Abstract
Chemotherapy is widely used for the treatment of ovarian cancer. Since chemotherapy resistance is the major cause of poor prognosis in patients with ovarian cancer, it is important to identify new methods to improve the efficacy of chemotherapy. Minichromosome maintenance complex component 2 (MCM2), which serves an essential role in DNA replication, has been recently identified as a novel proliferation marker with prognostic implications in multiple types of cancer. However, the role of MCM2 in ovarian cancer and its underlying molecular mechanisms remain unclear. Therefore, in the present study, the biological effects of MCM2 were investigated, particularly with respect to DNA damage and repair. In the present study, short hairpin RNA was employed to knockdown MCM2 expression in the A2780 ovarian cancer cell line. The sensitivity of A2780 cells to carboplatin was assessed by cell colony formation assay. The present results suggested that MCM2 knockdown inhibited the proliferation of tumor cells, induced G0/G1 phase arrest and did not exhibit effects on cell apoptosis. However, MCM2 knockdown significantly decreased the colony formation of A2780 cells treated with carboplatin. Furthermore, knockdown of MCM2 together with carboplatin treatment or UV irradiation increased the protein expression level of γ-H2A histone family member X and p53 compared with control cells. The present data suggested that the increased sensitivity to carboplatin may occur via the p53-dependent apoptotic response. Additionally, the present results suggested that knockdown of MCM2 may have therapeutic applications in enhancing the efficacy of carboplatin in patients with ovarian cancer.
Keywords: ovarian cancer, minichromosome maintenance complex component 2, carboplatin, DNA replication, DNA damage, p53
Introduction
Ovarian cancer is one of the most common gynecologic malignancies worldwide. According to recent global statistics, >230,000 new patients are diagnosed with this disease every year, and ovarian cancer accounts for ~140,000 mortalities annually (1). Although detection techniques and chemotherapy regimens for ovarian cancer have improved in recent years, the 5-year survival rate of patients with advanced stage ovarian cancer is ~30% (2). This low survival rate is primarily due to the number of patients diagnosed with ovarian cancer at late stages (3,4). Notably, cancer cells become more resistant to conventional chemotherapeutic agents at later stages (5). Therefore, the identification of novel therapeutic methods is critical to improve the prognosis of patients with ovarian cancer.
Minichromosome maintenance complex component 2 (MCM2) is one of the six proteins comprising the MCM complex, a stable heterohexamer crucial for the regulation of DNA replication (6,7). In eukaryotic cells, DNA synthesis is initiated from defined sites, called replication origins. During the G1 phase, replication origins interact with the origin recognition complex (ORC), which induces the sequential recruitment of cell division cycle 6, chromatin licensing and DNA replication factor 1 and MCM2-7 to form a prereplication complex (8–10). During the S phase, the activation of the MCM complex by cell cycle kinases triggers the initiation of the DNA replication (11). Additionally, the MCM complex restricts chromosome replication to only once per cell cycle (11,12). Previous studies have demonstrated that in yeast MCM mutations lead to chromosome loss, DNA damage and increased recombination (13,14). Consistent with previous studies on yeast, reduced expression levels of MCM2 in mice result in lymphomas (15,16). A large number of studies have confirmed that MCM2 is a reliable proliferation and prognostic marker of oral, gastric, colon and breast cancer, suggesting that it may represent a more reliable marker than traditional ones, such as marker of proliferation Ki-67 (MKI67) and proliferating cell nuclear antigen (17–20). A number of previous studies have reported that the expression level of MCM2 is significantly higher in ovarian adenocarcinomas compared with low malignant tumors (21,22) and a high level of MCM2 expression in malignant tumors is associated with higher grades, more advanced stages and poor prognosis (23,24).
Since the regulation of the expression levels of MCM2-7 is involved in ensuring proper genome replication preventing genome instability, the present study aimed to investigate whether agents that inhibit MCM2 gene expression may suppress cellular proliferation, influencing the cell cycle and leading to cell apoptosis. Moreover, the present study aimed to examine whether, under replication stress conditions, a reduction in MCM2 expression could sensitize cells to the chemotherapeutic drug carboplatin. In the present study, the expression level of MCM2 was knocked down in the human ovarian cancer cell line A2780. Collectively, the aim of the present study was to develop a novel therapeutic strategy for treating ovarian cancer.
Materials and methods
Cell culture
The human ovarian cancer cell line A2780 and the 293T cell line were obtained from The Chinese Academy of Sciences Committee and were verified by STR profiling. A2780 cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.). The 293T cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.). All cells were incubated at 37°C in a humidified incubator with 5% CO2.
Construction of the MCM2 short hairpin (sh)-RNA lentivirus vector and cell infection
RNA interference (RNAi) was used to downregulate the expression level of MCM2 in A2780 cells. Two shRNAs were used to target MCM2 and a scrambled shRNA was used as control (Table I; all from Sangon Biotech Co., Ltd). These shRNA oligos were annealed and ligated using AgeI and EcoRI restriction sites into the linearized pLKO.1-puro vector (Addgene) to generate pLKO.1-puro-MCM2 (shMCM2-1 and shMCM2-2) and pLKO.1-puro-control (shCON) recombinant vectors. The constructed shRNA-expressing vectors were then confirmed by DNA sequencing (Sangon Biotech Co., Ltd). The shRNA-expressing recombinant plasmids (2 µg) were transfected together with two helper plasmids, psPAX2 (1.5 µg) and pMD2.G (0.5 µg; both from Addgene) into 293T cells with Lipofectamine 2000™ transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Cell culture media containing lentiviral particles were collected 48 h after transfection. For cell infection, A2780 ovarian cancer cells were incubated in 6-well plates at a density of 4×105 cells/well. Subsequently, media containing 1×106 IFU/ml lentivirus in 8 µg/ml polybrene (Sigma-Aldrich; Merck KGaA) was added to A2780 cells for 24 h. Stably infected cells were selected using 1 µg/ml puromycin (Sigma-Aldrich; Merck KGaA) for 3–5 days and the RNAi knockdown efficiency was detected by western blot analysis.
Table I.
shRNA sequences.
| shRNA | Sequence (5′-3′) |
|---|---|
| shMCM2-1 | CCGGGCTCTTCATACTGAAGCAGTTCTCGAGAACTGCTTCAGTATGAAGAGCTTTTT |
| shMCM2-2 | CCGGCTATCAGAACTACCAGCGTATCTCGAGATACGCTGGTAGTTCTGATAGTTTTT |
| shCON | CCTAAGGTTAAGTCGCCCTCG |
MCM2, minichromosome maintenance complex component 2; shRNA, short hairpin RNA; shCON, control shRNA; shMCM2, shRNA targeting MCM2.
Western blot analysis
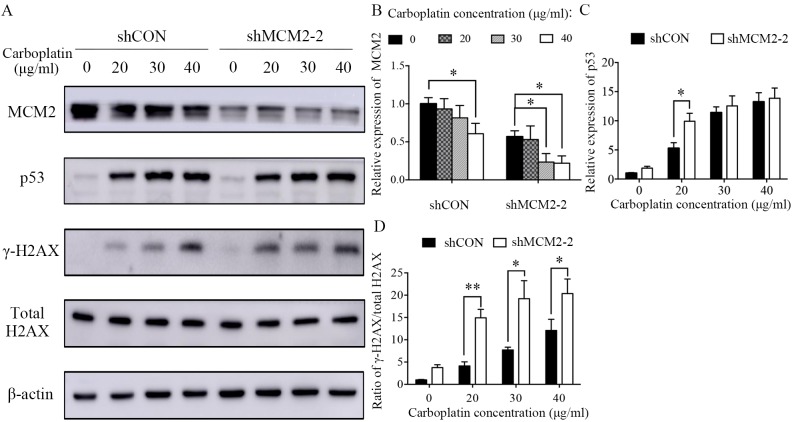
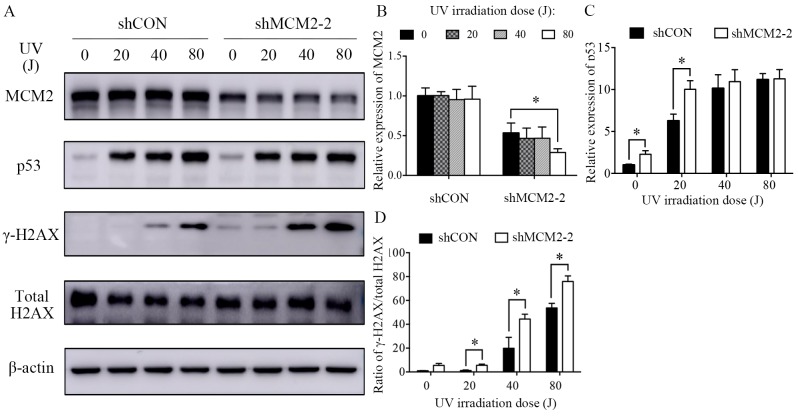
A2780 cells were treated with various concentrations of carboplatin (0, 20, 30 and 40 µg/ml) for 48 h at 37°C. Subsequently, the cells were lysed, and the total protein was extracted. Alternatively, A2780 cells were treated with various UV intensities (0, 20, 40 and 80 J). After 24 h, the cells were lysed, and the total protein was extracted. Cells were lysed for total protein extraction with a cell lysis buffer for Western and IP (cat. no. P0013J; Beyotime Institute of Biotechnology) containing protease inhibitor cocktail and PMSF (1%). The concentrations of total protein were quantified using a bicinchoninic protein assay kit (Beyotime Institute of Biotechnology). Extracted proteins were mixed with SDS-PAGE Sample Loading Buffer (cat. no. P0015; Beyotime Institute of Biotechnology) and boiled at 100°C for 10 min. The protein samples (20 µg/well) were separated by SDS-PAGE on 10–12% gels and transferred onto PVDF membranes (EMD Millipore). After blocking with 5% skimmed milk for 1 h at room temperature, the membrane was incubated at 4°C overnight with the following primary antibodies: MCM2 (1:1,000; cat. no. 3619; Cell Signaling Technology, Inc.), p53 (1:300; cat. no. 2527; Cell Signaling Technology, Inc.), γ-H2A histone family member X (γ-H2AX; 1:400; cat. no. 9718; Cell Signaling Technology, Inc.); H2AX (1:2,000; cat. no. ab124781; Abcam) and β-actin (1:3,000; cat. no. ab8224; Abcam). After washing with TBS-0.2% Tween-20 (TBST) three times, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse (1:4,000; cat. no. ab205719; Abcam) or anti-rabbit secondary antibodies (1:4,000; cat. no. ab205718; Abcam) for 1 h at room temperature. Then, the membranes were washed with TBST three times. Specific bands were visualized using an ECL detection kit (Thermo Fisher Scientific, Inc.). The protein bands were visualized by autoradiography. The band intensity was measured using the ImageJ software (version 1.48; National Institutes of Health). β-Actin was used to normalize the protein expression levels and the relative expression levels were subsequently calculated.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.) was used to assess cell proliferation. The cells were seeded in 96-well plates in triplicate at a density of ~1×103 cells/well and cultured in 100 µl RPMI-1640 for 24, 48, 72, 96 and 120 h. After incubation, the culture medium was replaced with 110 µl RPMI-1640 supplemented with CCK-8 reagent (10 µl CCK-8 in 100 µl RPMI-1640), and the cells were incubated at 37°C for 2 h. A microplate reader (BioTek Instruments, Inc.) was used to measure the optical density value at a wavelength of 450 nm. All experiments were performed in triplicate.
Cell cycle assay
Exponentially growing cells were seeded into 6-well plates at a density of 4×105 cells/well and synchronized in serum-free RPMI-1640 for 16–24 h. Then, the medium was replaced with RPMI-1640 containing 10% FBS and antibiotics. Following incubation for 24 h, cells were harvested, washed with ice-cold PBS, and fixed with precooled 70% ethanol overnight at 4°C. The next day, the fixed cells were washed with ice-cold PBS and stained with 500 µl propidium iodide (PI)/RNase Staining Buffer (BD Pharmingen; BD Biosciences) for 30 min at room temperature in the dark. Cell cycle analyses were performed with a flow cytometer (BD Biosciences). Cell cycle was analyzed using the ModFit software (version 4.1; Verity Software House, Inc.). The experiments were repeated three times.
Cell apoptosis assay
To examine apoptosis, the cells were harvested (4×105 cells/well) and washed twice with ice-cold PBS. Then, the cells were resuspended with 1X Annexin V binding buffer (BD Biosciences) and stained with PI and Annexin V-FITC according to the manufacturer's protocol (BD Biosciences). Cell apoptosis was detected by flow cytometry (BD Biosciences) and the assays were repeated three times. Cells negative for both PI and Annexin V were considered viable cells. PI-negative and Annexin V-positive cells were considered early apoptotic cells. PI-positive and Annexin V-positive cells were considered late-stage apoptotic cells. Cell apoptosis data were analyzed using BD CellQuest™ Pro software (version 6.1; BD Biosciences).
Colony formation assay
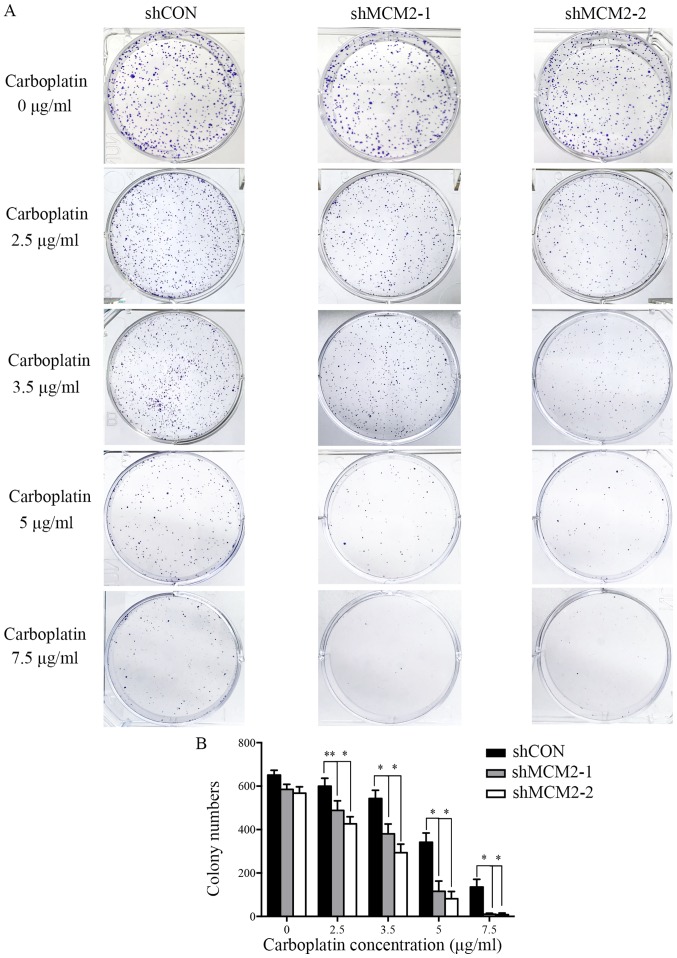
The control and knockdown group cells were seeded in 6-well plates at a density of 1,000 cells/well. Cells were then exposed to 0, 2.5, 3.5, 5 and 7.5 µg/ml carboplatin (Qilu Pharmaceutical Co., Ltd.) for 48 h at 37°C. Then, the medium was replaced with complete culture medium. Following incubation at 37°C for 10–14 days, the cells were fixed with 4% paraformaldehyde at room temperature for 15 min and then stained with 0.5% crystal violet dye at room temperature for 10 min. Colonies containing >50 cells were counted under a light microscope (magnification, ×100).
Statistical analysis
Statistical analyses were performed using SPSS 23.0 software (IBM Corp.). Data are presented as the mean ± standard deviation from three experiments. Two-tailed Student's t-test was used to evaluate the differences between two groups. One-way ANOVA was used to evaluate the differences between multiple groups, and Dunnett's test was used as the post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Lentivirus-mediated MCM2 knockdown in human ovarian cancer A2780 cells
To investigate the biological function of MCM2 in human ovarian cancer cells, shRNA targeting human MCM2 (shMCM2-1 and shMCM2-2) and a negative control (shCON) were infected into A2780 ovarian cancer cells to knockdown the expression of MCM2. The protein expression levels of MCM were investigated in stably infected A2780 cells by western blot analysis (Fig. 1), and the protein expression level of MCM2 was decreased by ~50% in shMCM2-1 (50.59±4%) and shMCM2-2 (49.1±3.6%) cells compared with cells infected with shCON (Fig. 1).
Figure 1.
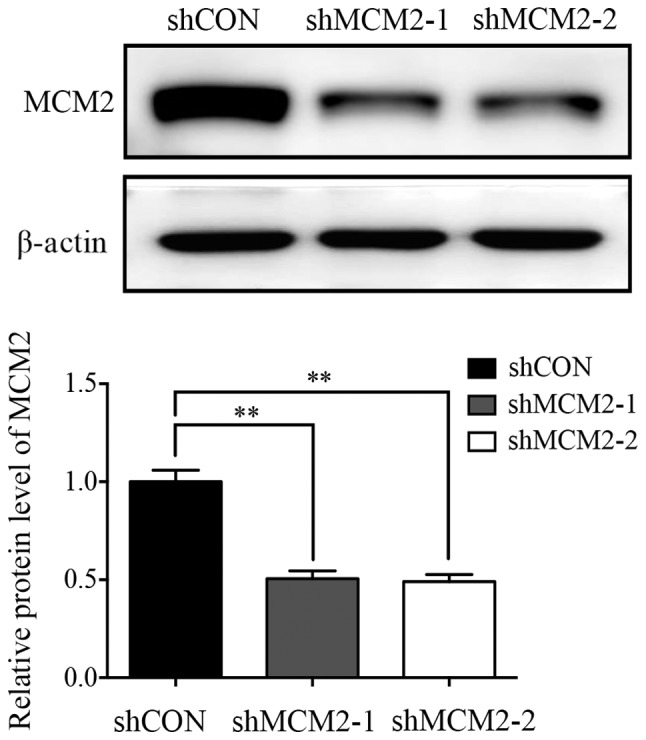
Expression of MCM2 in the ovarian cancer cell line A2780 after infection with MCM2 shRNA or control vector was determined by western blot assay. MCM2 protein expression was significantly decreased following infection with MCM2-shRNA compared with the control. Data are presented as the mean ± standard deviation from three experiments. **P<0.01 vs. shCON. MCM2, minichromosome maintenance complex component 2; shRNA, short hairpin RNA; shCON, control shRNA; shMCM2, shRNA targeting MCM2.
Effects of MCM2 knockdown on proliferation, cell cycle distribution and cell apoptosis in A2780 cells
Since MCM2 was shown to serve an important role in DNA replication in eukaryotes, and as a previous study reported that knockdown of MCM2 causes cell death (25), the present study investigated whether knockdown of MCM2 influenced cell proliferation, cell cycle and cell apoptosis. Therefore, CCK-8 assay was performed at different time points to measure cell viability. Notably, the proliferation of A2780 cells was significantly inhibited following knockdown of MCM2 compared with the control group at 4 and 5 days (Fig. 2A). To investigate the role of MCM2 in the regulation of the cell cycle, flow cytometry was performed. The present results suggested that inhibition of MCM2 in A2780 cells caused an increase in the G1 phase, whereas the number of cells in the G2 phase decreased compared with the control (Fig. 2B). The cell apoptosis assay suggested that cells infected with shMCM2 exhibited similar apoptotic rates compared with control cells (Fig. 2C). Therefore, MCM2 knockdown did not induce the apoptosis of A2780 cells. Notably, the present cell proliferation and apoptosis results were not in line with previous studies on hepatocellular carcinoma, lung carcinoma and esophageal cancer cells (25).
Figure 2.
Effect of MCM2 knockdown on A2780 cell proliferation, cell cycle and apoptosis. (A) Cell proliferation was assessed using the Cell Counting Kit-8 assay at 0, 1, 2, 3, 4 and 5 days after infection. Cell proliferation was inhibited by MCM2 knockdown at 4 and 5 days. (B) Cell cycle was analyzed by flow cytometry. Representative cell cycle histograms of A2780 cells are presented. Cells treated with MCM2 shRNA exhibited a higher number of cells in the G0/G1-phase. (C) Cell apoptosis was determined by flow cytometry. Infection with shMCM2 did not induce apoptosis in A2780 cells. A representative flow cytometry analysis is presented. Data are presented as the mean ± standard deviation from three experiments. *P<0.05 vs. shCON. MCM2, minichromosome maintenance complex component 2; shRNA, short hairpin RNA; shCON, control shRNA; shMCM2, shRNA targeting MCM2; OD, optical density.
Knockdown of MCM2 enhances carboplatin sensitivity in A2780 cells
Since the knockdown of MCM2 did not significantly influence cell proliferation at 1–3 days, and the downregulation of MCM2 did not affect cell apoptosis under normal circumstances, the present study investigated whether the effects of MCM2 knockdown would increase if replication forks were put under stress by supplementation with DNA-damaging drugs. To examine whether knockdown of MCM2 could sensitize A2780 cells to the common ovarian cancer chemotherapeutic drug carboplatin (5,26), a colony formation assay was performed. Control cells and MCM2 knockdown cells were treated with different concentrations of carboplatin. In the absence of carboplatin, all cells formed colonies (Fig. 3). Compared with the control group, the knockdown of MCM2 significantly decreased the colony formation of A2780 cells in response to various concentrations of carboplatin (Fig. 3). The present results suggested that the combined treatment of MCM2-shRNA and carboplatin significantly reduced the colony formation of A2780 cells compared with carboplatin treatment alone. Therefore, the present results support the hypothesis that the downregulation of MCM2 level can enhance the sensitivity of A2780 cells to carboplatin.
Figure 3.
MCM2-knockdown cells are hypersensitive to carboplatin. (A) Colony formation assays of A2780 cells expressing shMCM2 or shCON after exposure to various concentrations of carboplatin (0, 2.5, 3.5, 5 and 7.5 µg/ml). (B) Quantification of the colony formation assay. Data are presented as the mean ± standard deviation from three experiments. *P<0.05 and **P<0.01 vs. shCON. MCM2, minichromosome maintenance complex component 2; shRNA, short hairpin RNA; shCON, control shRNA; shMCM2, shRNA targeting MCM2.
Knockdown of MCM2 increases carboplatin and UV irradiation-induced double-strand breaks (DSBs) in A2780 cells
Next, the present study sought to identify the mechanism underlying carboplatin sensitivity in MCM2-knockdown cells. DNA damage and repair were investigated following treatment with two genotoxic agents, carboplatin and UV irradiation. Carboplatin treatment reduced the protein expression level of MCM2 in a dose-dependent manner (Fig. 4A and B). The present results suggested that MCM2 may serve a role in DNA damage and repair. However, the protein expression level of MCM2 was not affected in UV-irradiated cells (Fig. 5A and B). γ-H2AX is a sensitive marker of DNA DSBs (27). Therefore, to investigate the effects of MCM2 knockdown on DNA damage and repair, γ-H2AX expression following DNA damage was analyzed by western blotting, and γ-H2AX expression was normalized to the total protein expression level of H2AX. The present results suggested that the γ-H2AX/total H2AX ratio increased in MCM2-deficient cells compared with control cells after treatment with carboplatin and UV (Figs. 4A and 5A). The present results suggested that the ratio of γ-H2AX/total H2AX in cells treated with carboplatin and infected with shMCM2 was higher compared with control cells treated with carboplatin (Fig. 4A and D). Similarly, the UV irradiation experiment suggested that MCM2 knockdown significantly increased the ratio of γ-H2AX/total H2AX in A2780 cells exposed to UV irradiation at 20, 40 and 80 J (Fig. 5A and D). Therefore, the present results suggested that the enhanced sensitivity of MCM2-knockdown A2780 cells to carboplatin may have been caused by the accumulation of damaged DNA.
Figure 4.
Knockdown of MCM2 increases the protein expression levels of γ-H2AX and p53 after treatment with carboplatin. (A) A2780 cells were treated with various concentrations of carboplatin (0, 20, 30 and 40 µg/ml) for 48 h. Subsequently, the cells were lysed and the total protein was extracted. Protein expression levels of MCM2, p53, γ-H2AX and total H2AX were detected by western blotting. MCM2 knockdown increased DNA damage-associated markers in response to carboplatin. (B) Quantification of the protein expression levels of MCM2, (C) p53 and (D) the γ-H2AX/H2AX ratio. γ-H2AX was used as an indicator of double-strand breaks. Data are presented as the mean ± standard deviation from three experiments. *P<0.05 and **P<0.01 vs. the respective control. MCM2, minichromosome maintenance complex component 2; shRNA, short hairpin RNA; shCON, control shRNA; shMCM2, shRNA targeting MCM2; H2AX, H2A histone family member X.
Figure 5.
Knockdown of MCM2 increases the protein expression levels of γ-H2AX and p53 after treatment with UV. (A) A2780 cells were treated with various UV intensities (0, 20, 40 and 80 J) for 24 h. Subsequently, the cells were lysed and the total protein was extracted. Protein expression levels of MCM2, p53, γ-H2AX and total H2AX were detected by western blotting. MCM2 knockdown increased DNA damage-associated markers in response to UV irradiation. Quantification of the protein expression levels of (B) MCM2, (C) p53 and (D) the γ-H2AX/H2AX ratio. γ-H2AX was used as an indicator of double-strand breaks. Data are presented as the mean ± standard deviation from three experiments. *P<0.05 vs. the respective control. MCM2, minichromosome maintenance complex component 2; shRNA, short hairpin RNA; shCON, control shRNA; shMCM2, shRNA targeting MCM2; H2AX, H2A histone family member X.
DNA damage response in MCM2-deficient A2780 cells in vitro
The increased DNA damage in A2780 MCM2-knockdown cells after treatment with carboplatin and UV irradiation suggested that MCM2 may serve a role in mediating the DNA damage response. In a previous study, MCM2 was identified as a mediator of the DNA damage response when the replisome interacts with a genetic lesion (28). Therefore, the present study investigated the influence of MCM2 knockdown on the expression levels of proteins associated with the DNA damage repair pathway. The present results suggested that the protein expression levels of the tumor suppressor gene p53 were increased in MCM2 deficient cells compared with control cells after carboplatin treatment (Fig. 4A and C) and UV irradiation (Fig. 5A and C). An important DNA damage repair pathway involves p53 (29–31), which in turn serves an important role in cell cycle arrest and apoptotic response following cellular damage. The increased protein expression level of p53 suggested the activation of the p53-dependent apoptotic response.
Discussion
Carboplatin-based chemotherapy is the standard first-line treatment for patients with advanced stage ovarian cancer, tumor relapse or tumor metastasis. However, carboplatin-based chemotherapy resistance may occur and, in many cases, can result in treatment failure (26). For this reason, reversing carboplatin resistance in ovarian cancer is the major focus of multiple research studies. A previous study demonstrated that in the cisplatin-resistant ovarian carcinoma cell line PE01CDDP, the MCM2 expression level was increased by >2 folds compared with normal PE01 cells (32). Therefore, the present study hypothesized that MCM2 may be associated with chemotherapy resistance in patients with ovarian cancer.
MCM2-7 is a key factor during replication initiation and elongation (33). MCM2-7 was shown to interact with DNA during the G1 phase and is activated during the S phase, triggering DNA replication origin licensing and firing (33). It has been reported that the MCM2/5 active site functions as an ATP-dependent ‘gate’ of the MCM2-7 ring, and that the status of this gate regulates the function of MCM2-7 (34). Mutated MCM4 in mice causes a reduction in the overall levels of MCMs, leading to a high incidence of mammary adenocarcinomas (35). Additionally, when the expression levels of MCM2 are decreased to 33%, the average mouse lifespan is significantly reduced due to the occurrence of various types of cancer, in particular T- and B-cell lymphoma (16). Therefore, loss of MCM function in mice induces genome instability and cancer predisposition (16). However, increased expression levels of MCM2 have been detected in various types of cancer, and were identified to be associated with advanced stages and poor prognosis (21,36,37). Additionally, increased expression levels of MCM2 were identified to be associated with the upregulation of MKI67, an important marker of cell proliferation (21,36,37). These previous studies indicated that the precise control of MCM2 expression is important to maintain the stability of the genome. Therefore, in the present study, the expression level of MCM2 was knocked down in A2780 ovarian cancer cells. RNAi was used to downregulate the expression level of MCM2, and the biological effects of MCM2 were investigated in A2780 cells. The present results demonstrated that MCM2 knockdown exhibited a limited effect on A2780 cell proliferation and cell cycle, and did not affect cell apoptosis. The present findings suggested that targeted inhibition of MCM2 may not be an adequate therapy without additional treatments.
Therefore, the present study hypothesized that the acute downregulation of MCM2 may allow the formation of a limited, but sufficient, amount of active MCM2-7 complexes. The present results are consistent with various previous studies that have reported that normal DNA replication rates and cell proliferation in vitro are maintained also when the protein expression levels of MCM are significantly decreased (38–41). The present findings can be explained by the fact that the MCM2-7 complex is abundant in proliferating cells and is recruited to the chromatin at levels that are 3–20 folds higher than the levels required to unwind the DNA at the replication fork (42–45). Furthermore, MCM hexamers are homogenously distributed on DNA, and are not accumulated at the levels of the replication origins and ORCs (46,47). Previous studies have reported that MCM2-7 complexes can be found on DNA at significant distances from the ORCs (46,47). Moreover, each genomic site containing MCM2-7 bound to the DNA is a candidate replication origin that can potentially initiate replication.
Next, the present study aimed to identify the function of the excess levels of MCMs compared with the number of replication origins. The present results suggested that a partial reduction in MCM2 had limited effects on cell proliferation, cell cycle and apoptosis. Therefore, the present study investigated effects of MCM2 knockdown in the presence of replication stress. Thus, various concentrations of carboplatin were added to A2780 control cells and MCM2-knockdown cells. After exposure to carboplatin, the survival rate of MCM2-knockdown cells decreased significantly compared with the control cells.
Collectively, the results of the present and previous studies have suggested that under normal conditions, sufficient MCM complexes are recruited to the chromatin at various potential origins of replication (42,43). A previous study reported that following the initiation of replication at one replication origin, a signal is sent to inhibit the activation of additional replication origins (45). However, when cells experience replication stress and the replication forks are stalled, the dormant origins of replication may be activated to rescue stalled or damaged primary replication forks (45). In conclusion, cells maintain an excessive number of MCM2-7 complexes bound to dormant replication origins that can be activated during replication stress (41,45). The present data revealed that after decreasing the expression level of MCM2, A2780 cells exhibited significantly reduced proliferation following treatment with carboplatin, a replication inhibitor. Under limited DNA replication licensing conditions, a reduced number of potential replication origins are available, and cells may require the additional dormant replication origins used to rescue collapsed replication forks. This hypothesis could explain the hypersensitivity of MCM2-knockdown cells to replication inhibitors. An excessive number of potential DNA replication origins not only serves as a backup system in case of collapsed replication forks, but are also important in the maintenance of genome stability, since mice with reduced MCM levels or function exhibit high rates of cancer incidence (16). Additionally, a previous study reported that the increased number of potential DNA replication origins serves a role in suppressing genetic damage (48).
Additionally, in the present study, carboplatin treatment decreased the protein expression level of MCM2 and increased the level of p53 in a dose-dependent manner. According to a previous study, DNA DSB signals elicit ATM serine/threonine kinase-dependent phosphorylation of checkpoint kinase 2 (Chk2) (49). Activated Chk2 can transduce the signal via p53, which activates the transcription of the cell cycle inhibitor p21, leading to the downregulation of the protein expression level of MCM (49). However, the decrease in the protein expression level of MCM2 was not significant in UV-irradiated cells, suggesting that MCM2 may serve various roles in response to different forms of DNA damage. Moreover, both the carboplatin-treated and UV-irradiated cells showed higher protein expression levels of γ-H2AX and p53 in MCM2-knockdown cells compared with control cells. γ-H2AX is a sensitive marker of DNA DSB (27), and thus, the increased levels of γ-H2AX suggested that a higher frequency of DNA damage was present in MCM2-knockdown cells compared with control cells. The mechanism underlying MCM2 knockdown-mediated chemosensitivity in A2780 cells is unclear, but the present results suggested that the regulation of p53 was involved in this mechanism. The tumor suppressor gene p53 serves a pivotal role in the DNA damage response pathway, causing G1 cell cycle arrest, promoting DNA damage repair (29) or inducing cell apoptosis to protect genome stability (30,31). A previous study reported that defective apoptosis may contribute to resistance to platinum cytotoxicity, (50) and cells with p53 deletions (51) or mutations (52) are more resistant to platinum, possibly due to the inactivation of the p53-dependent apoptotic response. Additionally, a previous study suggested that p53 signaling may sensitize cells to cisplatin (52). Therefore, the sensitivity of MCM2-knockdown cells to carboplatin may be promoted by the activation of the p53-dependent apoptotic pathway. However, the specific mechanisms underlying MCM2 knockdown-mediated sensitivity of A2780 cells to carboplatin require further investigation.
In conclusion, the present data suggested that combined treatment with the chemotherapeutic agent carboplatin and MCM2 shRNA significantly enhanced the chemosensitivity of A2780 cells via upregulation of p53. Notably, the combined treatment was more effective than single treatment with either carboplatin or MCM2 shRNA alone. To the best of our knowledge, the present study is the first to suggest the inhibition of MCM2 as a strategy to potentiate the efficacy of carboplatin in ovarian cancer. However, only one cell type was used in the present study, and the reduction in the protein expression level of MCM2 following MCM2 was not optimal. Therefore, further experiments investigating various cell lines and overexpression models are required in order to identify the effects of MCM2 overexpression on cell proliferation, cell cycle and carboplatin resistance. The present findings suggested that the combination of MCM2 knockdown and carboplatin treatment may represent a novel therapeutic strategy to treat ovarian cancer. Additionally, MCM2 could represent a potential target for designing new drugs and for the development of novel therapeutic methods for ovarian cancer treatment.
Acknowledgements
The authors would like to thank Miss Qian Ma (Department of Oncology, Shanghai Medical College, Fudan University) for her assistance with the western blotting assay.
Funding
The present study was supported by a grant from The National Natural Science Foundation of China (grant nos. NSF-81772808, 81572552 and 81772774).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
MD performed the majority of the experiments, and wrote and revised the manuscript. AZ and SX constructed the recombinant lentiviral vector. JS performed the Cell Counting Kit-8 assay. HuZ and HoZ performed the cell cycle and apoptosis experiments. YW and JS performed the statistical analysis. RL and LG designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dahiya N, Morin PJ. MicroRNAs in ovarian carcinomas. Endocr Relat Cancer. 2010;17:F77–F89. doi: 10.1677/ERC-09-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 3.Wright JD, Shah M, Mathew L, Burke WM, Culhane J, Goldman N, Schiff PB, Herzog TJ. Fertility preservation in young women with epithelial ovarian cancer. Cancer. 2009;115:4118–4126. doi: 10.1002/cncr.24461. [DOI] [PubMed] [Google Scholar]
- 4.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 5.Fung-Kee-Fung M, Oliver T, Elit L, Oza A, Hirte HW, Bryson P. Optimal chemotherapy treatment for women with recurrent ovarian cancer. Curr Oncol. 2007;14:195–208. doi: 10.3747/co.2007.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsburg SL. Eukaryotic MCM proteins: Beyond replication initiation. Microbiol Mol Biol Rev. 2004;68:109–131. doi: 10.1128/MMBR.68.1.109-131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei M. The MCM complex: Its role in DNA replication and implications for cancer therapy. Curr Cancer Drug Targets. 2005;5:365–380. doi: 10.2174/1568009054629654. [DOI] [PubMed] [Google Scholar]
- 8.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 9.Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendez J, Stillman B. Perpetuating the double helix: Molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- 11.Parker MW, Botchan MR, Berger JM. Mechanisms and regulation of DNA replication initiation in eukaryotes. Crit Rev Biochem Mol Biol. 2017;52:107–144. doi: 10.1080/10409238.2016.1274717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Romanowski P, Madine MA. Mechanisms restricting DNA replication to once per cell cycle: The role of Cdc6p and ORC. Trends Cell Biol. 1997;7:9–10. doi: 10.1016/S0962-8924(97)30077-4. [DOI] [PubMed] [Google Scholar]
- 13.Hennessy KM, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 14.Gibson SI, Surosky RT, Tye BK. The phenotype of the minichromosome maintenance mutant mcm3 is characteristic of mutants defective in DNA replication. Mol Cell Biol. 1990;10:5707–5720. doi: 10.1128/MCB.10.11.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunnev D, Rusiniak ME, Kudla A, Freeland A, Cady GK, Pruitt SC. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene. 2010;29:3630–3638. doi: 10.1038/onc.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007;25:3121–3132. doi: 10.1634/stemcells.2007-0483. [DOI] [PubMed] [Google Scholar]
- 17.Yousef EM, Furrer D, Laperriere DL, Tahir MR, Mader S, Diorio C, Gaboury LA. MCM2: An alternative to Ki-67 for measuring breast cancer cell proliferation. Mod Pathol. 2017;30:682–697. doi: 10.1038/modpathol.2016.231. [DOI] [PubMed] [Google Scholar]
- 18.de Andrade BA, León JE, Carlos R, Delgado-Azañero W, Mosqueda-Taylor A, de Almeida OP. Expression of minichromosome maintenance 2, Ki-67, and geminin in oral nevi and melanoma. Ann Diagn Pathol. 2013;17:32–36. doi: 10.1016/j.anndiagpath.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 19.Czyzewska J, Guzińska-Ustymowicz K, Pryczynicz A, Kemona A, Bandurski R. Immunohistochemical evaluation of Ki-67, PCNA and MCM2 proteins proliferation index (PI) in advanced gastric cancer. Folia Histochem Cytobiol. 2009;47:289–296. doi: 10.2478/v10042-009-0042-y. [DOI] [PubMed] [Google Scholar]
- 20.Guzińska-Ustymowicz K, Pryczynicz A, Kemona A, Czyzewska J. Correlation between proliferation markers: PCNA, Ki-67, MCM-2 and antiapoptotic protein Bcl-2 in colorectal cancer. Anticancer Res. 2009;29:3049–3052. [PubMed] [Google Scholar]
- 21.Gakiopoulou H, Korkolopoulou P, Levidou G, Thymara I, Saetta A, Piperi C, Givalos N, Vassilopoulos I, Ventouri K, Tsenga A, et al. Minichromosome maintenance proteins 2 and 5 in non-benign epithelial ovarian tumours: Relationship with cell cycle regulators and prognostic implications. Br J Cancer. 2007;97:1124–1134. doi: 10.1038/sj.bjc.6603992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott IS, Heath TM, Morris LS, Rushbrook SM, Bird K, Vowler SL, Arends MJ, Coleman N. A novel immunohistochemical method for estimating cell cycle phase distribution in ovarian serous neoplasms: Implications for the histopathological assessment of paraffin-embedded specimens. Br J Cancer. 2004;90:1583–1590. doi: 10.1038/sj.bjc.6601660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudderidge TJ, Stoeber K, Loddo M, Atkinson G, Fanshawe T, Griffiths DF, Williams GH. Mcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinoma. Clin Cancer Res. 2005;11:2510–2517. doi: 10.1158/1078-0432.CCR-04-1776. [DOI] [PubMed] [Google Scholar]
- 24.Kodani I, Osaki M, Shomori K, Araki K, Goto E, Ryoke K, Ito H. Minichromosome maintenance 2 expression is correlated with mode of invasion and prognosis in oral squamous cell carcinomas. J Oral Pathol Med. 2003;32:468–474. doi: 10.1034/j.1600-0714.2003.00116.x. [DOI] [PubMed] [Google Scholar]
- 25.Feng D, Tu Z, Wu W, Liang C. Inhibiting the expression of DNA replication-initiation proteins induces apoptosis in human cancer cells. Cancer Res. 2003;63:7356–7364. [PubMed] [Google Scholar]
- 26.McGuire WP, III, Markman M. Primary ovarian cancer chemotherapy: Current standards of care. Br J Cancer. 2003;89(Suppl 3):S3–S8. doi: 10.1038/sj.bjc.6601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mah LJ, El-Osta A, Karagiannis TC. GammaH2AX: A sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–686. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 28.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bates S, Vousden KH. p53 in signaling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:12–18. doi: 10.1016/S0959-437X(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 30.Gottlieb TM, Oren M. p53 and apoptosis. Semin Cancer Biol. 1998;8:359–368. doi: 10.1006/scbi.1998.0098. [DOI] [PubMed] [Google Scholar]
- 31.Janus F, Albrechtsen N, Dornreiter I, Wiesmüller L, Grosse F, Deppert W. The dual role model for p53 in maintaining genomic integrity. Cell Mol Life Sci. 1999;55:12–27. doi: 10.1007/s000180050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macleod K, Mullen P, Sewell J, Rabiasz G, Lawrie S, Miller E, Smyth JF, Langdon SP. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005;65:6789–6800. doi: 10.1158/0008-5472.CAN-04-2684. [DOI] [PubMed] [Google Scholar]
- 33.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24:1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bochman ML, Schwacha A. The Mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 35.Shima N, Alcaraz A, Liachko I, Buske TR, Andrews CA, Munroe RJ, Hartford SA, Tye BK, Schimenti JC. A viable allele of Mcm4 causes chromosome instability and mammary adenocarcinomas in mice. Nat Genet. 2007;39:93–98. doi: 10.1038/ng1936. [DOI] [PubMed] [Google Scholar]
- 36.Yang C, Wen Y, Li H, Zhang D, Zhang N, Shi X, Jiang B, Ma X, Yang P, Tang H, et al. Overexpression of minichromosome maintenance 2 predicts poor prognosis in patients with gastric cancer. Oncol Rep. 2012;27:135–142. doi: 10.3892/or.2011.1473. [DOI] [PubMed] [Google Scholar]
- 37.Obermann EC, Went P, Zimpfer A, Tzankov A, Wild PJ, Stoehr R, Pileri SA, Dirnhofer S. Expression of minichromosome maintenance protein 2 as a marker for proliferation and prognosis in diffuse large B-cell lymphoma: A tissue microarray and clinico-pathological analysis. BMC Cancer. 2005;5:162. doi: 10.1186/1471-2407-5-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crevel G, Hashimoto R, Vass S, Sherkow J, Yamaguchi M, Heck MM, Cotterill S. Differential requirements for MCM proteins in DNA replication in Drosophila S2 cells. PLoS One. 2007;2:e833. doi: 10.1371/journal.pone.0000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsao CC, Geisen C, Abraham RT. Interaction between human MCM7 and Rad17 proteins is required for replication checkpoint signaling. EMBO J. 2004;23:4660–4669. doi: 10.1038/sj.emboj.7600463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oehlmann M, Score AJ, Blow JJ. The role of Cdc6 in ensuring complete genome licensing and S phase checkpoint activation. J Cell Biol. 2004;165:181–190. doi: 10.1083/jcb.200311044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ibarra A, Schwob E, Méndez J. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci USA. 2008;105:8956–8961. doi: 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei M, Kawasaki Y, Tye BK. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/MCB.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowles A, Chong JP, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the origin recognition complex and the replication licensing system in Xenopus. Cell. 1996;87:287–296. doi: 10.1016/S0092-8674(00)81346-X. [DOI] [PubMed] [Google Scholar]
- 44.Stoeber K, Tlsty TD, Happerfield L, Thomas GA, Romanov S, Bobrow L, Williams ED, Williams GH. DNA replication licensing and human cell proliferation. J Cell Sci. 2001;114:2027–2041. doi: 10.1242/jcs.114.11.2027. [DOI] [PubMed] [Google Scholar]
- 45.Woodward AM, Göhler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hyrien O, Marheineke K, Goldar A. Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays. 2003;25:116–125. doi: 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- 47.Masata M, Malínský J, Fidlerová H, Smirnov E, Raska I. Dynamics of replication foci in early S phase as visualized by cross-correlation function. J Struct Biol. 2005;151:61–68. doi: 10.1016/j.jsb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 48.Kunnev D, Freeland A, Qin M, Leach RW, Wang J, Shenoy RM, Pruitt SC. Effect of minichromosome maintenance protein 2 deficiency on the locations of DNA replication origins. Genome Res. 2015;25:558–569. doi: 10.1101/gr.176099.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwon HJ, Hong YK, Park C, Choi YH, Yun HJ, Lee EW, Kim BW. Widdrol induces cell cycle arrest, associated with MCM down-regulation, in human colon adenocarcinoma cells. Cancer Lett. 2010;290:96–103. doi: 10.1016/j.canlet.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 50.Minagawa Y, Kigawa J, Itamochi H, Kanamori Y, Shimada M, Takahashi M, Terakawa N. Cisplatin-resistant HeLa cells are resistant to apoptosis via p53-dependent and -independent pathways. Jpn J Cancer Res. 1999;90:1373–1379. doi: 10.1111/j.1349-7006.1999.tb00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asada N, Tsuchiya H, Tomita K. De novo deletions of p53 gene and wild-type p53 correlate with acquired cisplatin-resistance in human osteosarcoma OST cell line. Anticancer Res. 1999;19:5131–5137. [PubMed] [Google Scholar]
- 52.Kigawa J, Sato S, Shimada M, Takahashi M, Itamochi H, Kanamori Y, Terakawa N. p53 gene status and chemosensitivity in ovarian cancer. Hum Cell. 2001;14:165–171. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.