Abstract
Osteoarthritis (OA) is a type of degenerative joint disease that affects the health of the elderly. OA is characterized by articular cartilage degradation and joint inflammation. The present study aimed to investigate the role and mechanism of microRNA-let-7a (Let-7a) in OA by examining its role in lipopolysaccharide (LPS)-induced cartilage inflammatory injury in ATDC5 cells. ATDC5 cells were treated with various concentrations of LPS. The present results suggested that 5 and 10 µg/ml LPS significantly inhibited ATDC5 cell viability, and 5 µg/ml LPS was selected for further experiments. Reverse transcription-quantitative PCR (RT-qPCR) results suggested that treatment with LPS significantly induced the expression levels of multiple inflammatory factors, including tumor necrosis factor-α (TNF-α), interleukin (IL)-1β, IL-6 and IL-8, and increased the expression level of Let-7a in ATDC5 cells. IL-6 receptor (IL-6R) was identified to be a direct target of Let-7a using TargetScan and a dual-luciferase reporter assay. Subsequently, Cell Counting Kit-8 and flow cytometry analyses identified that Let-7a inhibitor could significantly promote cell viability and reduce cell apoptosis in ATDC5 cells treated with LPS, and these effects could be reversed by transfection with small interfering (si)RNA-IL-6R. ELISA was used to examine the expression of inflammatory factors in ATDC5 cells following treatment with LPS. Additionally, RT-qPCR and western blotting were performed to detect the mRNA and protein expression level of IL-6R and STAT3. The present results suggested that Let-7a inhibitor significantly reduced the expression level of TNF-α, IL-1β, IL-6 and IL-8 in ATDC5 cells, and this effect was reversed by transfecting siRNA-IL-6R. Moreover, RT-qPCR and western blot assay results suggested that Let-7a inhibitor significantly increased the expression level of IL-6R and phosphorylated STAT3, and these effects could be reversed by siRNA-IL-6R. Collectively, Let-7a inhibitor increased cell proliferation, reduced apoptosis and inhibited inflammatory response in ATDC5 cells treated with LPS. The present study provided a new potential therapeutic target for OA treatment.
Keywords: microRNA-let-7a, ATDC5 cells, inflammatory factor, STAT3
Introduction
Osteoarthritis (OA) is a chronic progressive bone and joint disease caused by articular cartilage degeneration and bone hyperplasia (1). An epidemiological study demonstrated that the incidence rates of OA are 44–70 and 60–70% in patients >55 and >65 years old, respectively (2). MicroRNAs (miRNAs) are a class of non-coding small RNAs that have been found to act as oncogenes or tumor suppressors during the development of tumors (3), and a previous study have shown that the expression of tumor-related miRNAs is associated with tumor occurrence and prognosis (4). miRNAs can regulate the proliferation, differentiation and apoptosis of tumor cells, affecting cell cycle and terminal differentiation (5,6). The occurrence of various types of tumors is associated with miRNA dysregulation (5,7). Therefore, investigating the regulatory mechanism of miRNAs is important for tumor pathogenesis and medical diagnosis (8,9). Importantly, an increasing number of studies have shown that miRNAs serve an important role in the occurrence and development of OA (10–12).
miRNA-let-7a (Let-7a) is the second identified miRNA (13). It has been reported that Let-7 is significantly downregulated in various tumor cells, such as ovarian cancer (14) and breast cancer (15). However, to the best of our knowledge, a limited number of studies examined the role of Let-7 in OA.
OA is an inflammatory disease characterized by articular cartilage degradation and joint inflammation (16–18), and apoptosis of chondrocytes is one of the main pathological features of OA (19,20). LPS-induced chondrocyte cell inflammatory injury has been widely used as an in vitro model to investigate OA (21–23). The aim of the present study was to investigate the role of let-7 in an in vitro model of OA induced by LPS. Additionally, the present study aimed to examine the effects of let-7 on chondrocyte cell proliferation and apoptosis. The identification of the mechanism associated with let-7 provided a theoretical basis for the development of new strategies for the prevention and treatment of OA.
Materials and methods
Cell culture
ATDC5 cells were purchased from the Cell Bank of Shanghai Institute of Cell Biology. Cells were cultured in 75 cm2 flasks with DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin, (Nanjing Sunshine Biotechnology Co., Ltd.) and 100 µg/ml streptomycin (Nanjing Sunshine Biotechnology Co., Ltd.). Cells were incubated at 37°C with 5% CO2. LPS treatment was performed when cell confluency reached 75%. Cells were treated for 5 h with LPS (Beyotime Institute of Biotechnology) at various concentrations (0, 1, 5 and 10 µg/ml). Cell counting Kit-8 (CCK-8; Sigma-Aldrich; Merck KGaA) was used to detect cell viability.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from 1×106 ATDC5 cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The concentration of RNA was detected using a NanoDrop 2000 (Thermo Fisher Scientific, Inc.). The RNA samples were stored at −80°C. Then, cDNA was synthesized using a miScript Reverse Transcription kit (Qiagen GmbH) according to the manufacturer's protocol. The QuantiFast SYBR Green PCR kit (Qiagen GmbH) was used to perform quantitative real-time polymerase chain reaction (RT-qPCR) under a CFX Connect Real-Time System (Bio-Rad Laboratories, Inc.). GAPDH was used as the internal control. The thermocycling conditions were as follows: 95°C for 10 min followed by 35 cycles of 95°C for 15 sec and 55°C for 40 sec. The 2−ΔΔCq method (24) was used to quantify the relative gene expression levels of the target genes. The following primers were purchased from GenScript Corporation: Let-7a forward, 5′-TGAGGTAGTAGGTTGTATAGTTAAA-3′ and reverse, 5′-AACGAGACGACGACAGACTTT-3′; interleukin 6 receptor (IL-6R) forward, 5′-TGGTGGATGTTCCCCCCGAG-3′ and reverse, 5′-TCCTGGGAATACTGGCACGG-3′; IL-1β forward, 5′-TGTGAAATGCCACCTTTTGA-3′ and reverse, 5′-TGAGTGATACTGCCTGCCTG-3′; IL-6 forward, 5′-CCGGAGAGGAGACTTCACAG-3′ and reverse, 5′-CAGAATTGCCATTGCACA-3′; TNF-α forward, 5′-GAACTGGCAGAAGAGGCACT-3′ and reverse, 5′-GGTCTGGGCCATAGAACTGA-3′; IL-8 forward, 5′-AGTGAGCTCATTGGCTGGCTTATCTTC-3′ and reverse, 5′-AGTAAGCTTGTTTCTTCCTGGCTCTTG-3′; STAT3 forward, 5′-AAGAGGCGGCAACAGATT-3′ and reverse, 5′-CGGTCTTGATGACGAGGG-3′; GAPDH forward, 5′-CTTTGGTATCGTGGAAGGACTC-3′ and reverse, 5′-GTAGAGGCAGGGATGATGTTCT-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The experiments were repeated three times.
Cell transfection
ATDC5 cells were seeded into six-well plates (1×106 cells/well) and cultured at 37°C for 24 h. Then, the cells were transfected with 100 nM miR-NC inhibitor (5′-UUCUCCGAACGUGUCACGU-3′; Guangzhou RiboBio Co., Ltd.), 100 nM Let-7a inhibitor (5′-AACUAUACAACCUCCUACCUCA-3′; Guangzhou RiboBio Co., Ltd.), 1 µg control-small interfering (si)RNA (cat. no. EYK-BVIS00414; Xiamen Yanke Biotechnology Co., Ltd.), 1 µg IL6R-siRNA (cat. no. XWCRH2387; Xiamen Yanke Biotechnology Co., Ltd.) or 100 nM Let-7a inhibitor + 1 µg IL6R-siRNA using Lipofectamine 3000 reagent (Invitrogen Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The transfection efficiency was detected after 48 h. The experiments were repeated three times.
CCK-8
ATDC5 cells were treated with 5 µg/ml LPS for 5 h after 48 h of transfection. In total, 100 µl cell suspension (1×104 cells/ml) was added to each well of the 96-well plates, and then the cells were cultured for 24 h. Subsequently, 10 µl CCK-8 reagent (Sigma-Aldrich; Merck KGaA) was added to each well. The absorbance at 450 nm was measured by an automatic enzyme-linked immune detector after 2 h of incubation. The CCK-8 experiments were repeated three times.
Flow cytometry analysis
ATDC5 cells were treated with 5 µg/ml LPS for 5 h after 48 h of transfection. Then, cells were digested using 0.25% trypsin, followed by washing with PBS. Cells were subsequently fixed with 70% ethanol overnight at 4°C. Apoptosis was detected using the Annexin V-FITC- propidium iodide kit (cat. no. 70-AP101-100; Multisciences Lianke Biotech Co., Ltd.) according to the manufacturer's protocol. Cell apoptosis rate was measured using a FACSCalibur flow cytometer (BD Biosciences) with Cell Quest software version 5.1 (BD Biosciences). The experiment was performed in triplicate.
ELISA
ATDC5 cells were treated with 5 µg/ml LPS for 5 h after 48 h of transfection. Then, the expression levels of TNF-α (cat. no. PT512; Beyotime Institute of Biotechnology), IL-1β (cat. no. PI301; Beyotime Institute of Biotechnology), IL-6 (cat. no. PI326; Beyotime Institute of Biotechnology) and IL-8 (cat. no. GD-QX2854; Shanghai Guduo Biological Technology Co., Ltd.) in the cell culture supernatant were detected using ELISA kits according to the manufacturer's protocol. Samples were diluted and cytokine standards were added to ELISA plates. Detection antibodies were added to the samples and incubated at room temperature for 1 h. Following incubation with streptavidin-horseradish peroxidase (HRP) conjugated secondary antibodies at room temperature for 20 min, plates were read at 450 nm. The experiment was repeated three times.
Western blot assay
ATDC5 cells were treated with 5 µg/ml LPS for 5 h after 48 h of transfection. Cells were washed with ice-cold PBS and then lysed with RIPA (Beyotime Institute of Biotechnology) buffer with 1% PMSF at 4°C for 1 h. Protein samples were collected by centrifugation for 5 min at 4°C at 12,000 × g. Protein concentration was determined using a bicinchoninic acid protein assay kit. Proteins (30 µg/lane) were separated by SDS-PAGE on 10% gels, electroblotted onto PVDF membranes and then blocked in 5% non-fat milk at room temperature for 2 h. Membranes were then incubated with primary antibodies: IL-6R (cat. no. ab83053; 1:1,000; Abcam), STAT3 (cat. no. ab119352; 1:1,000; Abcam), phosphorylated STAT3 (cat. no. ab76315; 1:1,000; Abcam) and GAPDH (cat. no. ab181602; 1:1,000; Abcam), overnight at 4°C and washed with PBS-0.1% Tween-20 (PBST) four times. Subsequently, the membranes were incubated with the HRP-conjugated anti-rabbit secondary antibody (cat. no. 7074; 1:2,000; Cell Signaling Technology, Inc.) for 2 h at room temperature, the membranes were then washed with PBST four times. Finally, ECL reagent (EMD Millipore) was used to visualize the protein bands using a FluorChem FC3 system (ProteinSimple). AlphaView 3.4.0 software (ProteinSimple) was used for the quantification of the protein bands. The experiments were repeated three times.
Dual-luciferase reporter assay
TargetScan version 7.2 (http://www.targetscan.org/vert_72/) was used to predict the potential targets of Let-7a, and an interaction between IL-6R and Let-7 was identified. To verify this prediction, the wild-type (WT) and mutant 3′-untranslated regions (UTRs) of IL-6R were cloned into a pmiR-RB-Report dual luciferase reporter gene plasmid vector (Guangzhou RiboBio Co., Ltd.). Cells were transfected with the reporter constructs and Let-7a mimic (5′-UGAGGUAGUAGGUUGUAUAGUU-3′; Guangzhou RiboBio Co., Ltd.) or miRNA-negative control (NC) mimic (5′-UUCUCCGAACGUGUCACGU-3′; Guangzhou RiboBio Co., Ltd.) using Lipofectamine 2000 (Life Technologies; Thermo Fisher Scientific, Inc.). Luciferase activity was assessed after 48 h using the Dual-Luciferase Reporter Assay system (Promega Corporation) according to the manufacturer's protocol. Luciferase activity was normalized to the Renilla luciferase activity. The experiment was repeated three times.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5 (GraphPad Software, Inc.). Data are presented as the mean ± SD. Comparisons between two groups were analyzed using Student's t-test. One-way ANOVA followed by Tukey's test was performed to compare multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
LPS inhibits ATDC5 cell viability
ATDC5 cells were treated with various concentrations of LPS (0, 1, 5 and 10 µg/ml) for 24 h, and CCK-8 assay was used to detect cell viability. The present results suggested that 5 and 10 µg/ml LPS could significantly inhibit ATDC5 cell viability (Fig. 1). Then, 5 µg/ml LPS was selected for further experiments.
Figure 1.
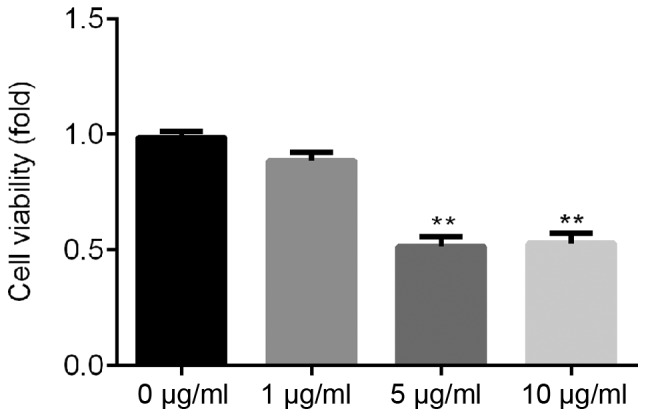
Effects of LPS on ATDC5 cell viability. Cell Counting Kit-8 was used to detect the effect of different concentrations of LPS (0, 1, 5, 10 µg/ml) on the viability of ATDC5 cells. Data are presented as the mean ± SD. **P<0.01 vs. 0 µg/ml LPS. LPS, lipopolysaccharide.
LPS significantly induces inflammatory response and increases the expression level of Let-7a
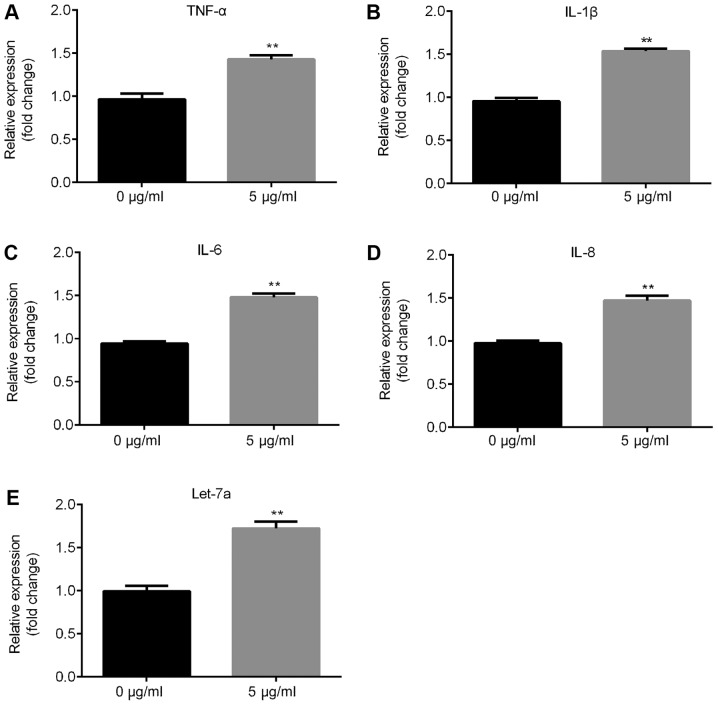
The present RT-qPCR results suggested that the mRNA expression levels of inflammatory factors, such as TNF-α, IL-1β, IL-6 and IL-8, significantly increased following treatment with 5 µg/ml LPS in ATDC5 cells compared with the control group (Fig. 2A-D) and Let-7a expression level was significantly increased (Fig. 2E).
Figure 2.
Effects of LPS on inflammatory response and Let-7a expression level. Reverse transcription-quantitative PCR was used to detect the effects of 5 µg/ml LPS on the expression level of the inflammatory response-associated factors (A) TNF-α, (B) IL-1β, (C) IL-6 and (D) IL-8. (E) Effects of LPS on the expression level of Let-7a. Data are presented as the mean ± SD. **P<0.01 vs. 0 µg/ml LPS. Let-7a, microRNA-let7a; LPS, lipopolysaccharide; TNF-α, tumor necrosis factor α; IL-, interleukin.
IL-6R is a target of Let-7a
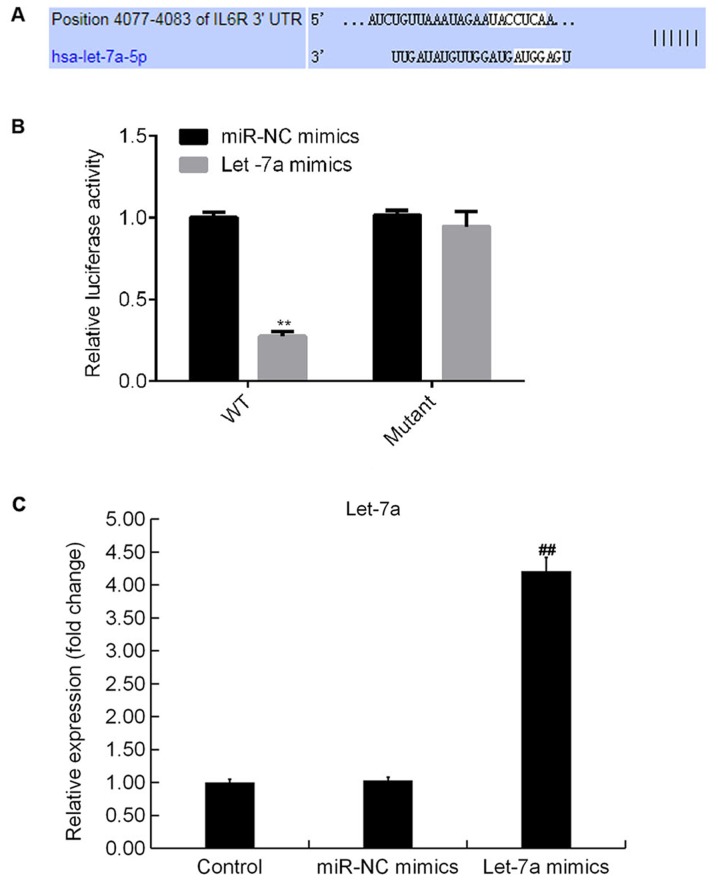
TargetScan analysis identified potential binding sites between Let-7a and IL-6R (Fig. 3A). The dual-luciferase reporter gene assay results suggested that compared with cotransfection of WT-3′UTR-IL-6R plasmid and mimic control, luciferase activity was significantly decreased following cotransfection with WT-3′UTR-IL-6R and Let-7a mimics. The present results suggested that IL-6R may be a direct target of Let-7a (Fig. 3B). In addition, Let-7a mimics significantly increased the expression level of Let-7a in ATDC5 cells (Fig. 3C).
Figure 3.
IL-6R is a target gene of Let-7a. (A) Binding sites between IL-6R and Let-7a, as predicted by TargetScan. (B) Dual-luciferase reporter assay was used to detect the association between IL-6R and Let-7a. (C) Expression level of Let-7a in ATDC5 cells in different conditions. Data are presented as the mean ± SD. **P<0.01 vs. miR-NC mimic; ##P<0.01 vs. Control. NC, negative control; IL-6R, interleukin 6 receptor; Let-7a, microRNA-let7a; miR, microRNA; WT, wild-type; UTR, untranslated region.
Let-7a inhibitor significantly promotes cell viability and reduces apoptosis
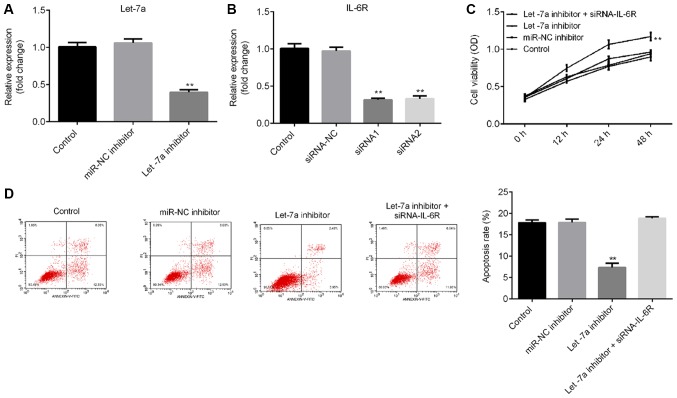
To investigate the effects of Let-7a on LPS-treated ATDC5 cells, ATDC5 cells were transfected with Let-7a inhibitor, miRNA-NC inhibitor or Let-7a inhibitor + IL-6R-siRNA for 48 h, and then the cells were treated with 5 µg/ml LPS for 5 h. RT-qPCR results indicated that Let-7a inhibitor significantly decreased Let-7a expression levels in ATDC5 cells (Fig. 4A), and the mRNA level of IL-6R in ATDC5 cells was significantly decreased following treatment with IL-6R-siRNA (Fig. 4B). The CCK-8 results and flow cytometry analysis suggested that Let-7a inhibitor significantly promoted cell viability (Fig. 4C) and decreased cell apoptosis (Fig. 4D) in LPS-treated ATDC5 cells. These effects were reversed by IL-6R knockdown.
Figure 4.
Effects of Let-7a inhibitor on cell proliferation and apoptosis in lipopolysaccharide-treated ATDC5 cells. (A) Expression level of Let-7a in ATDC5 cells in various conditions. (B) mRNA level of IL-6R in ATDC5 cells in various experimental groups. (C) Effects of Let-7a inhibitor on ATDC5 cell viability. (D) Effects of Let-7a inhibitor on ATDC5 cell apoptosis. Data are presented as the mean ± SD. **P<0.01 vs. control group. Let-7a, microRNA-let7a; IL-6R, interleukin 6 receptor; siRNA, small interfering RNA; OD, optical density; NC, negative control; miR, microRNA.
Let-7a inhibitor significantly reduces the protein expression levels of multiple inflammatory factors
ELISA results suggested that Let-7a inhibitor significantly reduced the expression levels of TNF-α, IL-1β, IL-6 and IL-8 in LPS-treated ATDC5 cells, and these effects were reversed by IL-6R knockdown (Fig. 5).
Figure 5.
Effects of Let-7a inhibitor on the expression of inflammatory factors in LPS-treated ATDC5 cells. ATDC5 cells were treated with 5 µg/ml LPS for 5 h after 48 h of transfection with miR-NC inhibitor, Let-7a inhibitor or Let-7a inhibitor + IL-6R-siRNA. ELISA was used to detect the expression of inflammatory factors, including (A) TNF-α, (B) IL-1β, (C) IL-6 and (D) IL-8. Data are presented as the mean ± SD. **P<0.01. Let-7a, microRNA-let7a; LPS, lipopolysaccharide; IL-6R, interleukin 6 receptor; siRNA, small interfering RNA; NC, negative control; TNF-α, tumor necrosis factor α; IL-, interleukin; miR, microRNA.
Let-7a inhibitor activates the STAT3 signaling pathway
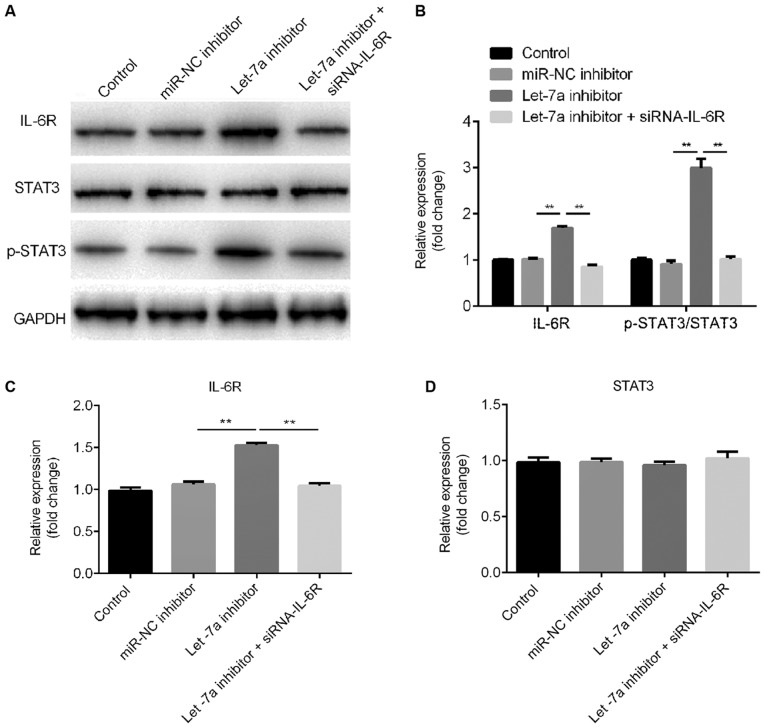
Western blot assay ad RT-qPCR results suggested that Let-7a inhibitor increased IL-6R protein and mRNA expression levels, respectively. These effects were reversed by IL-6R-siRNA (Fig. 6A-C). Let-7a inhibitor increased the protein expression level of phosphorylated STAT3 and this effect was reversed by IL-6R knockdown (Fig. 6A and B). Notably, Let-7a inhibitor and IL-6R-siRNA did not exhibit effects on the protein and mRNA expression levels of STAT3 (Fig. 6A and D).
Figure 6.
Effects of Let-7a inhibitor on the STAT3 signaling pathway. ATDC5 cells were treated with 5 µg/ml lipopolysaccharide for 5 h after 48 h of transfection with miR-NC inhibitor, Let-7a inhibitor or Let-7a inhibitor + IL-6R-siRNA. (A) Western blot assay was performed to quantify the protein expression level of p-STAT3 and IL-6R. (B) Densitometry analysis. Reverse transcription-quantitative PCR was used to detect the mRNA level of (C) IL-6R and (D) STAT3. Data are presented as the mean ± SD. **P<0.01. IL-6R, interleukin 6 receptor; p-, phosphorylated; siRNA, small interfering RNA; Let-7a, microRNA-let7a; NC, negative control; miR, microRNA.
Discussion
The deterioration of the main joint structure, cartilage, bone and synovium are a major feature of OA (25,26), which is the most common joint disease. Joint pain, stiffness and impaired movements are the major clinical symptoms of OA. The prevalence of OA increases with age. Notably, OA can cause chronic pain and reduce the quality of life of elderly patients (27).
LPS is one of the main components of the cell wall of Gram-negative bacteria (28). LPS can activate macrophages, triggering the inflammatory response and activating the innate immunity (28). In the present study, LPS-treated ATDC5 cells were used to establish an in vitro a model of chondrocyte inflammatory injury. The present results suggested that LPS was able to induce ATDC5 cells to synthesize a large amount of inflammatory cytokines, including TNF-α, IL-1β, IL-6 and IL-8, in line with previous studies (29,30).
The Let-7 family consists of miRNAs that are highly expressed in adult tissues (13). In the present paper, IL-6R was a identified as a direct target of Let-7a using a dual-luciferase reporter assay, and IL-6R was found to be negatively regulated by Let-7a. Consistent with a previous study, the present results suggested that Let-7a was involved in cell proliferation and apoptosis (31). In the present study, Let-7a inhibitor promoted cell viability and inhibited apoptosis in LPS-treated ATDC5 cells. Accumulating evidence demonstrated that pro-inflammatory cytokines, such as TNF-α and IL-1β, serve key roles in the pathogenesis of OA (32,33). TNF-α is a type of TNF released by macrophages, and acts as a trigger for the inflammatory response (34,35). IL-1β and IL-1α are the two types of IL-1, and these two cytokines can be synthesized by various cell types, such as monocyte macrophages and vascular endothelial cells (36). Notably, IL-1β is an important inflammatory mediator of the inflammatory process (37). IL-1β can not only induce the release of inflammatory mediators, including nitric oxide, prostaglandin E2 and matrix metalloproteinases, but also promote chondrocyte apoptosis, leading to articular cartilage damage (38,39). IL-6 is a cytokine produced by various cell types and it belongs to the interleukin family. IL-6 was identified to be able to transduce signals, activating and regulating immune cells, and mediating T and B cell activation, proliferation and differentiation, serving an important role in the inflammatory response (40,41). The inflammatory response involves lipid peroxidation and activation of multiple receptors, stimulating macrophages and other cells to secrete pro-inflammatory factors, such as IL-1, IL-2 and IL-8, activating a signaling cascade (34,35). In the present study, ELISA was used to detect the expression level of inflammatory factors in LPS-treated ATDC5 cells in various conditions. The present results suggested that Let-7a inhibitor significantly reduced the expression levels of TNF-α, IL-1β, IL-6 and IL-8 in LPS-treated ATDC5 cells, and these effects were reversed by IL-6R knockdown.
The IL-6/STAT3 signaling pathway is a key signal transduction pathway for the development and progression of malignant tumors (42–44). In addition, IL-6-mediated STAT3 activation is involved in cell proliferation and apoptosis (42–44). In the present study, the effects of Let-7a inhibitor on the STAT3 signaling pathway were investigated. Western blotting results suggested that Let-7a inhibitor significantly increased the protein expression level of phosphorylated STAT3 and this effect was reversed by IL-6R knockdown. The present results suggested that Let-7a inhibitor could activate the STAT3 signaling pathway.
Collectively, the present results suggested that Let-7a inhibitor could enhance cell proliferation, reduce apoptosis and inhibit inflammatory response in LPS-treated ATDC5 cells. The present study provided a novel potential therapeutic target and may facilitate the development of new approaches to improve the prevention and treatment of OA. However, the present study is a preliminary study, and further experiments are required to validate the role of Let-7a in OA. Importantly, it is necessary to investigate the expression level of Let-7a in patients with OA. In addition, the role of Let-7a in OA should be investigated using in vivo models. Moreover, the association between the expression level of Let-7a and the clinical features of patients with OA requires further investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by Youth Culture Program of the First Affiliated Hospital of Anhui Medical University (grant no. 2017kj06).
Availability of data and materials
All data sets used and/or generated during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CS and LZ contributed to the design of the study, data collection, statistical analysis and data interpretation. YH contributed to data collection, manuscript preparation and the literature search. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Allen KD, Golightly YM. State of the evidence. Curr Opin Rheumatol. 2015;27:276–283. doi: 10.1097/BOR.0000000000000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heijink A, Vanhees M, van den Ende K, van den Bekerom MP, van Riet RP, Van Dijk CN, Eygendaal D. Biomechanical considerations in the pathogenesis of osteoarthritis of the elbow. Knee Surg Sports Traumatol Arthrosc. 2016;24:2313–2318. doi: 10.1007/s00167-015-3518-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2016;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Hydbring P, Wang Y, Fassl A, Li X, Matia V, Otto T, Choi YJ, Sweeney KE, Suski JM, Yin H, et al. Cell-cycle-targeting micrornas as therapeutic tools against refractory cancers. Cancer Cell. 2017;31:576–590.e8. doi: 10.1016/j.ccell.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drusco A, Croce CM. MicroRNAs and cancer: A long story for short RNAs. Adv Cancer Res. 2017;135:1–24. doi: 10.1016/bs.acr.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7, miRNAs and their functional implications. Protein Cell. 2016;7:100–113. doi: 10.1007/s13238-015-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beyer C, Zampetaki A, Lin NY, Kleyer A, Perricone C, Iagnocco A, Distler A, Langley SR, Gelse K, Sesselmann S, et al. Signature of circulating microRNAs in osteoarthritis. Ann Rheum Dis. 2015;74:e18. doi: 10.1136/annrheumdis-2013-204698. [DOI] [PubMed] [Google Scholar]
- 11.Xue H, Tu Y, Ma T, Wen T, Yang T, Xue L, Cai M, Wang F, Guan M. miR-93-5p attenuates IL-1β-induced chondrocyte apoptosis and cartilage degradation in osteoarthritis partially by targeting TCF4. Bone. 2019;123:129–136. doi: 10.1016/j.bone.2019.03.035. [DOI] [PubMed] [Google Scholar]
- 12.Malemud CJ. MicroRNAs and osteoarthritis. Cells. 2018;7:E92. doi: 10.3390/cells7080092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 14.Lu L, Katsaros D, Risch HA, Canuto EM, Biglia N, Yu H. MicroRNA let-7a modifies the effect of self-renewal gene HIWI on patient survival of epithelial ovarian cancer. Mol Carcinog. 2016;55:357–365. doi: 10.1002/mc.22285. [DOI] [PubMed] [Google Scholar]
- 15.Liu K, Zhang C, Li T, Ding Y, Tu T, Zhou F, Qi W, Chen H, Sun X. Let-7a inhibits growth and migration of breast cancer cells by targeting HMGA1. Int J Oncol. 2015;46:2526–2534. doi: 10.3892/ijo.2015.2949. [DOI] [PubMed] [Google Scholar]
- 16.Johnson VL, Hunter DJ. The epidemiology of osteoarthritis. Best Pract Res Clin Rheumatol. 2014;28:5–15. doi: 10.1016/j.berh.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 17.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: A disease of the joint as an organ. Arthritis Rheumatism. 2012;64:1697–1707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burr DB, Gallant MA. Bone remodelling in osteoarthritis. Nat Rev Rheumatol. 2012;8:665–673. doi: 10.1038/nrrheum.2012.130. [DOI] [PubMed] [Google Scholar]
- 19.Kim HA, Lee YJ, Seong SC, Choe KW, Song YW. Apoptotic chondrocyte death in human osteoarthritis. J Rheumatol. 2000;27:455–462. [PubMed] [Google Scholar]
- 20.Yan S, Wang M, Zhao J, Zhang H, Zhou C, Jin L, Zhang Y, Qiu X, Ma B, Fan Q. MicroRNA-34a affects chondrocyte apoptosis and proliferation by targeting the SIRT1/p53 signaling pathway during the pathogenesis of osteoarthritis. Int J Mol Med. 2016;38:201–209. doi: 10.3892/ijmm.2016.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu Y, Long J, Guo W, Ye W. MicroRNA-195-5p inhibitor prevents the development of osteoarthritis by targeting REGγ. Mol Med Rep. 2019;19:4561–4568. doi: 10.3892/mmr.2019.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Li S, Zou Y. Knockdown of LncRNA H19 relieves LPS-induced damage by modulating miR-130a in osteoarthritis. Yonsei Med J. 2019;60:381–388. doi: 10.3349/ymj.2019.60.4.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying H, Wang Y, Gao Z, Zhang Q. Long non-coding RNA activated by transforming growth factor beta alleviates lipopolysaccharide-induced inflammatory injury via regulating microRNA-223 in ATDC5 cells. Int Immunopharmacol. 2019;69:313–320. doi: 10.1016/j.intimp.2019.01.056. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Goldring MB, Goldring SR. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann NY Acad Sci. 2010;1192:230–237. doi: 10.1111/j.1749-6632.2009.05240.x. [DOI] [PubMed] [Google Scholar]
- 26.Berenbaum F. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis) Osteoarthritis Cartilage. 2013;21:16–21. doi: 10.1016/j.joca.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Malfait AM. Osteoarthritis year in review 2015: Biology. Osteoarthritis Cartilage. 2016;24:21–26. doi: 10.1016/j.joca.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan Q, Zhang D, Liu C, Zhang C, Yuan D. Chikusetsusaponin V Inhibits LPS-activated inflammatory responses via SIRT1/NF-κB signaling pathway in RAW264.7 cells. Inflammation. 2018;41:2149–2159. doi: 10.1007/s10753-018-0858-8. [DOI] [PubMed] [Google Scholar]
- 29.Cheng Y, Yang C, Luo D, Li X, Le XC, Rong J. N-Propargyl caffeamide skews macrophages towards a resolving M2-Like phenotype against myocardial ischemic injury via activating Nrf2/HO-1 pathway and inhibiting NF-κB pathway. Cell Physiol Biochem. 2018;47:2544–2557. doi: 10.1159/000491651. [DOI] [PubMed] [Google Scholar]
- 30.De Santis R, Liepelt A, Mossanen JC, Dueck A, Simons N, Mohs A, Trautwein C, Meister G, Marx G, Ostareck-Lederer A, Ostareck DH. miR-155 targets caspase-3 mRNA in activated macrophages. RNA Biol. 2016;13:43–58. doi: 10.1080/15476286.2015.1109768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toledano H. The role of the heterochronic microRNA let-7 in the progression of aging. Exp Gerontol. 2013;48:667–670. doi: 10.1016/j.exger.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Amin AR. Regulation of tumor necrosis factor-α and tumor necrosis factor converting enzyme in human osteoarthritis. Osteoarthritis Cartilage. 1999;7:392–394. doi: 10.1053/joca.1998.0221. [DOI] [PubMed] [Google Scholar]
- 33.Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 2011;7:33–42. doi: 10.1038/nrrheum.2010.196. [DOI] [PubMed] [Google Scholar]
- 34.Calzascia T, Pellegrini M, Hall H, Sabbagh L, Ono N, Elford AR, Mak TW, Ohashi PS. TNF-alpha is critical for antitumor but not antiviral T cell immunity in mice. J Clin Invest. 2007;117:3833–3845. doi: 10.1172/JCI32567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu M, Gu M, Xu D, Lv Q, Zhang W, Wu Y. Protective effects of toll-like receptor 4 inhibitor eritoran on renal ischemia-reperfusion injury. Transplant Proc. 2010;42:1540–1544. doi: 10.1016/j.transproceed.2010.03.133. [DOI] [PubMed] [Google Scholar]
- 36.Striz I. Cytokines of the IL-1 family: Recognized targets in chronic inflammation underrated in organ transplantations. Clin Sci (Lond) 2017;131:2241–2256. doi: 10.1042/CS20170098. [DOI] [PubMed] [Google Scholar]
- 37.Logan RM, Stringer AM, Bowen JM, Yeoh AS, Gibson RJ, Sonis ST, Keefe DM. The role of pro-inflammatory cytokines in cancer treatment-induced alimentary tract mucositis: Pathobiology, animal models and cytotoxic drugs. Cancer Treat Rev. 2007;33:448–460. doi: 10.1016/j.ctrv.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Montaseri A, Busch F, Mobasheri A, Buhrmann C, Aldinger C, Rad JS, Shakibaei M. IGF-1 and PDGF-bb suppress IL-1β-induced cartilage degradation through down-regulation of NF-κB signaling: Involvement of Src/PI-3K/AKT pathway. PLoS One. 2011;6:e28663. doi: 10.1371/journal.pone.0028663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ying X, Peng L, Chen H, Shen Y, Yu K, Cheng S. Cordycepin prevented IL-β-induced expression of inflammatory mediators in human osteoarthritis chondrocytes. Int Orthop. 2014;38:1519–1526. doi: 10.1007/s00264-013-2219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lv SY, Wu Q, Liu JP, Shao J, Wen LL, Xue J, Zhang XS, Zhang QR, Zhang X. Levels of interleukin-1β, interleukin-18, and tumor necrosis factor-α in cerebrospinal fluid of aneurysmal subarachnoid hemorrhage patients may be predictors of early brain injury and clinical prognosis. World Neurosurg. 2018;111:e362–e373. doi: 10.1016/j.wneu.2017.12.076. [DOI] [PubMed] [Google Scholar]
- 41.Ataie-Kachoie P, Pourgholami MH, Richardson DR, Morris DL. Gene of the month: Interleukin 6 (IL-6) J Clin Pathol. 2014;67:932–937. doi: 10.1136/jclinpath-2014-202493. [DOI] [PubMed] [Google Scholar]
- 42.Bournazou E, Bromberg J. Targeting the tumor microenvironment: JAK-STAT3 signaling. JAKSTAT. 2013;2:e23828. doi: 10.4161/jkst.23828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rokavec M, Wu W, Luo JL. IL6-Mediated suppression of miR-200c directs constitutive activation of inflammatory signaling circuit driving transformation and tumorigenesis. Mol Cell. 2012;45:777–789. doi: 10.1016/j.molcel.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18:254–267. doi: 10.1038/cr.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data sets used and/or generated during the current study are available from the corresponding author on reasonable request.