Abstract
Context
The gut microbiome is a source of inflammatory factors such as lipopolysaccharide (LPS; endotoxin) that influence metabolic homeostasis. Rifaximin is a well-tolerated antibiotic that may reduce LPS.
Objective
We sought to develop a method to accurately assess postprandial endotoxemia and to determine whether rifaximin treatment improves metabolic homeostasis in obese humans with metabolic syndrome.
Design and Setting
Plasma LPS, adipose inflammation, glucose and lipid metabolism, and insulin sensitivity were evaluated in a clinical research setting.
Participants
Twelve obese human research participants with prediabetes or three features of metabolic syndrome participated.
Intervention
The research participants were randomized to placebo control or rifaximin soluble solid dispersion (80 mg/d) treatment groups and treated for 12 weeks.
Outcome Measures
We evaluated changes in insulin sensitivity with a euglycemic clamp; changes in lipid and glucose metabolism with oral lipid and glucose tolerance tests; changes in plasma LPS during the lipid tolerance test; and changes in adipose tissue and systemic inflammation by measuring inflammatory cytokines.
Results
Rifaximin treatment slightly worsened insulin sensitivity (P = 0.03), did not improve glucose or lipid homeostasis, and did not significantly improve adipose tissue inflammation. Our efforts to accurately assess plasma LPS using limulus amebocyte lysate assays revealed that the majority of LPS is masked from detection by limulus amebocyte lysate assays, but can be unmasked using a pretreatment step with protease. Unmasked LPS increases during the lipid tolerance test, but rifaximin treatment did not reduce this.
Conclusions
Rifaximin treatment did not lower plasma LPS or improve metabolic homeostasis in obese humans.
Keywords: rifaximin, lipopolysaccharide, insulin resistance, obesity
The gut microbiome influences metabolic homeostasis through a variety of mechanisms [1]. These include the generation of metabolites and substances such as lipopolysaccharide (LPS; endotoxin) that affect the function of the major organs involved in maintaining glucose and lipid homeostasis including adipose tissue [1–6]. Germ-free mice were shown to be resistant to high-fat diet-induced obesity [2], and subsequent studies in mice indicated that the gut microbiome contributes to obesity [4, 5, 7–9]. One mechanism promoting obesity is increased energy harvest from the gut microbiome [2, 4, 5].
In addition to promoting obesity, the gut microbiome has been shown to induce adipose inflammation, and LPS has been implicated in this process [3, 6]. During the assembly of chylomicrons, LPS from bacteria associates with chylomicrons [10], and there is a transient increase in plasma LPS levels after a fatty meal, which could lead to tissue inflammation from the lipoprotein-LPS complexes [11–13]. Furthermore, there are strong associations between insulin resistance, diabetes, fat intake, and plasma LPS levels [14–21]. These findings, often referred to as “metabolic endotoxemia” [22, 23], suggest that the gut microbiome may be a target for improving metabolic homeostasis, and studies in rodents have shown that antibiotics or prebiotic diets reduce plasma LPS and have beneficial effects on blood glucose [24–26].
Rifaximin is a poorly absorbed antibiotic in widespread use for the treatment of hepatic encephalopathy, Escherichia coli–induced traveler’s diarrhea, and recurrent Clostridium difficile infection. This drug is well-tolerated and patients are treated for long periods. In addition, rifaximin has been reported to reduce plasma LPS levels in patients with cirrhosis and hepatic encephalopathy [27–30]. We hypothesized that rifaximin may be beneficial in subjects with metabolic syndrome and insulin resistance through a similar reduction in LPS. Here we present a placebo-controlled study on the effects of rifaximin treatment on insulin sensitivity, adipose inflammation, and various measures of metabolic homeostasis. We developed a method of measuring plasma LPS levels during an oral lipid tolerance test (OLTT) that addresses important methodological concerns about measuring plasma LPS with limulus amebocyte lysate (LAL) assays [31], and used this to characterize endotoxemia and to determine whether rifaximin treatment lowers plasma LPS.
1. Materials and Methods
A. Human Research Participants and Study Design
Rifaximin soluble solid dispersion (SSD) is a formulation of rifaximin that is more gut-soluble and was supplied for this study along with matching placebo tablets by Salix Pharmaceuticals. Rifaximin SSD is given once daily at a dose of 80 mg, which is a lower dose than conventional rifaximin because of the improved solubility. Obese research participants with features of metabolic syndrome were recruited and assigned to placebo control or rifaximin treatment groups and treated for 12 weeks. Of the 12 subjects in this study, 10 were women, but only two were still menstruating; the others were postmenopausal. Of the two menstruating women, their periods were irregular, and it was not possible to time the euglycemic clamps described in the subsequent section to a specific phase of their cycle. The drug/placebo assignment was kept with the pharmacist and was unknown to the investigative team; however, pill counting during compliance visits ensured that participants were taking their assigned drugs. Carbohydrate and lipid metabolism as well as insulin sensitivity were assessed by oral glucose and lipid tolerance tests and euglycemic clamping before and after treatment as described [32]. Briefly, the oral glucose tolerance test (OGTT) was performed using a 75-g glucose challenge. Blood was collected at baseline and 30, 60, 90, and 120 minutes for measurement of glucose and insulin. The OLTT was performed using a high-fat breakfast shake that consisted of boost, corn oil, and cream; the high-fat shake consisted of 50% fat and was adjusted for 40% of daily estimated energy expenditure (see [32] for more detailed methods). Subcutaneous abdominal adipose tissue biopsies were obtained in the fasting state and immediately frozen at −80°C for analysis of mRNA expression. All subjects gave informed consent, and the protocols were approved by the institutional review board at the University of Kentucky.
B. Euglycemic Clamps
Peripheral insulin sensitivity was measured with a euglycemic clamp at two different insulin infusion rates. Upon arriving fasting at the Clinical Research Unit, a retrograde IV was inserted, the hand placed in a warming box, and a peripheral IV line was started for infusions. After a 30-minute period of baseline stabilization, an insulin infusion started at a rate of 0.25 mU/kg/min (low insulin) and a 20% glucose solution was infused at a variable rate to maintain euglycemia. Blood glucose was measured every 5 to 10 minutes, and blood insulin every 10 minutes during the final 30 minutes of the procedure. A steady state was generally attained at 2 hours. At the end of the low insulin infusion, the insulin infusion was increased to 1.0 mU/kg/min (high insulin), again with frequent blood glucose measurement and adjustment of the 20% glucose infusion. Glucose disposal [glucose infusion rate (GIR)] was determined during steady-state glucose infusion during the final 30 minutes of the procedure.
C. Plasma LPS Measurement by Conventional LAL Assays
For the measurement of LPS, we collected blood in citrate collection tubes during the OLTT and prepared plasma. The citrated plasma was diluted 1:5 in endotoxin free water (Lonza, Basel, Switzerland), heat inactivated at 70°C, and stored at −80°C. There is a methodological concern about using LAL assays to measure LPS levels in plasma during a lipid tolerance test because the substrate is not washed away from the LAL reagents before spectrometry. Therefore, substances that block light transmission such as triglycerides or absorb light at the same wavelength as the LAL chromophore will cause a false-positive signal [31]. To attempt to overcome this, we used the ToxinSensorTM Chromogenic LAL Endotoxin Assay kit (L00350; Genscript, Piscataway, NJ) because its high sensitivity would allow us to dilute interfering substances, and we included a control in which no LAL enzyme was added to determine assay interference. To assess this method, we measured plasma LPS in samples collected during an OLTT in a separate group of obese, insulin-resistant research participants at baseline [32]. We included a control reaction that was conducted without addition of the LAL lysate to determine whether the sample was causing a false-positive signal. The standard curve for the assay is shown in Supplementary Fig. 1A [33]. The raw absorbance data (A545) are presented in Supplementary Fig. 1B [33]. LPS did not increase with time during the OLTT using this method.
D. Plasma LPS Measured by a Modified LAL Assay to Unmask LPS
LPS can be masked from detection by proteins in the plasma; therefore, we heat inactivated the samples as described previously and also used endotoxin sample preparation kits, which use protease to eliminate the LPS binding proteins. The heat-inactivated, citrated plasma samples were pretreated with endotoxin sample preparation kits (BioDtech, Inc., Birmingham, AL) and then LPS was measured using the ToxinSensor kit (described above) according to the manufacturer’s instructions with and without the addition of LAL enzyme to control for any sample interference. We assessed this method on the placebo and rifaximin OLTT plasma before treatment. The raw absorbance data (A595) for all subjects before treatment are shown in Supplementary Fig. 1C [33]. This modified assay clearly shows an increase in LPS following a lipid-rich meal; this measurement clearly required steps to unmask LPS.
E. Adipose Tissue mRNA Expression
We isolated RNA using the RNeasy Lipid Tissue Mini Kit (Qiagen, 74804) with QIAzol supplied with the kit. We used real-time RT-PCR to measure gene expression of cytokines and adipokines according to [34]. The primer sequences are located in Supplementary Table 1 [35].
F. Plasma Cytokines and GLP-1
IL-6 and TNF-α were measured using the V-Plex Proinflammatory Panel I kit (K15049D [36], Mesoscale, Rockville, MD), and MCP1 was measured using the V-Plex Human MCP-1 Kit (K151NND [37], Mesoscale); assays were quantified using a QuickPlex SQ 120 (Mesoscale). GLP-1 and GIP were measured during the glucose or lipid tolerance tests as indicated using GLP-1 total [7–36, 9–36] ELISA (43-GPTHU-E01 [38], Alpco Diagnostics, Salem, NH), and Human GIP (Total) ELISA (Millipore [39], St. Charles, MO) assays.
G. Calculations
The dilution factor for samples analyzed by the conventional assay was 5, which is the dilution before heat inactivation (1:5). The dilution factor for plasma samples analyzed by the modified assay was 183, which was calculated by multiplying the dilution before heat inactivation (1:5) by the dilutions made during the protease treatment (1:11 in buffer 1; 1:3.33 in buffer 2). Homeostatic model assessment of insulin resistance (HOMA IR) was calculated by multiplying baseline glucose by baseline insulin and dividing by 405. The Matsuda index of insulin sensitivity (SI) was determined by the following formula: SI = 10,000/SQRT (baseline glucose × baseline insulin × (baseline glucose + 30 minutes glucose × 2 + 60 minutes glucose × 2 + 90 minutes glucose × 2 + 120 minutes glucose)/8 × (baseline insulin + 30 minutes insulin × 2 + 60 min insulin × 2 + 90 minutes insulin × 2 + 120 minutes insulin)/8). The insulinogenic index was calculated by the following formula: I = (30 minutes insulin − baseline insulin)/(30 minutes glucose – baseline glucose). The disposition index was determined by multiplying the Matsuda index by the insulinogenic index.
H. Statistics
Data are presented as mean SEM and were analyzed by paired, two-tailed Student t tests. Treatment differences were determined by calculating the change by each treatment and then performing an unpaired Student t test. Time course analyses of LPS levels during OLTTs were analyzed by two-way ANOVA. Statistical calculations were performed in GraphPad Prism, version 7.0.
2. Results
A. Plasma LPS During an OLTT
LPS has been reported to increase in plasma during an OLTT in humans [13], and therefore we sought to determine whether rifaximin affected baseline or postlipid LPS levels. As described in “Materials and Methods,” we developed two methods for detecting LPS during the OLTT that address important methodological concerns about sample interference by triglycerides [31], which increase after digestion of the lipid meal during an OLTT. Both the conventional and the modified LPS assays use a “no LAL” control to account for interference by triglycerides, and the assay is highly sensitive, which allows for dilution and minimizing of other interfering substances. We assessed the conventional assay using the baseline OLTT plasma from a cohort of obese subjects that we recently described [32]. These subjects had a body mass index of 37.3 ± 1.2, mean age of 53.4 ± 2.8, and were insulin resistant [32]. The conventional LAL assay did not detect an increase in LPS during an OLTT despite a substantial increase in triglyceride levels (Fig. 1).
Figure 1.
Measurement of plasma LPS during the OLTT using conventional LAL assays. (A) Plasma was collected from a cohort of obese research participants [32] at the indicated time after ingestion of lipid and triglyceride was measured. (B) LPS was measured using Genscript ToxinSensor assays in heat-inactivated plasma as described in “Materials and Methods.” The data represent mean ± SEM (n = 8) and were analyzed by repeated measures ANOVA.
We developed a modified assay that uses heat inactivation and protease pretreatment to remove LPS binding proteins that mask LPS from detection by the LAL assay. This modified assay was evaluated on the baseline OLTT plasma from the subjects in this study, and this revealed that unmasked LPS significantly increases during an OLTT (Supplementary Fig. 1B; P < 0.0001) [33]. Furthermore, analysis of the control reactions in which we omitted LAL showed that there was no assay interference because the signal was very low and there was no increase in absorbance in samples during the time course of the OLTT (Supplementary Fig. 1B) [33]. We therefore used this assay to assess postprandial plasma LPS during the OLTT of the subjects in this study, and did not perform conventional LPS assays. LPS detected by the modified assay is referred to as unmasked LPS.
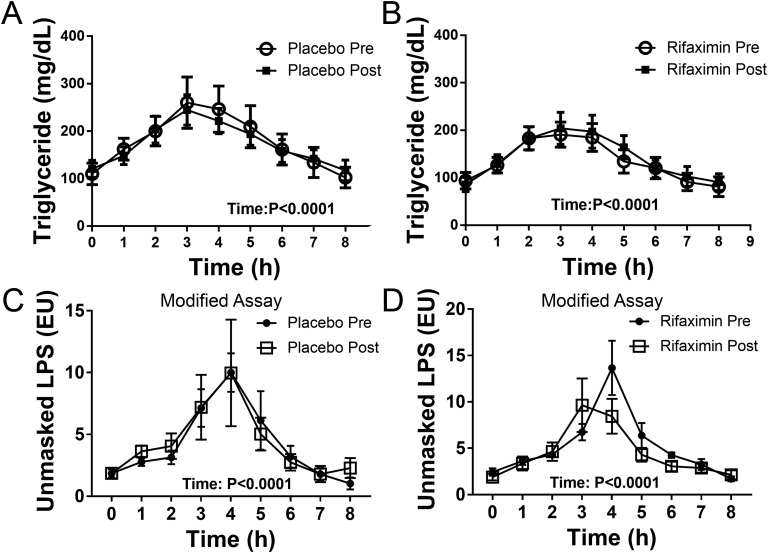
Triglyceride levels significantly increased and then decreased with time in the placebo and rifaximin treatment groups during the OLTT (Fig. 2A-B; P < 0.0001), but neither treatment changed the response. As shown in Fig. 2C-D, unmasked LPS levels in plasma rise and then fall during the OLTT (P < 0.0001). However, neither placebo nor rifaximin treatment significantly changed the unmasked LPS levels during the OLTT or at baseline (0 hours).
Figure 2.
Measurement of plasma LPS during the OLTT using a modified assay that uses protease pretreatment to unmask LPS. (A, B) Plasma was collected at the indicated time of the lipid tolerance test before and after placebo or rifaximin treatment as indicated, and triglyceride was measured. (C, D) LPS was measured using Genscript ToxinSensor assays in heat-inactivated, citrated plasma that was treated with an endotoxin sample preparation kit as described in “Materials and Methods.” The data represent mean ± SEM (n = 6) and were analyzed by repeated measures two-way ANOVA.
Although we did not find treatment differences in plasma LPS levels during the OLTT, the modified LAL assay using protease pretreatment to unmask LPS should be useful to others wishing to study postprandial endotoxemia. An important finding from these studies is that LPS increases in plasma of obese subjects during an OLTT, but it is masked from the LAL assay because we only detect it if we use protease pretreatment (compare Supplementary Fig. 1A and Supplementary Fig. 1B); see “Materials and Methods” for additional details. Furthermore, the conventional LAL assay detects a small level of free or unmasked LPS that is much lower than the level of masked LPS (compare Fig. 1B to Fig. 2C-D).
B. Metabolic Homeostasis
We performed OGTTs and euglycemic clamping before and after treatment to assess glucose homeostasis and insulin sensitivity. The results for each treatment and analysis of whether there was a difference in treatments are presented in Table 1. There was a substantial difference in treatment outcomes for the GIR obtained using high insulin because rifaximin treatment caused a slight decrease and placebo caused a slight increase in GIR (Table 1; treatment P = 0.03). However, the results of each treatment did not reach statistical significance (Table 1; placebo, P = 0.12; rifaximin, P = 0.14). Table 1 also shows that fasting insulin (P = 0.05) and HOMA IR (P = 0.05) significantly increased after rifaximin treatment, and there was a trend for an increase in fasting blood glucose; however, there was not an important difference in treatment outcomes for these results (Table 1). Together, these results suggest that rifaximin slightly reduced insulin sensitivity. Other plasma lipids were measured in addition to the triglyceride levels during the OLTT presented in Fig. 1, but did not significantly change (Table 1). Finally, we determined whether rifaximin treatment altered incretin secretion because there is evidence linking the gut microbiota to incretin secretion [40, 41]. As shown in Supplementary Fig. 2 [42], GLP-1 and GIP secretion was not significantly changed during the OGTT or the OLTT by rifaximin treatment.
Table 1.
Baseline Characteristics and Response to Treatments
| Treatmenta | Placebo | P | Rifaximin | P | Treatment | ||
|---|---|---|---|---|---|---|---|
| Pre | Post | Pre | Post | Difference P | |||
| No. (M/F) | 6 (1/5) | 6 (1/5) | |||||
| Age | 50 ± 2.9 | 50 ± 3 | |||||
| BMI | 36.1 ± 1.4 | 35.9 ± 1.7 | 0.54 | 38.5 ± 1.7 | 38.6 ± 1.9 | 0.81 | 0.53 |
| Weight (kg) | 97.2 ± 5.0 | 96.6 ± 5.8 | 0.60 | 105.1 ± 3.8 | 105.4 ± 4.6 | 0.80 | 0.62 |
| GIR (high insulin) | 120.1 ± 44.7 | 130.6 ± 43.6 | 0.12 | 116 ± 19.2 | 100.2 ± 17.4 | 0.14 | 0.03 b |
| S I (Matsuda) | 3.6 ± 2.2 | 2.8 ± 1.5 | 0.29 | 2.5 ± 0.4 | 2.1 ± 0.4 | 0.20 | 0.62 |
| HOMA-IR | 4.4 ± 1.6 | 4.9 ± 1.2 | 0.63 | 2.6 ± 0.2 | 3.5 ± 0.3 | 0.05 b | 0.67 |
| Insulinogenic index | 1.6 ± 0.2 | 2.0 ± 0.4 | 0.13 | 1.7 ± 0.5 | 1.7 ± 0.4 | 0.86 | 0.21 |
| Disposition index | 5.3 ± 3.2 | 4 ± 1.3 | 0.54 | 3.4 ± 0.4 | 3.1 ± 0.6 | 0.6 | 0.63 |
| Fasting glucose | 101 ± 5 | 101 ± 6 | 0.84 | 96.8 ± 3 | 101 ± 3 | 0.1c | 0.28 |
| Fasting insulin (uIU/mL) | 17.1 ± 6.0 | 19.0 ± 4.5 | 0.57 | 10.8 ± 1.1 | 14.4 ± 1.5 | 0.05 b | 0.67 |
| 2-h glucose | 137 ± 19 | 128 ± 13 | 0.50 | 126 ± 12 | 144 ± 20 | 0.38 | 0.26 |
| 2-h insulin | 213 ± 92 | 173 ± 82 | 0.24 | 178 ± 44 | 154 ± 45 | 0.51 | 0.75 |
| HbA1c | 6.0 ± 0.1 | 6.0 ± 0.1 | 0.81 | 5.7 ± 0.1 | 5.7 ± 0.1 | 0.62 | 0.78 |
| TG (mg/dL) | 166 ± 35 | 153 ± 30 | 0.73 | 126 ± 17 | 115 ± 16 | 0.35 | 0.95 |
| Cholesterol (mg/dL) | 204 ± 14 | 209 ± 13 | 0.48 | 191 ± 9 | 200 ± 12 | 0.45 | 0.79 |
| HDL (mg/dL) | 49 ± 5 | 44 ± 5 | 0.03 b | 55 ± 4 | 55 ± 3 | 0.89 | 0.14 |
| LDL (mg/dL) | 114 ± 18 | 132 ± 16 | 0.27 | 111 ± 10 | 122 ± 11 | 0.23 | 0.66 |
The results for each treatment were analyzed by a paired, two-tailed Student test. Treatment differences were determined by calculating the change (post-pre) caused by each treatment and performing an unpaired, two-tailed Student test.
Data represent mean ± SEM (n = 6).
b P ≤ 0.05.
P < 0.1.
C. Adipose and Systemic Inflammation
We measured plasma IL-4, IL-6, MCP-1, and TNF-α. IL-4 was below the limit of detection. Plasma TNF-α was lower after rifaximin treatment, but we did not detect a substantial difference between rifaximin and placebo treatment (Table 2; P = 0.44). We measured inflammatory and anti-inflammatory cytokine gene expression in subcutaneous white adipose tissue (SC WAT) by real-time RT-PCR. As shown in Table 2, we only detected a treatment difference for IL12B mRNA expression (P = 0.01); however, this was driven largely by increased IL12B mRNA expression after placebo treatment (P = 0.05). There was a trend for increased IL4 expression and an important decrease in TNF after rifaximin treatment, but we did not detect a substantial treatment difference between placebo and rifaximin treatment in the mRNA expression of either of these cytokines (Table 2). The reduction of adipose tissue TNF mRNA expression is consistent with the reduction in plasma TNF levels. Overall, these results suggest that rifaximin treatment does not improve systemic or SC WAT inflammation.
Table 2.
Systemic and Adipose Tissue Inflammation
| Genea | Placebo | P | Rifaximin | P | Treatment | ||
|---|---|---|---|---|---|---|---|
| Pre | Post | Pre | Post | Difference P | |||
| Plasma cytokines | |||||||
| IL-6 (pg/mL) | 1.02 ± 0.15 | 1.25 ± 0.22 | 0.09 | 1.01 ± 0.42 | 0.97 ± 0.31 | 0.78 | 0.16 |
| TNF-α (pg/mL) | 2.61 ± 0.18 | 2.62 ± 0.17 | 0.95 | 2.97 ± 0.26 | 2.83 ± 0.24 | 0.04 | 0.44 |
| MCP1 (pg/mL) | 159 ± 12.6 | 147 ± 4.8 | 0.343 | 132 ± 11 | 128 ± 10.4 | 0.325 | 0.50 |
| Adipose tissue gene expression b | |||||||
| IL4 | 0.90 ± 0.07 | 1.09 ± 0.19 | 0.25 | 1.01 ± 0.1 | 1.28 ± 0.21 | 0.07c | 0.69 |
| IL6 | 0.52 ± 0.17 | 0.55 ± 0.09 | 0.84 | 0.91 ± 0.49 | 0.88 ± 0.52 | 0.70 | 0.70 |
| CCL5 | 0.60 ± 0.14 | 0.49 ± 0.02 | 0.44 | 1.04 ± 0.39 | 0.87 ± 0.36 | 0.40 | 0.83 |
| ADIPOQ | 1.04 ± 0.08 | 0.99 ± 0.09 | 0.67 | 0.94 ± 0.12 | 1.03 ± 0.11 | 0.50 | 0.42 |
| IL1B | 0.61 ± 0.16 | 0.82 ± 0.29 | 0.50 | 1.39 ± 0.48 | 0.94 ± 0.32 | 0.19 | 0.14 |
| IL10 | 1.06 ± 0.28 | 1.12 ± 0.45 | 0.83 | 1.16 ± 0.36 | 1.23 ± 0.25 | 0.82 | 0.98 |
| IL12B | 1.14 ± 0.43 | 2.22 ± 0.57 | 0.05 | 1.28 ± 0.36 | 0.72 ± 0.19 | 0.13 | 0.01 |
| TNF | 1.30 ± 0.13 | 1.46 ± 0.28 | 0.58 | 2.00 ± 0.25 | 1.67 ± 0.25 | 0.03 | 0.12 |
| CCL2 | 1.11 ± 0.22 | 1.34 ± 0.30 | 0.14 | 1.30 ± 0.27 | 1.34 ± 0.30 | 0.90 | 0.33 |
| CD68 | 1.12 ± 0.23 | 0.98 ± 0.28 | 0.47 | 1.16 ± 0.25 | 1.28 ± 0.18 | 0.43 | 0.28 |
Boldface numbers indicate P < 0.05.
Gene expression was measured in SC WAT by real-time RT-PCR. The expression levels were normalized to the geometric mean of six housekeeping genes and the data, expressed as arbitrary units, are means ± SEM.
NCBI gene symbol.
P < 0.1.
D. Conclusions
The majority of plasma LPS is masked from detection by LAL assays, but can be detected with a protease pretreatment step that unmasks LPS. Unmasked LPS increases during an OLTT in obese human research participants. Rifaximin treatment did not reduce change baseline or plasma unmasked LPS during an OLTT. Consistent with this, rifaximin treatment did not improve glucose or lipid homeostasis or SC WAT inflammation. Rifaximin treatment slightly worsened insulin sensitivity and did not improve any measure of metabolic homeostasis.
3. Discussion
Many studies have drawn a link between the gut microbiome and metabolic homeostasis. One potential cause of chronic obesity-induced inflammation is the attachment of LPS to intestinally derived lipoproteins, which would then enter the circulation and activate TLR4 [22, 23]. We hypothesized that rifaximin would improve insulin sensitivity by lowering plasma LPS and adipose and systemic inflammation. Indeed, rifaximin has been demonstrated to lower plasma LPS in patients with hepatic encephalopathy, and the decrease in LPS correlates with the lowering of ammonia levels [43]. Furthermore, treatment of mice with antibiotics has clear effects on glucose tolerance [24]. After analyzing the effect of rifaximin SSD on plasma LPS in six subjects, it was apparent that plasma LPS was not being reduced by treatment; therefore, we would not be able to address our initial hypothesis. Furthermore, there was no evidence of improvement in insulin sensitivity, and there was therefore no justification for continuing the study. A limitation of the results presented, therefore, is the small sample size. Although not statistically significant, there were baseline differences between the groups in aspects of the OGTT such as HOMA IR. A larger study would be needed to normalize baseline differences and detect small changes in plasma or adipose inflammatory cytokines and relate these to the small reduction in insulin sensitivity. Finally, the unexpected lack of effect of rifaximin SSD on endotoxin levels in these obese, insulin resistant subjects raises the question of whether there is something different in patients with cirrhosis, who are responsive.
Consistent with our findings, a recent study of rifaximin treatment in subjects with nonalcoholic steatohepatitis found that rifaximin treatment worsened insulin resistance [44]. Our study did not identify any factors that would worsen insulin sensitivity such as an increase in adipose inflammation, an increase in any of the inflammatory cytokines that we measured in plasma, or plasma LPS (unmasked). Additional gut microbiota factors besides LPS that influence insulin sensitivity have been identified [45, 46]. It is thus possible that rifaximin treatment changed the gut microbiome in some manner to alter these or other factors to reduce insulin sensitivity. Together, these studies point to the complexity of the gut microbiota with regard to metabolic disease. This study attempts to modify the gut microbiota of obese humans with an antibiotic and then measure metabolic changes. Other studies have found that rifaximin reduces plasma LPS in patients with liver disease [28–30], but they used an assay that was different than the one we developed for this study (see the following section and “Materials and Methods”). It is also possible that rifaximin SSD, which was used in this study, has different effects on plasma LPS than rifaximin. Future studies may want to increase the dose of rifaximin SSD and consider effects on both the gut microbiome and plasma endotoxin.
Given the strong association between plasma LPS, adipose inflammation, and insulin sensitivity [14–21], strategies to reduce endotoxemia may be valuable therapies, but evaluation of such therapies will require a reliable LPS assay. We used the endotoxin sample preparation kit and high-sensitivity endotoxin detection assays to develop a method to detect unmasked LPS in the plasma. Importantly, we included a no-LAL enzyme control to demonstrate that the plasma did not interfere with the assay. Caution should be used when measuring plasma LPS; we recommend performing the no-LAL enzyme control because it is possible that plasma from subjects with hypertriglyceridemia may interfere with the assay. Using this method, we demonstrated that plasma LPS increased during an OLTT when we included a protease pretreatment step to unmask the LPS. However, we did not observe an increase in plasma LPS using the conventional method, where LPS is heavily masked.
Other studies have reported increased LPS in plasma following a lipid meal using a conventional assay [13]. A possible explanation is that we used a kit with higher sensitivity that allows for sample dilution to better reduce interfering substances, and we also diluted the samples prior to heat inactivation. The use of the two LAL assays presented here should be useful to other investigators who wish to quantify plasma LPS.
Acknowledgments
The authors thank Dorothy Ross, Brianna Harfmann, and Tamara Bennett for assistance with this study.
Financial Support: This study was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK R21 DK100258 (to P.A.K.) and CTSA Grant UL1TR001998. C.W.C. and J.M.E. are supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Clinical Trial Information: Clinicaltrials.gov no. NCT02124512 (registered 28 April 2014).
Author Contributions: B.S.F. and P.A.K. designed the study, analyzed data, and wrote the manuscript. B.Z. and T.B. performed laboratory studies. P.M.W. contributed to the statistical analysis of the data. C.W.C. and J.M.E. performed GLP-1 and GIP assays. All authors approved of the final version of the manuscript before submission. P.A.K. is the guarantor of this work, has access to all of the data, and takes responsibility for the data analysis.
Glossary
Abbreviations:
- GIR
glucose infusion rate
- HOMA IR
homeostatic model assessment of insulin resistance
- LAL
limulus amebocyte lysate
- LPS
lipopolysaccharide
- OGTT
oral glucose tolerance test
- OLTT
oral lipid tolerance test
- SC WAT
subcutaneous white adipose tissue
- SI
insulin sensitivity
- SSD
soluble solid dispersion
Additional Information
Disclosure Summary: The authors have nothing to declare.
Data Availability:
The data sets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.
References and Notes
- 1. Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22(10):1079–1089. [DOI] [PubMed] [Google Scholar]
- 2. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101(44):15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caesar R, Reigstad CS, Bäckhed HK, Reinhardt C, Ketonen M, Lundén GO, Cani PD, Bäckhed F. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 2012;61(12):1701–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104(3):979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. [DOI] [PubMed] [Google Scholar]
- 6. Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab. 2015;22(4):658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding S, Chi MM, Scull BP, Rigby R, Schwerbrock NM, Magness S, Jobin C, Lund PK. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS One. 2010;5(8):e12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fleissner CK, Huebel N, Abd El-Bary MM, Loh G, Klaus S, Blaut M. Absence of intestinal microbiota does not protect mice from diet-induced obesity. Br J Nutr. 2010;104(6):919–929. [DOI] [PubMed] [Google Scholar]
- 10. Vreugdenhil AC, Rousseau CH, Hartung T, Greve JW, van ’t Veer C, Buurman WA. Lipopolysaccharide (LPS)-binding protein mediates LPS detoxification by chylomicrons. J Immunol. 2003;170(3):1399–1405. [DOI] [PubMed] [Google Scholar]
- 11. Laugerette F, Vors C, Géloën A, Chauvin MA, Soulage C, Lambert-Porcheron S, Peretti N, Alligier M, Burcelin R, Laville M, Vidal H, Michalski MC. Emulsified lipids increase endotoxemia: possible role in early postprandial low-grade inflammation. J Nutr Biochem. 2011;22(1):53–59. [DOI] [PubMed] [Google Scholar]
- 12. Ghoshal S, Witta J, Zhong J, de Villiers W, Eckhardt E. Chylomicrons promote intestinal absorption of lipopolysaccharides. J Lipid Res. 2009;50(1):90–97. [DOI] [PubMed] [Google Scholar]
- 13. Erridge C, Attina T, Spickett CM, Webb DJ. A high-fat meal induces low-grade endotoxemia: evidence of a novel mechanism of postprandial inflammation. Am J Clin Nutr. 2007;86(5):1286–1292. [DOI] [PubMed] [Google Scholar]
- 14. Creely SJ, McTernan PG, Kusminski CM, Fisher M, Da Silva NF, Khanolkar M, Evans M, Harte AL, Kumar S. Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am J Physiol Endocrinol Metab. 2007;292(3):E740–E747. [DOI] [PubMed] [Google Scholar]
- 15. Ghanim H, Abuaysheh S, Sia CL, Korzeniewski K, Chaudhuri A, Fernandez-Real JM, Dandona P. Increase in plasma endotoxin concentrations and the expression of Toll-like receptors and suppressor of cytokine signaling-3 in mononuclear cells after a high-fat, high-carbohydrate meal: implications for insulin resistance. Diabetes Care. 2009;32(12):2281–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. [DOI] [PubMed] [Google Scholar]
- 17. Amar J, Burcelin R, Ruidavets JB, Cani PD, Fauvel J, Alessi MC, Chamontin B, Ferriéres J. Energy intake is associated with endotoxemia in apparently healthy men. Am J Clin Nutr. 2008;87(5):1219–1223. [DOI] [PubMed] [Google Scholar]
- 18. Harte AL, Varma MC, Tripathi G, McGee KC, Al-Daghri NM, Al-Attas OS, Sabico S, O’Hare JP, Ceriello A, Saravanan P, Kumar S, McTernan PG. High fat intake leads to acute postprandial exposure to circulating endotoxin in type 2 diabetic subjects. Diabetes Care. 2012;35(2):375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nowotny B, Zahiragic L, Krog D, Nowotny PJ, Herder C, Carstensen M, Yoshimura T, Szendroedi J, Phielix E, Schadewaldt P, Schloot NC, Shulman GI, Roden M. Mechanisms underlying the onset of oral lipid-induced skeletal muscle insulin resistance in humans. Diabetes. 2013;62(7):2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mehta NN, McGillicuddy FC, Anderson PD, Hinkle CC, Shah R, Pruscino L, Tabita-Martinez J, Sellers KF, Rickels MR, Reilly MP. Experimental endotoxemia induces adipose inflammation and insulin resistance in humans. Diabetes. 2010;59(1):172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shah R, Lu Y, Hinkle CC, McGillicuddy FC, Kim R, Hannenhalli S, Cappola TP, Heffron S, Wang X, Mehta NN, Putt M, Reilly MP. Gene profiling of human adipose tissue during evoked inflammation in vivo. Diabetes. 2009;58(10):2211–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saad MJ, Santos A, Prada PO. Linking gut microbiota and inflammation to obesity and insulin resistance. Physiology (Bethesda). 2016;31(4):283–293. [DOI] [PubMed] [Google Scholar]
- 23. Gomes JMG, Costa JA, Alfenas RCG. Metabolic endotoxemia and diabetes mellitus: A systematic review. Metabolism. 2017;68:133–144. [DOI] [PubMed] [Google Scholar]
- 24. Membrez M, Blancher F, Jaquet M, Bibiloni R, Cani PD, Burcelin RG, Corthesy I, Macé K, Chou CJ. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22(7):2416–2426. [DOI] [PubMed] [Google Scholar]
- 25. Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, Gibson GR, Delzenne NM. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50(11):2374–2383. [DOI] [PubMed] [Google Scholar]
- 26. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110(22):9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vlachogiannakos J, Saveriadis AS, Viazis N, Theodoropoulos I, Foudoulis K, Manolakopoulos S, Raptis S, Karamanolis DG. Intestinal decontamination improves liver haemodynamics in patients with alcohol-related decompensated cirrhosis. Aliment Pharmacol Ther. 2009;29(9):992–999. [DOI] [PubMed] [Google Scholar]
- 28. Kalambokis GN, Mouzaki A, Rodi M, Tsianos EV. Rifaximin improves thrombocytopenia in patients with alcoholic cirrhosis in association with reduction of endotoxaemia. Liver Int. 2012;32(3):467–475. [DOI] [PubMed] [Google Scholar]
- 29. Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, Fuchs M, Ridlon JM, Daita K, Monteith P, Noble NA, White MB, Fisher A, Sikaroodi M, Rangwala H, Gillevet PM. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8(4):e60042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gangarapu V, Ince AT, Baysal B, Kayar Y, Kılıç U, Gök Ö, Uysal Ö, Şenturk H. Efficacy of rifaximin on circulating endotoxins and cytokines in patients with nonalcoholic fatty liver disease. Eur J Gastroenterol Hepatol. 2015;27(7):840–845. [DOI] [PubMed] [Google Scholar]
- 31. Rood J, Smith SR. Triglyceride concentrations and endotoxemia. Am J Clin Nutr. 2008;88(1):248–249, author reply 249–250. [DOI] [PubMed] [Google Scholar]
- 32. Kern PA, Finlin BS, Ross D, Boyechko T, Zhu B, Grayson N, Sims R, Bland JS. Effects of KDT501 on metabolic parameters in insulin-resistant prediabetic humans. J Endocr Soc. 2017;1(6):650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finlin BS, Zhu B, Boyechko T, Westgate PM, Chia CW, Egan JM, Kern PA. Figure S1: Absorbance (A595) during the oral lipid tolerance test. Accessed 26 June 2019. https://figshare.com/articles/Figure_S1/8242694.
- 34. Finlin BS, Zhu B, Starnes CP, McGehee RE Jr, Peterson CA, Kern PA. Regulation of thrombospondin-1 expression in alternatively activated macrophages and adipocytes: role of cellular cross talk and omega-3 fatty acids. J Nutr Biochem. 2013;24(9):1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Finlin BS, Zhu B, Boyechko T, Westgate PM, Chia CW, Egan JM, Kern PA. Table S1: Primers. Accessed 26 June 2019. https://figshare.com/articles/Table_S1/8243102. [DOI] [PMC free article] [PubMed]
- 36. RRID:AB_2801398. http://antibodyregistry.org/search?q=AB_2801398.
- 37. RRID:AB_2801399. http://antibodyregistry.org/search?q=AB_2801399.
- 38. RRID:AB_2801400. http://antibodyregistry.org/search?q=AB_2801400.
- 39. RRID:AB_2801401. http://antibodyregistry.org/search?q=AB_2801401.
- 40. Hwang I, Park YJ, Kim YR, Kim YN, Ka S, Lee HY, Seong JK, Seok YJ, Kim JB. Alteration of gut microbiota by vancomycin and bacitracin improves insulin resistance via glucagon-like peptide 1 in diet-induced obesity. FASEB J. 2015;29(6):2397–2411. [DOI] [PubMed] [Google Scholar]
- 41. Wichmann A, Allahyar A, Greiner TU, Plovier H, Lundén GO, Larsson T, Drucker DJ, Delzenne NM, Cani PD, Bäckhed F. Microbial modulation of energy availability in the colon regulates intestinal transit. Cell Host Microbe. 2013;14(5):582–590. [DOI] [PubMed] [Google Scholar]
- 42. Finlin BS, Zhu B, Boyechko T, Westgate PM, Chia CW, Egan JM, Kern PA. Figure S2: Effect of rifaximin treatment on incretin secretion. Accessed 26 June 2019. https://figshare.com/articles/Figure_S2/8242811. [DOI] [PMC free article] [PubMed]
- 43. Kaji K, Takaya H, Saikawa S, Furukawa M, Sato S, Kawaratani H, Kitade M, Moriya K, Namisaki T, Akahane T, Mitoro A, Yoshiji H. Rifaximin ameliorates hepatic encephalopathy and endotoxemia without affecting the gut microbiome diversity. World J Gastroenterol. 2017;23(47):8355–8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cobbold JFL, Atkinson S, Marchesi JR, Smith A, Wai SN, Stove J, Shojaee-Moradie F, Jackson N, Umpleby AM, Fitzpatrick J, Thomas EL, Bell JD, Holmes E, Taylor-Robinson SD, Goldin RD, Yee MS, Anstee QM, Thursz MR. Rifaximin in non-alcoholic steatohepatitis: An open-label pilot study. Hepatol Res. 2018;48(1):69–77. [DOI] [PubMed] [Google Scholar]
- 45. Koh A, Molinaro A, Stahlman M, Khan MT, Schmidt C, Manneras-Holm L, Wu H, Carreras A, Jeong H, Olofsson LE, Bergh PO, Gerdes V, Hartstra A, de Brauw M, Perkins R, Nieuwdorp M, Bergstrom G, Backhed F. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell. 2018;175:947–961.e17. [DOI] [PubMed] [Google Scholar]
- 46. Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BA, Forslund K, Hildebrand F, Prifti E, Falony G, Le Chatelier E, Levenez F, Doré J, Mattila I, Plichta DR, Pöhö P, Hellgren LI, Arumugam M, Sunagawa S, Vieira-Silva S, Jørgensen T, Holm JB, Trošt K, Kristiansen K, Brix S, Raes J, Wang J, Hansen T, Bork P, Brunak S, Oresic M, Ehrlich SD, Pedersen O, Pedersen O; MetaHIT Consortium. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request.