Abstract
Post-operative cognitive dysfunction (POCD) is a complication of the central nervous system characterized by mental disorders, anxiety, personality changes and impaired memory. POCD occurs frequently after coronary artery bypass grafting (CABG) and can severely affect quality of life for patients. To date, the development of POCD biomarkers remains a challenge. Alterations in the expression of non-coding RNAs from brain tissue and peripheral blood have been linked to POCD. The present study aimed to detect the differential circular RNAs (circRNAs) in plasma exosomes of patients with POCD after CABG. The relative expression levels of circRNAs were analyzed using circRNA microarray analysis in the plasma exosomes of patients with POCD. Differentially altered circRNAs (P<0.05, fold change >1.5) were validated by reverse transcription-quantitative PCR in the plasma exosomes of patients with POCD. The target genes of the microRNAs were predicted using bioinformatics analysis. The functions and signaling pathways of these target genes were investigated by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes analyses. The microarray results indicated that the levels of nine circRNAs in patients with POCD were higher than those in the control subjects; and six circRNAs were at a lower level than those in control subjects. The RT-qPCR results from patients with POCD showed that only circRNA_089763 of the 15 circRNAs identified was significantly increased compared with control subjects. circRNA target gene prediction and functional annotation analysis showed significant enrichment in several GO terms and pathways associated with POCD. The present study provides evidence for the abnormal expression of POCD-induced circRNA_089763 in human plasma exosomes, as well as the involvement of POCD.
Keywords: circular RNA, post-operative cognitive dysfunction, exosomes, coronary artery bypass grafting
Introduction
Post-operative cognitive dysfunction (POCD) is a complication of the central nervous system after surgery that manifests as mental disorder, anxiety, personality changes and memory impairment (1,2). A number of risk factors are associated with the POCD including age, preoperative cognition and emotional state (3). A previous study noted that older men with APOE4 allele may be more vulnerable to POCD than older women with APOE4 allele (4). After coronary artery bypass grafting (CABG), the incidence of early cognitive dysfunction is high, at 30–60%, and POCD severely affects the post-operative quality of life of patients (5–7). Currently, the diagnosis of POCD is based on the description of symptoms from the patient, assessment of mental status and evaluation of clinical behaviors, which are complex and take time to evaluate (8,9). Although some biomarkers have been found to be associated with POCD such as S100B and NSE, the mechanism and genes which associated with these biomarkers remain to be elucidated (10).
Circular (circ) RNAs have recently emerged as a novel class of transcripts. circRNAs are noncoding RNA (ncRNA) molecules that regulate the stability or translational efficiency of target mRNAs through a competitive endogenous RNA (ceRNA) mechanism (11). The mammalian brain is the organ in which circRNAs are more abundant and first evidence of their functional significance started to emerge among many tissues and species (12). Several studies have reported that circRNAs are important regulatory factors in normal developmental, physiological and disease states, including cancer, mental disorders and cardiovascular diseases (13–15). However, to the best of our knowledge, no circRNAs have been reported to be associated with POCD.
Unlike tumor tissue, some tissues are not easy to obtain in clinical practice, including from the heart and brain. Recent studies have found that diseased tissues from the heart or brain can release exosomes, containing miRNAs, long non-coding RNAs (lncRNAs) and circRNAs, and that these exosomes can cross the blood-brain barrier and enter the circulation. These ncRNAs may be new biological markers that can be obtained with less invasive procedures from patients with cardiovascular and cerebral disease (16,17).
This present study aimed to identify circRNAs in the plasma exosomes of patients with POCD. These circRNAs were probed as potential specific biomarkers for the diagnosis of POCD.
Materials and methods
Patients
The present study was approved by the Medical Ethics Committee of The Affiliated Beijing Chaoyang Hospital of Capital Medical University. Written informed consent was obtained from each participant before enrollment. Patients, between 40 and 90 years of age, who were scheduled for first-time elective off-pump CABG surgery were recruited between June 2018 and October 2018. Surgeries were performed at The Affiliated Chaoyang Hospital of Capital Medical University. The exclusion criteria included pre-existing neurological disease, including a Mini-Mental State Examination (MMSE) score <24 points; a history of taking antidepressants or anti-anxiety drugs; anticipated difficulty with neuropsychological assessment, including severe visual or hearing impairment; deep hypothermic circulatory arrest during surgery; perioperative insulin therapy; and poor ventricular function (ejection fraction <30%).
Anesthesia protocol
Patients were given standard perioperative care and surgery was performed by the same team at The Affiliated Beijing Chaoyang Hospital of Capital Medical University. Anesthesia was administered according to a standard protocol, including induction with midazolam (0.05–0.1 mg/kg), fentanyl (5–8 µg/kg), etomidate (0.2–0.3 mg/kg) and vecuronium (1 mg/kg), and maintenance with fentanyl (0.1–0.15 µg/kg/min), midazolam (0.6–1 µg/kg/min), sevoflurane (0.5–1%) in oxygen and vecuronium (0.05 mg/kg, once every 30 min). Standard monitoring was performed for all patients, including electrocardiography, pulse oximetry, end-tidal carbon dioxide, nasopharyngeal temperatures, central venous and arterial blood pressure, arterial blood gas, cardiac output and bispectral index.
Neuropsychological assessment
Patients were first screened with the MMSE to exclude severe cognitive impairment. The enrolled patients completed neuropsychological tests 1 day before surgery and again 1 week after surgery. The neuropsychological evaluation test applied in this present study was based on a Chinese version of neuropsychological testing (18). The series of tests assessed several cognitive functions, including attention, motor skills, executive function, learning and memory. Concentration and the ability to ignore distracting stimuli was assessed with the Stroop Colour and Word Test (SCWT) (19), including SCWT-1, SCWT-2 and SCWT-3. The time required to complete the tests and the number of errors was recorded. Mental processing speed and visual scanning with the Trail Making Test (TMT) (20), including TMT-A and TMA-B, were measured and the time required to complete these tests was recorded. Concentration, processing speed and visual scanning abilities were tested with the Digit Symbol Substitution Test (21), and the correct matching quantity was measured. Short-term and long-term memory, and attention, were assessed by the Verbal Learning Test (VLT) (22), including VLT-A1, VLT-A2 and VLTA3. The total number of correct words immediately recalled and recalled after a 30 min delay were recorded. The Symbol Digits Modalities Test (SDMT) (23) was used to measure short-term memory, visuospatial skills and attention, and the number of errors recorded. These tests have been used to evaluate the POCD in a previous study (10). POCD was defined by a reduction in the post-operative test score compared with the pre-operative test score ≥20% in at least two of the eight tests, following the same criteria established by the previous study (10).
Data collection
Demographic and intraoperative data, including age, weight, sex, body mass index (BMI), ejection fraction (EF), New York Heart Association (NYHA) (24) class of heart failure and surgical time, were collected. Blood samples (10 ml) were collected 1 day before anesthesia and 7 days after surgery (at 08:00 a.m. following overnight fasting). Samples were collected in anticoagulant tubes, centrifuged at 4°C at 300 × g for 5 min and then at 1,200 × g and 4°C for 20 min. The plasma was stored at −80°C until further analysis.
Exosome isolation from plasma
The ultracentrifugation exosome isolation protocol was used to isolate exosomes from plasma. Briefly, plasma samples were centrifuged at 500 × g and 4°C for 5 min to remove the cells from the samples. The supernatant was transferred to a new polycarbonate tube and centrifuged at 2,000 × g for 10 min at 4°C. The supernatant was collected and transferred to a new polycarbonate tube. The samples were centrifuged at 10,000 × g for 30 min at 4°C to eliminate shed microvesicles (200–1,000 nm). The supernatants were collected and filtered using a 0.22 µm membrane filter (EMD Millipore). The samples were centrifuged at 100,000 × g for 2 h and 4°C. For RNA isolation, the exosome pellet was washed once with 1X PBS, followed by centrifugation 100,000 × g for 2 h at 4°C. The exosomes were resuspended in 1X PBS and stored at −80°C for further use.
Electron microscopy
Exosomes isolated from plasma were washed in PBS to further purify the sample, filtered using a 0.22-µm membrane filter (EMD Millipore), and ultracentrifuged at 100,000 × g for 2 h at 4°C to re-pellet the exosomes. The exosome pellet was resuspended and fixed in PBS containing 2% glutaraldehyde (4°C for 5 min) and then loaded onto former/carbon-coated electron microscopy grids. The samples were contrasted with 0.5% uranyl acetate to visualize the membranes and the samples were viewed with a Tecnai G2 Spirit 120 KV electron microscope (Thermo Fischer Scientific, Inc.).
circRNA microarray analysis
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and a RNeasy mini kit (74104; Qiagen, GmbH) were used for total RNA extraction according to the manufacturers' protocol. RNA quality and quantity were determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). Samples with an absorbance ratio >1.8 at 260 and 280 nm (A260/280), and at 260 and 230 nm (A260/230), an RNA concentration >60 ng and a total quantity >800 ng were defined as having sufficient quality and quantity for further experiments. Sample preparation and microarray hybridization was performed based on the standard protocol from Arraystar, Inc. Briefly, total RNA was digested with RNase R (Epicentre; Illumina, Inc.) to remove linear RNAs and enrich for circRNAs. The enriched circular RNAs were amplified and transcribed into fluorescent complementary RNA (cRNA) utilizing a random priming method (Arraystar Super RNA Labeling kit; Arraystar, Inc.). The labeled cRNAs were hybridized onto the Arraystar Rat circRNA Array (8×15K; Arraystar, Inc.). After washing the slides, the arrays were scanned with the Agilent G2505C Scanner (Agilent Technologies, Inc.). Agilent Feature Extraction software (version 11.0.1.1; Agilent Technologies, Inc.) was used to analyze the acquired array images. Quantile normalization and subsequent data processing was performed using the R software (3.4.2) limma package (v3.22.7, http://bioinf.wehi.edu.au/limma/). Differentially expressed circRNAs with statistical significance between patients with POCD and healthy individuals were identified through volcano plot filtering (25). Differentially expressed circRNAs between the two groups were identified through fold change filtering. Hierarchical clustering was performed to show the circRNA expression pattern among samples. circRNAs with a relative up- or downregulation of at least 1.5-fold signal density and P<0.05 were considered to be differentially expressed.
Reverse transcription-quantitative RT-q(PCR) validation of differentially expressed circRNAs
The expression levels of different circRNAs from the circRNA microarray analysis were validated using RT-qPCR. Complementary DNA templates were generated from 800 ng of total RNA and used for RT-qPCR. β-actin was used as an internal reference for template normalization. The primers for the different circRNAs were designed using Primer software (5.0, Premier, http://www.premierbiosoft.com/primerdesign; Table I). The ViiA 7 Real-time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for the amplification and detection of the circRNAs. The standard curve was prepared using a gradient dilution of the DNA template. Briefly, cDNA templates expressing the target and housekeeper genes were selected for the RT-qPCR reaction as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. The amplified PCR product and a 100-bp DNA ladder were electrophoresed on a 2% agarose gel and single specific bands were detected by ethidium bromide staining. After purification of the DNA of interest from the gel, a gradient dilution (10 dilutions, each decreasing by a factor of 10) was prepared and a standard curve was generated for the RT-qPCR assay. For RT-qPCR of the experimental groups, the target and housekeeping genes of each sample were subjected to the aforementioned RT-qPCR reaction. The concentrations of the target and housekeeping genes in each sample were directly generated by the machine using the standard curve as a reference. The relative level of the target gene=concentration of target gene/concentration of housekeeping gene.
Table I.
Sequences for all primers used.
| Gene name | Bidirectional primer sequence | Annealing temperature (°C) | Product length (bp) |
|---|---|---|---|
| β-actin (H) | F: 5′-GTGGCCGAGGACTTTGATTG-3′ | 60 | 73 |
| R: 5′-CCTGTAACAACGCATCTCATATT-3′ | |||
| hsa_circRNA_038651 | F: 5′-TACATCCAGGCCCACATCG-3′ | 60 | 80 |
| R: 5′-TTCATCACGCAGCGCTTG-3′ | |||
| hsa_circRNA_017502 | F: 5′-GCATCCGAACTGTCCCCTAC-3′ | 60 | 107 |
| R: 5′-ACCTGGCTCCTGTGTCATCAT-3′ | |||
| hsa_circRNA_089763 | F: 5′-GGTGGGCCATACGGTAGTATT-3′ | 60 | 88 |
| R: 5′-TTACTTCCTCTCTTTCTTCTTCCC-3′ | |||
| hsa_circRNA_091840 | F: 5′-TGTCTTCTTTGGTTCTGGGAG-3′ | 60 | 131 |
| R: 5′-CAATCTGAATGGTGGGGC-3′ | |||
| hsa_circRNA_049784 | F: 5′-CATCCCTGGCCGGTTCAA-3′ | 60 | 70 |
| R: 5′-AGTGGGGGTTTCTCTGTTTCCT-3′ | |||
| hsa_circRNA_000798 | F: 5′-ACTTGGGGTTTCTGTGGTTC-3′ | 60 | 70 |
| R: 5′-GAGCTCATGCCCTATGAGGA-3′ | |||
| hsa_circRNA_101688-1 | F: 5′-CTTATCCCCAGAGAGCAAACAA-3′ | 60 | 69 |
| R: 5′-GGCAGGAACATAACACCACG-3′ | |||
| hsa_circRNA_101688-2 | F: 5′-CAGATCACAGATTTTGGAACAGC-3′ | 60 | 131 |
| R: 5′-ACTCAGTCTGGGTCCTCACCAT-3′ | |||
| hsa_circRNA_002281-1 | F: 5′-CCAGGTCATAGTAATGGGAGC-3′ | 60 | 140 |
| R: 5′-GCTGTAGCTTCATGGTCAGAA-3′ | |||
| hsa_circRNA_002281-2 | F: 5′-GCCAGGTCATAGTAATGGGAG-3′ | 60 | 118 |
| R: 5′-TGAACGGTTTCGTAGAAAGGA-3′ | |||
| hsa_circRNA_100075 | F: 5′-CGATAACATTTAGCCTGGAACA-3′ | 60 | 74 |
| R: 5′-GTAGGTCTGATTGAAGCAAGCC-3′ | |||
| hsa_circRNA_003558-1 | F: 5′-CTTCTCCCCAGCCAATGTCG-3′ | 60 | 140 |
| R: 5′-CTGAAGGTAGGTGGTGAATAGGG-3′ | |||
| hsa_circRNA_003558-2 | F: 5′-TGGAGAGACAGTAAAGGAAAAGAC-3′ | 60 | 107 |
| R: 5′-GGTCAAGAATCTGAAGGTAGGTG-3′ | |||
| hsa_circRNA_101006-1 | F: 5′-GACTGGAGCAAGGTCGTCCT-3′ | 60 | 104 |
| R: 5′-AGAGCATGGGCCACTTTCTG-3′ | |||
| hsa_circRNA_101006-2 | F: 5′-GTACTGGCAAGACTGCAACACC-3′ | 60 | 84 |
| R: 5′-AGGCGATTACTCCGAGTCCC-3′ | |||
| hsa_circRNA_031757-1 | F: 5′-GGCTTTTGGCTGATGAGGATT-3′ | 60 | 62 |
| R: 5′-TAGCTGCACTATTACGAAGGGAC-3′ | |||
| hsa_circRNA_031757-2 | F: 5′-TGGCTGATGAGGATTGATGC-3′ | 60 | 125 |
| R: 5′-AGTCGAGCTTCATTGCAGAAT-3′ | |||
| hsa_circRNA_004954-1 | F: 5′-GGGAGCCCTGAATATACACG-3′ | 60 | 114 |
| R: 5′-CACCACCCGTTTCATTTTTAC-3′ | |||
| hsa_circRNA_004954-2 | F: 5′-ATACACGAGAAAGCCTGGAATG-3′ | 60 | 79 |
| R: 5′-GGCCTCAGCCACAGAATACAG-3′ | |||
| hsa_circRNA_012969-1 | F: 5′-TTTGACGGTGGATTTGGTTG-3′ | 60 | 96 |
| R: 5′-GTCCGGTGCATCTGACTTGA-3′ | |||
| hsa_circRNA_012969-2 | F: 5′-CAGGTTCTGAATCCCATGCT-3′ | 60 | 129 |
| R: 5′-CATCTTTCTTTGAGCCATAGGA-3′ |
circRNA, circular RNA; hsa, Homo sapiens. hsa_circRNA_102054 primer was not synthesized because hsa_circRNA_102054 and hsa_circRNA_102105 could not be distinguished.
Analysis of target genes
The miRDB V5 database (http://mirdb.org/miRDB/) was used to predict miRNA target genes in the human, mouse, and rat genomes. An additional database, targetscan7.1, was used for miRNA target predictions in human (http://www.targetscan.org/vert_71/) and rat (http://www.targetscan.org/mmu_71/). The overlapping results of two databases for each of these species are generally accepted.
Gene Ontology (GO) analysis
The GO project provides a controlled vocabulary to describe gene and gene product attributes in any organism (http://www.geneontology.org). The ontology covers three domains: Biological process, cellular component and molecular function. Fisher's exact test was used in Bioconductor topGO software (version 2.32.0, http://www.bioconductor.org/packages/release/bioc/html/topGO.html) to determine whether there was more overlap between the differentially expressed (DE) list and the GO annotation list than would be expected by chance. The P-value produced by topGO denotes the significance of GO term enrichment in the DE genes. The lower the P-value, the more significant the GO term (P≤0.05 is recommended).
Pathway analysis
Pathway analysis is a functional analysis mapping of genes to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The P-value (Expression Analysis Systemic Explorer-score, Fisher P-value or Hypergeometric P-value) denotes the significance of the pathway correlated to the conditions. The lower the P-value, the more significant the pathway (the recommended P-value cut-off is 0.05).
Constructing the circRNA-miRNA-mRNA co-expression network
The co-expression network of the circRNA- miRNA-mRNA was constructed using the analysis of circRNA-miRNA and the miRNA-gene networks. The related miRNA and mRNA of the DE circRNAs were screened and the circRNA-miRNA-mRNA network was constructed.
Statistical analysis
All statistical analyses were performed using SPSS 13.0 software (SPSS, Inc.). Normally distributed data are presented as the mean ± SD and were analyzed using the paired or unpaired t-test (including baseline analysis of age and operation time) or repeated measure analysis of covariance. Categorical data are expressed as percentages and frequencies, and were analyzed using the Pearson's χ2 test. Pearson's correlation analysis was used to illustrate the relationships among age, sex and weight. Regression analysis was used to investigate the factors contributing to the risk of POCD. P<0.05 was considered to indicate a statistically significant difference.
Results
Patients included and perioperative characteristics of patients
As shown in Fig. 1, 35 patients completed the cognitive tests and 12 patients (34.3%) developed POCD 7 days after surgery. The peri-operative characteristics of patients are shown in Table II. Patients were divided into two groups according to ‘cognitive tests’: POCD (n=12) and non-POCD (n=23). There were no statistically significant differences between the two groups in terms of sex, height, weight, BMI, EF, NYHA class of heart failure or surgical time (P>0.05). Patients with POCD were older than patients without POCD (P<0.05).
Figure 1.
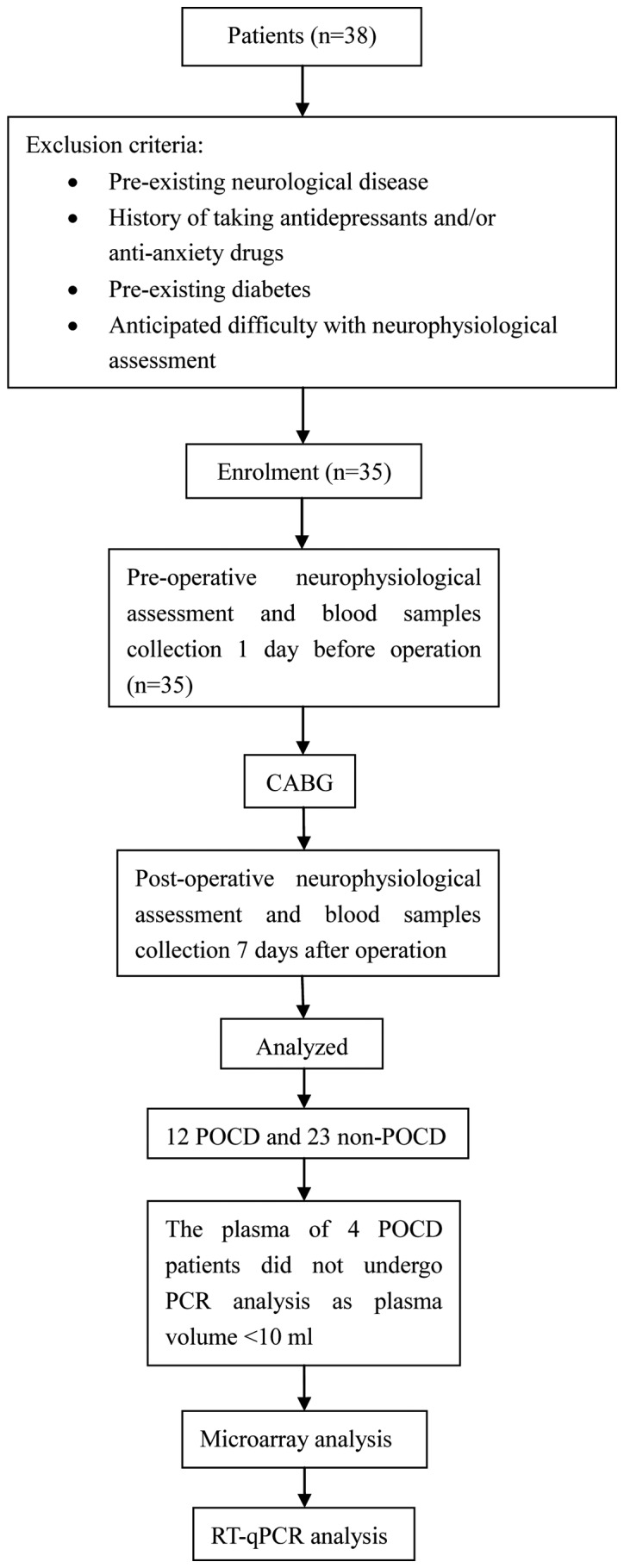
Experimental flow chart. POCD, post-operative cognitive dysfunction; CABG, coronary artery bypass grafting; RT-qPCR, reverse transcription-quantitative PCR.
Table II.
Characteristics of the patients.
| Characteristics | POCD group (n=12) | Non-POCD group (n=23) | P-value |
|---|---|---|---|
| Age, years | 67.67±8.50 | 60.78±8.95 | 0.04 |
| Female, n | 1/9 | 4/23 | 0.48 |
| Height, cm | 1.71±0.06 | 1.67±0.08 | 0.15 |
| Weight, kg | 74.33±7.41 | 71.65±12.93 | 0.52 |
| BMI, kg/cm2 | 25.36±2.24 | 25.50±3.86 | 0.91 |
| EF, % | 57.75±7.30 | 57.74±10.90 | 0.99 |
| NYHA | 0.78 | ||
| II | 5 | 10 | |
| III | 5 | 11 | |
| IV | 2 | 2 | |
| Surgical time, min | 266.67±43.99 | 238.04±51.54 | 0.12 |
POCD, post-operative cognitive dysfunction; BMI, body mass index; EF, ejection fraction; NYHA, New York Heart Association.
Exosome extraction results
As shown in Fig. 2, the red circle represents the plasma exosome. A transmission electron microscope image was taken of an exosome; the image shows the vesicular structure of the exocrine bodies and the size.
Figure 2.
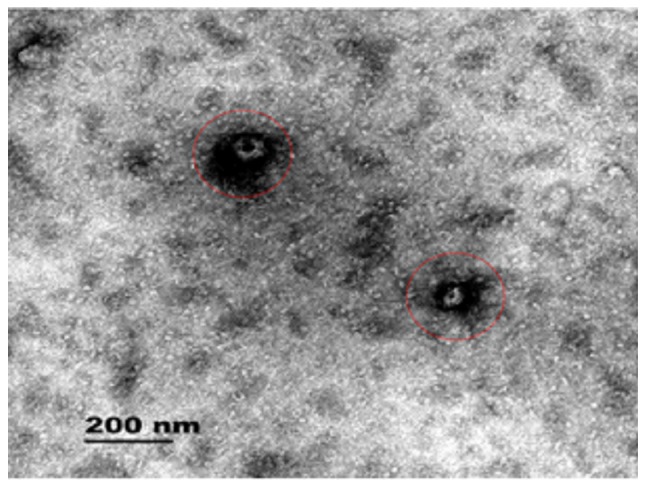
Plasma exosomes of patients after coronary artery bypass grafting. The red circles indicate the plasma exosome.
Expression profile of circRNAs in the plasma exosomes of patients with POCD using microarray analysis and secondary RT-qPCR validation
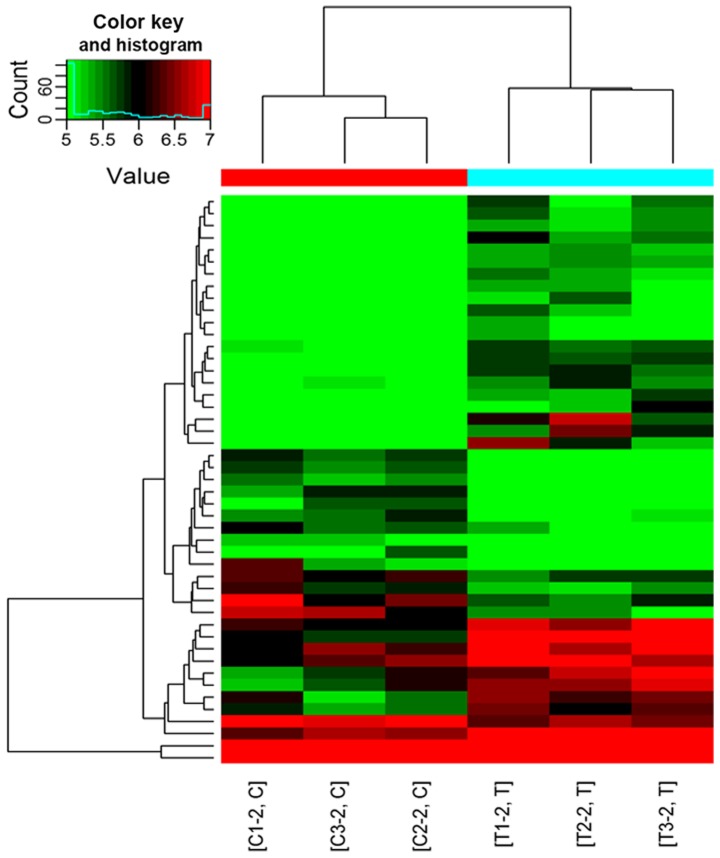
RNA concentration and purity was measured using a NanoDrop ND-1000 spectrophotometer. Samples with an A260/280 and A260/230 absorbance ratio >1.8, an RNA concentration >60 ng and total quantity >800 ng, were defined as having sufficient quality and quantity for further experimentation. The RNA purity, concentration and total quantity of 8 of the 12 samples from patients with POCD and 16 of the 23 samples from patients without POCD met these requirements and were used for subsequent microarray detection of the circRNA expression profile (3 samples from the POCD and 3 from the non-POCD group were analyzed by microarray, and 5 from each group analyzed by RTqPCR). As shown in Table III, circRNA microarray analysis of the plasma exosomes of patients with POCD compared with patients without POCD (P<0.05) indicated that 9 circRNAs were upregulated and 6 circRNAs were downregulated. As shown in Fig. 3, significantly different circRNAs were selected for hierarchical clustering analysis. The color key reflects the expression intensity of the circRNAs; with red indicating high expression, green indicating low expression and the color depth (ranging from black to color) indicating different expression intensities. The circRNA hierarchical clustering map revealed circRNA expression in all samples from both groups and indicated that the expression of a single circRNA was altered in all samples. RT-qPCR was used to validate the 15 circRNAs identified by microarray analysis (fold change ≥1.5; P<0.05). As shown in Table IV, of the 15 circRNAs analyzed by RT-qPCR only hsa_circRNA_089763 was confirmed to be differentially expressed in the plasma exosomes of patients with POCD compared to patients without POCD (P<0.05).
Table III.
Aberrantly expressed circRNAs in the plasma exosomes of patients with post-operative cognitive dysfunction from microarray analysis (fold change ≥1.5; P<0.05).
| A, Upregulated | |||
|---|---|---|---|
| circRNA | Fold change | P-value | Normalized intensity |
| has_circRNA_038651 | 1.88 | 0.0407 | 10.04 |
| has_circRNA_017502 | 1.71 | 0.0376 | 6.67 |
| has_circRNA_089763 | 2.39 | 0.0415 | 7.16 |
| has_circRNA_000798 | 3.39 | 0.0056 | 5.28 |
| has_circRNA_102054 | 1.76 | 0.0038 | 4.90 |
| has_circRNA_100075 | 1.60 | 0.0016 | 5.31 |
| has_circRNA_101688 | 1.61 | 0.0059 | 4.99 |
| has_circRNA_002281 | 2.10 | 0.0004 | 6.40 |
| has_circRNA_003558 | 1.61 | 0.0048 | 5.35 |
| B, Downregulated | |||
| circRNA | Fold change | P-value | Normalized intensity |
| hsa_circRNA_012969 | 1.51 | 0.0042 | 5.12 |
| hsa_circRNA_004954 | 1.89 | 0.0028 | 5.27 |
| hsa_circRNA_101006 | 1.76 | 0.0036 | 5.21 |
| hsa_circRNA_031757 | 1.86 | 0.0096 | 5.25 |
| hsa_circRNA_091840 | 1.51 | 0.0193 | 6.78 |
| hsa_circRNA_049784 | 1.61 | 0.0269 | 9.35 |
circRNA, circular RNA; hsa, Homo sapiens.
Figure 3.
Hierarchical clustering plot of differentially expressed circRNAs. Red indicates circRNAs with high expression levels and green indicates circRNAs with low expression levels. Each row represents a different circRNA and each column represents a sample. circRNA, circular RNA; T, patients with post-operative cognitive dysfunction; C, patients without post-operative cognitive dysfunction.
Table IV.
Expression profile of 15 circRNA in the plasma exosomes of patients with post-operative cognitive dysfunction using reverse transcription-quantitative PCR validation.
| circRNA | Fold change | Regulation | P-value | Cq value |
|---|---|---|---|---|
| hsa_circRNA_038651 | 6.1 | Up | 0.26321 | 34.0±0.8 |
| hsa_circRNA_017502 | 1.5 | Up | 0.20196 | 35.0±0.7 |
| hsa_circRNA_089763 | 2.1 | Up | 0.00055 | 34.7±0.7 |
| hsa_circRNA_000798 | – | – | – | >40 |
| hsa_circRNA_102054 | – | – | – | – |
| hsa_circRNA_100075 | – | – | – | >40 |
| hsa_circRNA_101688 | – | – | – | >40 |
| hsa_circRNA_002281 | – | – | – | >40 |
| hsa_circRNA_003558 | – | – | – | >40 |
| hsa_circRNA_012969 | – | – | – | >40 |
| hsa_circRNA_004954 | – | – | – | >40 |
| hsa_circRNA_101006 | – | – | – | >40 |
| hsa_circRNA_031757 | – | – | – | >40 |
| hsa_circRNA_091840 | 0.8 | Down | 0.66827 | 35.7±0.6 |
| hsa_circRNA_049784 | 0.8 | Down | 0.22230 | 32.1±0.3 |
circRNA, circular RNA; RT-qPCR; hsa, Homo sapiens.
Predicting the miRNAs regulated by circRNA_089763 and their corresponding target genes
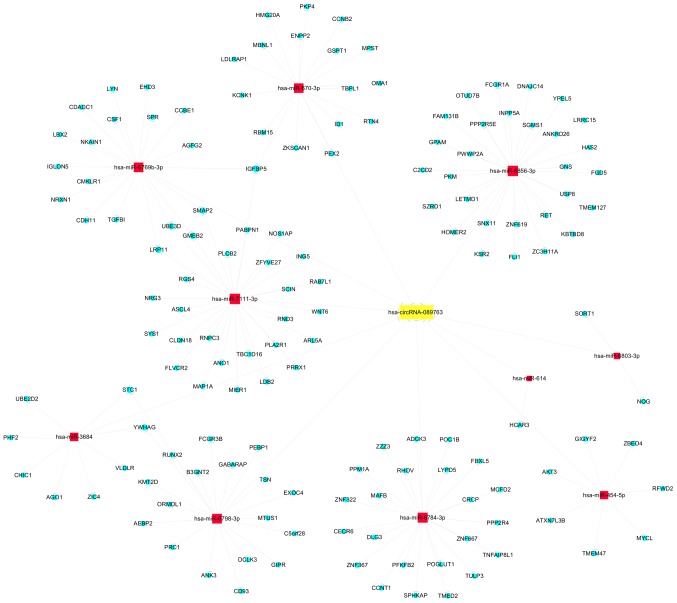
circRNAs act as miRNA sponges, binding and inhibiting miRNAs from regulating their target genes, and are also known as ceRNAs (11). As shown in Fig. 4, circRNA_089763 may sequester 10 miRNAs, including hsa-miR-670-3p, hsa-miR-6856-3p, hsa-miR-6769b-3p, hsa-miR-7111-3p, hsa-miR-6803-3p, hsa-miR-614, hsa-miR-6784-3p, hsa-miR-454-5p, hsa-miR-6798-3p and hsa-miR-3684. Each of these miRNAs regulates target genes. To identify these genes, prediction analysis was performed using the mirdb V5 database. As shown in Fig. 4, circRNA-089763 can indirectly regulate a number of target genes through endogenous competition. circRNA-089763 can bind miR-7111-3p, miR-6769b-3p and miR-670-3p simultaneously. These three microRNAs regulate the expression of insulin-like growth factor binding protein 5 (IGFBP5). A total of two other miRNAs, miR-3684 and miR-6798-3p, regulate the expression of stanniocalcin 1 (STC-1) and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein γ (YWHAG).
Figure 4.
Predicted miRNAs sequestered by circRNA_089763 and their corresponding target genes. miRNA/miR, microRNA; circRNA, circular RNA; hsa, Homo sapiens.
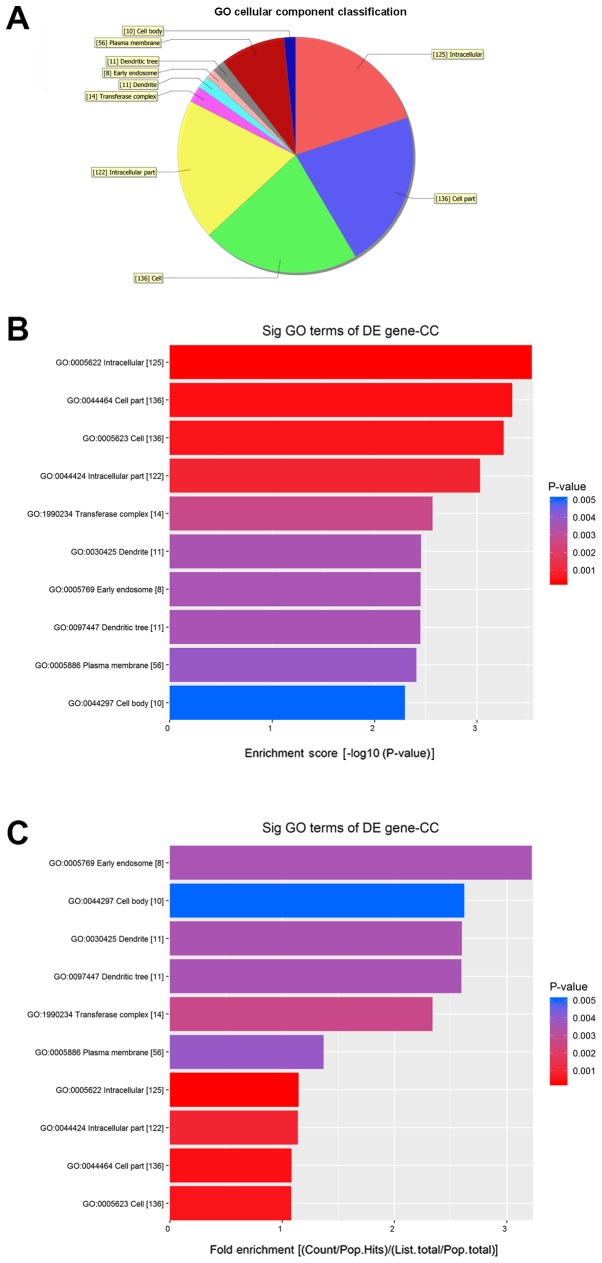
GO analysis results
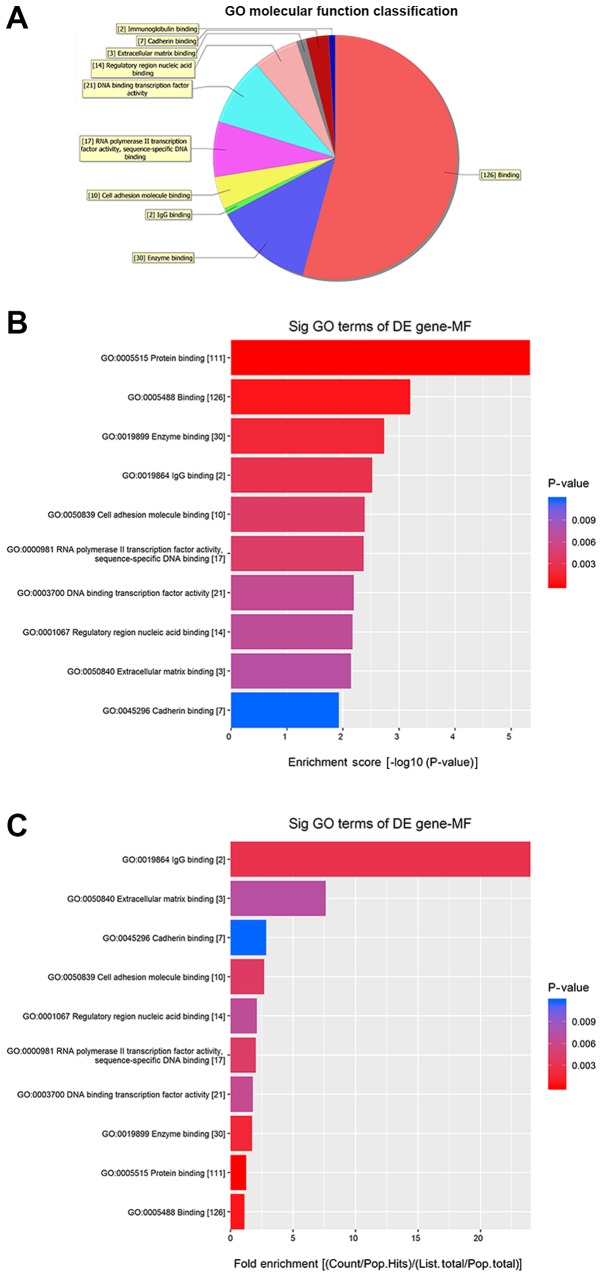
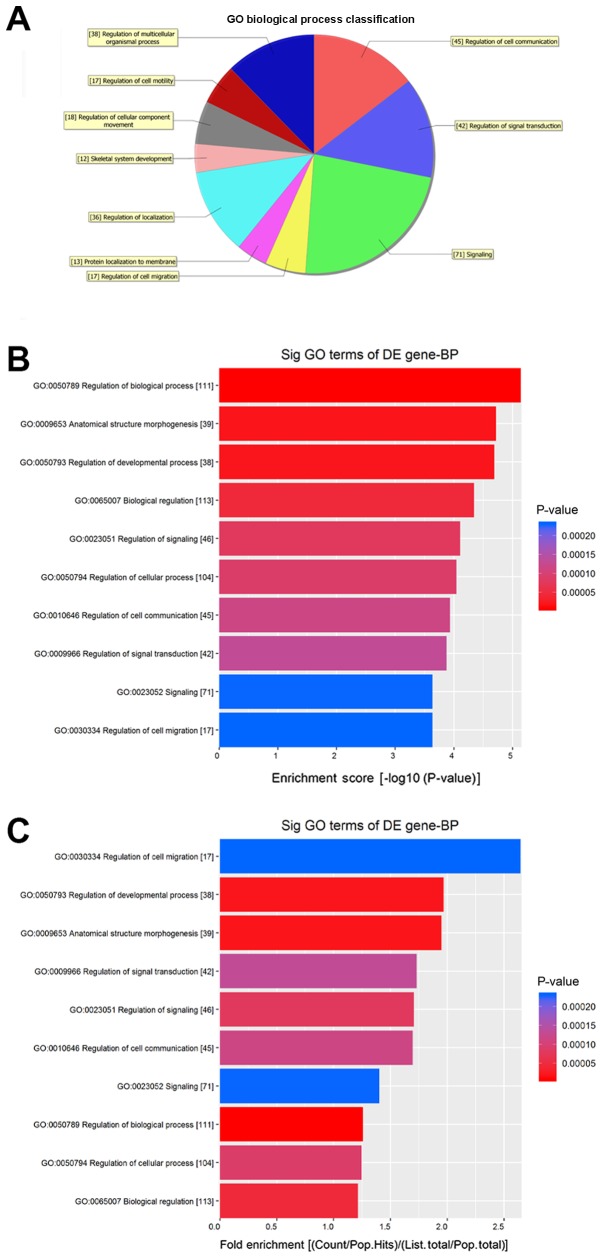
Fig. 5 shows the molecular functions (MFs) of neurons that may be regulated by the target genes of circRNA_089763 (P<0.05). Fig. 5A shows the classifications of these molecular functions. Fig. 5B orders the molecular functions based on their GO analysis enrichment scores. Fig. 5C shows the cellular processes, predicted using fold enrichment, regulated by the target genes. Fig. 6 shows the biological processes in neurons that may be regulated by the target genes of circRNA_089763 (P<0.05). Fig. 6A shows the classifications of the predicted biological processes and Fig. 6B orders the biological processes based on their GO analysis enrichment score. Fig. 6C shows the biological processes, predicted using fold enrichment, regulated by the target genes. Fig. 7 shows the cellular components that may be regulated by the target genes of circRNA_089763 (P<0.05). Fig. 7A shows the classifications of these cellular components and Fig. 7B orders the cellular components based on their GO analysis enrichment scores. Fig. 7C shows the cellular components, predicted using fold enrichment, regulated by the target genes.
Figure 5.
Molecular functions that were significantly enriched by the target genes of circular RNA_089763 based on GO analysis. (A) Classification of the predicted molecular functions. (B) The molecular functions ordered based on their GO analysis enrichment scores. (C) The cellular processes, predicted using fold enrichment, regulated by the target genes. GO, gene ontology; DE, differentially expressed; MF, molecular functions; sig, significant.
Figure 6.
Biological processes that were significantly enriched by the target genes of circular RNA_089763 based on GO analysis. (A) Classification of the predicted biological processes. (B) The biological processes ordered based on their GO analysis enrichment scores. (C) The biological processes, predicted using fold enrichment, regulated by the target genes. GO, gene ontology; DE, differentially expressed; BP, biological processes; sig, significant.
Figure 7.
Cellular component that were significantly enriched by the target gene of circular RNA_089763 based on GO analysis. (A) Classification of the predicted notable cellular components. (B) The cellular components ordered based on their GO analysis enrichment scores. (C) The cellular components, predicted using fold enrichment, regulated by the target genes. GO, gene ontology; DE, differentially expressed; CC, cellular components, sig, significant.
KEGG pathway analysis results
Table V shows the signaling pathways regulated by target genes of circRNA_089763 as revealed by KEGG pathway analysis (P<0.05). The identified pathways are involved in the regulation of central nervous system functions, including the ‘sphingolipid signaling pathway’, ‘FoxO signaling pathway’, ‘hippo signaling pathway’, ‘mRNA surveillance pathway’, ‘endocytosis’ and ‘glucagon signaling pathway’.
Table V.
Signaling pathways regulated by targeted genes of circular RNA_089763 (P<0.05).
| PathwayID | Definition | Fisher P-value | Enrichment_Score |
|---|---|---|---|
| hsa04666 | Fc gamma R-mediated phagocytosis-Homo sapiens (human) | 0.004481613 | 2.348566 |
| hsa04071 | Sphingolipid signaling pathway-Homo sapiens (human) | 0.01110983 | 1.954293 |
| hsa05230 | Central carbon metabolism in cancer-Homo sapiens (human) | 0.01217405 | 1.914565 |
| hsa04380 | Osteoclast differentiation-Homo sapiens (human) | 0.01462926 | 1.834777 |
| hsa05203 | Viral carcinogenesis-Homo sapiens (human) | 0.01612574 | 1.79248 |
| hsa04068 | FoxO signaling pathway-Homo sapiens (human) | 0.01621421 | 1.790104 |
| hsa04390 | Hippo signaling pathway-Homo sapiens (human) | 0.02684785 | 1.57109 |
| hsa04934 | Cushing syndrome-Homo sapiens (human) | 0.02684785 | 1.57109 |
| hsa03015 | mRNA surveillance pathway-Homo sapiens (human) | 0.02960263 | 1.52867 |
| hsa04144 | Endocytosis-Homo sapiens (human) | 0.03379667 | 1.471126 |
| hsa05165 | Human papillomavirus infection-Homo sapiens (human) | 0.03837845 | 1.415913 |
| hsa04922 | Glucagon signaling pathway-Homo sapiens (human) | 0.04051731 | 1.392359 |
| hsa04973 | Carbohydrate digestion and absorption-Homo sapiens (human) | 0.04185221 | 1.378282 |
| hsa05152 | Tuberculosis-Homo sapiens (human) | 0.04307338 | 1.365791 |
| hsa04928 | Parathyroid hormone synthesis, secretion and action-Homo sapiens (human) | 0.04352435 | 1.361268 |
| hsa05202 | Transcriptional misregulation in cancer-Homo sapiens (human) | 0.04842783 | 1.314905 |
hsa, Homo sapiens.
Discussion
Many patients develop POCD after heart surgery (26). POCD is a significant risk factor for a decline in quality of life after CABG (27). Thus, it is important to find methods for the prevention and treatment of POCD. POCD is difficult to accurately diagnose and few effective therapies are available; this is in part due to the fact that the mechanism of POCD remains unclear (28).
Because the pathophysiological mechanism of POCD is complex and the clinical efficacy of drugs is poor, a more comprehensive method to treat POCD is required (2). For many years, researchers have focused their work on protein-coding genes and the proteins encoded by these genes (10). Advances in big data research have revealed that only a small fraction of the genetic material encodes proteins. A large number of transcriptional products result from functionally diverse ncRNAs, including miRNA, lncRNAs and circRNAs. It is likely that the role of a single miRNA in the regulation of a specific target gene is relatively weak. However, a factor that could enrich multiple miRNAs that regulate the same specific target gene could greatly enhance the regulation of that gene.
circRNA molecules are different from traditional linear RNA, with 5′ and 3′ ends, in that they have a closed-ring structure that is unaffected by RNA exonucleases, has a more stable expression and is not easily degraded (29). Research has shown that circRNA molecules are rich in miRNA binding sites and that they act as an miRNA sponge. By functioning as ceRNAs, circRNAs can relieve the inhibitory effects of an miRNA on its target gene, thereby increasing the expression of that target gene (30,31). However, to the best of our knowledge, no circRNAs have been reported to be associated with POCD. Therefore, in the present study, the aim was to identify and investigate the potential role of circRNAs in POCD.
To facilitate the clinical diagnosis of cognitive function impairment, a number of studies have focused on biomarkers based on molecules found in the cerebrospinal fluid (CSF) and peripheral blood (32). However, it is difficult to obtain CSF from patients with POCD by lumbar puncture. Therefore, blood-based biomarkers for cognitive function impairment for routine testing would be more suitable. One approach could be to screen for plasma-specific circRNAs as biomarkers for POCD. Exosomes are membrane vesicles of 40–100 nm that are released from numerous cell types across the body. Exosomes contain various substances, including ncRNAs, lipids and a variety of proteins (33). It has been reported that exosomes may serve as vesicular carriers for intercellular communication in neurodegenerative disorders (34). A previous study described neuronal-derived exosomal proteins in serum from healthy people and patients with Alzheimer's disease (AD) (35). Accumulating evidence indicates that exosomes are important in the pathogenesis of AD (33,35,36). In the present study, candidate circRNAs were detected in the plasma exosomes of patients with POCD. These circRNAs were further investigated to determine whether they could provide information about regulatory factors involved in POCD or could be utilized as new biomarkers in the diagnosis and treatment of POCD.
The results in this present study showed that circRNAs from plasma exosomes from patients with POCD could be detected when there was sufficient quality and quantity of plasma exosomes. The RNA purity, concentration and total quantity from 8 out of 12 samples from patients with POCD and 16 out of 23 samples from patients without POCD met these requirements.
circRNA changes in the plasma exosomes of patients with POCD were examined and it was found that the expression of circRNA-089763 was significantly increased in patients with POCD. Different levels of change were detected for circRNAs when comparing the microarray and RT-qPCR method. However, the RT-qPCR results are more credible. Data obtained using a chip, for example in microarray experiments, provide a reference. This is due to the many unspecific factors in the hybridization of the chip and the number of objects measured by the chip may also cause distortion of the results. RT-qPCR results are generally taken as the standard (37).
As to the best of our knowledge, no previous studies had investigated the role of circRNAs in POCD, the regulation of target genes and signaling pathways required in-depth analysis. Structural prediction analysis indicated that the miRNAs sequestered by circRNA-089763 included hsa-miR-670-3p, hsa-miR-6856-3p, hsa-miR-6769b-3p, hsa-miR-7111-3p, hsa-miR-6803-3p, hsa-miR-614, hsa-miR-6784-3p, hsa-miR-454-5p, hsa-miR-6798-3p and hsa-miR-3684. Therefore, circRNAs, including circRNA-089763, could potentially induce the loss of function of multiple miRNAs. Using circRNAs to control the expression of a target gene would be expected to have a greater impact on gene function than using a single miRNA targeting the same gene. This hypothesis needs further testing in future studies.
In addition, according to the circRNA-miRNA-mRNA network analysis conducted in this present study, it was found that circRNA-089763 may function with miR-7111-3p, miR-6769b-3p and miR-670-3p simultaneously, and that these three miRNAs may regulate the IGFBP5 gene. IGFBP5 is involved in the regulation of amyloid-β and cognitive functions (38–40). circRNA-089763 may enrich miR-3684 and miR-6798-3p, two microRNAs that regulate STC-1 and YWHAG. Shahim et al (41) found that the level of STC-1 in cerebrospinal fluid is a potential biomarker in the differential diagnosis of dementia. Ramocki et al (42) reported that the YWHAG gene is involved in changes in cognitive and behavioral function. Therefore, it is hypothesized that the pathogenesis of POCD is associated with the abnormal expression levels of circRNA-089763 caused by peri-operative-related stimuli. circRNA-089763 may indirectly regulate cognitive function by affecting the expression of the IGFBP5, STC and YWHAG genes by sequestering the corresponding microRNAs. As proteins are the factors that carry out the functions of the target genes, it is important to investigate the association between circRNAs and POCD at the protein level in the future.
GO analysis of circRNA-089763 showed that its target genes regulated many molecular functions, including ‘protein binding’, ‘cell adhesion molecule binding’ and ‘DNA binding transcription factor activity’; biological processes, including ‘regulation of biological process’, ‘regulation of signaling’ and ‘regulation of developmental process’; and cellular components, including ‘dendritic tree’, ‘neuronal cell body’ and ‘postsynaptic density’. KEGG pathway analysis revealed that some of the signaling pathways regulated by the target genes of circRNA-089763 are involved in ‘sphingolipid signaling pathway’, ‘FoxO signaling pathway’, ‘hippo signaling pathway’, ‘mRNA surveillance pathway’, ‘endocytosis’ and ‘glucagon signaling pathway’. Many of these signaling pathways are closely associated with the occurrence and development of cognitive functions. Previous studies of signaling pathways are consistent with the GO and KEGG pathway analysis predictions in this present paper (43–51). For example, the sphingolipid signaling pathway was reported to play a role in AD and neurodegenerative diseases (43–45). The FoxO signaling pathway has also been shown to be involved in the pathogenesis of AD (46). Neuroinflammation has emerged as an important cause of cognitive decline during aging and in AD, and the glucagon signaling pathway has been shown to be related to neuroinflammation and cognitive function (47–50). Yang et al (51) reported a link between central autophagy and cognitive dysfunction.
Due to the multifaceted mechanisms underlying POCD, novel therapies should consider the multiple aspects of molecular imbalance. As miRNAs are involved in several pathophysiological aspects of POCD, they could serve as novel targets for POCD therapies. circRNAs may be used as steady-state regulators in locations associated with cognitive function or POCD. Although overexpression of circRNAs acting as miRNA sponges would increase the translation of their target genes, a decrease in circRNA expression would result in the inappropriate silencing of an array of downstream target genes. To prevent circRNA dysfunction in the body, artificial circRNA mimics could be developed to restore normal transcriptional regulation.
Due to the limited number of clinical CABG patients, the number of samples in this present study was low. However, this present study is the basis for further research. To increase the sample size, other surgical diseases will be considered in future studies to further investigate whether circRNA_089763 is elevated in the circulating blood of patients with POCD. The genes and pathways related to circRNA-089763 require further verification. In addition, a unified world standard for POCD is still lacking, the diagnostic criteria used in this present study are based on the criteria used in a previous study (10).
In summary, the present study found that POCD resulted in the abnormal expression of circRNA-089763 in patients who underwent CABG. circRNA-089763 may be a novel biomarker that could be utilized for the treatment of POCD after CABG.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Projects of the National Natural Science Foundation of China (grant no. 81271478) and the Department of Science & Technology of Sichuan Province (grant no. 14JC0093).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
MW, XW and SG designed and performed the experiments, analyzed the data and wrote the manuscript. MW, PS, YL and XZ provided the patient samples, conceived the study and revised the manuscript. MW, JY, XW and XA performed the experiments, analyzed the data and revised the manuscript. XW and SG designed the experiments, provided reagents and revised the manuscript.
Ethics approval and consent to participate
The study was performed in accordance with the Declaration of Helsinki, and was approved by the Ethics Committee of Beijing Chaoyang Hospital (no. 2017-221-1). The participants provided written informed consent to participate in this study.
Patient consent for publication
Consent for publication was obtained from all participants.
Competing interests
The authors declare that they have no competing interests.
References
- 1.O'Brien H, Mohan H, Hare C, Reynolds J, Kenn R. Mind over matter? The hidden epidemic of cognitive dysfunction in the older surgical patient. Ann Surg. 2017;265:677–691. doi: 10.1097/SLA.0000000000001900. [DOI] [PubMed] [Google Scholar]
- 2.Skvarc D, Berk M, Byrne L, Dean O, Dodd S, Lewis M, Marriott A, Moore EM, Morris G, Page RS, Gray L. Post-operative cognitive dysfunction: An exploration of the inflammatory hypothesis and novel therapies. Neurosci Biobehav Rev. 2018;84:116–133. doi: 10.1016/j.neubiorev.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Kulason K, Nouchi R, Hoshikawa Y, Noda M, Okada Y, Kawashima R. Indication of cognitive change and associated risk factor after thoracic surgery in the elderly: A pilot study. Front Aging Neurosci. 2017;9:396. doi: 10.3389/fnagi.2017.00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schenning KJ, Murchison CF, Mattek NC, Kaye JA, Quinn JF. Sex and genetic differences in postoperative cognitive dysfunction: A longitudinal cohort analysis. Biol Sex Differ. 2019;10:14. doi: 10.1186/s13293-019-0228-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boodhwani M, Rubens FD, Wozny D, Rodriguez R, Alsefaou A, Hendry PJ, Nathan HJ. Predictors of early neurocognitive deficits in low-risk patients undergoing on-pump coronary artery bypass surgery. Circulation. 2006;114:I461–I466. doi: 10.1161/CIRCULATIONAHA.105.001354. [DOI] [PubMed] [Google Scholar]
- 6.Liu YH, Wang DX, Li LH, Wu XM, Shan GJ, Su Y, Li J, Yu QJ, Shi CX, Huang YN, Sun W. The effects of cardiopulmonary bypass on the number of cerebral microemboli and the incidence of cognitive dysfunction after coronary artery bypass graft surgery. Anesth Analg. 2009;109:1013–1022. doi: 10.1213/ane.0b013e3181aed2bb. [DOI] [PubMed] [Google Scholar]
- 7.Bishawi M, Hattler B, Almassi GH, Spertus JA, Quin JA, Collins JF, Grover FL, Shroyer AL. Preoperative factors associated with worsening in health-related quality of life following coronary artery bypass grafting in the Randomized On/Off Bypass (ROOBY) trial. Am Heart J. 2018;198:33–38. doi: 10.1016/j.ahj.2017.12.014. [DOI] [PubMed] [Google Scholar]
- 8.Needham MJ, Webb CE, Bryden DC. Postoperative cognitive dysfunction and dementia: What we need to know and do. Br J Anaesth. 2017;119:i115–i125. doi: 10.1093/bja/aex354. [DOI] [PubMed] [Google Scholar]
- 9.Rundshagen I. Postoperative cognitive dysfunction. Dtsch Arztebl Int. 2014;111:119–125. doi: 10.3238/arztebl.2014.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva FP, Schmidt AP, Valentin LS, Pinto KO, Zeferino SP, Oses JP, Wiener CD, Otsuki DA, Tort AB, Portela LV, et al. S100B protein and neuron-specific enolase as predictors of cognitive dysfunction after coronary artery bypass graft surgery: A prospective observational study. Eur J Anaesthesiol. 2016;33:681–689. doi: 10.1097/EJA.0000000000000450. [DOI] [PubMed] [Google Scholar]
- 11.Deng L, Liu G, Zheng C, Zhang L, Kang Y, Yang F. Circ-LAMP1 promotes T-cell lymphoblastic lymphoma progression via acting as a ceRNA for miR-615-5p to regulate DDR2 expression. Gene. 2019;701:146–151. doi: 10.1016/j.gene.2019.03.052. [DOI] [PubMed] [Google Scholar]
- 12.Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. 2017;14:1028–1034. doi: 10.1080/15476286.2016.1255398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C, Yao M, Li C, Shan K, Yang H, Wang J, Liu B, Li XM, Yao J, Jiang Q, Yan B. Silencing Of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics. 2017;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han B, Chao J, Yao H. Circular RNA and its mechanisms in disease: From the bench to the clinic. Pharmacol Ther. 2018;187:31–44. doi: 10.1016/j.pharmthera.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Floris G, Zhang L, Follesa P, Sun T. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol. 2017;54:5156–5165. doi: 10.1007/s12035-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, Zheng Q, Li Y, Wang P, He X, Huang S. ExoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otero-Ortega L, Laso-García F, Gómez-de Frutos M, Fuentes B, Diekhorst L, Díez-Tejedor E, Gutiérrez-Fernández M. Role of exosomes as a treatment and potential biomarker for stroke. Transl Stroke Res. 2018;10:241–249. doi: 10.1007/s12975-018-0654-7. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Zhang S, Zhang J, Zhu J, He H, Zhang Y, Zhang W, Tian D. Construct validity and reliability of the test your memory chinese version in older neurology outpatient attendees. Int J Ment Health Syst. 2018;12:64. doi: 10.1186/s13033-018-0240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seo EH, Lee DY, Choo IH, Kim SG, Kim KW, Youn JC, Jhoo JH, Woo JI. Normative study of the stroop color and word test in an educationally diverse elderly population. Int J Geriatr Psychiatry. 2008;23:1020–1027. doi: 10.1002/gps.2027. [DOI] [PubMed] [Google Scholar]
- 20.Płotek W, Łyskawa W, Kluzik A, Grześkowiak M, Podlewski R, Żaba Z, Drobnik L. Evaluation of the trail making test and interval timing as measures of cognition in healthy adults: Comparisons by age, education, and gender. Med Sci Monit. 2014;20:173–181. doi: 10.12659/MSM.889776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forn C, Belloch V, Bustamante JC, Garbin G, Parcet-Ibars MA, Sanjuan A, Ventura N, Avila C. A symbol digit modalities test version suitable for functional MRI studies. Neurosci Lett. 2009;456:11–14. doi: 10.1016/j.neulet.2009.03.081. [DOI] [PubMed] [Google Scholar]
- 22.Geffen GM, Butterworth P, Geffen LB. Test-retest reliability of a new form of the auditory verbal learning test (AVLT) Arch Clin Neuropsychol. 1994;9:303–316. doi: 10.1016/0887-6177(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 23.Benedict RH, DeLuca J, Phillips G, LaRocca N, Hudson LD, Rudick R, Multiple Sclerosis Outcome Assessments Consortium Validity of the symbol digit modalities test as a cognition performance outcome measure for multiple sclerosis. Mult Scler. 2017;23:721–733. doi: 10.1177/1352458517690821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Criteria Committee of the New York Heart Association, corp-author. Nomenclature and criteria for diagnosis of diseases of the heart and great vessels. In: Dolgin M, editor. 9th. Little, Brown & Co; Boston: 1994. pp. 253–256. [Google Scholar]
- 25.Li W, Freudenberg J, Suh YJ, Yang Y. Using volcano plots and regularized-chi statistics in genetic association studies. Comput Biol Chem. 2014;48:77–83. doi: 10.1016/j.compbiolchem.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Gilman S. Cerebral disorders after open-heart operations. N Engl J Med. 1965;272:489–498. doi: 10.1056/NEJM196503112721001. [DOI] [PubMed] [Google Scholar]
- 27.Nijs K, Vandenbrande J, Vaqueriza F, Ory JP, Yilmaz A, Starinieri P, Dubois J, Jamaer L, Arijs I, Stessel B. Neurological outcome after minimal invasive coronary artery surgery (NOMICS): Protocol for an observational prospective cohort study. BMJ Open. 2017;7:e017823. doi: 10.1136/bmjopen-2017-017823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cascella M, Bimonte S. The role of general anesthetics and the mechanisms of hippocampal and extra-hippocampal dysfunctions in the genesis of postoperative cognitive dysfunction. Neural Regen Res. 2017;12:1780–1785. doi: 10.4103/1673-5374.219032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Rossum D, Verheijen BM, Pasterkamp RJ. Circular RNAs: Novel regulators of neuronal development. Front Mol Neurosci. 2016;9:74. doi: 10.3389/fnmol.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji MH, Yuan HM, Zhang GF, Li XM, Dong L, Li WY, Zhou ZQ, Yang JJ. Changes in plasma and cerebrospinal fluid biomarkers in aged patients with early postoperative cognitive dysfunction following total hip-replacement surgery. J Anesth. 2013;27:236–242. doi: 10.1007/s00540-012-1506-3. [DOI] [PubMed] [Google Scholar]
- 33.Iranifar E, Seresht BM, Momeni F, Fadaei E, Mehr MH, Ebrahimi Z, Rahmati M, Kharazinejad E, Mirzaei H. Exosomes and microRNAs: New potential therapeutic candidates in alzheimer disease therapy. J Cell Physiol. 2018;234:2296–2305. doi: 10.1002/jcp.27214. [DOI] [PubMed] [Google Scholar]
- 34.Frühbeis C, Fröhlich D, Kuo WP, Amphornrat J, Thilemann S, Saab AS, Kirchhoff F, Möbius W, Goebbels S, Nave KA, et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang TT, Liu CG, Gao SC, Zhang Y, Wang PC. The serum exosome derived microRNA-135a, −193b, and −384 were potential alzheimer's disease biomarkers. Biomed Environ Sci. 2018;31:87–96. doi: 10.3967/bes2018.011. [DOI] [PubMed] [Google Scholar]
- 36.Ding M, Shen Y, Wang P, Xie Z, Xu S, Zhu Z, Wang Y, Lyu Y, Wang D, Xu L. Exosomes isolated from human umbilical cord mesenchymal stem cells alleviate neuroinflammation and reduce amyloid-beta deposition by modulating microglial activation in alzheimer's disease. Neurochem Res. 2018;43:2165–2177. doi: 10.1007/s11064-018-2641-5. [DOI] [PubMed] [Google Scholar]
- 37.Zhou M, Wang MH, Wang XB, Liu KZ, Wan YQ, Li M, Liu L, Zhang C. Abnormal expression of microRNAs induced by chronic unpredictable mild stress in rat hippocampal tissues. Mol Neurobiol. 2018;55:917–913. doi: 10.1007/s12035-016-0365-6. [DOI] [PubMed] [Google Scholar]
- 38.Barucker C, Sommer A, Beckmann G, Eravci M, Harmeier A, Schipke CG, Brockschnieder D, Dyrks T, Althoff V, Fraser PE, et al. Alzheimer amyloid peptide aβ42 regulates gene expression of transcription and growth factors. J Alzheimers Dis. 2015;44:613–624. doi: 10.3233/JAD-141902. [DOI] [PubMed] [Google Scholar]
- 39.Fernandes J, Vieira AS, Kramer-Soares JC, Da Silva EA, Lee KS, Lopes-Cendes I, Arida RM. Hippocampal microRNA-mRNA regulatory network is affected by physical exercise. Biochim Biophys Acta Gen Subj. 2018;1862:1711–1720. doi: 10.1016/j.bbagen.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 40.Jing XP, Peng QH, Hu R, Zou HW, Wang HZ, Yu XQ, Zhou JW, Degen A, Wang ZS. Dietary supplements during the cold season increase rumen microbial abundance and improve rumen epithelium development in tibetan sheep. J Anim Sci. 2018;96:293–305. doi: 10.1093/jas/skx032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahim P, Blennow K, Johansson P, Svensson J, Lista S, Hampel H, Andersson LC, Zetterberg H. Cerebrospinal fluid stanniocalcin-1 as a biomarker for alzheimer's disease and other neurodegenerative disorders. Neuromolecular Med. 2017;19:154–160. doi: 10.1007/s12017-016-8439-1. [DOI] [PubMed] [Google Scholar]
- 42.Ramocki MB, Bartnik M, Szafranski P, Kolodziejska KE, Xia Z, Bravo J, Miller GS, Rodriguez DL, Williams CA, Bader PI, et al. Recurrent distal 7q11.23 deletion including HIP1 and YWHAG identified in patients with intellectual disabilities, epilepsy, and neurobehavioral problems. Am J Hum Genet. 2010;87:857–865. doi: 10.1016/j.ajhg.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noel A, Ingrand S, Barrier L. Inhibition of GSK3β by pharmacological modulation of sphingolipid metabolism occurs independently of ganglioside disturbance in a cellular model of Alzheimer's disease. Exp Neurol. 2015;271:308–318. doi: 10.1016/j.expneurol.2015.06.021. [DOI] [PubMed] [Google Scholar]
- 44.Mielke MM, Lyketsos CG. Alterations of the sphingolipid pathway in Alzheimer's disease: New biomarkers and treatment targets? Neuromolecular Med. 2010;12:331–340. doi: 10.1007/s12017-010-8121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ariga T, Jarvis WD, Yu RK. Role of sphingolipid-mediated cell death in neurodegenerative diseases. J Lipid Res. 1998;39:1–16. [PubMed] [Google Scholar]
- 46.Wang X, Wang Z, Chen Y, Huang X, Hu Y, Zhang R, Ho MS, Xue L. FoxO mediates APP-induced AICD-dependent cell death. Cell Death Dis. 2014;5:e1233. doi: 10.1038/cddis.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin L, Chong T, Rodriguez R, Pugazhenthi S. Glucagon-like peptide-1-mediated modulation of inflammatory pathways in the diabetic brain: Relevance to alzheimer's disease. Current Alzheimer Res. 2016;13:1346–1355. doi: 10.2174/1567205013666160401114751. [DOI] [PubMed] [Google Scholar]
- 48.Tramutola A, Arena A, Cini C, Butterfield DA, Barone E. Modulation of GLP-1 signaling as a novel therapeutic approach in the treatment of Alzheimer's disease pathology. Expert Rev Neurother. 2017;17:59–75. doi: 10.1080/14737175.2017.1246183. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Chen S, Xu Z, Chen S, Yao W, Gao X. GLP-1 receptor agonists downregulate aberrant GnT-III expression in Alzheimer's disease models through the Akt/GSK-3β/β-catenin signaling. Neuropharmacology. 2018;131:190–199. doi: 10.1016/j.neuropharm.2017.11.048. [DOI] [PubMed] [Google Scholar]
- 50.An FM, Chen S, Xu Z, Yin L, Wang Y, Liu AR, Yao WB, Gao XD. Glucagon-like peptide-1 regulates mitochondrial biogenesis and tau phosphorylation against advanced glycation end product-induced neuronal insult: Studies in vivo and in vitro. Neuroscience. 2015;300:75–84. doi: 10.1016/j.neuroscience.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 51.Yang N, Li L, Li Z, Ni C, Cao Y, Liu T, Tian M, Chui D, Guo X. Protective effect of dapsone on cognitive impairment induced by propofol involves hippocampal autophagy. Neuroscience Lett. 2017;649:85–92. doi: 10.1016/j.neulet.2017.04.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.