Abstract
Gastric cancer (GC) remains a leading cause of cancer-associated mortality globally. Emerging evidence suggests that microRNAs (miRs) function as oncogenes or tumor suppressors, contributing to various aspects of cancer progression, including invasion and metastasis. In the present study, the specific role of miR-299-3p in the invasion of GC cells was investigated. The expression level of miR-299-3p was measured using reverse transcription-quantitative PCR and in situ hybridization in human GC tissues. Effects of miR-299-3p on GC cell invasion were determined by Transwell assay. Bioinformatics and luciferase reporter assays were performed to identify and verify the downstream effectors of miR-299-3p. miR-299-3p expression analysis in clinical GC samples revealed a significant downregulation of miR-299-3p compared with non-tumor tissues. Inhibition of miR-299-3p promoted the invasive abilities of GC cells, whereas its overexpression significantly suppressed cell invasion. Bioinformatics and luciferase reporter assays identified heparanase (HPSE) as a direct target of miR-299-3p, the ectopic expression of which reversed the impairment in cell invasion induced by miR-299-3p upregulation. Furthermore, HPSE expression was negatively associated with miR-299-3p levels in human GC tissues. Overall, the present study indicated that miR-299-3p functions as a tumor suppressor by directly targeting HPSE, highlighting its potential as a target for the treatment of GC.
Keywords: miR-299-3p, gastric cancer, cell invasion, heparanase
Introduction
Gastric cancer (GC) is one of the most common malignancies, and the second leading cause of cancer-associated mortality globally, with 951,600 newly diagnosed cases and 723,100 deaths reported annually (1–3). Tumor invasion and metastasis, accounting for the majority of cancer-associated mortality, are essential for tumor progression (4). It has been shown that the invasion of cancer cells into the lymphatic vessels or peritoneal cavity is significantly correlated with secondary growth in distant organs and tumor recurrence, serving as an independent predictor of survival in patients with GC (5,6). Therefore, it is crucial to understand the molecular mechanisms underlying cell motility and metastasis in GC.
MicroRNAs (miRNAs/miRs) are a class of non-coding RNAs (21–25 nucleotides) that regulate gene expression post-transcriptionally (7). Accumulating evidence has indicated that miRNAs can function as oncogenes or tumor suppressors, and are frequently dysregulated in cancers (8,9). Previous studies have suggested that miR-299-3p is involved in a variety of processes of carcinogenesis. For example, miR-299-3p functioned as a tumor suppressor by inhibiting proliferation and invasion in human colon carcinoma (10). A similar inhibitory role of miR-299-3p was reported in breast cancer, where miR-299-3p suppressed the invasive behavior of cancer cells by targeting Oct4 (11). Additionally, Zheng et al (12) reported a role for miR-299-3p in the chemoresistance of lung cancer, as miR-299-3p promoted sensitivity to doxorubicin by directly targeting ATP-binding cassette E1. Recently, tumor-suppressive roles and prognostic values of miR-299-3p were further reported in hepatocellular carcinoma and thyroid cancer (13,14); however, the role of miR-299-3p in the progression of GC remains largely unknown.
In the present study, the potential role of miR-299-3p in the invasion of human GC cells was explored. miR-299-3p expression analysis revealed that miR-299-3p was significantly downregulated in GC tissues. The inhibition of miR-299-3p promoted the invasion of HGC-27 cells in vitro, whereas the overexpression of miR-299-3p induced opposing effects. Bioinformatics and luciferase reporter assays revealed that miR-299-3p targeted the mRNA 3′-untranslated region (3′-UTR) of heparanase (HPSE), an enzyme involved in the remodeling of the extracellular matrix (ECM) (15,16). A rescue experiment confirmed that HPSE was the downstream effector of miR-299-3p in regulating GC cell invasion.
Materials and methods
Tissue samples
A total of 30 GC specimens and 25 adjacent normal tissues (median age, 52 years; range, 42–67 years; female to male ratio, 7:23) were collected at Danyang People's Hospital Affiliated to Nantong University (Danyang, Jiangsu, China). The patients were diagnosed with GC and underwent surgical resection between August 2008 and July 2013. The selection criteria for patients were as follows: i) Pathologically confirmed patients with gastric cancer; ii) the patients had no history of other cancers; and iii) the patients had no preoperative chemotherapy or radiotherapy. The tumor was staged according to the 7th editions international union against cancer TNM staging standard (17). Histologic classification was based on Lauren's criteria (18) as intestinal-type (11 cases), diffuse-type (11 cases) or mixed-type (8 cases). Consent forms were signed by all patients. The procedures were approved by the Research Ethics Committee of Nantong University. The clinicopathological information of the patients is presented in Table SI.
Bioinformatics
Prediction of miRNA targets and sequence conservation analysis was performed using miRDB (http://mirdb.org/) (19) and TargetScan (version 7.2; http://www.targetscan.org/vert_72/). miRNA expression data and clinical information of GC samples were obtained from The Cancer Genome Atlas database Stomach Adenocarcinoma database (release date, September 2014) (http://cancergenome.nih.gov). Based on the selection criteria that samples with both prognostic information and expression data of miR-299-3p, 307 GC samples were included in the present study. Survival analysis was performed using the R package Survival (version 2.43–3; http://CRAN.R-project.org/package=survival) in R (version 3.5.2). The log-rank test was used to assess the statistical significance between stratified groups.
Immunohistochemistry (IHC)
Samples were fixed in 10% buffered formalin at room temperature for 1 day and embedded in paraffin. Paraffin sections (thickness, 4 µm) were deparaffinized, rehydrated in a graded alcohol series and washed in distilled water. Following antigen retrieval with citrate buffer (pH 6.0), endogenous peroxidase was quenched by incubating with hydrogen peroxide blocking reagent (cat. no. ab64218, Abcam) for 5–10 min at room temperature and sections were blocked with goat serum (cat. no. ab7481, Abcam) for 2 h at room temperature. Primary antibody against HPSE (1:200; cat. no. ab85543; Abcam) was then applied at 4°C overnight. Following washing, the sections were incubated with peroxidase-conjugated secondary antibodies against rabbit IgG (1:2,000; cat. no. ab205718, Abcam) for 1 h at room temperature. Color was developed with DAB solution (Dako; Agilent Technologies GmbH). The sections were counterstained with hematoxylin for 10 sec at room temperature prior to coverslipping. The sections were viewed under a light microscope (Olympus Corporation; magnification, ×40). For each section, ≥5 random fields were examined. The staining intensity of HPSE was scored as 0 (absent), 1 (weak), 2 (moderate) and 3 (strong) in a double-blinded manner. HPSE expression was categorized as low (score of 0–1) or high (score of 2–3).
miRNA in situ hybridization (ISH)
To evaluate the expression of miR-299-3p in human GC and adjacent control tissues, ISH was performed using a locked nucleic acid-conjugated miR-299-3p-specific probe against the mature miR-299-3p sequence from Exiqon (Qiagen GmbH; cat. no. 339400), according to the manufacturer's protocol. Briefly, deparaffinized sections were treated with Pepsin protease (cat. no. S3002; Dako; Agilent Technologies, Inc.) at 37°C for 30 min. miR-299-3p specific probe were then hybridized at 48°C for 2 h, followed by a series of signal amplification and washing steps. Hybridization signals were detected by DAB staining as described in the IHC section, followed by counterstaining with hematoxylin for 10 sec at room temperature. Slides were examined using an Olympus BX50 microscope (magnification, ×40; Olympus Corporation), with three random fields captured per sample. The staining intensity of miR-299-3p was scored as 0 (absent), 1 (weak), 2 (moderate) and 3 (strong) in a double-blinded manner. miR-299-3p expression was categorized as low (score of 0–1) or high (score of 2–3).
Cell culture and transfection
The human GC HGC-27 and SGC-7901 cell lines were obtained from the Shanghai Institute of Biochemistry and Cell Biology. Cells were maintained in RPMI 1640 supplemented with 10% FBS (both Gibco; Thermo Fisher Scientific, Inc.), 1% penicillin and 1% streptomycin. miR-299-3p inhibitor (anti-miR-299-3p) (UAUGUGGGAUGGUAAACCGCUU; cat. no. INH0292), non-targeting inhibitor control (NC) (UUGUACUACACAAAAGUACUG, Product ID: INH9002), miR-299-3p mimics (miR-299-3p) (UAUGUGGGAUGGUAAACCGCUU; cat. no. MIM0292) and scramble mimics (Ctrl) (UUGUACUACACAAAAGUACUG; cat. no. MIM9002) were synthesized by Shanghai GenePharma Co., Ltd. Full-length HPSE cDNA was purchased from GeneCopoeia, Inc. and subcloned into the eukaryotic expression vector pcDNA3.0 (Invitrogen; Thermo Fisher Scientific, Inc.). The empty vector pcDNA3.0 was used as a negative control. A total of 1 µg plasmid was transfected into sub-confluent cells using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. The miRNA mimics and inhibitor were transfected at a final concentration of 50 and 100 nM, respectively. Experiments were performed 48 h after transfection.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from human tissues, HGC-27 cells and SGC-7901 cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. cDNA was synthesized from 10 ng total RNA using a TaqMan™ microRNA RT kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) at 42°C for 50 min, followed by heat inactivation for 10 min at 70°C. The levels of miR-299-3p and U6 small nuclear RNA were measured by qPCR using TaqMan assays (cat. no. A25576; Thermo Fisher Scientific, Inc.), following the manufacturer's protocols. The thermocycling conditions were as follows: Initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 60 sec. The primer sequences were as follows: miR-299-3p forward, 5′-ACACTCCAGCTGGGTATGTGGGATGGTAAAC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. All reactions were performed in duplicate in a 12-µl reaction volume. The relative expression levels of targets were calculated using 2−∆∆Cq method (20).
Cell invasion assay
Transwell 24-well plates with 8-µm pore size chamber inserts (Corning Inc.) were used to determine cell invasion. The upper chamber inserts were coated with diluted Matrigel (0.3 mg/ml; BD Biosciences). Cells (2×105) suspended in serum-free media were seeded into the upper chamber, with the lower chamber containing 20% FBS as a chemoattractant. Following incubation for 48 h, non-invading cells in the upper chamber were gently removed. Cells migrating to the lower surface were fixed with 4% formaldehyde for 15 min at room temperature and stained with 0.1% crystal violet, and five random fields in each chamber were photographed under a microscope (Olympus Corporation; magnification, ×200) and counted. The mean value was calculated from three independent experiments.
Luciferase assay
The HPSE 3′-UTR region containing putative miR-299-3p binding sites was amplified by RT-qPCR with Phusion Taq DNA polymerase (Thermo Fisher Scientific, Inc.) from cDNA generated from RNA extracted from HGC-27 and inserted into the pGL3 control vector (Promega Corporation). Amplification was performed as follows: Initial denaturation at 95°C for 5 min, followed by 30 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 1 min, followed by a final elongation step at 72°C for 5 min. A mutant (MUT) 3′-UTR of HPSE was also generated by RT-qPCR. The primer sequences used for PCR amplification were as follows: HPSE 3′-UTR forward, TGCTCTAGAACTGCCGTTACCGTGAAACT; HPSE 3′-UTR reverse, CCGGAATTCGACTGACCAACACCCTCTCC; HPSE 3′-UTR MUT forward, TAAGGAAATGTATATTCCCACTATACTAGACGTTCAAACAGGC; HPSE 3′-UTR MUT reverse, CTGGCCTGTTTGAACGTCTAGTATAGTGGGAATATACATTTCC. A total of 80 ng pGL3 reporter plasmid (Promega Corporation), 5 ng Renilla luciferase plasmid (Promega Corporation) and 50 nM miR-299-3p or control mimics were co-transfected into HGC-27 cells when the cell reach 80–90% confluence using Lipofectamine 2000 reagent, following the manufacturer's instructions. Luciferase activity was measured 48 h following transfection using a Dual Luciferase Assay (Promega Corporation), according to the manufacturer's protocols. Relative luciferase activity was determined as the ratio of firefly to Renilla luciferase activity. All transfection assays were conducted in triplicate.
Western blot assay
Whole cell lysates were prepared using RIPA buffer (Bio-Rad Laboratories, Inc.) containing the protease inhibitor PMSF (Invitrogen; Thermo Fisher Scientific, Inc.). The protein concentrations were determined using a bicinchoninic acid assay (Beyotime Institute of Biotechnology). A total of 20 µg protein per lane was loaded and resolved by 10% SDS-PAGE and then transferred onto PVDF membranes (EMD Millipore). The membranes were blocked with 5% (w/v) non-fat milk for 1 h at room temperature, followed by incubation with primary antibodies overnight at 4°C. Following washing, the membranes were incubated with anti-rabbit horseradish peroxidase-conjugated secondary antibody in blocking solution for 1 h at room temperature. Immunoreactive proteins were detected using enhanced chemiluminescence (Bio-Rad Laboratories, Inc.). The relative band intensity was quantified using ImageJ software (National Institutes of Health). The following antibodies were used: Anti-HPSE (1:500; cat. no. ab85543; Abcam), anti-GAPDH (1:2,000; cat. no. 2118; Cell Signaling Technology, Inc.) and goat anti-rabbit immunoglobulin G (1:5,000; cat. no. 7074; Cell Signaling Technology, Inc.).
Statistical analysis
All data are presented as the mean ± standard deviation. Statistical analysis was performed by unpaired Student's t-test or one-way ANOVA followed by post hoc Tukey's multiple comparison tests as indicated using GraphPad Prism software 6.0 (GraphPad Software, Inc.). Survival analysis was performed using the Kaplan-Meier method with a Log-rank statistical test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-299-3p is downregulated in GC tissues
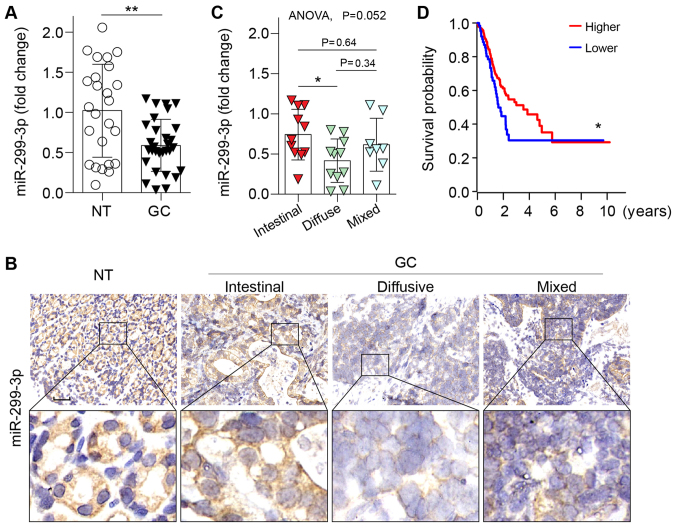
To investigate the potential involvement of miR-299-3p in GC, its expression was evaluated in human GC tissues via RT-qPCR analysis. The expression levels of miR-299-3p were significantly downregulated in GC than in non-tumor tissues (P<0.01; Fig. 1A). In addition, the expression of miR-299-3p was analyzed by an ISH assay in GC and adjacent non-tumor tissues. A moderate miR-299-3p signal was evident in the adjacent control tissues, and a normal structure of the stomach mucosa was observed. Conversely, a large number of malignant cells with low miR-299-3p expression were observed in GC tissues (Fig. 1B). Of note, reduced ISH staining of miR-299-3p was more evident in diffusive GC (Fig. 1B). This observation was further revealed by miR-299-3p expression analysis according to histological subtype, with the most notably downregulated expression in the diffuse subtype (Fig. 1C). The Cancer Genome Atlas database was subsequently explored, and it was revealed that patients with GC with reduced miR-299-3p expression exhibited significantly shorter overall survival (Fig. 1D). Collectively, these data suggested that miR-299-3p may serve a tumor-suppressive role in the development of GC.
Figure 1.
Downregulation of miR-299-3p in GC tissues. (A) Relative expression of miR-299-3p in 30 primary GC compared with 25 adjacent NT tissues. The expression of miR-299-3p was normalized to U6. **P<0.01. (B) Representative in situ hybridization of miR-299-3p in non-tumor tissue and different histological subtypes of GC specimens. Scale bar, 50 µm. (C) Relative expression of miR-299-3p in different subtypes of GC classified according to the Lauren classification as intestinal-type (n=11), diffuse-type (n=11) or mixed-type (n=8). ANOVA with Tukey's post hoc test; *P<0.05. (D) Kaplan-Meier analysis of the overall survival of patients with GC with low and high expression of miR-299-3p; data were obtained from The Cancer Genome Atlas GC database. *P<0.05. miR-299-3p, microRNA-299-3p; GC, gastric cancer; NT, non-tumor.
miR-299-3p inhibits the invasion of GC cell lines
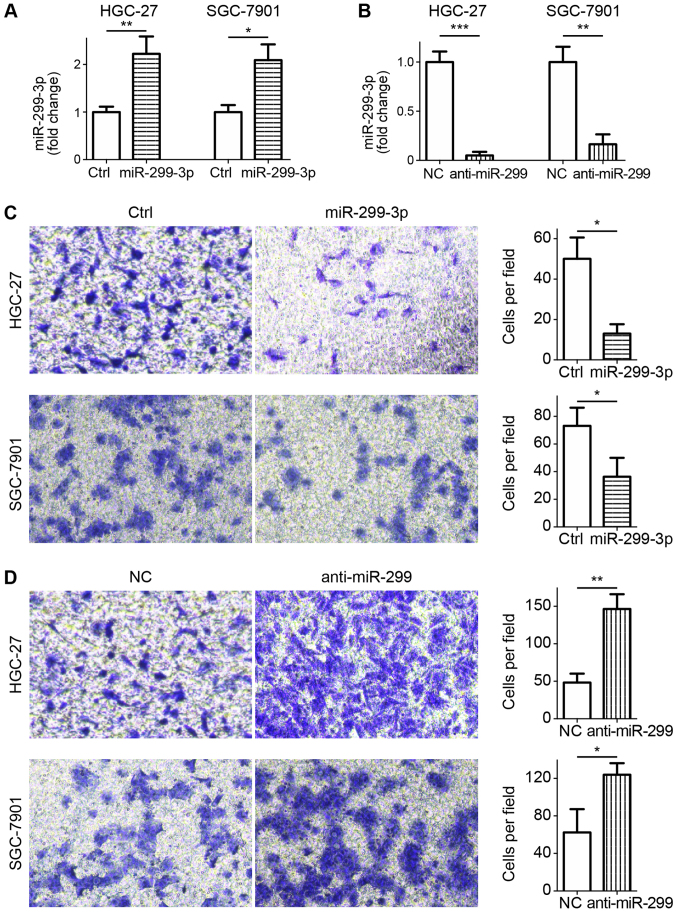
To assess the role of miR-299-3p in the invasion of GC cells, ectopic miR-299-3p mimic or miR-299-3p inhibitor was transfected into HGC-27 and SGC-7901 cells, leading to the efficient up- or downregulation of miR-299-3p, respectively (Fig. 2A and B). Matrigel invasion assays revealed that the ectopic expression of miR-299-3p significantly attenuated HGC-27 and SGC-7901 cell invasion (Fig. 2C). Conversely, miR-299-3p inhibition led to an enhanced invasion capacity, as determined by the increased number of cells moving through the Matrigel layer (Fig. 2D). The results indicated that miR-299-3p negatively regulated the invasion of GC cells.
Figure 2.
Effects of miR-299-3p on cell invasion in vitro. (A) HGC-27 and SGC-7901 cells were transfected with miR-299-3p or Ctrl. *P<0.05, **P<0.01. (B) HGC-27 and SGC-7901 cells were transfected with anti-miR-299-3p or NC. **P<0.01, ***P<0.001. (C) Cells were seeded into Matrigel-coated invasion chambers 24 h post-transfection with miR-299-3p or Ctrl, and allowed to invade for 24 h. Magnification, ×200. Right panel, quantification of the invasion of cells through the Matrigel layer. *P<0.05. (D) Cells were seeded into Matrigel-coated invasion chambers 24 h post-transfection with anti-miR-299-3p or NC, and allowed to invade for 24 h. Magnification, ×200. Right panel, quantification of the invasion of cells through the Matrigel layer. *P<0.05, **P<0.01. miR-299-3p, microRNA-299-3p; Ctrl, scramble mimics; NC, non-targeting inhibitor control.
miR-299 directly targets HPSE
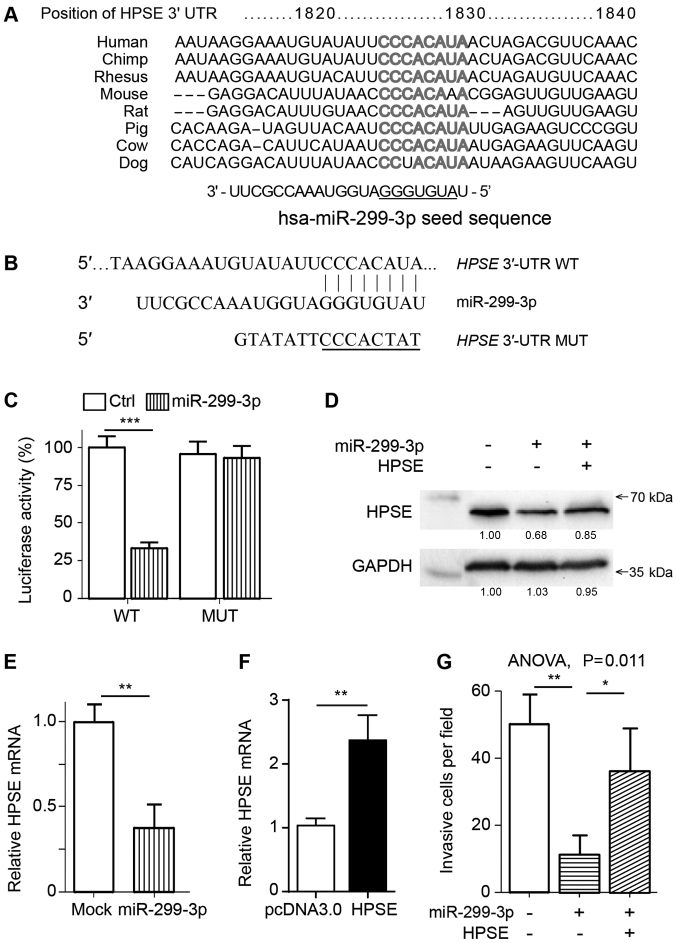
To identify the molecular mechanisms via which miR-299-3p inhibited tumor cell invasion, the two widely used online miRNA target prediction programs, TargetScan and miRDB (21,22), were cross-referenced to identify putative mRNA targets of miR-299-3p. It was observed that HPSE, an enzyme previously implicated in the remodeling of the ECM and cancer invasion and metastasis (15,23), was identified by the two programs to contain an miR-299-3p target site in its 3′-UTR (Fig. 3A and B). Additionally, sequence analysis revealed that the seven consecutive bases corresponding to the seed region of miR-299-3p in the HPSE 3′-UTR were almost completely conserved in humans and other mammals (Fig. 3A). The 3′-UTR of HPSE was cloned into a luciferase reporter vector, and it was revealed that miR-299-3p mimics significantly decreased the luciferase activity; this effect was eliminated when the miR-299-3p binding sites were mutated (Fig. 3C). Consistent with the reporter assay, the overexpression of miR-299-3p induced downregulated expression of the endogenous HPSE at the protein and mRNA levels (Fig. 3D and E). Collectively, these data suggested that HPSE is a direct target of miR-299-3p, as it directly repressed HPSE expression and targeted the seed sequence in its 3′-UTR.
Figure 3.
HPSE is a functional target of miR-299-3p. (A) Alignment of human miR-299-3p and HPSE mRNA 3′-UTR sequences among mammals. The sequences were obtained from the TargetScan database. The underlined sequence indicates the seed region of miR-299-3p. (B) Sequence of miR-299-3p binding site within human HPSE mRNA 3′-UTR, and schematic reporter constructs containing the WT HPSE 3′-UTR or MUT nucleotides. (C) Analysis of relative luciferase reporter activities in HGC-27 cells following transfection, as indicated. (D) Western blot assay revealing HPSE levels in HGC-27 cells following transfection with miR-299-3p mimic or HPSE plasmid. The relative band intensity was indicated under each lane. GAPDH was used as a loading control. (E) RT-qPCR analysis of HPSE mRNA expression in HGC-27 cells following treatment with miR-299-3p or Ctrl mimics. Expression was normalized to GAPDH. (F) RT-qPCR analysis of HPSE expression following transfection of HGC-27 cells with HPSE plasmid. Expression was normalized to GAPDH. (G) Transwell analysis of HGC-27 cells following transfection with miR-299-3p mimic or HPSE plasmid. ANOVA with Tukey's post hoc test; *P<0.05, **P<0.01, ***P<0.001. miR-299-3p, microRNA-299-3p; HPSE, heparanase; 3′-UTR, 3′-untranslated region; WT, wildtype; MUT, mutant; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; Ctrl, scramble mimic.
To determine whether the function of miR-299-3p in the invasion of cells involved the suppression of HPSE, a rescue experiment was conducted by co-transfecting an HPSE overexpression vector and miR-299-3p into HGC-27 cells. Co-transfection restored the expression of HPSE, as determined by western blot analysis 48 h post-transfection (Fig. 3D and F). HGC-27 cells co-transfected with miR-299-3p and HPSE exhibited enhanced invasive abilities compared with miR-299-3p-overexpressing cells (Fig. 3G). The results indicated that restoring HPSE expression could rescue the cell invasion deficit induced by miR-299-3p, suggesting that HPSE is a mediator of miR-299-3p's anti-invasive effects.
HPSE expression is negatively associated with miR-299-3p expression in human GC tissues
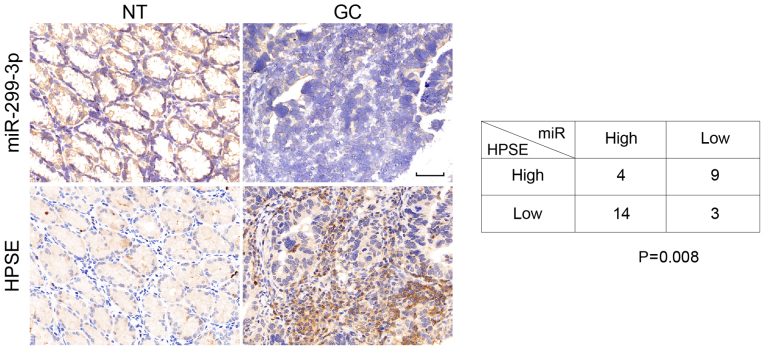
Finally, to investigate the clinical association between HPSE and miR-299-3p expression in patients with GC, the levels of miR-299-3p were determined by ISH, and HPSE via IHC in serial sections of human GC tissue specimens. Intensive miR-299-3p staining was observed in the cytoplasm of tumor-adjacent cells, where the expression of HPSE was absent (Fig. 4). Conversely, GC samples exhibited weak miR-299-3p signals but moderate diffuse HPSE staining (Fig. 4). Statistical analysis revealed a significant inverse association between miR-299-3p and HPSE expression in GC samples (P=0.008; Fig. 4). These observations were consistent with the findings that miR-299-3p targeted HPSE in GC cells, indicating the potential clinical relevance of these results.
Figure 4.
Inverse association between miR-299-3p and HPSE expression in human GC tissues. In situ hybridization demonstrating cytoplasmic expression of miR-299-3p in NT, but not in the GC tissues. Immunohistochemistry revealed diffuse staining of HPSE in GC, but not in NT tissues. Scale bar, 50 µm. Right panel, expression status of miR-299-3p and HPSE in 30 GC clinical samples. miR-299-3p, microRNA-299-3p; HPSE, heparanase; NT, non-tumor; GC, gastric cancer.
Discussion
GC is one of the most malignant cancers worldwide, with limited chemotherapy options (1–3). Tumor invasion and metastasis are the predominant causes of cancer mortality (4). Understanding the mechanisms underlying tumor cell dissemination and developing strategies to inhibit cancer cell invasion may have significant impacts on patient outcomes. miRNAs have emerged as important therapeutic targets for the treatment of cancer (24). In the present study, novel insight into the molecular mechanisms of miR-299-3p in GC invasion was provided. Specifically, reduced expression of miR-299-3p was observed in GC tissues, and downregulation of miR-299-3p in GC cells enhanced cell invasion through HPSE.
It has been suggested that miR-299-3p acts as a tumor-suppressive miRNA, regulating tumor cell proliferation, migration, invasion and cell senescence. For example, the overexpression of miR-299-3p in laryngeal cancer cells inhibited cell growth by targeting human telomerase reverse transcriptase, a catalytic subunit of telomerase, crucial to the immortality of cancer cells (25). Furthermore, Jong et al (26) conducted miRNA profiling and observed that miR-299-3p was one of the most upregulated miRNAs in senescent cells compared with normal cells; the inhibition of miR-299-3p delayed premature senescence and overcame H2O2-induced senescence by augmenting IGF1 signaling. Previous studies have also revealed that miR-299-3p impedes cell migration and invasion in human breast and colon cancer (10,11). In the present study, overexpression of miR-299-3p inhibited the invasion of GC cells, which was consistent with the results of previous studies, indicating a general tumor-suppressive function of miR-299-3p. These observations provided rationale for further investigation of the role of miR-299-3p as a diagnostic biomarker and therapeutic target for a variety of human cancers.
HPSE is an endonuclease that participates in the degradation and remodeling of ECM by cleaving heparan sulfate (HS) covalently attached to HS proteoglycans, a process involved in angiogenesis, tissue repair and lipid metabolism (16,27). It has been demonstrated that HPSE is overexpressed in various types of malignancies, including liver cancer, melanoma and ovarian cancer (23,28,29). Due to its critical functions in tumor invasion and metastasis, clinical trials with HPSE inhibitors, such as HS mimics, have been under intense investigation (16,30–32); however, the precise molecular mechanisms responsible for the regulation of HPSE are yet to be fully determined. In the present study, it was revealed that miR-299-3p induced its effects on GC cells by inhibiting HPSE expression via direct interactions with the HPSE 3′-UTR. An inverse association was also demonstrated between the levels of miR-299-3p and HPSE in clinical GC specimens. As high expression levels of HPSE have been reported to be a reliable indicator of poor prognosis in a variety of cancers, including melanoma and ovarian cancer (28,29), the close association between miR-299-3p and HPSE suggests prognostic implications for miR-299-3p in GC. However, requires further investigation with larger populations.
Of note, miR-299-3p was recently revealed by transcriptomic profiling to be the third-most downregulated miRNA in cancer-associated fibroblasts (CAFs) in lung adenocarcinoma (33). Due to the notable tumor-promoting properties of CAFs, the downregulation of miR-299-3p may modulate the niche, favoring cancer cell invasion and proliferation. Importantly, high levels of HPSE have been reported to facilitate the formation of a pre-metastatic niche (15,34–36). It is therefore proposed that the miR-299-3p/HPSE axis may also exist in CAFs, meriting further investigations.
In conclusion, the present study identified miR-299-3p/HPSE as a novel mechanism influencing the invasion of GC cells. These findings warrant further investigation into how miR-299-3p may be used as a biomarker and potential target to monitor and suppress the progression of GC.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was funded by Danyang Science and Technology Development Special Fund Project (Grant ID: SF201506) to XZ.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XZ and ZG designed the study. XZ, MH and ZG performed all the experiments and analyzed the data. The manuscript was drafted by ZG with input from the other authors. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Research Ethics Committee of Nantong University (Nantong, China). Informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Guan X. Cancer metastases: Challenges and opportunities. Acta Pharm Sin B. 2015;5:402–418. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuoka T, Yashiro M. Rho/ROCK signaling in motility and metastasis of gastric cancer. World J Gastroenterol. 2014;20:13756–13766. doi: 10.3748/wjg.v20.i38.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu J, Xue Z, Zhang S, Guo X, Zhai L, Shang S, Zhang Y, Lu H. Integrated analysis of the prognostic role of the lymph node ratio in node-positive gastric cancer: A meta-analysis. Int J Surg. 2018;57:76–83. doi: 10.1016/j.ijsu.2018.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Song F, Yang D, Liu B, Guo Y, Zheng H, Li L, Wang T, Yu J, Zhao Y, Niu R, et al. Integrated microRNA network analyses identify a poor-prognosis subtype of gastric cancer characterized by the miR-200 family. Clin Cancer Res. 2014;20:878–889. doi: 10.1158/1078-0432.CCR-13-1844. [DOI] [PubMed] [Google Scholar]
- 8.Tang H, Deng M, Tang Y, Xie X, Guo J, Kong Y, Ye F, Su Q, Xie X. miR-200b and miR-200c as prognostic factors and mediators of gastric cancer cell progression. Clin Cancer Res. 2013;19:5602–5612. doi: 10.1158/1078-0432.CCR-13-1326. [DOI] [PubMed] [Google Scholar]
- 9.Wu JG, Wang JJ, Jiang X, Lan JP, He XJ, Wang HJ, Ma YY, Xia YJ, Ru GQ, Ma J, et al. miR-125b promotes cell migration and invasion by targeting PPP1CA-Rb signal pathways in gastric cancer, resulting in a poor prognosis. Gastric Cancer. 2015;18:729–739. doi: 10.1007/s10120-014-0421-8. [DOI] [PubMed] [Google Scholar]
- 10.Wang JY, Jiang JB, Li Y, Wang YL, Dai Y. MicroRNA-299-3p suppresses proliferation and invasion by targeting VEGFA in human colon carcinoma. Biomed Pharmacother. 2017;93:1047–1054. doi: 10.1016/j.biopha.2017.07.030. [DOI] [PubMed] [Google Scholar]
- 11.Göhring AR, Reuter S, Clement JH, Cheng X, Theobald J, Wölfl S, Mrowka R. Human microRNA-299-3p decreases invasive behavior of cancer cells by downregulation of Oct4 expression and causes apoptosis. PLoS One. 2017;12:e0174912. doi: 10.1371/journal.pone.0174912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng D, Dai Y, Wang S, Xing X. MicroRNA-299-3p promotes the sensibility of lung cancer to doxorubicin through directly targeting ABCE1. Int J Clin Exp Pathol. 2015;8:10072–10081. [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Qi M, Yang Q, Li JY. miR-299-3p functions as a tumor suppressor in thyroid cancer by regulating SHOC2. Eur Rev Med Pharmacol Sci. 2019;23:232–240. doi: 10.26355/eurrev_201901_16769. [DOI] [PubMed] [Google Scholar]
- 14.Dang S, Zhou J, Wang Z, Wang K, Dai S, He S. miR-299-3p functions as a tumor suppressor via targeting Sirtuin 5 in hepatocellular carcinoma. Biomed Pharmacother. 2018;106:966–975. doi: 10.1016/j.biopha.2018.06.042. [DOI] [PubMed] [Google Scholar]
- 15.Sanderson RD, Elkin M, Rapraeger AC, Ilan N, Vlodavsky I. Heparanase regulation of cancer, autophagy and inflammation: New mechanisms and targets for therapy. FEBS J. 2017;284:42–55. doi: 10.1111/febs.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Washington K. 7th edition of the AJCC cancer staging manual: Stomach. Ann Surg Oncol. 2010;17:3077–3079. doi: 10.1245/s10434-010-1362-z. [DOI] [PubMed] [Google Scholar]
- 18.Lauren P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma. An Attempt at a Histo-Clinical Classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 19.Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43 (Database Issue) 2015:D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:18. doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen XP, Luo JS, Tian Y, Nie CL, Cui W, Zhang WD. Downregulation of heparanase expression results in suppression of invasion, migration, and adhesion abilities of hepatocellular carcinoma cells. Biomed Res Int. 2015;2015:241983. doi: 10.1155/2015/241983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li M, Li J, Ding X, He M, Cheng SY. microRNA and cancer. AAPS J. 2010;12:309–317. doi: 10.1208/s12248-010-9194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Chen SM, Chen C, Zhang ZX, Dai MY, Zhang LB, Wang SB, Dai Q, Tao ZZ. microRNA-299-3p inhibits laryngeal cancer cell growth by targeting human telomerase reverse transcriptase mRNA. Mol Med Rep. 2015;11:4645–4649. doi: 10.3892/mmr.2015.3287. [DOI] [PubMed] [Google Scholar]
- 26.Jong HL, Mustafa MR, Vanhoutte PM, AbuBakar S, Wong PF. MicroRNA 299-3p modulates replicative senescence in endothelial cells. Physiol Genomics. 2013;45:256–267. doi: 10.1152/physiolgenomics.00071.2012. [DOI] [PubMed] [Google Scholar]
- 27.Zcharia E, Jia J, Zhang X, Baraz L, Lindahl U, Peretz T, Vlodavsky I, Li JP. Newly generated heparanase knock-out mice unravel co-regulation of heparanase and matrix metalloproteinases. PLoS One. 2009;4:e5181. doi: 10.1371/journal.pone.0005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Wen W, Wu H, Chen Y, Ren G, Guo W. Heparanase expression correlates with poor survival in oral mucosal melanoma. Med Oncol. 2013;30:633. doi: 10.1007/s12032-013-0633-5. [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, Chan H, Wei L, Pan Z, Zhang J, Li L. Overexpression of heparanase in ovarian cancer and its clinical significance. Oncol Rep. 2013;30:2279–2287. doi: 10.3892/or.2013.2701. [DOI] [PubMed] [Google Scholar]
- 30.Ferro V, Dredge K, Liu L, Hammond E, Bytheway I, Li C, Johnstone K, Karoli T, Davis K, Copeman E, Gautam A. PI-88 and novel heparan sulfate mimetics inhibit angiogenesis. Semin Thromb Hemost. 2007;33:557–568. doi: 10.1055/s-2007-982088. [DOI] [PubMed] [Google Scholar]
- 31.Vreys V, David G. Mammalian heparanase: What is the message? J Cell Mol Med. 2007;11:427–452. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao BY, Wang Z, Hu J, Liu WF, Shen ZZ, Zhang X, Yu L, Fan J, Zhou J. PI-88 inhibits postoperative recurrence of hepatocellular carcinoma via disrupting the surge of heparanase after liver resection. Tumour Biol. 2016;37:2987–2998. doi: 10.1007/s13277-015-4085-8. [DOI] [PubMed] [Google Scholar]
- 33.Negrete-Garcia MC, Ramírez-Rodriguez SL, Rangel-Escareño C, Muñoz-Montero S, Kelly-García J, Vázquez-Manríquez ME, Santillán P, Ramírez MM, Ramírez-Martínez G, Ramírez-Venegas A, Ortiz-Quintero B. Deregulated microRNAs in cancer-associated fibroblasts from front tumor tissues of lung adenocarcinoma as potential predictors of tumor promotion. Tohoku J Exp Med. 2018;246:107–120. doi: 10.1620/tjem.246.107. [DOI] [PubMed] [Google Scholar]
- 34.Putz EM, Mayfosh AJ, Kos K, Barkauskas DS, Nakamura K, Town L, Goodall KJ, Yee DY, Poon IK, Baschuk N, et al. NK cell heparanase controls tumor invasion and immune surveillance. J Clin Invest. 2017;127:2777–2788. doi: 10.1172/JCI92958. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Waning DL, Guise TA. Molecular mechanisms of bone metastasis and associated muscle weakness. Clin Cancer Res. 2014;20:3071–3077. doi: 10.1158/1078-0432.CCR-13-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, Sanderson RD. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J Biol Chem. 2013;288:10093–10099. doi: 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.