Abstract
Increasing evidence has suggested the crucial role of the dysregulation of microRNAs (miRNAs) in osteosarcoma (OS) progression. MicroRNA (miR)-330-5p has been reported to exert tumor suppressive effects in various types of human cancer. However, the role of miR-330-5p in the development of OS and the underlying mechanism remain to be clarified. In the present study, miR-330-5p expression was found to be significantly decreased in OS tissues and cell lines. In addition, low miR-330-5p expression was highly correlated with the overall survival and clinical stage of OS. Overexpression of miR-330-5p inhibited the viability, migration and invasion, and promoted the apoptosis of OS cells, as well as induced cell cycle arrest at the G2/M phase. Subsequently, the proto-oncogene survivin was identified as a functional target of miR-330-5p, and this was validated using a luciferase reporter assay. It was also demonstrated that survivin expression was markedly increased in OS tissues, and that it was negatively correlated with the expression of miR-330-5p. Furthermore, overexpression of survivin significantly abrogated the tumor-suppressive effect induced by miR-330-5p on OS cells. In conclusion, these results revealed that the miR-330-5p/survivin axis has a significant tumor-suppressive effect on OS, and may serve as a diagnostic and therapeutic target for the treatment of OS.
Keywords: osteosarcoma, microRNA-330-5p, survivin
Introduction
Osteosarcoma (OS) is a primary bone malignancy that mainly occurs in young adults and adolescents (1). In spite of recent developments in combinational chemotherapy and surgical techniques, the long-term survival of OS patients remains unsatisfactory, with metastatic and recurrent disease observed (2). Thus, there is an urgent need for exploring the pathogenesis of OS, and identifying novel and efficient biomarkers for predicting the prognosis of OS patients.
MicroRNAs (miRNAs) are a type of highly conserved noncoding RNAs with a length of 18–25 nucleotides, which regulate gene expression by binding to the 3′-untranslated regions (UTRs) of their target mRNA in order to repress transcription or induce mRNA degradation (3,4). Currently, growing evidence suggests that the involvement of miRNAs in multiple cellular processes, including cell apoptosis, proliferation and metastasis (5,6). Dysregulated miRNAs may contribute to the development of numerous types of human cancer, including OS. For instance, an evident increase in miR-378 was reported in OS tissues and cells, while miR-378 overexpression promoted the proliferation of OS cells by targeting Krüppel-like factor 9 (7). Liu et al (8) reported decreased expression of miR-935 in OS tissues, and restoration of this expression evidently restricted cell proliferation and invasion. In addition, the study by Yang et al (9) demonstrated that miR-183 suppressed the LDL receptor-related protein 6/Wnt/β-catenin signaling and, thereby, inhibited MG63 cell growth, migration and invasion in vivo and in vitro. Furthermore, Dong et al (10) reported that miR-223 was markedly decreased in the serum of OS patients, suggesting that miR-223 may serve as a potential diagnostic and prognostic biomarker of OS.
The aberrant expression of miR-330-5p has been reported in certain types of cancer, including glioblastoma (11) and pancreatic cancer (12). A study by Wang et al (13) revealed that high miR-330-5p expression was correlated with worse prognosis in patients with breast cancer. It was also reported that miR-330-5p was significantly decreased in cutaneous malignant melanoma (CMM) tissues, and forced expression of miR-330-5p suppressed CMM cell proliferation and invasion (14). In addition, Wei et al (15) demonstrated that miR-330-5p functioned as an oncogene in non-small cell lung cancer (NSCLC) through activating the mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) signaling pathway. Nevertheless, the biological function and clinical value of miR-330-5p in OS remain to be investigated.
In the present study, the expression levels of miR-330-5p in OS tissues and cell lines were investigated, and the correlation between miR-330-5p expression and the clinicopathological characteristics of patients was then analyzed. The study also investigated the effects of miR-330-5p expression on the proliferation, invasion, apoptosis and cell cycle distribution of OS cells. In addition, the regulatory mechanisms of miR-330-5p on OS cells, as well as the potential relationship between miR-330-5p and proto-oncogene survivin (also known as baculoviral IAP repeat-containing protein 5) were investigated. The study findings provide novel insights into the role of miR-330-5p in the development of OS.
Materials and methods
Patients and samples
A total of 63 surgically resected OS tissue specimens were acquired from patients with OS at the Department of Traumatic Orthopaedics at Anhui Provincial Hospital, Anhui Medical University (Hefei, China) between January 2012 and December 2016. The patients were assigned into two groups according to the presence or absence of metastasis, as determined by radiology. The clinicopathological data of the patients are shown in Table I. The clinical stage of the patients was classified according to the Tumor Node Metastasis (TNM) Classification of Malignant Tumors (Sixth edition) from the Union for International Cancer Control (10). In addition, 20 osteochondroma (a benign bone lesion) tumor tissue samples from amputees were selected and served as the Control group; the Control group included 9 males and 11 females, whose ages ranged between 10 and 57 with a mean age of 25.93±10.57. The present study was approved by the Institutional Ethics Review Board of Anhui Provincial Hospital, Anhui Medical University, and informed consent was obtained from adult participant or from the legal guardians of participants <18 years old prior to participation in this study. The samples were immediately snap-frozen in liquid nitrogen and stored at −80°C until further use.
Table I.
Association between miR-330-5p expression and clinicopathological features of osteosarcoma patients.
| miR-330-5p expression | ||||
|---|---|---|---|---|
| Clinicopathological feature | No. of cases | High (n=25) | Low (n=38) | P-value |
| Sex | 0.961 | |||
| Male | 33 | 13 | 20 | |
| Female | 30 | 12 | 18 | |
| Age (years) | 0.493 | |||
| ≥18 | 41 | 15 | 26 | |
| <18 | 22 | 10 | 12 | |
| Tumor location | 0.065 | |||
| Tibia/femur | 39 | 12 | 27 | |
| Other | 24 | 13 | 11 | |
| Clinical stage | 0.028a | |||
| IIA | 45 | 14 | 31 | |
| IIB/III | 18 | 11 | 7 | |
| Tumor size (cm) | 0.274 | |||
| ≥8 | 28 | 9 | 19 | |
| <8 | 35 | 16 | 19 | |
| Distant metastasis | 0.011a | |||
| Present | 19 | 3 | 16 | |
| Absent | 44 | 22 | 22 | |
P<0.05. miR, microRNA.
Cell culture
The OS-derived human cell lines HOS, U2OS and MG63, and the conditionally immortalized human fetal osteoblastic cell line hFOB1.19 were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). The 293T cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). HOS, U2OS, MG63 and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). The hFOB1.19 cells were maintained in a 1:1 mixture of Ham's F12 medium and DMEM supplemented with 2.5 ml glutamine (without phenol red) and 10% FBS. All cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and miRNA was extracted using the mirVana RNA isolation kit (Ambion; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. RNA concentration was measured using NanoDrop ND-1000 (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). RT reactions for miRNA detection were performed using a miRNA Reverse Transcription kit (Qiagen, Inc., Valencia, CA, USA), whereas RT reactions for mRNA detection were conducted using the Takara PrimeScript™ First Strand cDNA Synthesis (Takara Biotechnology Co., Ltd., Dalian, China). Subsequently, an ABI 7500 system (Thermo Fisher Scientific, Inc.) was used to conduct qPCR using a standard protocol described in the SYBR Green PCR kit (Toyobo Life Science, Osaka, Japan). Briefly, the amplification protocol was set as follows: Initial denaturation at 95°C for 5 min; followed by 30 cycles of denaturation at 94°C for 15 sec, annealing at 55°C for 30 sec and extension at 70°C for 30 sec. U6 and GAPDH were used as the internal references and to normalize expression data for miR-330-5p and survivin levels, respectively. The sequences of the primers were as follows: miR-330-5p, forward 5′-TCTCTGGGCCTGTGTCTTAGGC-3′, reverse 5′-GCTATCTCAGGGCTTGTTGCTTCAGTCCTCCTGGG-3′; U6, forward 5′-TGCGGGTGCTCGCTTCGCAGC-3′, reverse 5′-CCAGTGCAGGGTCCGAGGT-3′; survivin, forward 5′-GCACTTTCTTCGCAGTTTC-3′, reverse 5′-GTGAGGTGTGCTGTTCGAGA-3′; GAPDH, forward 5′-AGGTCGGTGTGAACGGATTTG-3′, reverse 5′-TGTAGACCATGTAGTTGAGGTCA-3′. The relative expression levels were calculated using the 2−ΔΔCq method (16).
Cell transfection
miR-330-5p mimics (5′-TCTCTGGGCCTGTGTCTTAGGC-3′), negative control mimics (mimics NC; 5′-GCCTAAGACACAGGCCCAGAGA-3′), miR-330-5p inhibitor and inhibitor NC were purchased from GenePharma Co., Ltd. (Shanghai, China). In addition, the coding domain sequences of survivin mRNA were amplified by PCR and inserted into a pcDNA3.0 overexpression vector (pcDNA-survivin; Invitrogen; Thermo Fisher Scientific, Inc.). Cells were cultured to 80% confluence and subsequently transfected with miR-330-5p mimics (50 nM), control mimics (50 nM), miR-330-5p inhibitor (50 nM) or control inhibitor (50 nM) using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to a previous study (11). pcDNA-survivin (2 µg) and the control empty plasmid pcDNA3.0 (2 µg) was transfected into OS cells with Lipofectamine 2000, following manufacturer's protocol. Transfected cells were incubated in a 37°C incubator with 5% CO2 for 24, 48 or 72 h. For cell viability assay, cell apoptosis assay and cell cycle distribution assay, cells were harvested for further experiments after 48 h of transfection at 37°C. Cell invasion assay and wound-healing assay were performed after 24 h of transfection at 37°C, and images of the stained cells were captured at the next 24 h.
Cell viability assay
For the cell viability assay, U2OS and MG63 cells were seeded in a 96-well plate at the density of 5×103 cells per well and transfected with the miR-330-5p mimics, miR-330-5p inhibitor or pcDNA-survivin. After 24, 48 or 72 h of incubation, the relative viability of the cells was determined using a Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology, Jiangsu, China). Briefly, 10 µl CCK-8 solution was added to each well and incubated at 37°C in a 5% CO2 cell incubator for 90 min. Next, the absorbance rates were measured at 450 nm with a SpectraMax M5 reader (Molecular Devices, LLC, Shanghai, China). All experiments were performed in triplicate.
Cell apoptosis assay
After 48 h of transfection, an Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection kit (Abcam, Cambridge, UK) was used to detect cell apoptosis, according to the manufacturer's protocol. Following harvesting and washing twice with PBS, the cells were stained with Annexin V-FITC and propidium iodide (PI), and incubated for 15 min at room temperature in the dark. Cell apoptosis was subsequently detected using a FACScan flow cytometer (Beckman Coulter, Inc., Brea, CA, USA).
Cell cycle distribution assay
After 48 h of transfection, U2OS and MG63 cells were harvested, washed twice with PBS and fixed with 70% ethanol at 4°C overnight, washed with PBS twice and incubated with 100 µl RNaseA at 37°C in dark for 25 min. The cells were washed with PBS and stained with PI staining solution (50 µg/ml; containing 1 mg/ml RNase A and 0.1% Triton X-100 in PBS). Finally, the cell cycle distribution was assessed with a BD FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). Experiments were performed in triplicate.
Cell invasion Transwell assays
A cell invasion assay was conducted in a 24-well plate with Transwell chamber inserts (pore size, 8 mm; Corning Incorporated, Corning, NY, USA). Transfected cells (1×105 cells) were placed into the upper chamber with Matrigel (BD Biosciences), respectively. The lower chamber was loaded with medium supplemented with 10% FBS. Following incubation for 48 h, the cells on the upper surface of the membrane were removed, while the invading cells on the bottom surface of the Transwell chambers were fixed with methanol for 20 min at room temperature and stained with 0.1% crystal violet for 30 min. Images of the stained cells were captured, and the number of cells was counted under an inverted microscope (Olympus Corporation, Tokyo, Japan). The results were averaged among three independent experiments.
Wound-healing assay
U2OS and MG63 cells (2×105 per well) were plated onto 6-well plates and cultured until confluence of ~90% was reached. After 24 h of transfection, the plates were scrapped with a 10 µl pipet tip, and the medium was replaced with fresh serum-free medium. Initial images were acquired as a reference, and further images of the previously photographed region were collected after 48 h. The percentage of wound-healing rate=[(width at 0 h-width at 48 h)/width at 0 h] ×100.
Bioinformatics analysis and luciferase reporter assay
The miRNA target prediction websites, including PicTar (https://pictar.mdc-berlin.de), TargetScan (http://targetscan.org) and miRDB (http://www.mirdb.org) were used to search for the putative targets of miR-330-5p. Luciferase complexes were constructed by ligating oligonucleotides containing the wild-type (wt) or mutated (mut) putative target site of the survivin 3′-untranslated region (3′UTR) into the multi-cloning site of the pGL3 luciferase reporter vector (Promega Corporation, Madison, WI, USA). Subsequently, 293T cells (1×105 cells/well) were plated in a 24-well plate and co-transfected with 80 ng of pGL3-survivin-3′-UTR (wt) or pGL3-survivin-3′-UTR (mut) and 50 nmol/l of the mimics or inhibitors (including miR-330-5p mimics, mimics NC, miR-330-5p inhibitor and inhibitor NC) using Lipofectamine® 2000, according to the manufacturer's protocol. Cells were transfected for 48 h at 37°C, then luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega Corporation). To correct for differences in transfection and harvesting efficiencies, Renilla luciferase activity was used to normalize the firefly luciferase activity.
Western blot analysis
Total protein was extracted from the cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China), and the total cellular protein concentration was measured using a BCA assay kit (Pierce; Thermo Fisher Scientific, Inc.). Next, total protein samples (40 µg) were separated by 8% SDS-PAGE and subsequently transferred to polyvinylidene difluoride membranes (GE Healthcare, Chicago, IL, USA) by electroblotting. Primary antibodies against survivin (cat no. ab76424; 1:5,000; Abcam, Cambridge, MA, USA) and β-actin (cat no. sc-58673; 1:2,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) were then probed with the proteins on the membrane overnight at 4°C. Following further incubation with secondary antibodies (cat no. 7076; 1:10,000; Cell Signaling Technology, Inc., Danvers, MA, USA) at 37°C for 1 h. Protein bands were visualized with an Enhanced Chemiluminescence kit (GE Healthcare). Finally, ImageJ software (version 1.46; National Institutes of Health, Bethesda, MD, USA) was used to assess the intensity of the bands of interest; the endogenous control β-actin was used to normalize the expression of the selected genes.
Statistical analysis
Statistical analysis was performed using the SPSS program (version 18.0; SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard deviation. Student's t-test or one-way analysis of variance followed by Tukey's post hoc test was used to analyze the difference among two or more than two sample groups, respectively. Spearman's analysis was used to calculate the correlation between the expression of miR-330-5p and survivin. The survival rate of patients was determined using the Kaplan-Meier method, and differences between groups were examined using the log-rank test. P<0.05 was considered to denote a difference that was statistically significant.
Results
Expression of miR-330-5p and its diagnostic value in OS tissues and cells
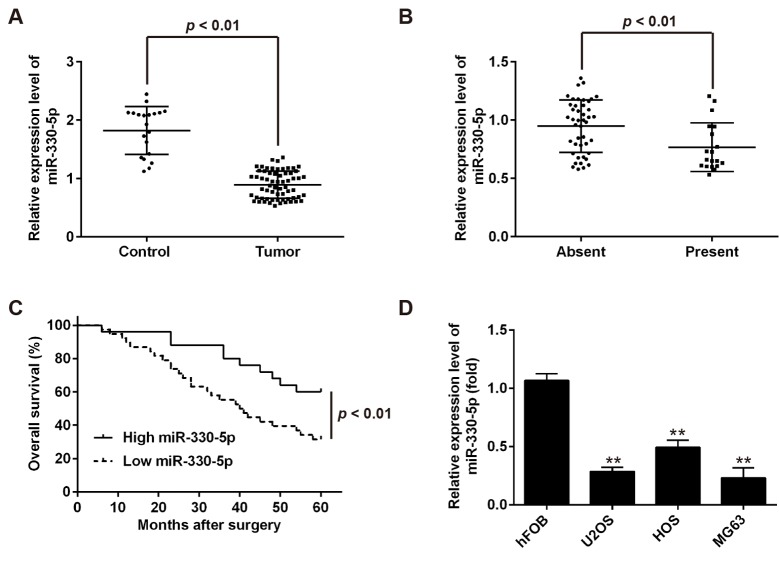
To investigate the potential role of miR-330-5p in OS, the expression of miR-330-5p was first determined in 63 OS tumor and 20 benign osteochondroma (control) tissues by RT-qPCR assay. As shown Fig. 1A, compared with the control group, miR-330-5p was significantly downregulated in OS tissues. Furthermore, the expression of miR-330-5p was markedly lower in tumors with distant metastasis as compared with that in tumors without distant metastasis (Fig. 1B), indicating that miR-330-5p downregulation is associated with OS metastasis. To further determine the clinical value of miR-330-5p, the patients were divided into the high miR-330-5p expression group (n=25) and low expression group (n=38), using the median value of miR-330-5p expression in patients OS as a cutoff point (0.997). The results demonstrated that low miR-330-5p expression was associated with the clinical stage and distant metastasis, but not with age, gender, tumor location and tumor size in the OS patients (Table I). Compared with the patients in the high miR-330-5p expression group, patients in the low miR-330-5p expression group had a significantly lower 5-year overall survival rate (Fig. 1C). Taken together, these findings indicated that miR-330-5p may serve as an effective biomarker for the prognosis of patients with OS.
Figure 1.
miR-330-5p is downregulated in OS tissues and cell lines. (A) miR-330-5p expression was measured by RT-qPCR in tumor tissues (n=63) and control osteochondroma tissues (n=20). (B) miR-330-5p expression was measured in OS tissues with or without distant metastasis by RT-qPCR. (C) Kaplan-Meier survival curve demonstrated the association between the overall survival of osteosarcoma patients and the miR-330-5p levels. (D) miR-330-5p expression was detected in the HOS, U2OS and MG63 cells, and normal hFOB1.19 cells were used as the control. Data represent the mean ± standard deviation of three independent experiments. **P<0.01 vs. hFOB1.19 cells. miR, microRNA; OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
miR-330-5p expression levels were also measured in the HOS, U2OS and MG63 OS cell lines. In accordance with the results in OS tissues, the expression levels of miR-330-5p were significantly downregulated in the HOS, U2OS and MG63 cells, as compared with those in normal hFOB1.19 cells (Fig. 1D). U2OS and MG63 cells were selected for use in further experiments since they exhibited the lowest expression of miR-330-5p.
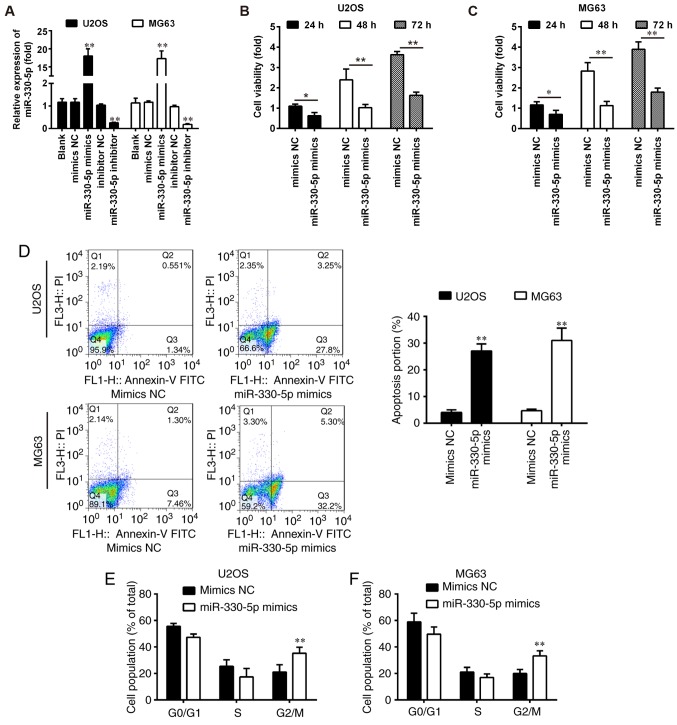
Overexpression of miR-330-5p inhibits OS cell viability, promotes cell apoptosis and induces cell cycle arrest
The downregulation of miR-330-5p in OS tissues suggested that miR-330-5p may function as a tumor suppressor in OS progression. To test this hypothesis, miR-330-5p mimics or inhibitor were transfected into U2OS and MG63 cells. Following miR-330-5p transfection, the expression level of miR-330-5p in the OS cell lines was significantly increased when compared with that in the control groups (Fig. 2A). Conversely, miR-330-5p expression levels were significantly decreased in miR-330-5p inhibitor-transfected cells compared with inhibitor NC-transfected cells (Fig. 2A). Next, the effect of miR-330-5p overexpression on cell viability was examined by a CCK-8 assay. The results revealed that overexpression of miR-330-5p significantly suppressed the viability of U2OS and MG63 cells, as compared with that in the mimics NC group (Fig. 2B and C). Furthermore, the overexpression of miR-330-5p significantly promoted the apoptosis of U2OS and MG63 cells (Fig. 2D). Cell cycle analysis was further performed to demonstrate the underlying mechanisms by which miR-330-5p affected the cell viability. Flow cytometric analysis revealed that the overexpression of miR-330-5p in U2OS and MG63 cells significantly increased the number of cells at G2/M phase (Fig. 2E and F). Taken together, these results suggested that miR-330-5p inhibited cell proliferation by inducing apoptosis and cell cycle arrest at G2/M phase.
Figure 2.
Overexpression of miR-330-5p suppressed cell viability, promoted cell apoptosis and induced cell cycle arrest. (A-F) U2OS and MG63 cells were transfected with 50 nM miR-330-5p mimics, inhibitors, mimics NC or inhibitor NC, and used for subsequent analyses. (A) Transfection efficiency was assessed by reverse transcription-quantitative polymerase chain reaction after transfection for 48 h; **P<0.01 vs. Blank group. (B) U2OS and (C) MG63 cell viability was measured by Cell Counting Kit-8 assay at 24, 48 or 72 h. (D) Cell apoptosis was detected by flow cytometry after transfection for 48 h. Flow cytometry was also performed to observe the cell cycle distribution in (E) U2OS and (F) MG63 cells after transfection for 48 h. Data represent the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01 vs. mimics NC group. miR, microRNA; NC, negative control.
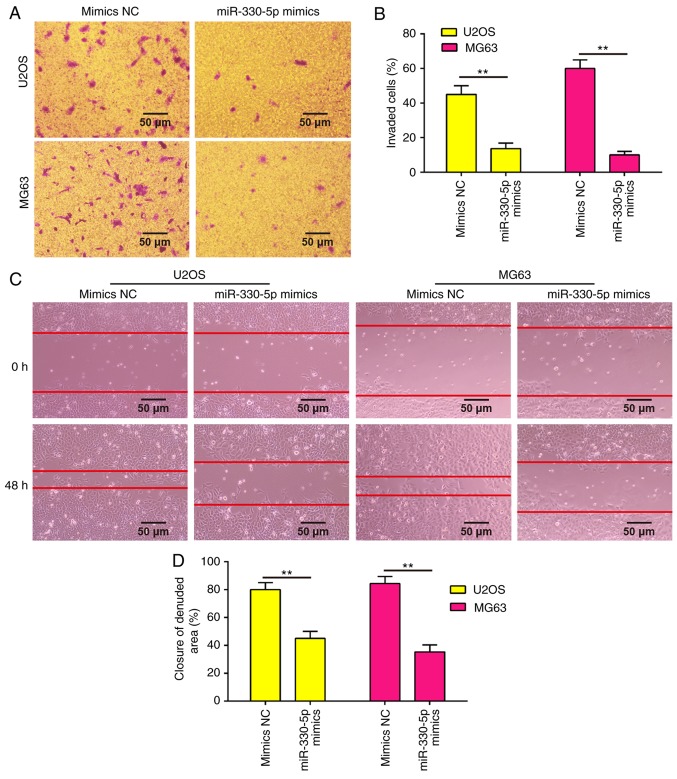
Overexpression of miR-330-5p inhibits the invasion and migration abilities of OS cells
Since the metastatic ability of OS is a critical factor in the poor prognosis of patients, the present study examined whether miR-330-5p is able to modulate the metastatic ability of OS cells using Transwell invasion and wound healing assays. As shown in Fig. 3A and B, overexpression of miR-330-5p in U2OS and MG63 cells significantly decreased the cell invasion ability as compared with the mimics NC group. Similarly, the wound healing assay demonstrated that overexpression of miR-330-5p clearly decreased the cell migration distance as compared with that of cells transfected with negative controls (Fig. 3C and D). Collectively, these data suggested that miR-330-5p suppressed the metastatic ability of OS cells.
Figure 3.
Overexpression of miR-330-5p suppressed cell invasion and migration. U2OS and MG63 cells were transfected with the 50 nM miR-330-5p mimics or mimics-NC for 48 h, and then used for analysis. (A) Wound healing assay and (B) quantified migration abilities of U2OS and MG63 cells. (C) Transwell invasion assay and (D) quantified invasion abilities of U2OS and MG63 cells. Data represent the mean ± standard deviation of three independent experiments. **P<0.01. miR, microRNA; NC, negative control.
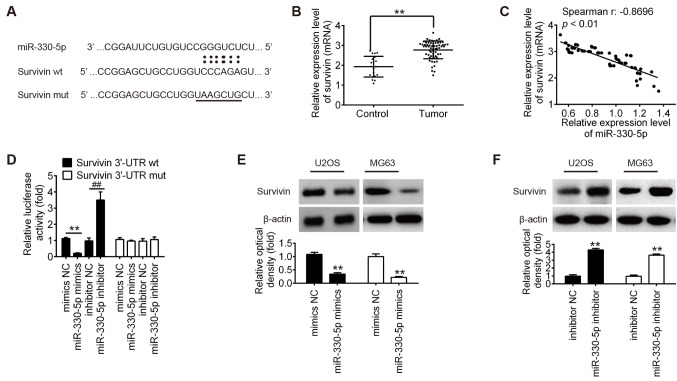
Survivin is a direct target of miR-330-5p
Several studies have reported that miR-330-5p is involved in tumor progression via targeting the 3′-UTR of its target genes (11,14), including MUC1, ITGA5 and Pdia3 (17). To identify new potential target genes of miR-330-5p, the databases PicTar, TargetScan and miRBase were searched for candidate genes. Bioinformatics analysis revealed a miR-330-5p binding site in the 3′-UTR of survivin (Fig. 4A). In addition, it was observed that the mRNA level of survivin was significantly upregulated in OS tissues compared with the control group (Fig. 4B), and miR-330-5p expression was inversely correlated with survivin expression in OS tissues (r=−0.8696; P<0.01; Fig. 4C). To validate whether the 3′-UTR of survivin is a functional target of miR-330-5p, a luciferase reporter assay was performed. Compared with the NC group, introduction of miR-330-5p mimics markedly inhibited the relative luciferase activity of cells co-transfected with the survivin-3′-UTR wt, while miR-330-5p inhibitor significantly increased the luciferase activity of the wt group. However, the luciferase activity of the reporters containing the mut binding site exhibited no evident changes (Fig. 4D). To further investigate whether miR-330-5p regulates survivin expression, the effect of miR-330-5p overexpression or inhibition on survivin protein expression was examined in OS cells. It was clearly observed that overexpression of miR-330-5p inhibited the protein expression of survivin in the U2OS and MG63 cells, whereas knockdown of miR-330-5p promoted survivin expression (Fig. 4E and F). These results demonstrated that miR-330-5p was able to suppress the expression of the proto-oncogene survivin in OS cells by directly targeting the survivin 3′-UTR.
Figure 4.
Survivin is a direct target of miR-330-5p. (A) Schematic of the survivin 3′-UTR containing the miR-330-5p binding site. (B) Survivin expression was measured by reverse transcription-quantitative polymerase chain reaction in OS tissues (n=63) and osteochondroma tissues (n=20). (C) Survivin expression was negatively associated with miR-330-5p expression in OS tissues (r=−0.8696, P<0.01). (D) 293T cells were co-transfected with 80 ng survivin-3′-UTR wt or mut reporter plasmids, and with 50 nM miR-330-5p mimics, mimics NC, miR-330-5p inhibitor or inhibitor NC for 48 h, and relative luciferase activity was measured. (E) U2OS and (F) MG63 cells were transfected with the 50 nM miR-330-5p mimics, mimics-NC, miR-330-5p inhibitor or inhibitor NC for 48 h, and then survivin protein expression was determined by western blot analysis. Data represent the mean ± standard deviation of three independent experiments. **P<0.01. miR, microRNA; OS, osteosarcoma; NC, negative control; 3′-UTR, 3′-untranslated region; wt, wild-type; mut, mutant.
miR-330-5p suppresses cell growth and induces cell apoptosis through targeting survivin
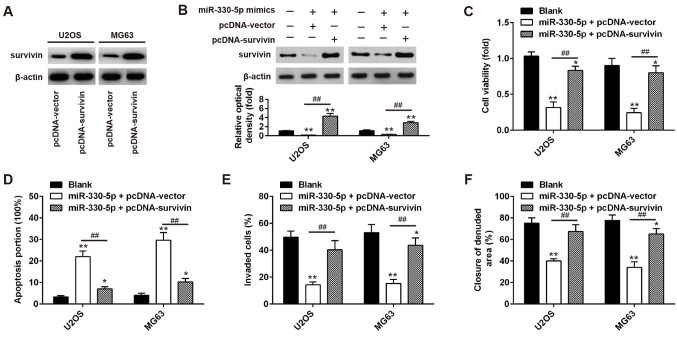
First, the overexpression efficiency of pcDNA-survivin transfection in OS cells was determined (Fig. 5A); the expression levels of survivin in was significantly higher compared with empty vector-transfected cells, which indicated successful transfection of the overexpression vector. To investigate whether miR-330-5p exerts an antitumor effect through regulating survivin, U2OS and MG63 cells were co-transfected with miR-330-5p mimics and pcDNA-survivin plasmid, and cell viability, apoptosis, invasion and migration was assessed. As shown in Fig. 5B, the transfection of cells with survivin expression vectors significantly upregulated survivin expression and restored the decreased survivin expression in miR-330-5p mimics-transfected cells. CCK-8 assay and flow cytometry assays were then conducted to determine the cell viability and apoptosis, respectively. The results demonstrated that overexpression of survivin significantly increased the viability and inhibited apoptosis in miR-330-5p mimics + pcDNA-vector-transfected cells (Fig. 5C and D). Furthermore, the inhibitory effects of miR-330-5p overexpression on the invasion and migration abilities of cells were reversed by survivin overexpression (Fig. 5E and F). These results suggest that miR-330-5p functions as a tumor suppressor in OS, at least partially, through regulating survivin expression.
Figure 5.
miR-330-5p inhibited cell viability and induced cell apoptosis by targeting survivin. (A) U2OS and MG63 cells were transfected with 2 µg pcDNA-survivin or pcDNA-vector for 48 h, and the overexpression efficiency was detected by western blot analysis. (B) U2OS and MG63 cells were co-transfected with the 2 µg pcDNA-survivin and 50 nM miR-330-5p mimics for 48 h, and then used for analysis. Western blot analysis was used to detect the survivin protein level after 48 h of co-transfection. (C) Viability of U2OS and MG63 cells was measured by Cell Counting Kit-8 assay after 48 h of co-transfection. (D) Apoptosis was detected by flow cytometry after 48 h of co-transfection. (E) Migration ability of U2OS and MG63 cells, as measured by the wound healing assay after 48 h of co-transfection. (F) Invasion ability of U2OS and MG63 cells, as measured by the Transwell invasion assay after 48 h of co-transfection. Data represent the mean ± standard deviation of three independent experiments. *P<0.05 and **P<0.01 vs. Blank group in U2OS or MG63 cells; ##P<0.01 vs. miR-330-5p + pcDNA-vector group in U2OS or MG63 cells. miR, microRNA; NC, negative control.
Discussion
In the present study, the results indicated that miR-330-5p was significantly downregulated in OS tissues and cell lines, and its low expression was closely associated with the clinical stage and overall survival of OS patients. Subsequently, functional experiments in OS cell lines revealed that overexpression of miR-330-5p inhibited the cell viability, migration and invasion, promoted cell apoptosis and induced cell cycle arrest. Mechanism research demonstrated that the underlying mechanism of miR-330-5p was associated with targeting and inhibiting survivin, a well-known oncogenic gene. Overall, the current study provided an insight into the possible involvement of the miR-330-5p/survivin axis in the development and progression of OS.
Recent publications have demonstrated that miR-330-5p functions as a tumor suppressor or an oncogene in the context of different cancer types. For instance, Kong et al (18) reported that miR-330-5p was significantly decreased in NSCLC, while overexpression of miR-330-5p markedly inhibited NSCLC cell growth and promoted cell apoptosis. Lee et al (19) also demonstrated that miR-330 functioned as a tumor suppressor and induced the apoptosis of prostate cancer cells through targeting E2F1. By contrast, miR-330 was found to be upregulated in glioblastoma and to function as an oncogenic factor by enhancing proliferation and invasion, and inhibiting apoptosis through the activation of ERK and phosphoinositide 3-kinase/protein kinase B pathways (20,21). However, the expression and role of miR-330-5p in OS remain unknown. In the current study, miR-330-5p was significantly downregulated in OS tissues, with a markedly lower expression detected in metastatic tissues, indicating that decreased miR-330-5p was associated with OS metastasis. The study also identified that low miR-330-5p expression in OS patients was significantly associated with the clinical stage and distant metastasis, as well as poorer prognosis. These findings suggested that miR-330-5p may serve as an effective biomarker for the prognosis of patients with OS. Next, the role of miR-330-5p in malignant progression of OS was examined. In OS cells, overexpression of miR-330-5p significantly inhibited the cell proliferation, promoted cell apoptosis and induced cell cycle arrest at G2/M phase in vitro. It was also observed that the migration and invasion of OS cells were suppressed by miR-330-5p overexpression, which suggests that miR-330-5p overexpression exerts an antitumor effect in OS.
Survivin, the most important member of the inhibitor of apoptosis family, is known to serve important roles in tumor cell proliferation and invasion, therapeutic resistance and poor prognosis (22,23). For instance, a previous study reported that survivin was apparently overexpressed in ovarian cancer, and its overexpression induced cell proliferation and angiogenesis (24). Wang and Ye (25) reported that interfering with the expression of survivin was able to inhibit the cell proliferation, migration and invasion, and promote apoptosis in breast cancer cells. These previous findings suggested that survivin exerts an oncogenic role in tumorigenesis. Recently, Chen et al (26) demonstrated that miR-34a and miR-203 repressed the proliferation of human OS cells by downregulating the expression of survivin. In the present study, the results identified survivin as a direct and functional target of miR-330-5p, and miR-330-5p was found to directly negatively regulate survivin expression in OS cells. Furthermore, it was demonstrated that survivin was highly expressed in OS tissues, and a significant negative correlation was observed between miR-330-5p and survivin expression. Notably, overexpression of survivin partially reversed the inhibitory effects of miR-330-5p in OS cells. These data suggest that miR-330-5p exerts its tumor suppressive role by targeting survivin.
In conclusion, the data of the present study revealed that miR-330-5p functions as a tumor suppressor, and participates in the inhibition of cell proliferation and invasion, as well as promotes apoptosis and induces cell cycle arrest in OS cells by directly regulating the expression of survivin. These findings indicated that miR-330-5p may be a promising therapeutic target for the treatment of OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
HW and LL performed the experiments, contributed to data analysis and wrote the paper. SF designed the study and contributed to data analysis and experimental materials. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All participants provided informed consent for the use of human specimens for clinical research. The present study was approved by the Anhui Provincial Hospital, Anhui Medical University (Hefei, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kobayashi E, Hornicek FJ, Duan Z. MicroRNA involvement in osteosarcoma. Sarcoma. 2012;2012:359739. doi: 10.1155/2012/359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruland OS, Bauer H, Alvegaard T, Smeland S. Treatment of osteosarcoma. The Scandinavian Sarcoma Group experience. Cancer Treat Res. 2009;152:309–318. doi: 10.1007/978-1-4419-0284-9_16. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: A review. J Physiol Biochem. 2011;67:129–139. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 6.Sotillo E, Thomas-Tikhonenko A. Shielding the messenger (RNA): microRNA-based anticancer therapies. Pharmacol Ther. 2011;131:18–32. doi: 10.1016/j.pharmthera.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peng N, Miao Z, Wang L, Liu B, Wang G, Guo X. MiR-378 promotes the cell proliferation of osteosarcoma through down-regulating the expression of Kruppel-like factor 9. Biochem Cell Biol. 2018;96:515–521. doi: 10.1139/bcb-2017-0186. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Li Q, Zhao X, Cui B, Zhang L, Wang Q. MicroRNA-935 inhibits proliferation and invasion of osteosarcoma cells by directly targeting High mobility group box 1. Oncol Res. 2018 Feb 22; doi: 10.3727/096504018X15189093975640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Wang L, Wang Q, Li L, Fu Y, Sun J. MiR-183 inhibits osteosarcoma cell growth and invasion by regulating LRP6-Wnt/β-catenin signaling pathway. Biochem Biophys Res Commun. 2018;496:1197–1203. doi: 10.1016/j.bbrc.2018.01.170. [DOI] [PubMed] [Google Scholar]
- 10.Dong J, Liu Y, Liao W, Liu R, Shi P, Wang L. miRNA-223 is a potential diagnostic and prognostic marker for osteosarcoma. J Bone Oncol. 2016;5:74–79. doi: 10.1016/j.jbo.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng L, Ma J, Ji H, Liu Y, Hu W. miR-330-5p suppresses glioblastoma cell proliferation and invasiveness through targeting ITGA5. Biosci Rep. 2017;37(pii):BSR20170019. doi: 10.1042/BSR20170019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Trehoux S, Lahdaoui F, Delpu Y, Renaud F, Leteurtre E, Torrisani J, Jonckheere N, Van Seuningen I. Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells. Biochim Biophys Acta. 2015;1853:2392–2403. doi: 10.1016/j.bbamcr.2015.05.033. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Chen SH, Kong P, Zhang LY, Zhang LL, Zhang NQ, Gu H. Increased expression of miR-330-3p: A novel independent indicator of poor prognosis in human breast cancer. Eur Rev Med Pharmacol Sci. 2018;22:1726–1730. doi: 10.26355/eurrev_201803_14587. [DOI] [PubMed] [Google Scholar]
- 14.Su BB, Zhou SW, Gan CB, Zhang XN. MiR-330-5p regulates tyrosinase and PDIA3 expression and suppresses cell proliferation and invasion in cutaneous malignant melanoma. J Surg Res. 2016;203:434–440. doi: 10.1016/j.jss.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 15.Wei CH, Wu G, Cai Q, Gao XC, Tong F, Zhou R, Zhang RG, Dong JH, Hu Y, Dong XR. MicroRNA-330-3p promotes cell invasion and metastasis in non-small cell lung cancer through GRIA3 by activating MAPK/ERK signaling pathway. J Hematol Oncol. 2017;10:125. doi: 10.1186/s13045-017-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Kim BK, Yoo HI, Choi K, Yoon SK. miR-330-5p inhibits proliferation and migration of keratinocytes by targeting Pdia3 expression. FEBS J. 2015;282:4692–4702. doi: 10.1111/febs.13523. [DOI] [PubMed] [Google Scholar]
- 18.Kong R, Liu W, Guo Y, Feng J, Cheng C, Zhang X, Ma Y, Li S, Jiang J, Zhang J, et al. Inhibition of NOB1 by microRNA-330-5p overexpression represses cell growth of non-small cell lung cancer. Oncol Rep. 2017;38:2572–2580. doi: 10.3892/or.2017.5927. [DOI] [PubMed] [Google Scholar]
- 19.Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT, Goan YG, Lu PJ. MicroRNA-330 acts as tumor suppressor and induces apoptosis of prostate cancer cells through E2F1-mediated suppression of Akt phosphorylation. Oncogene. 2009;28:3360–3370. doi: 10.1038/onc.2009.192. [DOI] [PubMed] [Google Scholar]
- 20.Qu S, Yao Y, Shang C, Xue Y, Ma J, Li Z, Liu Y. MicroRNA-330 is an oncogenic factor in glioblastoma cells by regulating SH3GL2 gene. PLoS One. 2012;7:e46010. doi: 10.1371/journal.pone.0046010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Yao Y, Xue Y, Ma J, Shang C, Wang P, Liu L, Liu W, Li Z, Qu S, Li Z, Liu Y. MiR-330-mediated regulation of SH3GL2 expression enhances malignant behaviors of glioblastoma stem cells by activating ERK and PI3K/AKT signaling pathways. PLoS One. 2014;9:e95060. doi: 10.1371/journal.pone.0095060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luk SU, Xue H, Cheng H, Lin D, Gout PW, Fazli L, Collins CC, Gleave ME, Wang Y. The BIRC6 gene as a novel target for therapy of prostate cancer: Dual targeting of inhibitors of apoptosis. Oncotarget. 2014;5:6896–6908. doi: 10.18632/oncotarget.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kusner LL, Ciesielski MJ, Marx A, Kaminski HJ, Fenstermaker RA. Survivin as a potential mediator to support autoreactive cell survival in myasthenia gravis: A human and animal model study. PLoS One. 2014;9:e102231. doi: 10.1371/journal.pone.0102231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang HX, Chen G, Li GL, Jiang YJ. Expression and significance of Survivin and Smac in ovarian mucinous tumors. Zhonghua Bing Li Xue Za Zhi. 2010;39:387–390. (In Chinese) [PubMed] [Google Scholar]
- 25.Wang H, Ye YF. Effect of survivin siRNA on biological behaviour of breast cancer MCF7 cells. Asian Pac J Trop Med. 2015;8:225–228. doi: 10.1016/S1995-7645(14)60320-5. [DOI] [PubMed] [Google Scholar]
- 26.Chen X, Chen XG, Hu X, Song T, Ou X, Zhang C, Zhang W, Zhang C. MiR-34a and miR-203 Inhibit survivin expression to control cell proliferation and survival in human osteosarcoma cells. J Cancer. 2016;7:1057–1065. doi: 10.7150/jca.15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.