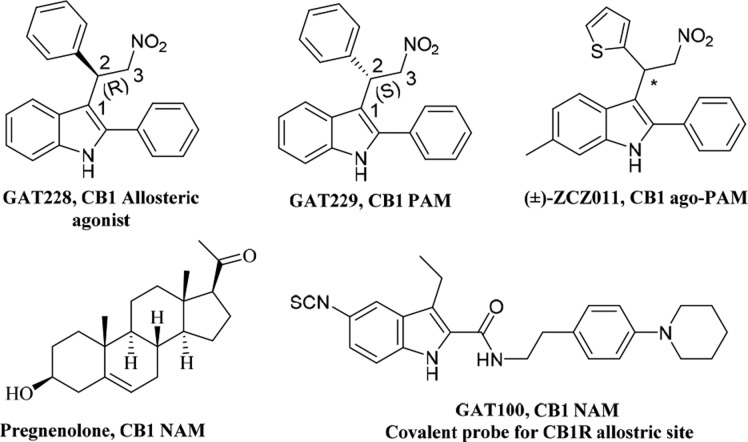
Abstract
Positive allosteric modulation of the cannabinoid 1 receptor (CB1R) has demonstrated distinct therapeutic advantages that address several limitations associated with orthosteric agonism and has opened a promising therapeutic avenue for further drug development. To advance the development of CB1R positive allosteric modulators, it is important to understand the molecular architecture of CB1R allosteric site(s). The goal of this work was to use Force-Biased MMC Simulated Annealing to identify binding sites for GAT228 (R), a partial allosteric agonist, and GAT229 (S), a positive allosteric modulator (PAM) at the CB1R. Our studies suggest that GAT228 binds in an intracellular (IC) TMH1–2–4 exosite that would allow this compound to act as a CB1 allosteric agonist as well as a CB1 PAM. In contrast, GAT229 binds at the extracellular (EC) ends of TMH2/3, just beneath the EC1 loop. At this site, this compound can act as CB1 PAM only. Finally, these results were successfully validated through the synthesis and biochemical evaluation of a focused library of compounds.
Keywords: CB1 cannabinoid receptor, PAM binding site, MMC simulated annealing, structure−activity relationship
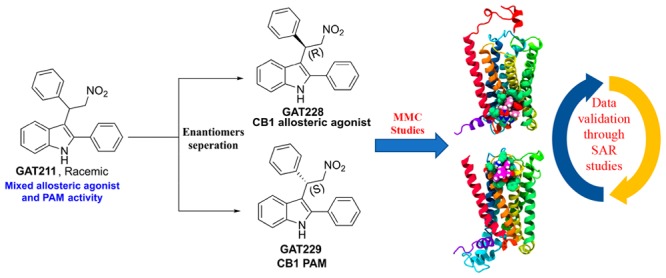
The cannabinoid 1 receptor (CB1R) is one of the most widely expressed metabotropic G protein-coupled receptors (GPCRs) in brain. Its participation in various (patho)physiological processes has made CB1 (in)activation a viable therapeutic modality for treating prevalent unsolved medical problems including neurological/neurodegenerative diseases, chronic and neuropathic pain, substance-use disorders, obesity, and diabetes.1 Naturally occurring CB1R agonists, including the endocannabinoids anandamide (AEA) and 2-arachidonoylglycerol (2-AG) and the principal psychoactive cannabis constituent, Δ9-tetrahydrocannabinol (Δ9-THC), activate the receptor by engaging its orthosteric site.2 Orthosterically acting agonists or antagonists have been met with significant challenges in clinical advancement due to their association with undesirable central nervous system (CNS) side effects. From the alternative approaches pursued to modulate the CB1R activity, allosteric modulation that targets topographically distinct site(s) has met with significant success so far in preclinical settings.3,4,26 Positive allosteric modulators (PAMs) enhance orthosteric ligand binding or receptor activity, whereas negative allosteric modulators (NAMs) reduce orthosteric ligand binding or receptor activity. Endogenous ligands such as LipoxinA4 has been shown to act as a CB1R PAM,5 whereas pregnenolone has been demonstrated to have CB1R NAM activity.6 A small molecule GAT211 (Figure 1) was the first well-characterized CB1R ago-PAM.7 It exhibited agonism through allosteric site to varying degrees in the absence of orthosteric ligand and also potentiated CB1R orthosteric activation.7 It displayed both PAM and agonist activity in multiple cell lines expressing human recombinant CB1R and in mouse-brain membranes rich in native CB1R.14 GAT211 was instrumental in establishing the therapeutic role of CB1R PAMs in treating several diseases in preclinical studies.8−11 Enantiomeric resolution of GAT211 led to identification of R-(+)-enantiomer GAT228 as a partial allosteric agonist with a weak PAM activity. The S-(−)-enantiomer GAT229 was a pure CB1 PAM and lacked intrinsic activity in the biological systems examined. Our current preclinical studies suggest that CB1R allosteric agonists, pure PAMs, and ago-PAMs can provide distinct therapeutic advantages depending on the disease to be treated. To facilitate drug discovery and development of CB1R allosteric modulators of this structural class, it is essential to understand the molecular architecture of CB1R allosteric site(s). Although CB1R has been crystallized recently in the active and inactive form, it does not provide understanding of the therapeutically relevant allosteric site(s).12−15 The goal of the present work was to use Force-Biased MMC Simulated Annealing to identify binding sites for the allosteric agonist GAT228 and pure PAM GAT229.
Figure 1.
Representative CB1R allosteric modulators
Transmembrane helices (TMHs) in all Class A GPCRs have varying lipid face sizes depending on their tilt and also their relative position in the TMH bundle. Due to this architecture, distinctive crevices exist on the lipid face of all Class A GPCRs. Some of these crevices are large enough to form binding exosites for lipid components like cholesterol or for allosteric modulators. Hanson and co-workers described a consensus IC binding site for cholesterol in a TMH1–2–4 exosite at the β2-AR (PDB code 2RH1) that is present in 26% of human Class A GPCRs.16 Another cholesterol IC exosite has been identified at TMH1–7-Hx8 in the β2-AR (PDB code 2RH1) and the 5HT-2B (PDB code 4IB4) receptors.17 On the extracellular (EC) side, cholesterol has been found to bind at TMH2–3 and the EC1 loop in the α2A-AR (PDB code 4EIY).17
We have shown that the CB1 biased NAM, pregnenolone, binds in the IC TMH1–7-Hx8 exosite that allows the modulator to limit the movement of TMH7, a movement important for β-arrestin biased signaling.6 Recent CB1R crystal structures reveal that at the CB1R IC TMH1–2–4 lipid facing crevice, there are several aromatic residues (H2.41,F2.42 and W4.50) in the TMH1–2–4 region 1 (PDB code 5UO9; see Figure 2).18 In the CB1R* state complexed with Gi protein (PDB code 6N4B; see Figure 2),18 a fourth aromatic residue moves into the TMH1–2–4 region, F4.46. This residue changes its position to occupy the TMH1–2–4 exosite by undergoing a rotameric change (γ1g+ → trans). In addition, TMH2 comes closer to TMH4 on the IC side of the bundle. This conformation of F4.46 allows it to form an aromatic stack with W4.50.18
Figure 2.
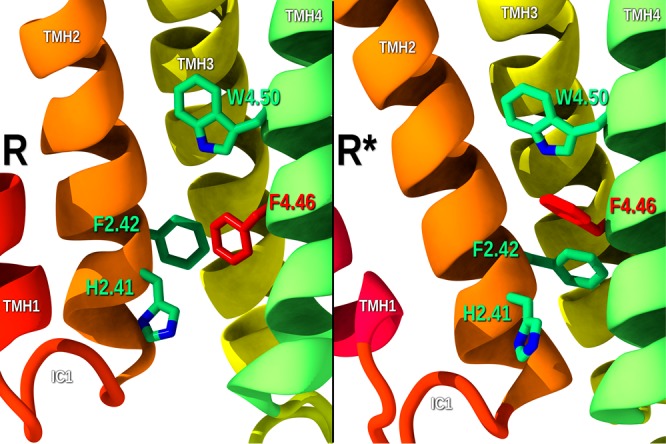
Location of four key aromatic residues in the IC TMH1–2–4 exosite region is shown. While H2.41, F2.42, and W4.50 have similar orientations in R and R*, F4.46 points into the orthosteric site in the inactive state (R), but points into the TMH1–2–4 exosite in the CB1 activated state in R*.
Results and Discussion
The GAT228 and GAT229 structures were broken into fragments as shown in Figure 3. All fragments were prepared with partial charges using Amber 2002, a point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations.19 Four MMC runs were performed in which our CB1R* model was immersed in a box filled with copies of one of these fragments (for details, see Figure S1). Analysis of the MMC runs for GAT229 revealed that, while each fragment bound to multiple positions on the CB1R*, there was only one region in which all fragments clustered in the correct spatial proximity for the S stereoisomer. This was at the extracellular end of TMH2/3, just beneath the EC1 loop. Y2.59, a polar residue that faces lipid, was found to attract the nitro group of the AS stereoisomer fragment, while D2.63 consistently was found interacting with the N–H of the indole fragment (Figure S1).
Figure 3.
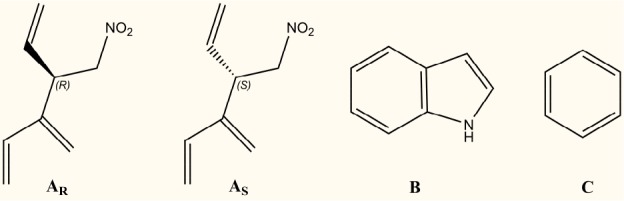
Fragments of GAT228 and GAT229 created for MMC studies.
GAT228 AGO-PAM Binding at IC TMH1–2–4 Exosite
Figure 4 illustrates the binding site identified for GAT228 by MMC. This site is in the IC TMH1–2–4 exosite, which accommodates cholesterol in many Class A GPCRs. It is important to note that CB1 does not have the cholesterol consensus sequence motif identified for 26% of human Class A GPCRs16 at the TMH1–2–4 exosite. So, it is less likely that the CB1 TMH1–2–4 exosite binds cholesterol, leaving it available for other ligands. The principal GAT228 interactions at this site includes a strong dual hydrogen bond between the GAT228 NO2 group and R(148) (R(148) NH1 to nitro O1, N–O distance, 2.8 Å; N–H–O, 169° and R(148) NH2 to nitro O2, N–O distance, 3.1 Å; N–H–O, 173°), while R(148) also is involved in a cation−π interaction with the indole ring of GAT228 (distance from R(148) Cz to indole five-membered ring centroid, 3.8 Å; distance from R(182) Cz to indole six-membered ring centroid, 4.6 Å).20 GAT228 also has a hydrogen bond between the indole N–H and H2.41 (H2.41 ND to nitro O2, N–O distance, 2.9 Å; N–H–O, 175°). There are also several aromatic–aromatic interactions: an aromatic tilted T-stack between the phenyl ring attached to stereocenter and F4.46 (tilted T, centroid–centroid distance 5.0 Å, angle 83°); an offset parallel aromatic interaction between F4.46 and the indole ring phenyl substituent (centroid–centroid distance 4.2 Å, angle, 15°); and an aromatic tilted T-stack between W4.50 and the indole ring phenyl substituent (centroid–centroid distance 5.0 Å, angle 79°). The total interaction energy for GAT228 at this site is −37.18 kcal/mol, and Glide score was −6.5 kcal/mol (see interaction energy table, Table S1).
Figure 4.
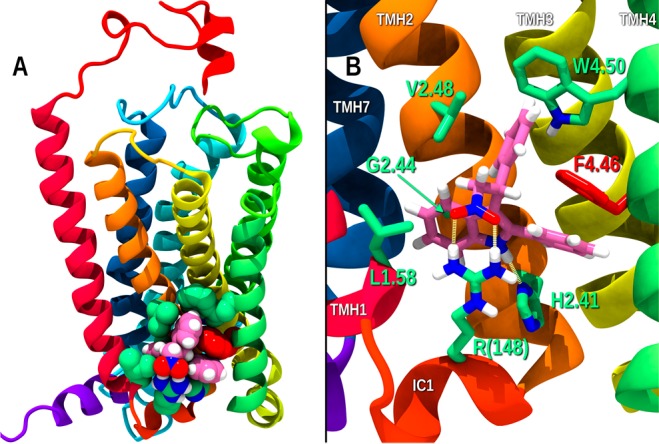
(A) Binding site identified for GAT228 by MMC. This site is in the IC TMH1–2–4 exosite. Interacting receptor residues are shown contoured at their VDW radii and colored green. Atoms in GAT228 are also contoured at their VDW radii and colored magenta. (B) GAT228 docked in the TMH1–2–4 exosite is shown.
How Could a Ligand Bound in an Exosite Be an Agonist?
Like all Class A GPCRs, CB1 has charged residues at the IC side of TMH3 (R3.50) and TMH6 (D6.30) that form a salt bridge (called the “ionic lock”) that keeps CB1 in its inactive state, unable to couple to G protein. When the ionic lock is broken, either by ligand binding or as consequence of a mutation, CB1 assumes its activated state conformation (R*). We reported several years ago that a missense mutation (F4.46(238)L) in the rat Cnr1 gene that encodes for CB1 represents a gain of function mutation that results in an adolescent-like phenotype.21 Cell based assays found that the F4.46L mutant was highly constitutively active and consistent with these results, molecular dynamics calculations revealed the toggle switch for CB1 activation, and the ionic lock was more frequently broken in the F4.46L mutant compared to WT. When GAT228 binds in the IC TMH1–2–4 exosite (see Figure 4), it forms an extended aromatic cluster, interacting directly with F4.46 (holding the F4.46 in its activated state γ1 = trans conformation). At the same time, GAT228 can interact with H2.41 and W4.50.
Figure 5 provides a view of the inactive (R; Figure 5A) and activated state (R*; Figure 5B) CB1R models. The view is from the IC side of the TMH bundle looking toward EC. In the inactive state, F4.46 is in a γ1 = g+ and points into the orthosteric TMH2–3–4 pocket (Figure 5A). When CB1R activates, F4.46 undergoes a γ1 g+ → trans change and F4.46 leaves the orthosteric pocket and faces into the TMH1–2–4 exosite (Figure 5B). In this latter case, TMH2 and TMH3 (on IC end) can move toward TMH4 and away from TMH6, stretching the R3.50/D6.30 ionic lock until it breaks and activates CB1 (see Figure 5B). Thus, by its binding at the TMH1–2–4 exosite, GAT228 binding can promote the F4.46 γ1 g+ → trans rotameric change that may cause activation of CB1, making this ligand a nonorthosteric agonist. Because GAT228 promotes a receptor active state conformation, it can also act as a PAM. In this case, F4.46 will be in its R* conformation (γ1 = trans) because an orthosteric agonist like CP55940 has activated CB1. GAT228 binding to the exosite in this situation will stabilize the activated state of CB1, thus acting as a PAM.
Figure 5.
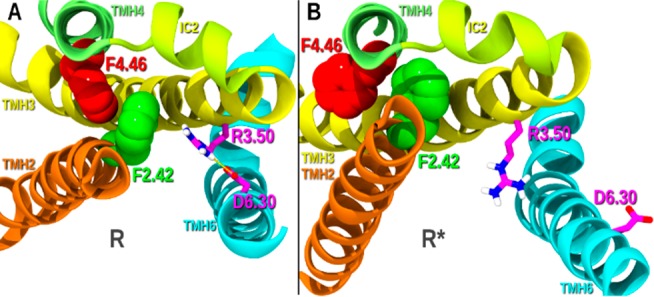
View of the inactive (R; Figure 6A) and activated states (R*; Figure 6B) CB1 models. The view is from the IC side of the TMH bundle looking toward EC. (A) In the inactive state, F4.46 is in a γ1 = g+, and it points into the orthosteric TMH2–3–4 pocket. (B) When CB1 activates, F4.46 undergoes a γ1 g+ → trans change, causing F4.46 to leave the orthosteric pocket and face into the TMH1–2–4 exosite.
To study an allosteric ligand that affects the binding and activity of an agonist, it is important to study its effect on the agonist bound CB1 activated (R*) state. Recently, Saleh and colleagues reported multiple adjunct binding sites in the Ago-PAM activity of 2-phenylindole modulators such as GAT228 using metadynamics studies in the ligand binding pocket of the CB1 inactive (R) state crystal structure (PDB code 5U09)22 in the absence or presence of the CB1 agonist, CP55940. Since the authors did not see the receptor convert to R* during their simulations, the GAT228 site they identified is a site that may influence the low-affinity state for CP55940 that does not signal because it remains the R state. Therefore, the GAT228 binding site identified may be less relevant to the pharmacology of GAT228.
GAT229 PAM Binding at TMH2–3-EC1 Site
Figure 6 illustrates the EC TMH2–3-EC1 binding site identified for GAT229 by MMC. GAT229 (magenta) can bind in this site in the presence of CP55940 (lavender). Figure 6 shows that when GAT229 interacts at its EC TMH2–3-EC1 site, its indole ring N–H hydrogen bonds with D2.63 (N–O distance, 3.0 Å; N–H–O, 141°), while D2.63 maintains its interaction with K(373). In addition, the GAT229 NO2 group hydrogen bonds with Y2.59 (O–O distance, 2.9 Å; O–H–O, 137°); the EC1 loop residue R(182) has a cation−π interaction with the indole ring (distance from R(182) Cz to indole five-membered ring centroid, 4.1 Å; distance from R(182) Cz to indole six-membered ring centroid, 3.8 Å) and Y2.59 has an offset parallel aromatic stack with the GAT229 indole ring phenyl substituent (centroid–centroid distance 4.1 Å, angle 11°). This last aromatic interaction is made stronger by the creation of a triple stack as F3.27 has a tilted-T aromatic stacking interaction with Y2.59 (centroid–centroid distance 5.3 Å, angle 80°). The net interaction energy of GAT229 in this complex is −32.73 kcal/mol, and its Glide score was −4.5 kcal/mol (see Table S2 for interaction energy table).
Figure 6.
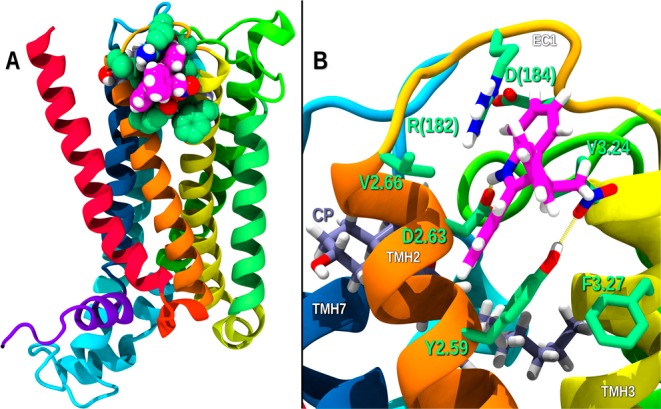
(A) EC TMH2–3-EC1 binding site identified for GAT229 by MMC. Receptor residues that interact with GAT229 are shown contoured at their VDW radii and colored green. Atoms in GAT229 are also contoured at their VDW radii and colored magenta. (B) Key interactions of GAT229 at its EC site.
As discussed in the Introduction, the α2A-AR (PDB code 4EIY) structure illustrates another cholesterol binding site in Class A GPCRs that is on the extracellular (EC) side, where cholesterol binds at the EC ends of TMH2–3 and the EC1 loop. As shown in Figure 6, our MMC results place GAT229 in this same site. Positive allosteric interactions typically stabilize the receptor/orthosteric ligand complex in its activated state conformation. Although there are clear changes on the IC side of GPCRs when they are activated (ionic lock broken and TMH6 straightening), there are also changes in EC loop conformations that take place during activation. CB1 EC2 loop mutation studies performed in the Kendall lab indicate that the EC2 loop moves toward the TMH bundle when CB1 activates.23 CB1 EC3 loop mutations in our lab have indicated that the EC3 loop residue K(373) interacts directly with D2.63 when CB1 activates.24 In its docking position, GAT229 stabilizes the EC3 loop conformation in the activated state and therefore should act as a CB1 PAM.
Chemistry and Structure–Activity Relationship Studies
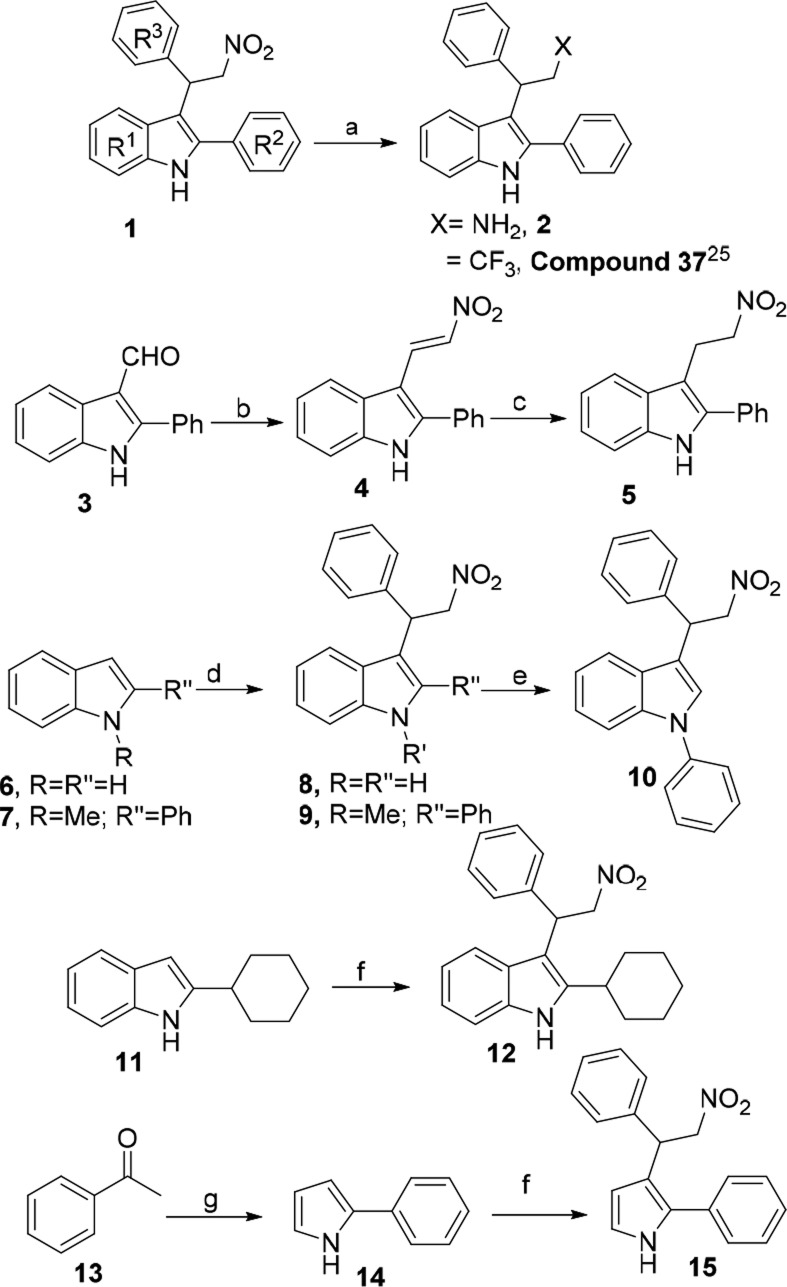
To validate MMC results, we designed and synthesized (Scheme 1Scheme 1) a focused library of GAT211 analogs (Table 1) in which the key pharmacophoric groups were removed and evaluated for their CB1R PAM activity using our previous published protocols.7
Scheme 1. Synthesis of Focused Library of GAT211 Analogs.
Reagents and conditions (a) NaBH4, NiCl2·6H2O, THF, MeOH, −5 0C-0 0C, 81%; (b) CH3NO2, NH4OAc, 115 0C, 4 h, 82%; (c) NaBH4, CH3OH, H2O, rt, 2 h, 72%; (d) β-nitrostyrene, Et4NBr, dioxane:water (2:1), 105 0C, 68%; (e) iodobenzene, K3PO4, DMSO, 150 0C, MW, 90 min, 25%; (f) β-nitrostyrene, CF3CO2NH4, 10% eq. EtOH, reflux, 115 °C, 9−12 h, 61−91%; (g) (i) NH2OH·HCl, NaOAc, 80% aq. EtOH, reflux, 2 h, 90%; (ii) acetylene, KOH, DMSO, 150 °C, 4 h, 44%.
Table 1. CB1 PAM Activity of All GAT211 Analogsa.
| cAMP |
β-arrestin2 |
|||
|---|---|---|---|---|
| compound | EC50 (nM) | Emax (%) | EC50 (nM) | Emax (%) |
| 1, GAT211 | 230(140–370) | 110 ± 6.8 | 940 (540–1,800) | 46 ± 9.5 |
| 2 | >10,000 | 12 | >10,000 | 12 |
| compound 37(25) | NA | NA | moderate PAM activity @ 1μM | NA |
| 5 | 5,528 | 85 | >10,000 | 0 |
| 8 | >10,000 | 0 | >10,000 | 0 |
| 9 | >10,000 | 0 | >10,000 | 0 |
| 10 | >10,000 | 0 | >10,000 | 0 |
| 12 | 992 | 121 | >10,000 | 0 |
| 15 | >10,000 | 0 | >10,000 | 0 |
Compounds were evaluated by Eurofins DiscoverX in independent experiments using the HitHunter cAMP assay and PathHunter β-arrestin assay. PAM activity was assessed using EC20 CP55,940.7
The amine analog of GAT211 2 was synthesized by the nitro-reduction of 1 (GAT211). Accordingly, nitro compound 1 was treated with NiCl2·6H2O and NaBH4 in methanol to yield amine 2 in 81% yield. α,β-Unsaturated nitro compound, 4 was synthesized from 2-phenyl-1H-indole-3-carbaldehyde, 3 by treating with nitromethane in the presence of ammonium acetate. The double bond of 4 was selectively reduced with NaBH4 to get compound 5 in 72% yield. However, 3-(2-nitro-1-phenylethyl)-1H-indole 8 and 1-methyl-3-(2-nitro-1-phenylethyl)-2-phenyl-1H-indole 9 were synthesized using Et4NBr and β-nitro styrene under reflux condition from 6 and 7, respectively, in good yields. N-Arylation of 8 was carried out using iodobenzene under microwave irradiation in DMSO to yield N-arylated analog 10. The C2-cyclohexyl analog 12 was synthesized from 2-cyclohexyl-1H-indole 11 using literature known protocol in 63% yield. 2-Phenyl-1H-pyrrole 14 was synthesized from acetophenone 13 using Trofimov reaction. Finally, 14 was treated with β-nitro styrene using CF3CO2NH4 to yield Michael adduct 15 in 91% yield27 (for details see Supporting Information).
Significance of the NO2 Group
Replacement of nitro group of GAT211 (cAMP: EC50 = 230 nM and Emax = 110%; β-arrestin2: EC50 = 940 nM and Emax = 46%) with amine (−NH2, 2) and −CF3 (compound 37)25 leads to dramatic loss in potency and efficacy in both cAMP and β-arrestin2 assays. These data strongly support the MMC studies prediction that a strong hydrogen bond interaction exist between the −NO2 group and R(148) for GAT228 and Y2.59 amino acid for GAT229, respectively (Figures 4B and 6B), and it is important for PAM activity. The CF3 group does not serve as efficient bioisosteric replacement to the nitro group.
Significance of C2 Phenyl Ring
MMC results suggested the C2 aromatic ring is important for allosteric activity due to an aromatic tilted T-stack between C-2 phenyl ring of GAT228 and F4.46, and triple stack as F3.27 has a tilted-T aromatic stacking interaction with Y2.59 for GAT229, respectively (Figures 4B and 6B). Both analogs 8 and 10 lacking the C2 phenyl ring (R2 = H) were found to be inactive thus signifying the importance of C2 aromatic ring. Interestingly, when C2-phenyl group of GAT211 was replaced by cyclohexyl ring, the resulting compound 12 showed slight improvement in cAMP potency as well as in efficacy (cAMP: EC50 = 992 nM and Emax = 121%) when compared to 8 in which the C2 phenyl ring was absent. This may be due to the van der Waals interaction of cyclohexyl group with key amino acid, F4.46.
We also observed considerable loss in efficacy and potency when the phenyl ring (R3) of 1 was replaced with hydrogen (R3 = H, 5), which leads to the interruption of the predicted π–π interaction.
Significance of Indole Ring
The pyrrole analog of GAT211 (R1 = H, 15), in which the phenyl ring of indole moiety is absent, showed dramatic drop in CB1R PAM activity in both assays, suggesting that cation−π interaction is important between indole ring of GAT228 and R(148) and indole ring of GAT229 and R(182) (Figures 4B and 6B). Finally, to check the importance of the N–H group of the indole ring, we synthesized N-methyl analog 9 and N-phenyl analog 10. As expected these compounds were found to be inactive/less active in both assays through repeated testing. Either the disruption of key hydrogen bonding between indole N–H and H2.41 (GAT228) and D2.63 (GAT229) (Figures 4B and 6B) or the introduction of a sterically hindered N-aryl group might account for the loss of the activity.
Conclusion
The MMC studies performed here deciphered the molecular basis for the observed distinct pharmacological effects exhibited by these enantiomers of GAT211. This work identified two distinct CB1R allosteric sites that are novel and had not been identified before. These results were further validated successfully through SAR studies. These findings hold the potential for successfully navigating future optimizations of a 2-phenylindole class of CB1R PAM for improved potency and efficacy.
Glossary
ABBREVIATIONS
- CB1
cannabinoid receptor type 1
- PAM
positive allosteric modulator
- ago-PAM
allosteric agonist and positive allosteric modulator
- EC
extracellular
- IC
intracellular
- TMHs
transmembrane helices
- MMC
Metropolis Monte Carlo
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.9b00256.
Synthetic procedures, analytical data, and MMC method (PDF)
Author Contributions
G.A.T., P.H.R., D.P.H., and S.G. designed and conducted experiments, analyzed data, and contributed to the writing of the manuscript. S.G., P.M.K., and P.C.S. synthesized the analogs.
This work was supported by NIH/National Eye Institute, RO1 EY024717 (to G.A.T.), RO1 DA003934 (to P.H.R.) and KO5 DA021358 (to P.H.R.).
The authors declare no competing financial interest.
Supplementary Material
References
- Di Marzo V. Targeting the endocannabinoid system: to enhance or reduce? Nature reviews. Nat. Rev. Drug Discovery 2008, 7, 438–55. 10.1038/nrd2553. [DOI] [PubMed] [Google Scholar]
- Lu H. C.; Mackie K. An Introduction to the Endogenous Cannabinoid System. Biol. Psychiatry 2016, 79, 516–25. 10.1016/j.biopsych.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janero D. R.; Thakur G. A. Leveraging allostery to improve G protein-coupled receptor (GPCR)-directed therapeutics: cannabinoid receptor 1 as discovery target. Expert Opin. Drug Discovery 2016, 11, 1223–1237. 10.1080/17460441.2016.1245289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley C. W.; Emmitte K. A.; Hopkins C. R.; Bridges T. M.; Gregory K. J.; Niswender C. M.; Conn P. J. Practical Strategies and Concepts in GPCR Allosteric Modulator Discovery: Recent Advances with Metabotropic Glutamate Receptors. Chem. Rev. 2016, 116, 6707–41. 10.1021/acs.chemrev.5b00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni A. R.; Garai S.; Janero D. R.; Thakur G. A.. Design and Synthesis of Cannabinoid 1 Receptor (CB1R) Allosteric Modulators: Drug Discovery Applications. In Methods in Enzymology; Reggio P. H., Ed. Academic Press: New York, 2017; Vol. 593, Chapter 13, pp 281−315. [DOI] [PubMed] [Google Scholar]
- Pamplona F. A.; Ferreira J.; Menezes de Lima O. Jr; Duarte F. S.; Bento A. F.; Forner S.; Villarinho J. G.; Bellocchio L.; Wotjak C. T.; Lerner R.; Monory K.; Lutz B.; Canetti C.; Matias I.; Calixto J. B.; Marsicano G.; Guimaraes M. Z.; Takahashi R. N. Anti-inflammatory lipoxin A4 is an endogenous allosteric enhancer of CB1 cannabinoid receptor. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 21134–9. 10.1073/pnas.1202906109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee M.; Vitiello S.; Bellocchio L.; Hebert-Chatelain E.; Monlezun S.; Martin-Garcia E.; Kasanetz F.; Baillie G. L.; Panin F.; Cathala A.; Roullot-Lacarriere V.; Fabre S.; Hurst D. P.; Lynch D. L.; Shore D. M.; Deroche-Gamonet V.; Spampinato U.; Revest J. M.; Maldonado R.; Reggio P. H.; Ross R. A.; Marsicano G.; Piazza P. V. Pregnenolone can protect the brain from cannabis intoxication. Science 2014, 343, 94–8. 10.1126/science.1243985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprairie R. B.; Kulkarni P. M.; Deschamps J. R.; Kelly M. E. M.; Janero D. R.; Cascio M. G.; Stevenson L. A.; Pertwee R. G.; Kenakin T. P.; Denovan-Wright E. M.; Thakur G. A. Enantiospecific Allosteric Modulation of Cannabinoid 1 Receptor. ACS Chem. Neurosci. 2017, 8, 1188–1203. 10.1021/acschemneuro.6b00310. [DOI] [PubMed] [Google Scholar]
- Laprairie R. B.; Bagher A. M.; Rourke J. L.; Zrein A.; Cairns E. A.; Kelly M. E. M.; Sinal C. J.; Kulkarni P. M.; Thakur G. A.; Denovan-Wright E. M. Positive allosteric modulation of the type 1 cannabinoid receptor reduces the signs and symptoms of Huntington’s disease in the R6/2 mouse model. Neuropharmacology 2019, 151, 1–12. 10.1016/j.neuropharm.2019.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitjavila J.; Yin D.; Kulkarni P. M.; Zanato C.; Thakur G. A.; Ross R.; Greig I.; Mackie K.; Straiker A. Enantiomer-specific positive allosteric modulation of CB1 signaling in autaptic hippocampal neurons. Pharmacol. Res. 2018, 129, 475–481. 10.1016/j.phrs.2017.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slivicki R. A.; Xu Z.; Kulkarni P. M.; Pertwee R. G.; Mackie K.; Thakur G. A.; Hohmann A. G. Positive Allosteric Modulation of Cannabinoid Receptor Type 1 Suppresses Pathological Pain Without Producing Tolerance or Dependence. Biol. Psychiatry 2018, 84, 722–733. 10.1016/j.biopsych.2017.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur G. A.; Kulkarni P. M.. Allosteric Modulators of CBl Cannabinoid Receptors. US 10246414 B2, 2013.
- Hua T.; Vemuri K.; Pu M.; Qu L.; Han G. W.; Wu Y.; Zhao S.; Shui W.; Li S.; Korde A.; Laprairie R. B.; Stahl E. L.; Ho J.-H.; Zvonok N.; Zhou H.; Kufareva I.; Wu B.; Zhao Q.; Hanson M. A.; Bohn L. M.; Makriyannis A.; Stevens R. C.; Liu Z.-J. Crystal Structure of the Human Cannabinoid Receptor CB1. Cell 2016, 167, 750–762. 10.1016/j.cell.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua T.; Vemuri K.; Nikas S. P.; Laprairie R. B.; Wu Y.; Qu L.; Pu M.; Korde A.; Jiang S.; Ho J.-H.; Han G. W.; Ding K.; Li X.; Liu H.; Hanson M. A.; Zhao S.; Bohn L. M.; Makriyannis A.; Stevens R. C.; Liu Z.-J. Crystal structures of agonist-bound human cannabinoid receptor CB1. Nature 2017, 547, 468. 10.1038/nature23272. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shao Z.; Yin J.; Chapman K.; Grzemska M.; Clark L.; Wang J.; Rosenbaum D. M. High-resolution crystal structure of the human CB1 cannabinoid receptor. Nature 2016, 540, 602–606. 10.1038/nature20613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni P. M.; Kulkarni A. R.; Korde A.; Tichkule R. B.; Laprairie R. B.; Denovan-Wright E. M.; Zhou H.; Janero D. R.; Zvonok N.; Makriyannis A.; Cascio M. G.; Pertwee R. G.; Thakur G. A. Novel Electrophilic and Photoaffinity Covalent Probes for Mapping the Cannabinoid 1 Receptor Allosteric Site(s). J. Med. Chem. 2016, 59, 44–60. 10.1021/acs.jmedchem.5b01303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M. A.; Cherezov V.; Griffith M. T.; Roth C. B.; Jaakola V.-P.; Chien E. Y. T.; Velasquez J.; Kuhn P.; Stevens R. C. A Specific Cholesterol Binding Site Is Established by the 2.8 Å Structure of the Human β2-Adrenergic Receptor. Structure 2008, 16, 897–905. 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gater D. L.; Saurel O.; Iordanov I.; Liu W.; Cherezov V.; Milon A. Two classes of cholesterol binding sites for the beta2AR revealed by thermostability and NMR. Biophys. J. 2014, 107, 2305–12. 10.1016/j.bpj.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna Kumar K.; Shalev-Benami M.; Robertson M. J.; Hu H.; Banister S. D.; Hollingsworth S. A.; Latorraca N. R.; Kato H. E.; Hilger D.; Maeda S.; Weis W. I.; Farrens D. L.; Dror R. O.; Malhotra S. V.; Kobilka B. K.; Skiniotis G. Structure of a Signaling Cannabinoid Receptor 1-G Protein Complex. Cell 2019, 176, 448–458. 10.1016/j.cell.2018.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y.; Wu C.; Chowdhury S.; Lee M. C.; Xiong G.; Zhang W.; Yang R.; Cieplak P.; Luo R.; Lee T.; Caldwell J.; Wang J.; Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- Trujillo C.; Rodriguez-Sanz A. A.; Rozas I. Aromatic Amino Acids-Guanidinium Complexes through Cation-pi Interactions. Molecules 2015, 20, 9214–28. 10.3390/molecules20059214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M.; Kasanetz F.; Lynch D. L.; Friemel C. M.; Lassalle O.; Hurst D. P.; Steindel F.; Monory K.; Schafer C.; Miederer I.; Leweke F. M.; Schreckenberger M.; Lutz B.; Reggio P. H.; Manzoni O. J.; Spanagel R. Enhanced Functional Activity of the Cannabinoid Type-1 Receptor Mediates Adolescent Behavior. J. Neurosci. 2015, 35, 13975–88. 10.1523/JNEUROSCI.1937-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh N.; Hucke O.; Kramer G.; Schmidt E.; Montel F.; Lipinski R.; Ferger B.; Clark T.; Hildebrand P. W.; Tautermann C. S. Multiple Binding Sites Contribute to the Mechanism of Mixed Agonistic and Positive Allosteric Modulators of the Cannabinoid CB1 Receptor. Angew. Chem., Int. Ed. 2018, 57, 2580–2585. 10.1002/anie.201708764. [DOI] [PubMed] [Google Scholar]
- Ahn K. H.; Bertalovitz A. C.; Mierke D. F.; Kendall D. A. Dual role of the second extracellular loop of the cannabinoid receptor 1: ligand binding and receptor localization. Mol. Pharmacol. 2009, 76, 833–42. 10.1124/mol.109.057356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu J.; Shore D. M.; Kapur A.; Trznadel M.; Makriyannis A.; Reggio P. H.; Abood M. E. Novel insights into CB1 cannabinoid receptor signaling: a key interaction identified between the extracellular-3 loop and transmembrane helix 2. J. Pharmacol. Exp. Ther. 2013, 345, 189–97. 10.1124/jpet.112.201046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni P. M.; Ranade A.; Garai S.; Thakur G. A. Micro-wave-accelerated Conjugate Addition of 2-Arylindoles to Substi-tuted β-Nitrostyrenes in the Presence of Ammonium Trifluoroace-tate: An Efficient Approach for the Synthesis of a Novel Class of CB1 Cannabinoid Receptor Allosteric Modulators. J. Heterocycl. Chem. 2017, 54, 2079–2084. 10.1002/jhet.2861. [DOI] [Google Scholar]
- Tseng C.-C.; Baillie G.; Donvito G.; Mustafa M. A.; Juola S. E.; Zanato C.; Massarenti C.; Dall’Angelo S.; Harrison W. T. A.; Lichtman A. H.; Ross R. A.; Zanda M.; Greig I. R. The Trifluoromethyl Group as a Bioisosteric Replacement of the Aliphatic Nitro Group in CB1 Receptor Positive Allosteric Modulators. J. Med. Chem. 2019, 62, 5049–5062. 10.1021/acs.jmedchem.9b00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.