular carcinoma. J Am Coll Surg. 2000;191:381–388. doi: 10.1016/S1072-7515(00)00700-6. [DOI] [PubMed] [Google Scholar]
- 36.Poon RT, Fan ST, Lo CM, Ng IO, Liu CL, Lam CM, Wong J. Improving survival results after resection of hepatocellular carcinoma: A prospective study of 377 patients over 10 years. Ann Surg. 2001;234:63–70. doi: 10.1097/00000658-200107000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosch J, Forns X. Therapy. Statins and liver disease: From concern to ‘wonder’ drugs? Nat Rev Gastroenterol Hepatol. 2015;12:320–321. doi: 10.1038/nrgastro.2015.78. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Gao C, Fang L, Zhao HC, Yao SK. Metformin and reduced risk of hepatocellular carcinoma in diabetic patients: A meta-analysis. Scand J Gastroenterol. 2013;48:78–87. doi: 10.3109/00365521.2012.719926. [DOI] [PubMed] [Google Scholar]
- 39.Forones NM, Carvalho AP, Giannotti-Filho O, Lourenço LG, Oshima CT. Cell proliferation and apoptosis in gastric cancer and intestinal metaplasia. Arq Gastroenterol. 2005;42:30–44. doi: 10.1590/S0004-28032005000100008. [DOI] [PubMed] [Google Scholar]
- 40.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 41.Altieri DC. Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 42.Choudhary GS, Al-Harbi S, Almasan A. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods Mol Biol. 2015;1219:1–9. doi: 10.1007/978-1-4939-1661-0_1. [DOI] [PubMed] [Google Scholar]
- 43.Newell P, Toffanin S, Villanueva A, Chiang DY, Minguez B, Cabellos L, Savic R, Hoshida Y, Lim KH, Melgar-Lesmes P, et al. Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J Hepatol. 2009;51:725–733. doi: 10.1016/j.jhep.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villanueva A, Toffanin S, Llovet JM. Linking molecular classification of hepatocellular carcinoma and personalized medicine: Preliminary steps. Curr Opin Oncol. 2008;20:444–453. doi: 10.1097/CCO.0b013e328302c9e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang J, Ouyang C, Chen X, Fu B, Lu Y, Hong Q. Effect of Jak2 kinase inhibition on Stat1 and Stat3 activation and apoptosis of tubular epithelial cells induced by ATP depletion/recovery. J Nephrol. 2008;21:919–923. [PubMed] [Google Scholar]
- 46.Quentmeier H, Geffers R, Jost E, Macleod RA, Nagel S, Rohrs S, Romani J, Scherr M, Zaborski M, Drexler HG. SOCS2: Inhibitor of JAK2V617F-mediated signal transduction. Leukemia. 2008;22:2169–2175. doi: 10.1038/leu.2008.226. [DOI] [PubMed] [Google Scholar]
- 47.Herbst A, Kolligs FT. Wnt signaling as a therapeutic target for cancer. Methods Mol Biol. 2007;361:63–91. doi: 10.1385/1-59745-208-4:63. [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Pi C, Wang G. Inhibition of PI3K/Akt/mTOR pathway by apigenin induces apoptosis and autophagy in hepatocellular carcinoma cells. Biomed Pharmacother. 2018;103:699–707. doi: 10.1016/j.biopha.2018.04.072. [DOI] [PubMed] [Google Scholar]
- 49.Zhao L, Li C, Liu F, Zhao Y, Liu J, Hua Y, Liu J, Huang J, Ge C. A blockade of PD-L1 produced antitumor and antimetastatic effects in an orthotopic mouse pancreatic cancer model via the PI3K/Akt/mTOR signaling pathway. Onco Targets Ther. 2017;10:2115–2126. doi: 10.2147/OTT.S130481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X, Zeng J, Wang L, Zhang X, Liu Z, Zhang H, Dong J. Overexpression of microRNA-133b is associated with the increased survival of patients with hepatocellular carcinoma after curative hepatectomy: Involvement of the EGFR/PI3K/Akt/mTOR signaling pathway. Oncol Rep. 2017;38:141–150. doi: 10.3892/or.2017.5699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.