Abstract
Background: Estrogen receptor-positive, progesterone receptor-positive, and HER2-positive breast cancers (triple-positive breast cancers, TPBCs) account for 5% to 10% of all breast cancers. The clinical and molecular features of TPBCs remain elusive. In this study, we aim to analyze the multiomics landscape and responsiveness of TPBCs to trastuzumab.
Methods: We employed five cohorts. The first cohort was from the Surveillance, Epidemiology, and End Results database (n=32,056) and was used to determine the clinical characteristics of TPBC. The second, third and fourth cohorts were from The Cancer Genome Atlas (n=162), GSE2603 (n=37) and GSE2109 (n=30) datasets, respectively, and were used to examine the genomic features and molecular classification of TPBC. The fifth cohort comprised TPBC patients treated at Fudan University Shanghai Cancer Center (FUSCC, n=171) and was used to investigate an immunohistochemistry-defined luminal A-like subgroup of TPBC.
Results: Patients with TPBC had a significantly better prognosis than those with ER-PR-HER2+ breast cancer. Genomic analysis revealed that TPBCs showed a lower TP53 mutation rate (30% vs. 69%, P < 0.001) and lower levels of HER2 mRNA and protein expression than ER-PR-HER2+ breast cancers. More than 40% of TPBCs were classified as the luminal A intrinsic subtype, with an even lower HER2 expression level. Based on the immunohistochemical detection of CDCA8, BCL2 and STC2, we identified a luminal A-like subgroup of TPBCs in the FUSCC cohort (CDCA8-negative, BCL2- and/or STC2-positive). Patients with luminal A-like TPBC had a better prognosis and benefited less from trastuzumab than those with TPBC of other subtypes.
Conclusions: TPBCs consist of clinically and genomically heterogeneous subgroups that may require different therapeutic strategies. The luminal A-like subgroup of TPBCs is associated with a better prognosis and reduced benefit from trastuzumab.
Keywords: triple-positive breast cancer, multiomics, molecular classification, luminal A, trastuzumab
Introduction
Breast cancer is increasingly recognized as a heterogeneous disease that exhibits substantial differences in terms of pathological features, biological behavior and gene expression profiles. It is widely accepted that breast cancers can be classified into four molecular subtypes (luminal A, luminal B, HER2-enriched and basal-like) according to the gene expression patterns 1, 2. In clinical practice, a simplified classification based on the detection of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2) and Ki-67 by immunohistochemistry (IHC) and/or fluorescence in situ hybridization has been adopted as a substitute 3. The classification results help guide treatment decisions.
Approximately 15-20% of breast cancers overexpress HER2, and nearly half of these HER2+ breast cancers also express hormone receptors (HRs) 4, 5. Previous retrospective studies reported a better prognosis for HR+HER2+ breast cancers than for HR-HER2+ breast cancers 6. However, few studies have focused specifically on the clinical features and prognosis of ER+PR+HER2+ breast cancers (triple-positive breast cancers, TPBCs). As for treatment, although HER2-targeted therapy (e.g., trastuzumab) can improve the prognosis of patients with HER2+ breast cancer regardless of HR status, the benefit may be lower in HR+ patients than in HR- patients 7.
Compared with the surrogate IHC-based classification, the intrinsic molecular classification based on gene expression profiling might better reveal the molecular essence and indicate treatment sensitivity 8-10. Although TPBC was all categorized as the luminal B subtype according to the surrogate IHC classification 3, it is heterogeneous according to the intrinsic molecular classification. Previous studies have demonstrated that HR+HER2+ breast cancers comprised at least luminal A, luminal B and HER2-enriched intrinsic subtypes, and the intrinsic molecular classification was associated with the tumor sensitivity to HER2-targeted therapy 11, 12. Certain subgroups of TPBCs, such as luminal A, may not be sensitive to HER2-targeted therapy.
In this study, we employed five well-characterized cohorts of breast cancer patients to comprehensively study the clinicopathologic and molecular features of TPBCs. In addition, we developed and validated a clinically practical method based on IHC to identify a luminal A-like subgroup of TPBCs, which may benefit less from trastuzumab.
Methods
Study Cohorts
Our study comprised five cohorts. The first cohort included 32,056 patients with HER2-positive breast cancer identified from the Surveillance, Epidemiology and End Results (SEER) database. We used this cohort to examine the clinicopathologic features and prognoses of HER2-positive breast cancers according to ER and PR status. The second cohort comprised 162 patients with HER2-positive breast cancer identified from The Cancer Genome Atlas (TCGA). Based on the somatic mutation, copy number, RNA-seq and reverse-phase protein array (rppa) protein expression data, we unraveled the genomic landscape and examined the HER2 expression level of HER2-positive breast cancers according to ER and PR status. The third and fourth cohorts were from two publicly available microarray datasets (GSE2603 and GSE2109), which included 37 and 30 patients with TPBC, respectively. We used the TCGA cohort and these two cohorts to study the intrinsic molecular classification of TPBCs and to select the genes that can be used to identify luminal A subtype TPBCs. The fifth cohort was a prospective observational cohort, which included 171 consecutive TPBC patients treated at Fudan University Shanghai Cancer Center (FUSCC) between 2007 and 2014.
Detailed information about our study cohort, bioinformatics and immunohistochemical analysis methods can be found in the Supplementary File.
Statistical analysis
Student's t test was used to compare differences in continuous variables, while Pearson's chi-square test and Fisher's exact test were used to compare differences in categorical variables. For multiple testing adjustment, a false discovery rate was calculated using the R function “p.adjust”. In the SEER and TCGA cohorts, breast cancer-specific survival (BCSS) was defined as the interval from diagnosis to death due to breast cancer. Overall survival (OS) was defined as the interval from diagnosis to death from any cause. Disease-free survival (DFS) was defined as the interval from diagnosis to the first recurrence or death. In the FUSCC cohort, relapse-free survival (RFS) was defined as the time from diagnosis to the first relapse, incidence of contralateral breast cancer, or death from any cause. The survival curves were plotted using the Kaplan-Meier method, and the survival differences between groups were compared using the log-rank test. Multivariate survival analyses were performed using the Cox proportional hazards model, and the results were reported as hazard ratios (HRs) with 95% confidence intervals (CIs).
Statistical analyses were performed using R version 3.5.3 (R Foundation for Statistical Computing, Vienna, Austria). All tests were two-sided, and a P value < 0.05 was considered statistically significant.
Results
Clinicopathologic features and prognosis of triple-positive breast cancers
We outlined the demographic, tumor, and treatment characteristics of patients with HER2-positive breast cancers in the SEER cohort according to ER and PR status (Table S1). TPBCs accounted for 50.8% of HER2-positive breast cancers, while ER+PR-HER2+, ER-PR+HER2+ and ER-PR-HER2+ breast cancers accounted for 18.1%, 1.9% and 29.2%, respectively. Compared with ER-PR-HER2+ patients (≥ 50 years, 69.1%), TPBC (≥ 50 years, 62.8%) or ER-PR+HER2+ patients (≥ 50 years, 64.2%) tended to be younger, while ER+PR-HER2+ patients (≥ 50 years, 74.7%) tended to be older. Tumors of TPBC and ER+PR-HER2+ breast cancer were less frequently of a higher grade (grade 3 or undifferentiated, 48.9% and 58.6%, respectively) than those of ER-PR-HER2+ breast cancer (grade 3 or undifferentiated, 75.5%). The tumor size of TPBC and ER+PR-HER2+ breast cancer was relatively smaller (T1, 52.8% and 51.4%, respectively) than that of ER-PR-HER2+ breast cancer (T1, 46.6%). Patients with TPBC or ER+PR-HER2+ breast cancer were less likely to be diagnosed with node-positive disease (38.4% and 38.1%, respectively) than those with ER-PR-HER2+ breast cancer (42.3%). In addition, the rate of lumpectomy in patients with TPBC or ER+PR-HER2+ breast cancer was higher (51.2% and 46.9%) than that in patients with ER-PR-HER2+ breast cancer (42.7%).
We next compared the survival difference among the four groups. Compared with patients with ER-PR-HER2+ breast cancer, those with TPBC or ER+PR-HER2+ breast cancer had significantly better BCSS and OS (all log-rank P < 0.001) (Figure 1). After adjusting for patient age, race, T category, N category, tumor grade and surgery type in multivariate analyses, TPBC (BCSS, HR = 0.49, 95% CI: 0.43-0.57, P < 0.001; OS, HR = 0.67, 95% CI: 0.60-0.76, P < 0.001) and ER+PR-HER2+ breast cancer (BCSS, HR = 0.79, 95% CI: 0.67-0.93, P = 0.004; OS, HR = 0.82, 95% CI: 0.72-0.94, P = 0.005) were still associated with better BCSS and OS than ER-PR-HER2 breast cancer (Table 1). The survival difference between ER-PR+HER2+ breast cancer and ER-PR-HER2+ breast cancer was not statistically significant in either univariate or multivariate analysis.
Figure 1.
(A) BCSS and (B) OS of HER2-positive breast cancers according to ER and PR status. P values are calculated using the log-rank test. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant. Abbreviations: BCSS: breast cancer-specific survival; OS: overall survival.
Table 1.
Multivariate analyses of breast cancer-specific survival (BCSS) and overall survival (OS) using Cox proportional hazards models in the SEER cohort.
| BCSS | OS | |||||
|---|---|---|---|---|---|---|
| Variables | HR (95% CI) | P | HR (95% CI) | P | ||
| Age at diagnosis | ||||||
| ≤ 50 years | Reference | - | Reference | - | ||
| > 50 years | 1.83 (1.59-2.10) | < 0.001 | 2.64 (2.32-3.01) | < 0.001 | ||
| Race | ||||||
| White | Reference | - | Reference | - | ||
| Black | 1.29 (1.10-1.52) | 0.002 | 1.15 (1.00-1.32) | 0.052 | ||
| Others | 0.56 (0.44-0.71) | < 0.001 | 0.58 (0.48-0.70) | < 0.001 | ||
| Grade | ||||||
| 1 or 2 | Reference | - | Reference | - | ||
| 3 or UD | 1.43 (1.24-1.65) | <0.001 | 1.23 (1.10-1.37) | < 0.001 | ||
| T category | ||||||
| T1 | Reference | - | Reference | - | ||
| T2-4 | 2.67 (2.27-3.13) | < 0.001 | 2.01 (1.79-2.26) | < 0.001 | ||
| N category | ||||||
| N0 | Reference | - | Reference | - | ||
| N1-3 | 2.63 (2.28-3.04) | < 0.001 | 1.75 (1.57-1.95) | < 0.001 | ||
| HER2+ subgroup | ||||||
| ER-PR-HER2+ | Reference | - | Reference | - | ||
| ER-PR+HER2+ | 0.77 (0.51-1.17) | 0.222 | 0.88 (0.63-1.24) | 0.477 | ||
| ER+PR-HER2+ | 0.79 (0.67-0.93) | 0.004 | 0.82 (0.72-0.94) | 0.005 | ||
| ER+PR+HER2+ | 0.49 (0.43-0.57) | < 0.001 | 0.67 (0.60-0.76) | < 0.001 | ||
| Surgery | ||||||
| Lumpectomy | Reference | - | Reference | - | ||
| Mastectomy | 1.54 (1.34-1.78) | < 0.001 | 1.42 (1.27-1.58) | < 0.001 | ||
Abbreviations: BCSS: breast cancer-specific survival; OS: overall survival; HR: hazard ratio; CI: confidence interval; UD: undifferentiated.
In summary, according to ER and PR status, HER2-positive breast cancers can be divided into four groups with different clinicopathologic features. TPBCs and ER+PR-HER2+ breast cancers were associated with a better prognosis than ER-PR-HER2+ breast cancers.
Genomic analyses revealed a lower HER2 expression level in TPBCs than in ER-PR-HER2+ breast cancers
To provide deeper insight into the biological nature of TPBCs, we investigated the genomic landscape of HER2-positive breast cancers according to ER and PR status using the TCGA data. Whole-exome sequencing data were available for 147 of the 162 HER2-positive breast cancers. Overall, 12,236 somatic mutations were identified, including 11,106 single-nucleotide variants (SNVs) and 1,130 insertions or deletions (indels) (Figure 2A). TPBCs harbored a median of 31.5 nonsynonymous somatic mutations per tumor, while ER+PR-HER2+, ER-PR+HER2+ and ER-PR-HER2+ breast cancers harbored 50.5, 40 and 40.5 nonsynonymous somatic mutations per tumor, respectively. TP53 (40%) was the most frequently mutated gene, followed by PIK3CA (29%), MUC4 (10%), MUC16 (7%) and CDH1 (7%). Of interest, TPBCs showed a significantly lower TP53 mutation rate than ER-PR-HER2+ breast cancers (30% vs. 69%, P < 0.001). In addition, MUC16, GATA3 and ERBB3 mutations were strongly associated with the ER+PR-HER2+ phenotype (Figure 2B).
Figure 2.
The genomic landscape of HER2-positive breast cancers from the TCGA dataset according to ER and PR status. (A) 162 HER2-positive samples are classified into four groups according to the IHC-based ER and PR status. Clinical and molecular features are annotated below. The mutation data are available for 147 of 162 HER2-positive cases. (B) Known cancer-related genes mutated in at least 4% of the patients withmutation data. The non-synonymous mutation rates of these genes in each group are listed on the right. (C) Known cancer-related genes located in significant GISTIC 2.0 peaks with a q value < 0.05 (The CNA events are defined according to the discrete copy number calls provided by GISTIC 2.0: -2 = homozygous deletion; -1 = hemizygous deletion; 0 = neutral; 1 = gain; 2 = amplification). Homozygous and hemizygous deletion are collectively called gene loss. The amplification or loss rates of these genes in each group are listed on the right. Differences in the rates of gene mutation, amplification and loss are compared between the ER-PR-HER+ group and each of the other three groups. P values are calculated using the chi-square test or Fisher's exact test. ***, P < 0.001; **, P < 0.01; *, P < 0.05. Abbreviations: I/E: Indeterminate or Equivocal; TPBC: triple-positive breast cancer; IHC: immunohistochemistry; FISH: fluorescence in situ hybridization.
We next examined the somatic copy number alterations (CNAs) of reported cancer-related genes (Figure 2C) 13, 14. ERBB2 was the most frequently affected gene by somatic CNAs (60%). Of note, TPBCs showed significantly lower rates of ERBB2 and MYC amplification than ER-PR-HER2+ breast cancers (ERBB2: 53% vs. 85%, P = 0.001; MYC: 24% vs. 47%, P = 0.013). The frequency of CCND1 amplification and FANCA loss was higher in TPBCs than in ER-PR-HER2+ breast cancers (CCND1: 26% vs. 6%, P = 0.013; FANCA, 72% vs. 50%, P = 0.023). In addition, ER+PR-HER2+ breast cancers also exhibited a lower ERBB2 amplification rate (59% vs. 85%, P = 0.017).
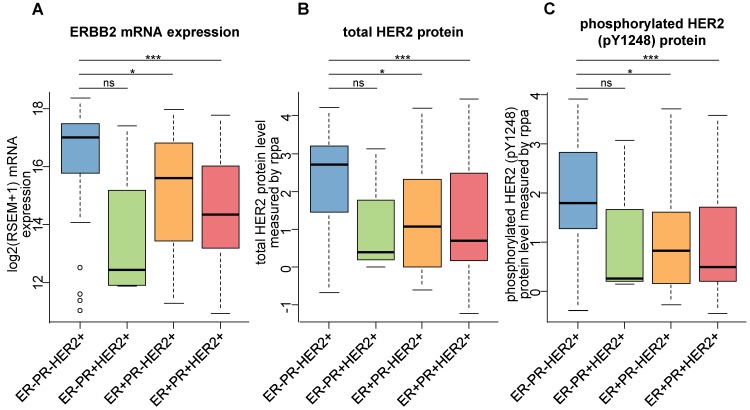
The different ERBB2 amplification rates among the four groups inspired us to concentrate on the HER2 expression levels of HER2-positive breast cancers according to ER and PR status. We found that the ERBB2 mRNA expression was significantly lower in TPBCs than in ER-PR-HER2+ breast cancers (Figure 3A). Besides, both total HER2 protein and phosphorylated HER2 protein levels were significantly lower in TPBCs than in ER-PR-HER2+ breast cancers (Figure 3B-C). All these data suggested a relatively low level of HER2 expression in TPBCs compared with ER-PR-HER2+ breast cancers.
Figure 3.
Low levels of HER2 expression and activation in TPBC compared with ER-PR-HER2+ breast cancers. (A) ERBB2 mRNA expression in HER2-positive breast cancers according to ER and PR status. (B) Total HER2 protein levels in HER2-positive breast cancers according to ER and PR status. (C) Phosphorylated HER2 (pY1248) protein levels in HER2-positive breast cancers according to ER and PR status. P values are calculated using Student's t test. ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant. Abbreviation: TPBC: triple-positive breast cancer; rppa: reverse-phase protein array.
In summary, genomic analyses revealed different molecular features of TPBCs from ER-PR-HER2+ breast cancers. In particular, TPBCs showed a lower HER2 expression level than ER-PR-HER2+ breast cancers.
Intrinsic molecular classification of TPBCs
Intrinsic molecular subtyping of breast cancer is essential for understanding the biological features of this disease and for making treatment choices. However, few studies have examined the intrinsic subtyping and the molecular essence of TPBCs. Thus, we explored the distribution of PAM50 intrinsic subtypes of TPBCs from the TCGA, GSE2603 and GSE2109 cohorts (Figure 4A). Surprisingly, the luminal A intrinsic subtype accounted for more than 40% of TPBCs in all three cohorts. The percentages of luminal A intrinsic subtype in TPBCs (TCGA, 50.6%; GSE2603, 40.5%; GSE2109, 43.3%) were much higher than those in ER+PR-HER2+ (TCGA, 16.0%; GSE2603, 15.4%; GSE2109, 0%) and ER-PR-HER2+ (TCGA, 0%; GSE2603, 0%; GSE2109, 9.1%) breast cancers. Based on the intrinsic molecular classification results, we further explored the HER2 expression levels in different intrinsic subtypes using the TCGA data. The ERBB2 mRNA expression, total HER2 protein level and phosphorylated HER2 protein level were all lower in the luminal A subtype than in the other subtypes (Figure 4B).
Figure 4.
Subtyping of TPBCs according to PAM50 intrinsic classification. (A) PAM50 intrinsic classification of TPBCs from the TCGA, GSE2603 and GSE2109 datasets. (B) Low HER2 expression level of the luminal A subtype among TPBCs. P values are calculated using Student's t test. ***, P < 0.001; **, P < 0.01; *, P < 0.05. Abbreviations: TPBC: triple-positive breast cancer; LA: luminal A; OS: other subtypes; rppa: reverse-phase protein array.
We also compared the prognosis of TPBCs of the luminal A subtype and the other subtypes using the TCGA cohort. We observed a tendency of better DFS and OS in TPBCs of the luminal A subtype than in those of the other subtypes, but the difference did not reach statistical significance due to the small sample size (DFS: log-rank P = 0.106, OS: log-rank P = 0.088; Figure S1). The differences in clinicopathologic characteristics between the luminal A subtype and the other subtypes were not significant (Table S2).
In summary, TPBCs comprised considerable luminal A intrinsic subtype breast cancers, which may be associated with a relatively good prognosis and a low HER2 expression level compared with the other intrinsic subtypes.
Identification of luminal A-like TPBCs and its clinical implications
The PAM50 intrinsic subtyping should be regarded as an important reference to the prognostic evaluation and therapeutic decision-making in patients with TPBC. We aimed to develop a clinically feasible method to identify luminal A subtype TPBCs using immunohistochemical markers (Figure S2). We first identified the differentially expressed genes between the luminal A subtype and the other subtypes using the TCGA dataset. These genes were further filtered and validated in the GSE2603 and GSE2109 datasets (Tables S3-S4).
We next tested the relationship between the mRNA expression and protein expression of the remaining genes and retained those with a correlation coefficient of > 0.5. Finally, we conducted receiver operating characteristic (ROC) analysis to assess the accuracy of using the mRNA expression of retained genes to identify the luminal A subtype. According to the area under the curve (AUC) in the TCGA dataset, we selected STC2, BCL2 and CDCA8 (Tables S5-S6) as the immunohistochemical markers. STC2 and BCL2 were highly expressed in the luminal A subtype compared with the other subtypes, while CDCA8 was expressed at a relatively low level in the luminal A subtype (Figure 5A). A significant positive correlation was observed between each gene's protein expression and mRNA expression (Figure S3). The AUC of using BCL2 expression to identify the luminal A subtype was 0.761 in TCGA, 0.758 in GSE2603 and 0.810 in GSE2109. The AUC of using STC2 expression to identify the luminal A subtype was 0.751 in TCGA, 0.739 in GSE2603 and 0.882 in GSE2109. The AUC of using CDCA8 expression to identify the luminal A subtype was 0.918 in TCGA, 0.820 in GSE2603 and 0.955 in GSE2109 (Figure 5B). We also investigated whether the expression of ESR1, PGR and ERBB2 could be used to identify luminal A subtype TPBCs and found that the accuracy was far inferior to that of our selected genes (Figures S4-S5).
Figure 5.
Identification of luminal A subtype TPBCs by the mRNA expression of BCL2, STC2 and CDCA8. (A) Expression of BCL2, STC2 and CDCA8 between TPBCs of the luminal A subtype and those of the other subtypes in the TCGA, GSE2603 and GSE2109 cohorts. P values are calculated using Student's t test. ***, P < 0.001; **, P < 0.01; *, P < 0.05. (B) ROC curves of using the mRNA expression of BCL2, STC2 and CDCA8 to identify luminal A subtype TPBCs. Abbreviations: TPBC: triple-positive breast cancer; LA: luminal A; OS: other subtypes; ROC: receiver operating characteristic; AUC: area under the curve; CI: confidence interval.
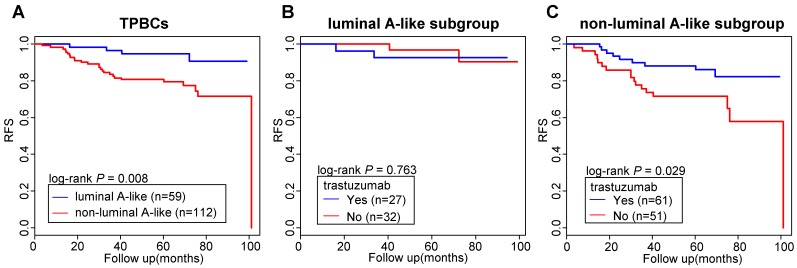
Next, we validated the clinical implications of these three genes in the FUSCC cohort by performing immunohistochemical detection to identify a luminal A-like subgroup of TPBCs. According to the immunohistochemical staining results (Figure S6), we classified 171 TPBC tumors into a luminal A-like subgroup (n=59) and a non-luminal A-like subgroup (n=112) (Table S7). Tumors were classified into the luminal A-like subgroup if they were CDCA8-negative and positive for at least one of BCL2 and STC2, while those showing other immunohistochemical staining results were placed into the non-luminal A-like subgroup. Patients with luminal A-like TPBC showed significantly better RFS than those with non-luminal A-like TPBC (log-rank P = 0.008) (Figure 6A). In the multivariate survival analysis, the luminal A-like subgroup was still associated with better relapse-free survival (HR = 0.33, 95% CI: 0.11-0.97, P = 0.045) (Table S8).
Figure 6.
Clinical implications of luminal A-like TPBCs. (A) Difference in relapse-free survival between luminal A-like and non-luminal A-like TPBCs. (B) Comparison of relapse-free survival between patients treated with trastuzumab and those not treated with trastuzumab in the luminal A-like subgroup. (C) Comparison of relapse-free survival between patients treated with trastuzumab and those not treated with trastuzumab in the non-luminal A-like subgroup. P values are calculated using the log-rank test. Abbreviations: TPBC: triple-positive breast cancer; RFS: relapse-free survival.
Of note, in the non-luminal A-like subgroup, patients treated with trastuzumab had significantly better RFS than those not treated with trastuzumab (log-rank P = 0.029), while in the luminal A-like subgroup, there was no difference in RFS between patients treated with trastuzumab and those not treated with trastuzumab (log-rank P = 0.763) (Figure 6B-C, Figure S7).
In summary, we found that the mRNA expression of STC2, BCL2 and CDCA8 can be used to identify luminal A subtype TPBCs. Based on this result, we developed an IHC-based method incorporating these three genes to identify a luminal A-like subgroup of TPBCs and demonstrated that patients with luminal A-like TPBC had a better prognosis and benefited less from trastuzumab therapy.
Discussion
In this study, we systemically studied the clinical and molecular features of TPBCs. We demonstrated that patients with TPBC had a significantly better prognosis than those with ER-PR-HER2+ breast cancer. We also found that TPBCs exhibited a relatively low HER2 expression level compared with ER-PR-HER2+ breast cancers. We further unraveled the intrinsic molecular classification of TPBCs and found that a considerable percentage of TPBCs were classified as the luminal A subtype that showed an even lower HER2 expression level. Finally, we developed a practical method based on the immunohistochemical detection of STC2, BCL2 and CDCA8 to identify a luminal A-like subgroup of TPBCs that was associated with a relatively good prognosis and reduced benefit from trastuzumab.
Compared with HER2-negative breast cancers, HER2-positive breast cancers are more aggressive and have been associated with a poorer prognosis before the introduction of HER2-targeted therapy. HER2-positive breast cancers can be further classified into ER+PR+HER2+, ER+PR-HER2+, ER-PR+HER2+ and ER-PR-HER2+ breast cancers according to ER and PR status. Taking advantage of the SEER database, we investigated the clinicopathologic characteristics and prognoses of these four groups. We found that compared with ER-PR-HER2+ breast cancers, TPBCs showed a lower tumor stage and grade and were independently associated with longer BCSS and OS. These results indicated that TPBCs were less aggressive than ER-PR-HER2+ breast cancers.
We next examined the molecular features of TPBCs and compared them with those of other HER2-positive subgroups. We revealed that TPBCs may have different driver events from ER-PR-HER2+ breast cancers, including a lower TP53 mutation rate, a lower ERBB2 amplification rate and a higher CCND1 amplification rate. Among these events, we concentrated on ERBB2 amplification, which has been associated with tumor sensitivity to HER2-targeted therapy 15. We further compared the differences in HER2 mRNA and protein expression between TPBCs and ER-PR-HER2+ breast cancers. All these analyses suggested a lower HER2 expression level in TPBCs.
As a clinically defined entity, TPBC is heterogeneous in terms of its intrinsic molecular subtypes. Previous studies have shown that nearly 30% of HR+/HER2+ breast cancers were classified as the luminal A subtype according to intrinsic molecular classification 12, 16. By contrast, the luminal A intrinsic subgroup accounted for more than 40% of the TPBCs in all three cohorts with intrinsic molecular classification results in our study. These luminal A subtype TPBCs showed even lower levels of HER2 mRNA and protein expression than TPBCs of the other intrinsic subtypes. This result suggested that luminal A subtype TPBCs might be driven primarily by HR signaling pathways rather than the HER2 signaling pathway 5, 17. In addition, patients with luminal A subtype TPBC tended to have a better prognosis. Thus, we inferred that luminal A subtype TPBCs represent a special subgroup of HER2-positive breast cancers with a particularly low level of HER2 expression and a favorable prognosis. This subgroup of TPBCs might benefit less from HER2-targeted therapy.
The identification of luminal A subtype TPBCs may be essential to guide individualized treatment of TPBC patients. By analyzing the gene expression profiling data, we demonstrated that the mRNA expression of STC2, BCL2 and CDCA8 can be used to identify luminal A subtype TPBCs. According to previous studies, both STC2 and BCL2 are estrogen-responsive genes that are upregulated in luminal breast cancers and correlate with a better prognosis 18-22. CDCA8 is a critical regulator of mitosis and cell division and is associated with cancer growth and progression 23. All these three markers have been detected by IHC in the previous studies 20, 24, 25. We detected these three markers by IHC in 171 TPBCs from the FUSCC cohort and identified a luminal A-like subgroup of TPBCs. Patients with luminal A-like TPBC had a better prognosis and benefited less from adjuvant trastuzumab. These results suggested that it might be possible for some patients with luminal A-like TPBC to be treated with de-escalated trastuzumab therapy. Vici et al also explored the efficacy of trastuzumab in TPBCs 26. They found that increased expression of ER was associated with reduced trastuzumab benefit and that this benefit tended to disappear in patients whose tumors expressed ER in > 50% of cells. However, our data suggested that the accuracy of using ER expression to identify the luminal A intrinsic subtype was far inferior to that of our selected markers.
Our study has some limitations. First, we did not perform intrinsic molecular subtyping of the TPBCs in the FUSCC cohort; therefore, we were unable to examine the accuracy of our IHC-based method for identifying the luminal A intrinsic subtype. Nevertheless, based on the treatment and follow-up data, we observed that patients with luminal A-like TPBC identified by our IHC-based method had a relatively better prognosis and benefited less from trastuzumab therapy. These results demonstrated the clinical implications of our IHC-based method. Second, we were unable to obtain data on HER2-targeted therapy from the SEER and TCGA datasets; therefore, we could not directly compare the efficacy of HER2-targeted therapy between TPBCs and other HER2-positive subgroups or between TPBCs of the luminal A intrinsic subtype and TPBCs of the other subtypes. Third, since there are only a very small number of patients in the FUSCC cohort who did not receive chemotherapy due to old age, we are unable to analyze whether it is possible for patients with luminal A-like TPBC to be treated with de-escalated chemotherapy.
In conclusion, compared with ER-PR-HER2+ breast cancers, TPBCs are less aggressive and show a lower HER2 expression level. A considerable proportion of TPBCs are luminal A subtype breast cancers according to the intrinsic molecular classification. Evaluating the expression of STC2, BCL2 and CDCA8 via IHC can help identify a luminal A-like subgroup of TPBCs, which has a relatively better prognosis and benefits less from trastuzumab therapy.
Supplementary Material
Supplementary materials and methods, figures and tables.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81874112, 81874113, 81572583 and 81502278), the Training Plan of Excellent Talents in Shanghai Municipality Health System (2017YQ038), the “Chen Guang” project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (17CG01), Shanghai Pujiang Program (18PJD007), the Training Plan of Excellent Talents of Fudan University Shanghai Cancer Center (YJYQ201602), the Municipal Project for Developing Emerging and Frontier Technology in Shanghai Hospitals (SHDC12010116), the Cooperation Project of Conquering Major Diseases in Shanghai Municipality Health System (2013ZYJB0302), the Innovation Team of Ministry of Education (IRT1223), and the Shanghai Key Laboratory of Breast Cancer (12DZ2260100). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Ethics approval and consent to participate
Our study was approved by the independent ethics committee/institutional review board at FUSCC Ethical Committee. The data from the SEER, TCGA, GSE2603, GSE2109 datasets are publicly available and therefore do not require informed patient consent.
Authors' contributions
YZJ and ZMS outlined the manuscript; all authors contributed to the literature search, data collection, data analysis, and data interpretation. SZ, XYL and XJ provided the figures and drafted the manuscript, with additional input from all authors. All authors approved the final manuscript. SZ, XYL and XJ contributed equally to this work.
Abbreviations
- AUC
area under the curve
- BCSS
breast-cancer-specific survival
- CC
correlation coefficient
- CNA
copy number alteration
- ER
estrogen receptor
- FUSCC
Fudan University Shanghai Cancer Center
- HER2
human epidermal growth factor receptor 2
- IHC
immunohistochemistry
- OS
overall survival
- PR
progesterone receptor
- RFS
relapse-free survival
- ROC
receiver operating characteristic
- SEER
Surveillance, Epidemiology, and End Results
- TCGA
The Cancer Genome Atlas
- TPBC
triple-positive breast cancer
References
- 1.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA. et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 2.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10869–74. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ. et al. Strategies for subtypes-dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Annals of oncology: official journal of the European Society for Medical Oncology. 2011;22:1736–47. doi: 10.1093/annonc/mdr304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 5.Vici P, Pizzuti L, Natoli C, Gamucci T, Di Lauro L, Barba M. et al. Triple positive breast cancer: a distinct subtype? Cancer Treat Rev. 2015;41:69–76. doi: 10.1016/j.ctrv.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Untch M, Gelber RD, Jackisch C, Procter M, Baselga J, Bell R. et al. Estimating the magnitude of trastuzumab effects within patient subgroups in the HERA trial. Annals of oncology: official journal of the European Society for Medical Oncology. 2008;19:1090–6. doi: 10.1093/annonc/mdn005. [DOI] [PubMed] [Google Scholar]
- 7.Pogue-Geile KL, Kim C, Jeong JH, Tanaka N, Bandos H, Gavin PG. et al. Predicting degree of benefit from adjuvant trastuzumab in NSABP trial B-31. J Natl Cancer Inst. 2013;105:1782–8. doi: 10.1093/jnci/djt321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guarneri V, Dieci MV, Bisagni G, Frassoldati A, Bianchi GV, De Salvo GL, De-escalated therapy for HR+/HER2+ breast cancer patients with Ki67 response after 2 weeks letrozole: results of the PerELISA neoadjuvant study. Annals of oncology: official journal of the European Society for Medical Oncology; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu XY, Ma D, Xu XE, Jin X, Yu KD, Jiang YZ. et al. Genomic Landscape and Endocrine-Resistant Subgroup in Estrogen Receptor-Positive, Progesterone Receptor-Negative, and HER2-Negative Breast Cancer. Theranostics. 2018;8:6386–99. doi: 10.7150/thno.29164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X, Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer cell; 2019. [DOI] [PubMed] [Google Scholar]
- 11.Llombart-Cussac A, Cortes J, Pare L, Galvan P, Bermejo B, Martinez N. et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): an open-label, single-group, multicentre, phase 2 trial. The Lancet Oncology. 2017;18:545–54. doi: 10.1016/S1470-2045(17)30021-9. [DOI] [PubMed] [Google Scholar]
- 12.Carey LA, Berry DA, Cirrincione CT, Barry WT, Pitcher BN, Harris LN. et al. Molecular Heterogeneity and Response to Neoadjuvant Human Epidermal Growth Factor Receptor 2 Targeting in CALGB 40601, a Randomized Phase III Trial of Paclitaxel Plus Trastuzumab With or Without Lapatinib. J Clin Oncol. 2016;34:542–9. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.An O, Dall'Olio GM, Mourikis TP, Ciccarelli FD. NCG 5.0: updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings. Nucleic acids research. 2016;44:D992–9. doi: 10.1093/nar/gkv1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R. et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–83. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lesurf R, Griffith OL, Griffith M, Hundal J, Trani L, Watson MA. et al. Genomic characterization of HER2-positive breast cancer and response to neoadjuvant trastuzumab and chemotherapy-results from the ACOSOG Z1041 (Alliance) trial. Annals of oncology: official journal of the European Society for Medical Oncology. 2017;28:1070–7. doi: 10.1093/annonc/mdx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim HK, Park KH, Kim Y, Park SE, Lee HS, Lim SW, Discordance of the PAM50 Intrinsic Subtypes Compared with IHC-Based Surrogate in Breast Cancer Patients: Potential Implication of Genomic Alterations of Discordance. Cancer Res Treat; 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Viale G, de Snoo FA, Slaets L, Bogaerts J, van 't Veer L, Rutgers EJ. et al. Immunohistochemical versus molecular (BluePrint and MammaPrint) subtyping of breast carcinoma. Outcome results from the EORTC 10041/BIG 3-04 MINDACT trial. Breast Cancer Res Treat. 2018;167:123–31. doi: 10.1007/s10549-017-4509-9. [DOI] [PubMed] [Google Scholar]
- 18.Esseghir S, Kennedy A, Seedhar P, Nerurkar A, Poulsom R, Reis-Filho JS. et al. Identification of NTN4, TRA1, and STC2 as prognostic markers in breast cancer in a screen for signal sequence encoding proteins. Clinical cancer research: an official journal of the American Association for Cancer Research. 2007;13:3164–73. doi: 10.1158/1078-0432.CCR-07-0224. [DOI] [PubMed] [Google Scholar]
- 19.Bouras T, Southey MC, Chang AC, Reddel RR, Willhite D, Glynne R. et al. Stanniocalcin 2 is an estrogen-responsive gene coexpressed with the estrogen receptor in human breast cancer. Cancer research. 2002;62:1289–95. [PubMed] [Google Scholar]
- 20.Dawson SJ, Makretsov N, Blows FM, Driver KE, Provenzano E, Le Quesne J. et al. BCL2 in breast cancer: a favourable prognostic marker across molecular subtypes and independent of adjuvant therapy received. Br J Cancer. 2010;103:668–75. doi: 10.1038/sj.bjc.6605736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perillo B, Sasso A, Abbondanza C, Palumbo G. 17beta-estradiol inhibits apoptosis in MCF-7 cells, inducing bcl-2 expression via two estrogen-responsive elements present in the coding sequence. Mol Cell Biol. 2000;20:2890–901. doi: 10.1128/mcb.20.8.2890-2901.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang KT, Kim K, Chang JH, Oh S, Kim YA, Lee JY. et al. BCL2 Regulation according to Molecular Subtype of Breast Cancer by Analysis of The Cancer Genome Atlas Database. Cancer Res Treat. 2018;50:658–69. doi: 10.4143/crt.2017.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai C, Miao CX, Xu XM, Liu LJ, Gu YF, Zhou D. et al. Transcriptional activation of human CDCA8 gene regulated by transcription factor NF-Y in embryonic stem cells and cancer cells. J Biol Chem. 2015;290:22423–34. doi: 10.1074/jbc.M115.642710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer HA, Tolle A, Jung M, Fritzsche FR, Haendler B, Kristiansen I. et al. Identification of stanniocalcin 2 as prognostic marker in renal cell carcinoma. Eur Urol. 2009;55:669–78. doi: 10.1016/j.eururo.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Chang JL, Chen TH, Wang CF, Chiang YH, Huang YL, Wong FH. et al. Borealin/Dasra B is a cell cycle-regulated chromosomal passenger protein and its nuclear accumulation is linked to poor prognosis for human gastric cancer. Exp Cell Res. 2006;312:962–73. doi: 10.1016/j.yexcr.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Vici P, Pizzuti L, Sperduti I, Frassoldati A, Natoli C, Gamucci T. et al. "Triple positive" early breast cancer: an observational multicenter retrospective analysis of outcome. Oncotarget. 2016;7:17932–44. doi: 10.18632/oncotarget.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods, figures and tables.