Abstract
Background and Aim: We have previously shown that high-mobility group box 1 (HMGB1) is an independent biomarker for shortened survival of prostate cancer (PCa) patients. However, the specific role of HMGB1 in tumor development and progression remains largely unknown. In this study, we investigated the molecular mechanisms of HMGB1 in PCa tumorigenesis.
Methods: Gain-of-function and loss-of-function experiments were used to determine the biological functions of HMGB1 both in vitro and in vivo. Bioinformatic analysis, immunoprecipitation, and immunofluorescence assays were applied to discern and examine the relationship between HMGB1 and its potential targets. Specimens from 64 patients with PCa were analyzed for the expression of HMGB1 and its relationship with Brahma-related gene 1 (BRG1) was examined by immunohistochemistry.
Results: The results demonstrated that ectopic expression of HMGB1 facilitated growth and metastasis of PCa by enhancing Akt signaling pathway and promoting epithelial-mesenchymal transition (EMT), while silencing of HMGB1 showed the opposite effects. Mechanistically, HMGB1 exerted these functions through its interaction with BRG1 which may augment BRG1 function and activate the Akt signaling pathway thereby promoting EMT. Importantly, both HMGB1 and BRG1 expression was markedly increased in human PCa tissues.
Conclusions: Taken together, these findings indicate that upregulation of HMGB1 promotes PCa development via activation of Akt and accelerates metastasis through regulating BRG1-mediated EMT. HMGB1 could be used as a novel potential target for the treatment of PCa.
Keywords: HMGB1, epithelial-mesenchymal transition, Brahma-related gene 1, Akt pathway, prostate cancer
Introduction
Prostate cancer (PCa) is the second most common human malignancy in men worldwide 1. Although the recent advances in diagnosis and surgical resection can cure a majority of patients with localized disease, nearly 20-30% of treated men inevitably progress to castration-resistant prostate cancer (CRPC) followed by metastasis 2, the leading cause of poor prognosis and mortality 3. However, the underlying molecular mechanisms in PCa metastasis remain unclear. Therefore, there is an urgent need to detect innovative diagnostic and therapeutic methods based on the biological and molecular mechanisms of metastatic spread of PCa.
High-mobility group box 1 (HMGB1), a member of HMG1-type polypeptides, was initially identified as a DNA chaperone that is involved not only in inflammation but also in cancer 4, 5. Aberrant expression and release of HMGB1 are associated with various human carcinomas including colon, breast, lung, cervical, and liver cancers 6-10. During tumor progression and treatment, HMGB1 has been shown to play different roles in facilitating both cell survival and death by modulating various signaling pathways, including inflammation, gene transcription, autophagy, metastasis, metabolism, and apoptosis 11. Several signaling pathways mediated by HMGB1 have been implicated in carcinogenesis including PI3K/Akt signaling and epithelial-mesenchymal transition (EMT) process 12, 13. Our previous studies have revealed that HMGB1 is up-regulated in human PCa 14. However, the precise role of intracellular HMGB1 in metastasis during PCa tumorigenesis remains largely unknown.
In this study we explored the role of HMGB1 in growth, invasion, and metastasis of PCa in vitro as well as tumorigenesis in vivo. Our results illustrated that HMGB1 exerts these functions through constitutively activating the PI3K/Akt signaling pathway, modulating the expression of Brahma-related gene 1 (BRG1, also known as SMARCA4) in PCa cells, and regulating EMT via BRG1. Thus, our study has revealed a potential mechanism by which HMGB1 mediates the development and progression of PCa.
Materials & Methods
Patients and clinical samples
Paraffin specimens were collected from radical prostatectomy (RP) performed at the Nanfang Hospital between 2012 and 2016; 14 benign prostatic hyperplasia (BPH) tissues were used as controls. Characteristics of patients were retrospectively obtained from the review of medical records. Among the selected patients, the median age was 69 years (range: 30-89 years). The clinical stages and Gleason Scores (GS) were reassessed based on the American Joint Committee on Cancer (AJCC) 2002 and the World Health Organization (WHO) classification system. The detailed clinicopathological data are shown in Table S2. All patients signed the informed consent to participate in the study according to the ethical protocols of the Ethics Committee of Nanfang Hospital, Southern Medical University.
Cell lines and culture conditions
Human immortalized prostate epithelial cell line RWPE-1 and four PCa cell lines PC-3, DU145, 22Rv1, and LNCaP were purchased from Stem Cell Bank, Chinese Academy of Sciences. Cells were grown in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS, Hyclone). RWPE-1 cells were cultured in Keratinocyte Serum Free Medium (KSFM) (Gibco, No. 10744-019) supplemented with 5 ng/mL epidermal growth factor (EGF) (Gibco, No. 10450-013), and 1% antibiotic-antimycotic solution (Gibco, No. 15140-122). All cell lines were maintained at 37 °C in 5% CO2.
RNAi and gene transfection
Three small interfering RNAs (siRNAs) targeting HMGB1 (si-HMGB1-1, 2, 3) and negative control (NC) siRNA with no specific target were synthesized by RiboBio Co. (Guangzhou, China). The siRNA sequences that specifically target HMGB1 are shown below: si-h-HMGB1_001: GTTGGTTCTAGCGCAGTTT; si-h-HMGB1_002:GGACAAGGCCCGTTATGAA; si-h-HMGB1_003: GAGGCCTCCTTCGGCCTTC.
Stable cell lines expressing HMGB1 or sh-HMGB1 were generated using the lentivirus vector (GeneChem Bio-Medical Biotechnology, Shanghai, People's Republic of China) according to the manufacturer's instruction. The sequence of short hairpin RNA (shRNA) against HMGB1 was 5'-GGACAAGGCCCGTTATGAA-3'. Stable cell lines overexpressing HMGB1 or sh-HMGB1 were selected for 10 days with 5 μg/mL puromycin. The siRNA had previously been shown to knockdown BRG1 expression efficiently and specifically 15.
RNA extraction and Quantitative real-time PCR (qRT-PCR) assays
Total RNA was extracted from cells with RNAiso Plus reagent (TaKaRa) and cDNA was reverse- transcribed by using PrimeScript RT reagent Kit (TaKaRa). The RT-PCR analysis was conducted using the SYBR Green PCR Master Mix (TaKaRa) with Applied Bio-systems 7500 Fast Real-Time RCR System (Applied Biosystems, Foster City, CA, USA). Each measurement was performed in triplicate, and the results were normalized to the internal control of GAPDH. The relative expression of the target gene was determined by 2-△△Ct method. The specific primers were as indicated: HMGB1: 5'-AAAGCGGACAAGGCCCGTTAT-3' (forward) and 5'-AAGAGGAAGAAGGCCGAAGGAG-3' (reverse). BRG1: 5'-CAGATCCGTCACAGGCAAAAT-3' (forward) and 5'-TCTCGATCCGCTCGTTCTCTT-3' (reverse). Primers set for GAPDH was 5'-CCAGGTGGTCTCCTCTGACTTC-3' (forward) and 5'-GTGGT CGTTGAGGGCAATG-3' (reverse).
Cell viability
Cell viability was conducted using cell counting kit-8 (CK-04, Dojindo) according to the manufacturer's instructions. Briefly, PCa cells at 2 × 103 per well were seeded into 96-well plates. Then, 10% v/v CCK-8 was added to each plate and the cells were incubated at 37 ℃ for 2 h. Subsequently, the optical density (OD) at 450nm at predetermined time points was measured by a microplate reader (EXL800, BioTek Instruments).
Colony formation
PCa cells (300 cells/well) were seeded into six‐well plates and incubated for two weeks to allow colony formation. Then, the colonies were fixed with 4% paraformaldehyde and stained with Giemsa. Each group was replicated in three wells, and the colony formation experiment was performed with three replicates.
5-ethynyl-2'-deoxyuridine assay (EdU) incorporation Assay
Cell-Light ™ EdU staining kit (RiboBio, Guangzhou, China) was utilized to determine cell proliferation activity based on the manufacturer's instructions. Images were taken with a microscope (Olympus, Tokyo, Japan) at 200×. The proportion of EdU positively stained cells (with red fluorescence) to Hoechst-stained cells (with blue fluorescence) in per well was calculated.
Cell-cycle analysis
Cell-cycle analysis was executed as described previously 16. In brief, human PCa cells were digested using 0.25% Trypsin/EDTA solution and fixed with ice-cold 70% ethanol at 4 ℃ overnight. Subsequently, cells were stained with 50 μg/mL propidium iodide (PI) (keygentec. Nanjing, China). DNA content of cells in each group was determined by flow cytometry (FACS Calibur, Becton Dickinson). Synchronization in the cell cycle analysis was not used; all assays were repeated for three times.
Cell scratch assay
PCa cells were seeded at a density of 1 × 106 cells/well in 6-well plates and grown to 75%-90%. Linear wounds were scratched with a 10 μL plastic pipette tip and the cell debris was washed away with PBS twice. Subsequently, cells were incubated in FBS-free culture medium. Wounds of the scraped area were monitored and captured at indicated time points with an inverted microscope (Olympus IX71) at 100× magnification. Distance from each side of the scratch was quantified at 3 distinctive fields within the same scratch.
Transwell migration assay
Cell migration assay was conducted utilizing Transwell inserts (Costar, Corning, Cambridge, MA, USA) with a chamber (8.0μm pore size). 5 × 104 cells were resuspended in serum-free medium and seeded into the upper compartment of the insert. Then, 500μL complete medium was added to the lower chamber. After 36 h, the cells were fixed with 4% paraformaldehyde and stained with Giemsa (Boster Ltd., Wuhan, China). Subsequently, cells on the top surface of the membrane were removed, and those on the bottom surface were photographed using an inverted microscope (Olympus DP72) at 200× magnification. Five randomly chosen visual fields were captured and counted using Image J software. The number of cells migrating through the chambers in each group was documented as the average value.
Transwell invasion assay
For invasion assay, the Transwell membrane was pre-coated with Matrigel matrix (40111ES08; Yisheng Biotechnology Co., Ltd., Shanghai, China). Subsequently, 6 × 104 PCa cells were seeded in the upper section of the Boyden chambers. After 36 h of culture at standard conditions, the superficial cell layer was wiped off with cotton balls before visualization. The next steps were the same as in the migration assay.
Mice xenograft and tumor metastasis
All experimental animal procedures were authorized by the Animal Care and Use Committee of Southern Medical University and animals were raised under Specific Pathogen Free (SPF) conditions. Xenograft tumor models were generated by subcutaneous injection of 2×106 PC3/sh-Ctrl, PC3/sh- HMGB1, 22Rv1/vector, and 22Rv1/Lv-HMGB1 cells (n = 6 per group), on the axillae of 4 to 6 weeks BALB/c nude mice obtained from the Animal Center of Southern Medical University, Guangzhou, China. Tumor size was measured every 5 days and the volume was calculated using the formula: volume = (length × width2)/2 16. To assess metastasis, 5 × 106 cells in 100μL of PBS were injected via the tail veins of mice (n = 6 for each group). After 40 days, lung metastases of tumor nodules were observed under the microscope. Image J software was used to calculate the lung colonization of foci in three randomly selected fields. The presence of cancer cells in the tumor tissues was verified with H&E (hematoxylin and eosin) staining and immunohistochemistry (IHC) staining using antibodies against HMGB1 and Ki-67 (Abcam, #16667).
Western blot analysis
Cells were extracted using RIPA lysis buffer containing protease inhibitors (#KGP250, KeyGEN BioTECH, Nanjing, China) according to the manufacturer's protocol. After extraction, equal amounts of proteins in the cell lysates were separated by SDS/PAGE gels (4-12%, Bio-Rad) and electrically transferred onto polyvinylidene fluoride (PVDF, Millipore) membranes. Subsequently, membranes were blocked with 5% no fat milk or bovine serum albumin (BSA) and incubated for 16 h at 4 ℃ with the following specific primary antibodies: rabbit anti- HMGB1 (#ab18256, Abcam) and rabbit anti-BRG1 (#ab110641, Abcam); EMT marker (#9782, Cell signaling, MA, USA), rabbit anti-cyclin D1 (#2978), rabbit anti-c-myc (#5605), rabbit anti-p27 Kip1(#3686), rabbit anti-p21 Waf1/Cip1(#2947), rabbit anti-CDK4 (#12790), rabbit anti-Akt (#4691), rabbit anti-phospho- AktSer473(#4060) and rabbit anti-phospho-AktThr308(#13038). These antibodies were purchased from Cell Signaling Technology except for mouse anti-β-actin (#60008-1-Ig, Proteintech Group) and rabbit anti-α- tubulin (#ab18251, Abcam). Subsequently, all membranes were immersed in horseradish peroxidase- linked secondary anti-rabbit IgG or anti-mouse IgG antibodies (Cell Signaling Technology) for 1 h at room temperature. The bands were visualized using the enhanced chemiluminescence (ECL) detection system (Pierce Biotechnology, Rockford, IL, USA). The intensity of protein bands was quantified with the Image J software.
Immunofluorescence assays
Cells were fixed with 4% paraformaldehyde (E672002, Sangon Biotech) for 15 min at room temperature followed by incubation with 0.5% Triton solution (A110694, Sangon Biotech) for 10 min to permeabilize the cell membrane. Then, cells were incubated with Tris-buffered saline containing 5% bovine serum albumin (BSA) for 30 min. Subsequently, samples were incubated with rabbit anti-HMGB1 (#ab18256, Abcam) and mouse anti- BRG1 (#sc17796, Santa Cruz) at 4 ℃ overnight. Finally, the fluorescent secondary antibody Alexa Fluor 488-conjugated goat anti-rabbit IgG (#4412S, Cell Signaling Technology) and Alexa Fluor 594-conjugated goat anti-mouse IgG (#8890, Cell Signaling Technology) were used to detect primary antibodies. DAPI (E607303, Sangon Biotech) was applied for nuclear staining. Fluorescence images were visualized and collected under inverted confocal microscopy (DM5000B, Leica).
Plasmid transfection
The full-length human wild-type Akt and BRG1 cDNAs were inserted into PENTER-3FLAG-SV40- Kana expression vector obtained from Shangdong Vigene Biosciences Co. All constructs were validated by sequencing. PC-3/sh-HMGB1 and LNCaP/sh- HMGB1 cells were transiently transfected with pUSEamp-myr-akt or pUSEamp empty vector and BRG1 or empty vector by using Lipofectamine™ 3000 Transfection Reagent following the manufacturer's instructions (L3000015, Thermo Fisher Scientific, USA). Plasmid expression was verified by Western blotting.
Co-immunoprecipitation
Co-immunoprecipitation of proteins was executed as previously described 17 with modifications. In brief, PC-3 and LNCaP cell protein supernatants were pre-treated with 50 μL A/G beads (Selleck Chemicals, Houston, U.S.A) before immunoprecipitation and then with 5 μg control IgG (Santa Cruz Biotechnology), HMGB1, or BRG1 antibodies overnight at 4 °C. After further incubation with 50 μL A/G beads at 4 °C for 6 h, the immune-precipitates were eluted with ice-cold PBS containing 0.2% NP-40 for 5 times. Subsequently, these immunoprecipitated proteins were electrophoresed on SDS-PAGE, and visualized by silver staining (Byeotime, Shanghai, China). LC-MS/MS 18 was used to analyze the digested gels. The immunoprecipitated protein complexes were subsequently isolated by metal boiling in 2 × SDS-PAGE sample buffer for 10 min and utilized for immunoblotting with both anti-HMGB1 (#ab18256, Abcam) and anti-BRG1 (#ab110641, Abcam) antibodies.
Immunohistochemical analysis and evaluation
Tissue microarray (TMA) and IHC were employed to investigate protein expression in tissues as described previously 16, 19, 20. For IHC, the specimens were incubated with antibodies against HMGB1 (1:500), and BRG1 (1:50). Positive cells were scored as follows: 0 (no staining, 0%), 1 (staining range, 1-25%), 2 (staining range, 26-50%), 3 (staining range, 51-75%), or 4 (staining range, 76-100%). Intensity scores were recorded as: 0 (no staining), 1 (weakly staining, light yellow), 2 (moderately staining, yellowish brown), and 3 (strongly staining, brown). The multiplication of the above two scores was recorded as follows: - (0-1), +(2-4), ++ (5-8), +++ (9-12). A score of ≥2 was regarded as upregulated, while <2 score was considered as low expression. In case of discrepancies (>5%), the results were reevaluated. All evaluations were performed by three independent senior pathologists using the same microscope.
Statistical analysis
All statistical analyses were conducted using SPSS version 19.0 software (SPSS, Chicago, IL, USA). Numerical data were expressed as means ± SD. Differences between variables were analyzed by two-tailed Student t-test or One-way analysis of variance (ANOVA) for continuous variable groups. When ANOVA was significant, post hoc testing of differences between groups was carried out using the LSD test. Kaplan-Meier survival curve was plotted from the mice survival data and examined by the log-rank test. Correlations between the protein abundance and clinicopathological factors in PCa tumor tissues and BPH tissues were confirmed by Pearson's chi-square test or Fisher exact test for categorical/binary measures. HMGB1 and BRG1 expressions were explored by Spearman's correlation. P<0.05 was considered as statistically significant.
Results
HMGB1 accelerates PCa cell proliferation in vitro
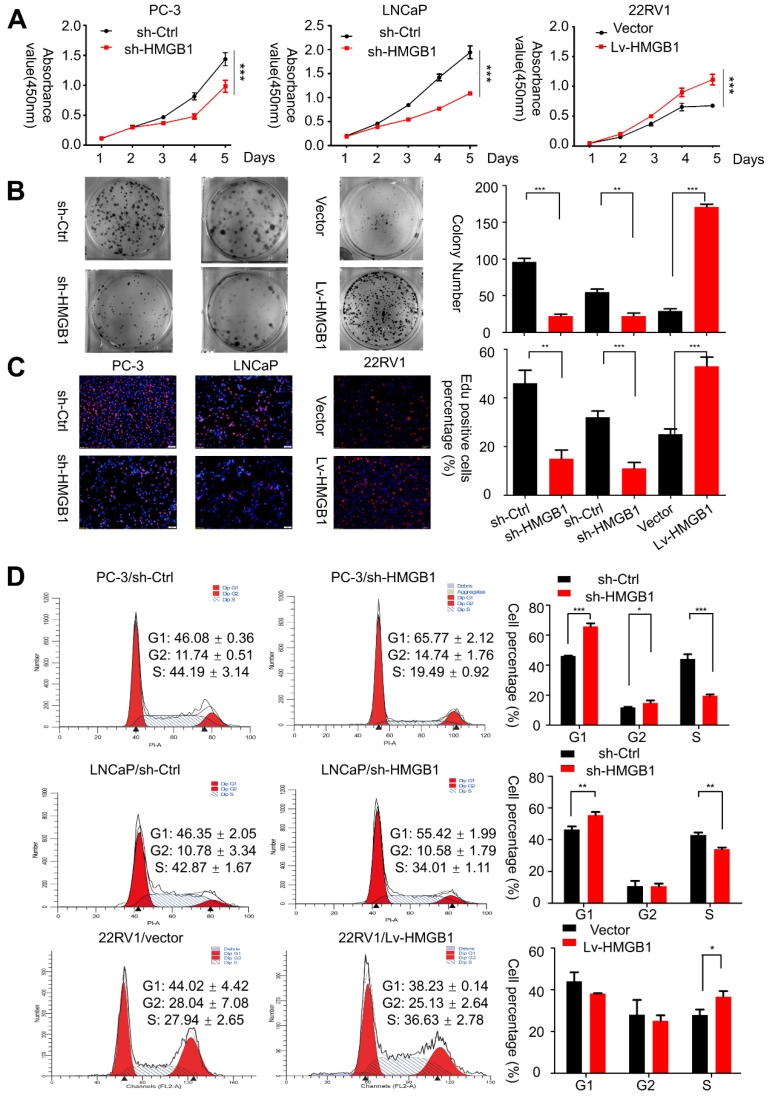
To verify whether HMGB1 is essential for PCa oncogenesis, we first analyzed the endogenous protein expression of HMGB1 in four tumor-derived PCa cell lines (PC-3, DU145, 22Rv1, and LNCaP) by Western blotting. PC-3 and LNCaP cells exhibited relatively higher levels of HMGB1 protein than other cells, while 22Rv1 cells expressed HMGB1 at a much lower level (Figure S1A). Hence, we chose PC-3, LNCaP, and 22Rv1 cells for the subsequent studies. First, three siRNAs targeting HMGB1 were designed to silence its expression in PC-3 and LNCaP cell lines. We chose si-HMGB1#2 for the subsequent experiments due to the highest inhibitory efficiency (Figure S1B). Then, we established stable silenced HMGB1 in PC-3 and LNCaP cells as well as ectopic HMGB1 in 22Rv1 cells by using lentiviral vectors. Western blot and RT-PCR analyses revealed that sh-HMGB1 significantly silenced intracellular HMGB1 protein compared with the control (sh-Ctrl), while the HMGB1 overexpression group showed a dramatically increased level of HMGB1 protein (Figure S1C). CCK-8 assay demonstrated that the proliferation of HMGB1-silenced cells was remarkably decreased but was increased in ectopic-HMGB1-expressing cells (Figure 1A). Consistent with this, the colony-forming ability was notably suppressed in PC-3/sh-HMGB1 and LNCaP/sh-HMGB1 cells compared with sh-Ctrl cells but was dramatically increased in 22Rv1/Lv-HMGB1 cells (Figure 1B).
Figure 1.
HMGB1 promoted the aggressive behavior and proliferation ability of PCa cells in vitro. (A and B) CCK-8 proliferation assay and colony-formation assay were used for detecting the proliferation ability in HMGB1-knockdown or -overexpressing PCa cell lines. (C) Representative micrographs (left) and quantification (right) of EdU incorporation. (D) Cell-cycle analysis revealed that knocking down HMGB1 expression in PC-3 and LNCaP cells increased the percentage of cells in the G1 phase and decreased the percentage in S phase, while ectopic expression of HMGB1 decreased the percentage of cells in the G1 phase and increased the percentage of cells in S phase, *P<0.05, **P<0.01, ***P<0.001.
Then immunofluorescent staining for EdU incorporation assays showed increased DNA synthesis in 22Rv1 cells with ectopic expression of HMGB1, whereas blocking of HMGB1 significantly suppressed DNA synthesis (Figure 1C). Additionally, flow cytometry analysis was conducted to confirm whether the change of proliferation is attributed to alterations in the cell cycle profile. Results showed that HMGB1 knockdown in PC-3 and LNCaP cells induced G1 arrest while ectopic expression of HMGB1 in 22Rv1 cells accelerated cell cycle progression into the S phase (Figure 1D). These results indicated that HMGB1 enhances cell growth at least partially by inducing G1/S transition in PCa cells.
HMGB1 facilitates PCa cell invasion and migration in vitro
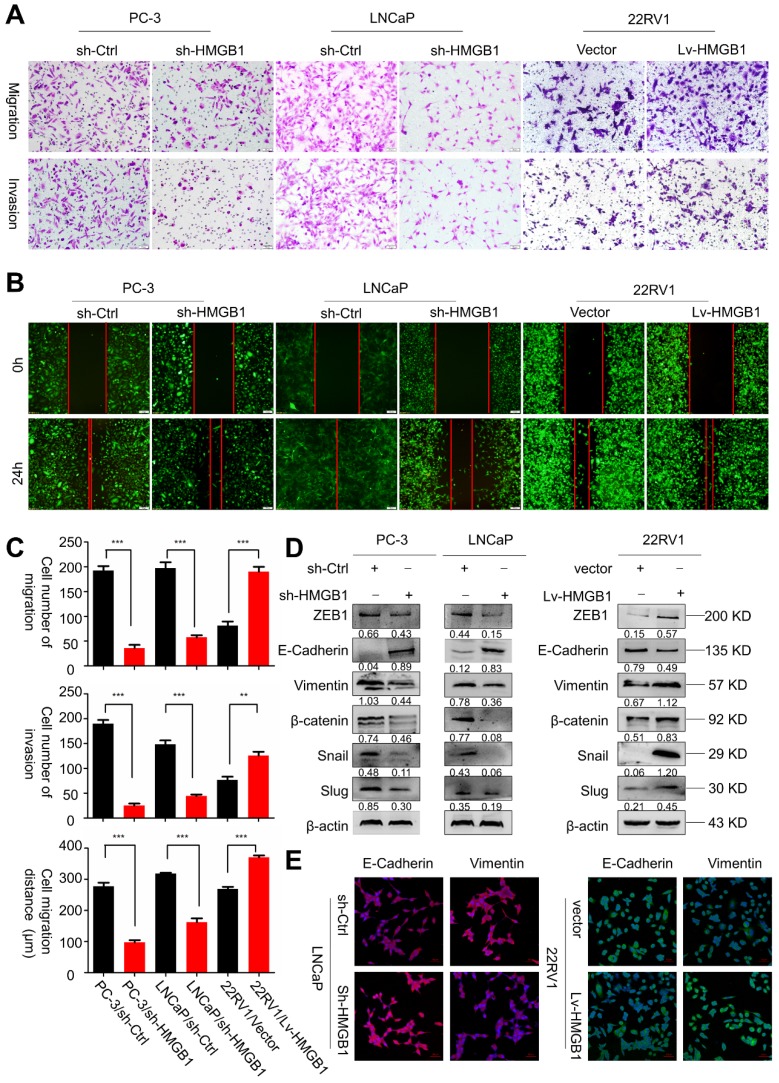
Next, we sought to determine the role of HMGB1 in tumor invasion and metastasis. Results of migration and invasion in vitro assays showed that depleted HMGB1 suppressed cell migration and invasion of PC-3 and LNCaP cells (Figure 2A-B). Opposite effects on cell migration and invasion were observed in ectopic-HMGB1-expressing 22Rv1 cells (Figure 2C). Since EMT has been widely regarded as a crucial process in tumor invasion and metastasis 21, we hypothesized that HMGB1 affected PCa cell migration and invasion through regulating the EMT progression. Western blotting and immunofluorescence analyses were performed to detect the protein expression of EMT-related markers. As anticipated, results demonstrated that HMGB1 depletion reduced the abundance of mesenchymal markers vimentin, β-catenin, Snail, Slug, and ZEB1, but increased protein abundance of epithelial marker E-cadherin in PC-3 and LNCaP cells. In contrast, overexpression of HMGB1 displayed the opposite effect (Figure 2D-E). These results suggested that HMGB1 promotes invasion and tumor metastasis of PCa cells by regulating EMT.
Figure 2.
HMGB1 facilitated migration and invasion of PCa cells in vitro. (A-B) HMGB1 knockdown markedly attenuated cell migration and invasion in PC-3 and LNCaP cells, while up-regulation of HMGB1 increased the migration and invasion ability of 22Rv1 cells as measured by Transwell migration and wound healing assays. The invasive capability was determined by using Matrigel invasion chambers. (C) Graphical illustration of statistical results of transfection of HMGB1-shRNA or ectopic-HMGB1 on cell invasion and migration. Data are presented as mean ± SD (n = 3). Migrated cells were counted using Image J software and represent mean values per field from at least three fields. Experiments were carried in duplicate. *P<0.05; **P<0.01; ***P<0.001 compared with the control (D) EMT markers were detected by Western blotting in both PCa cell lines with HMGB1 knocked down or overexpressed. β-actin served as the loading control. (E) Immunostaining of mesenchymal and epithelial markers in LNCaP/sh-Ctrl and LNCaP/sh-HMGB1 or 22Rv1/vector and 22Rv1/Lv-HMGB1 cells as indicated, Scale bar = 50 μm.
HMGB1 promotes the growth and metastasis of human PCa cells in vivo
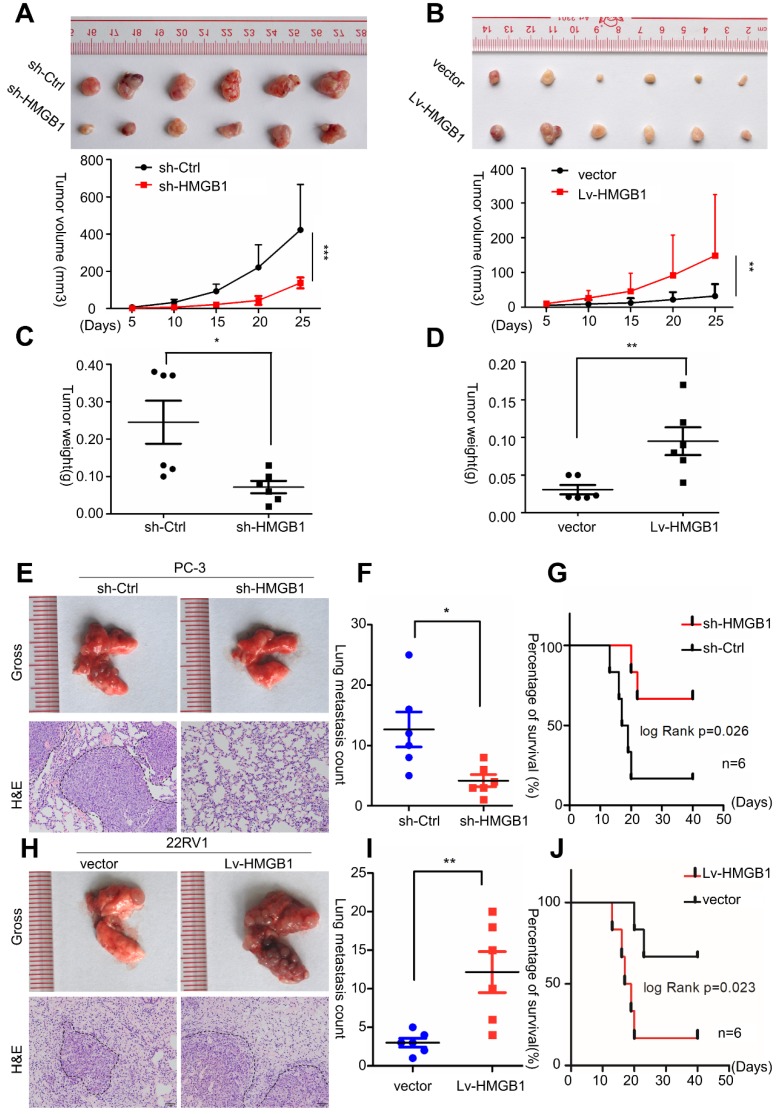
To validate the potential impact of HMGB1 depletion on PCa cell proliferation in vivo, PC-3/sh-HMGB1 cells and PC-3/sh-Ctrl cells were injected subcutaneously in nude mice. Tumors in mice implanted in PC-3/sh-HMGB1 cells grew slower in comparison with control cells. HMGB1 knockdown cells exhibited significantly smaller tumor volume and weight than control cells 25 days after injection (Figure 3A-C). The 22Rv1 cells with HMGB1 overexpression, on the other hand, showed rapid growth speed than the control vector cells (Figure 3B-D). H&E staining showed the histopathological features of the tumor tissues. The expression of Ki-67 proliferation antigen was dramatically weaker in tumor tissues with sh-HMGB1 cells and stronger in xenografts with HMGB1 overexpression as seen by IHC staining (Figure S2). These results provided evidence that HMGB1 is a remarkable determinant for PCa cell growth.
Figure 3.
HMGB1 enhanced tumor growth and metastasis in vivo. Cells were injected into the hindlimbs of nude mice (n = 6). (A and B) Representative images of the gross tumors are shown (upper panels), Tumor growth curves were measured during the growth of the tumors (lower panels). Tumors derived from PC-3 cells expressing sh-HMGB1 grew significantly slower than those from cells with sh-Ctrl, whereas ectopic expression of HMGB1 in 22Rv1 cells dramatically enhanced tumor growth. (C and D) Final tumor weights were measured in each group. (E and H) Gross and microscopic illustrations of lung metastases in mice injected with PC-3/sh-Ctrl and PC-3/sh-HMGB1 cells or 22Rv1/Vector and 22Rv1/Lv-HMGB1 cells (n = 6). The lung sections were stained with H&E (200×). (F and I) The number of metastatic nodules in individual mice was counted under the microscope, *P<0.05, **P<0.01. (G and J) Kaplan-Meier overall survival curves for mice with PCa stratified by HMGB1 knockdown and negative control (log-rank test, n = 6, P = 0.026), as well as overexpressed HMGB1and negative control (log-rank test, n = 6, P = 0.023), respectively.
To investigate the effects of HMGB1 on PCa metastasis in vivo, tail vein xenograft model was generated. The tumor presence was validated by histological examination (Figure 3E-H). Results demonstrated that mice injected with HMGB1 shRNA cells produced less lung colonization and peritoneal colonization compared to those with the control cells (Figure 3E-F). Moreover, we found that knockdown of HMGB1 could prolong the survival time of mice compared with the control group (Figure 3G) while overexpression of HMGB1 exhibited the reverse effects (Figure 3H-J). These findings corroborated our previous data 14 and demonstrated that HMGB1 has a key role in PCa cell growth and metastasis in vivo.
HMGB1 promotes PCa cell growth and proliferation through the PI3K/Akt pathway
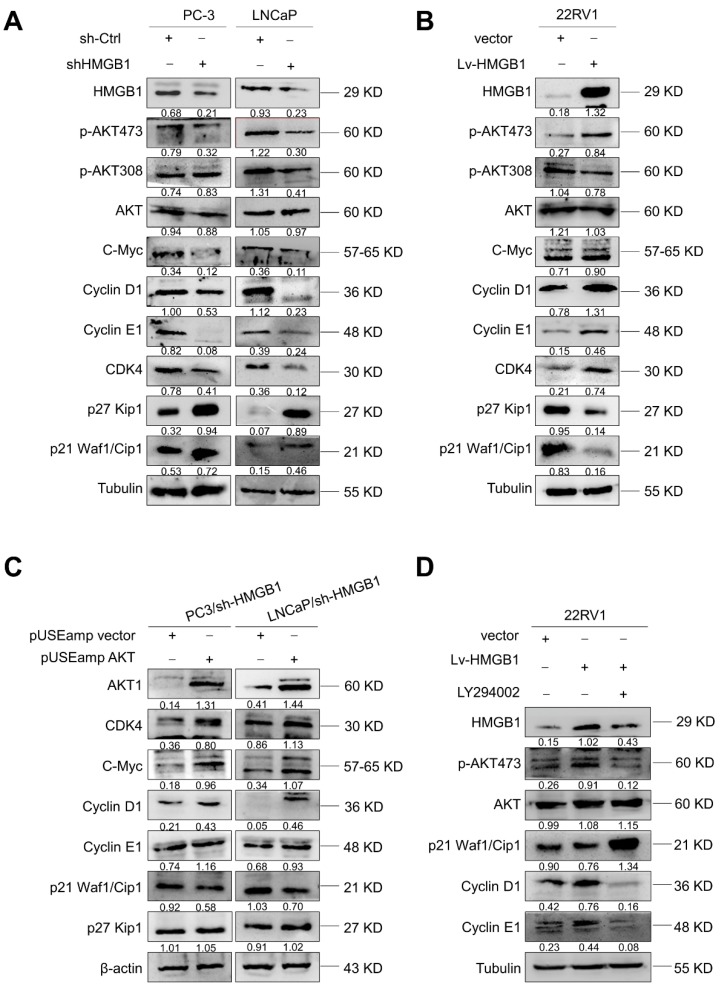
Since HMGB1 promoted G1/S transition of PCa cells, we further investigated the key cell-cycle regulation signaling networks. Indeed, we observed an increase in p21 Waf1/Clip1 and p27 Kip1 levels and a decrease in phosphorylated AktSer-473, CDK4, C-myc, cyclin D1, and cyclin E levels with HMGB1 deletion in PC-3 and LNCaP cells (Figure 4A). Conversely, overexpression of HMGB1 exhibited the opposite effects (Figure 4B). To further address whether the HMGB1 was required for Akt-mediated promotion of PCa proliferation, we transiently transfected active Akt1 into PC-3/sh-HMGB1 and LNCaP/sh-HMGB1 cells and measured the proliferation. CCK-8 and colony formation assays showed that the HMGB1-depleted cells regained high proliferative ability upon upregulation of Akt (Figure S3A-B). Also, the ratio of cells in the G1 phase reduced concurrently with a high proportion in S phase (Figure S3C). Subsequently, Western blotting was performed to evaluate the effect of myc-Akt overexpression on the levels of cell-cycle-related proteins in PC-3-sh-HMGB1 and LNCaP-sh-HMGB1 cells. The up-regulated expression of Akt in HMGB1-depleted cells increased the expression of CDK4, c-myc, cyclin D1, and cyclin E, together with decreased expression of p21 Waf1/Clip1, whereas the level of p27 Kip1 was not altered (Figure 4C). These data indicated the existence of an underlying mechanism, other than the Akt pathway, in HMGB1-mediated downregulation of p27 Kip1 expression. Our results also demonstrated that treatment with PI3K inhibitor LY294002 partially restored p21 Waf1/Clip1 expression and decreased p-AktSer-473, cyclin D1, and cyclin E levels in 22Rv1 cells with ectopic expression of HMGB1 (Figure 4D). Functional assays also demonstrated that inhibition of Akt signaling blocked the HMGB1-mediated up-regulation of PCa cell proliferation and cell cycle progression (Figure S3D-F). Taken together, these results further indicated that Akt pathway might involve in HMGB1-induced PCa cell proliferation.
Figure 4.
HMGB1 activated Akt signaling pathway and increased key cell-cycle regulatory proteins. (A) Western blotting revealed that HMGB1 knockdown in PC-3 and LNCaP cells caused a decrease in p-Akt Ser473, CDK4, c-myc, cyclin D1, and cyclin E, along with an increase in p27, p21. (B) Expression of these proteins showed an opposite effect in 22Rv1 cells with ectopic-HMGB1. (C) Introduction of Akt into PC-3 and LNCaP HMGB1-shRNA cells restored the levels of CDK4, c-myc, cyclin D1, and cyclin E together with decreased expression of p21; the p27 level showed less change. (D) 22Rv1/Lv-HMGB1 cells were treated with Akt inhibitor LY294002 (50 μM) for 24 h. Inhibition of the Akt signaling suppressed the promoting effect of HMGB1 overexpression on the Akt pathway.
HMGB1-mediated EMT in PCa cells is regulated by BRG1-induced activation of the Akt signaling pathway
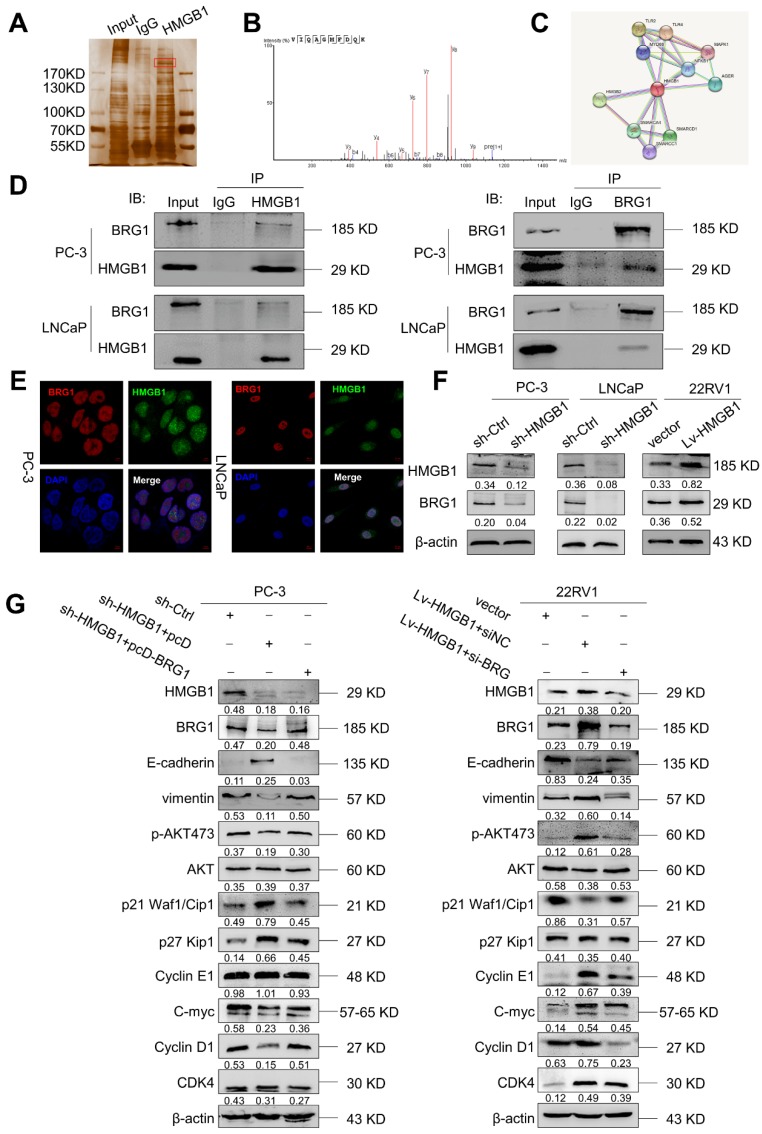
To elucidate the potential molecular mechanism of HMGB1 in PCa cells, we immunoprecipitated the HMGB1 protein with an anti-HMGB1 antibody and identified the proteins that may directly interact with HMGB1 by LC-MS/MS. Among the HMGB1- interacting proteins identified by mass spectrometry (Figure 5A-B, Table S1), BRG1 had been reported to drive the progression of PCa 22, 23. In addition, the single protein function partner network of HMGB1 in String analysis also indicated that BRG1 may interact with HMGB1 (Figure 5C). To validate the protein- protein interaction between HMGB1 and BRG1, co-immunoprecipitation (Co-IP) with an antibody against HMGB1 was carried out. After immunoprecipitation with HMGB1 conjugated beads, BRG1 was found in the lysates from both PC-3 and LNCaP cells (Figure 5D). It has been reported that ZEB1/BRG1 transcriptionally regulates E-cadherin expression and EMT that is implicated in the initial stages of tumor invasion 24. Thus, we hypothesized that HMGB1 may interact with BRG1 protein to induce EMT. As expected, in a reciprocal Co-IP with BRG1 conjugated beads, HMGB1 precipitated with BRG1 (Figure 5D). Moreover, HMGB1 and BRG1 co-localized in both cell lines in the cell nucleus as observed by immunofluorescent staining (Figure 5E) indicating the physical interaction between HMGB1 and BRG1 in PC-3 and LNCaP cells. To confirm whether HMGB1 regulates BRG1 expression, immunoblotting was conducted to detect the abundance of BRG1 in PCa cells when HMGB1 expression was altered. Notably, BRG1 expression was downregulated in PC-3 and LNCaP cells with HMGB1 knockdown, while the level of BRG1 was dramatically upregulated in 22Rv1 cells with HMGB1 overexpression (Figure 5F). To validate the role of the HMGB1-BRG1 axis in HMGB1-mediated EMT, we examined whether BRG1 overexpression and silencing would reverse the effects of HMGB1 knockdown and ectopic expression in PCa cells. When we induced BRG1 siRNA in HMGB1 overexpressing cells, immunoblotting showed reversed EMT markers and inhibition of phosphorylation of Aktser473 accompanied by suppression of cell cycle-related proteins (Figure 5G). These data demonstrated that HMGB1 triggers EMT in PCa partly by enhancing BRG1 activity.
Figure 5.
BRG1 is an interactive factor of HMGB1. (A) Immunoprecipitation and silver staining were performed by using PC-3 cell lysate with the anti-HMGB1 antibody. (B) The spectrogram of the differential protein band was identified as SMARCA4 also known as BRG1 (Quadrupole Mass Spectrometer). (C) The single protein function partner network of HMGB1 in String analysis. (D) Coimmunoprecipitation was performed to validate the interaction between HMGB1 and BRG1 in PC-3 and LNCaP cells. (E) Confocal immunofluorescence analysis showed the presence and localization of HMGB1 and BRG1 in the nuclei of PC-3 and LNCaP cells, Scale bar = 10 μm. (F) HMGB1 regulated BRG1 protein expression in PCa cells. Knockdown of HMGB1 attenuated BRG1 protein expression in PC-3 and LNCaP cells, whereas ectopic expression of HMGB1 facilitated BRG1 protein expression in 22Rv1 cells. (G) HMGB1 mediated Akt pathway and EMT via BRG1 in PCa cells. BRG1 overexpression in HMGB1 deficient PC-3 cells rescued the phosphorylation of Akt and EMT, while knockdown of BRG1 in ectopic HMGB1-overexpressing 22Rv1 cells attenuated these effects.
HMGB1-mediated EMT in PCa cells is regulated by BRG1-induced activation of the Akt signaling pathway
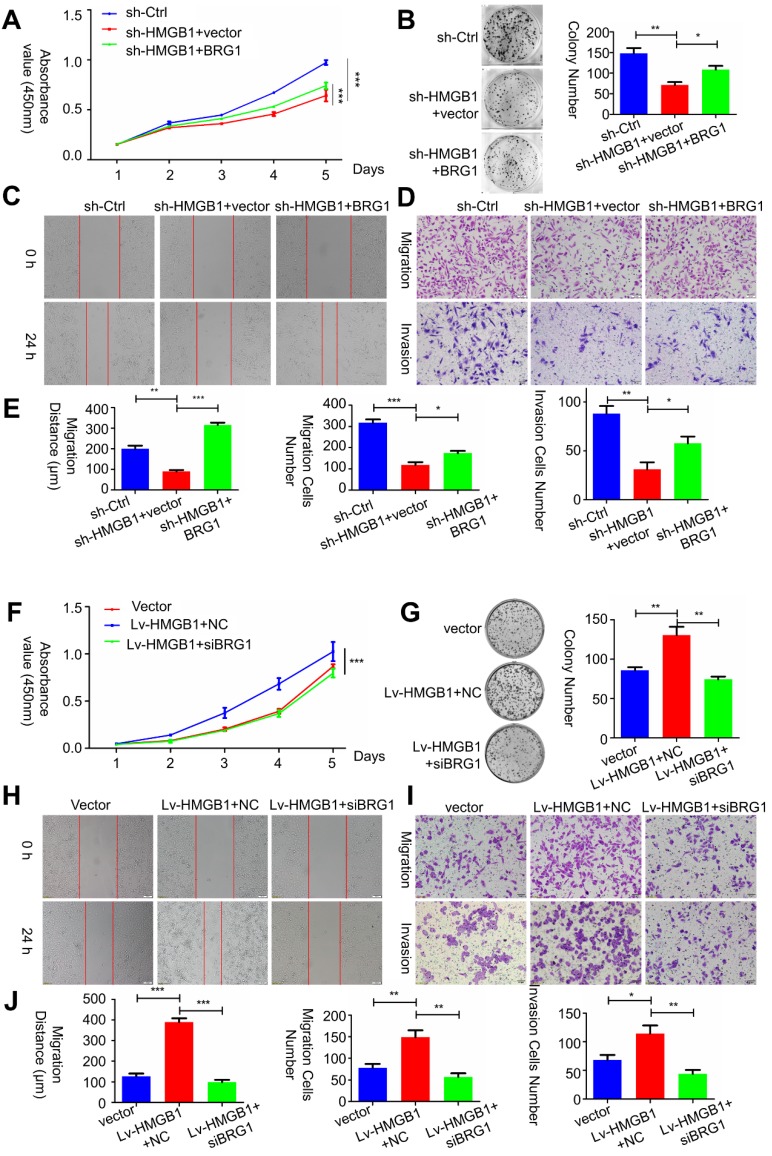
To determine whether HMGB1-mediated BRG1 affected the cell viability as well as migration and invasion of PCa cells, co-transfection of HMGB1 and si-BRG1 or HMGB1-shRNA and BRG1 was performed. Consist with EMT marker reversal, BRG1 overexpression in HMGB1-knockdown cells impaired the migration and invasion of PC-3 cells (Figure 6 A-E). In contrast, depletion of BRG1 remarkably impaired aggressive behavior of the HMGB1- overexpressing cells (Figure 6F-J). Moreover, Western blot analysis results demonstrated that blocking Akt activation excessively increased the expression of E-cadherin, but not vimentin (Figure S4), indicated that this aberrant signal triggered EMT process may partly contribute to the activation of Akt signaling pathway. Collectively, these results suggested that HMGB1 mediates EMT of PCa cells via inducing BRG1 expression.
Figure 6.
HMGB1mediated carcinogenic effects in PCa cells via BRG1. (A-C) Effects of co-transfection of HMGB1-shRNA and BRG1 or HMGB1 and si-BRG1 on cell proliferation, invasion, and migration by CCK-8, colony formation, scratch-wound-healing, and Matrigel invasion assays. (D and J) Graphical illustration of statistical results of co-transfection of HMGB1-shRNA and BRG1 or HMGB1 and si-BRG1 on cell migration and invasion. *P<0.05, **P<0.01, ***P<0.001, n = 3.
HMGB1 and BRG1 are co-expressed in PCa cells and PCa tumors
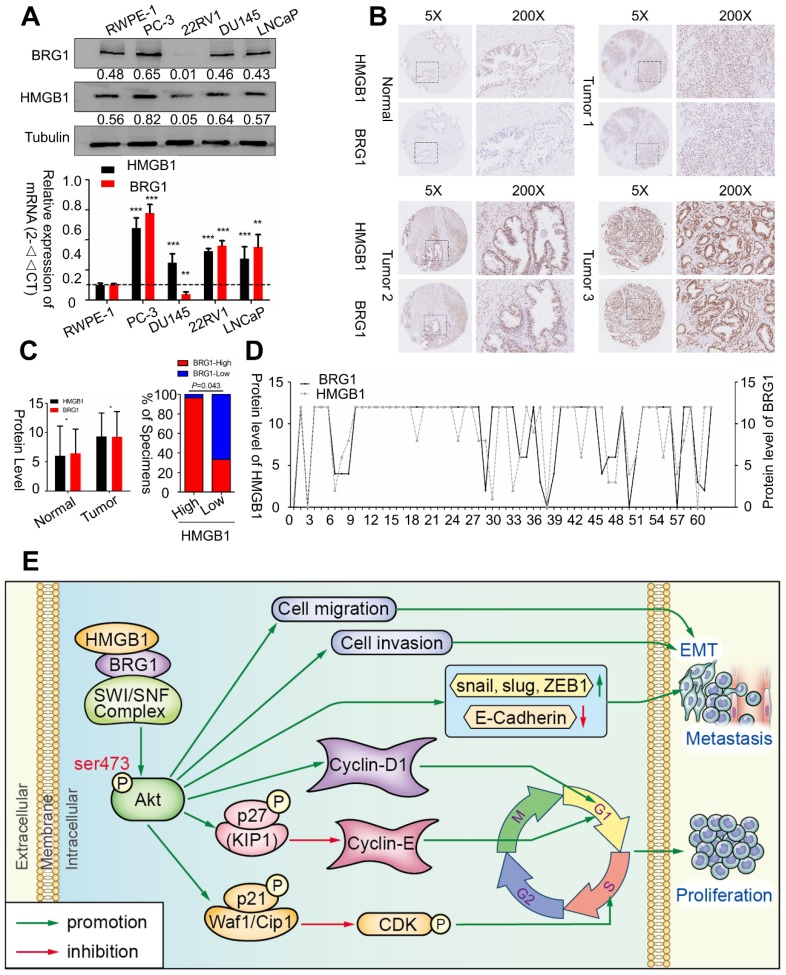
To confirm whether BRG1 is expressed in different PCa cell lines and in human PCa tissues, we evaluated BRG1 and HMGB1 proteins together with mRNA expression in control RWPE-1 cells and four different PCa cell lines by Western blotting and qRT-PCR. High expression of both BRG1 and HMGB1 was detected in PC-3, DU145, and LNCaP cells (Figure 7A). Furthermore, IHC of 64 paraffin- embedded human PCa tissues and 14 benign prostatic hyperplasia (BPH) or normal prostate samples showed that expression of both HMGB1 (P = 0.008, Table 1) and BRG1 (P = 0.045, Table 1) was dramatically increased in PCa tumors than the corresponding BPH tissues (Figure 7B-C). We then analyzed the correlation between the expression of these two proteins and clinicopathological variables. Chi‑square analysis demonstrated that expression of HMGB1 and BRG1 was significantly associated with tumor Gleason score/International Society of Urological Pathology (ISUP) Grade Group (PHMGB1 = 0.01 and PBRG1 = 0.006) and clinical stage (PHMGB1 = 0.002 and PBRG1 = 0.045), but not associated with patient's age, T stage, regional lymph nodes, or distant metastasis (Table 2). Furthermore, our results showed a spatial and protein level correlation between HMGB1 and BRG1 in prostate carcinoma tissues (r2 =0.574, P<0.001; Figure 7D) implying that HMGB1 modulates BRG1 function in prostate carcinoma progression.
Figure 7.
BRG1 was positively correlated with HMGB1 in PCa cells and tissues. (A) HMGB1 and BRG1 protein and mRNA expression in the normal prostate epithelial cell line (RWPE-1) and five PCa cell lines were detected by Western blot (α-tubulin was used as loading control) and qRT-PCR. (B) BRG1 and HMGB1 expressions in 64 paraffin-embedded PCa tissue samples were detected by immunohistochemistry staining. BRG1 and HMGB1 were spatially correlated. BRG1 and HMGB1 expression levels in PCa tissue specimens were positively correlated. Four Representative immunohistochemical staining photographs of normal tissue (Normal) and tumor tissue samples (Tumor 1, Tumor 2 and Tumor 3) are shown as indicated. (C) Left panel: quantification of BRG1 and HMGB1 expression levels in PCa tumors (n = 64) which were significantly higher than the normal tissues (n = 14). Magnification: 200×. *P < 0.05 compared to normal tissue; Right panel: percentage of PCa specimens showing low or high HMGB1 expression relative to the level of BRG1. (D) Spearman's correlation analysis showed a positive relationship between the BRG1 and HMGB1 levels in 62 human PCa tissues. r = 0.574, P < 0.001. (E) Schematic representation of the proposed mechanism of HMGB1 in PCa cells.
Table 1.
Expression of HMGB1 and BRG1 in BPH and prostate cancer tissues.
| HMGB1 protein expression |
P value |
BRG1 protein expression |
P value |
|||||
|---|---|---|---|---|---|---|---|---|
| Type | N | Positive (%) | Negative (%) | N | Positive (%) | Negative (%) | ||
| BPH | 14 | 8 (57.1) | 6 (42.9) | 0.008 | 14 | 10 (71.4) | 4 (28.6) | 0.045 |
| PCa | 62 | 56 (90.3) | 6 (9.7) | 64 | 60 (93.8) | 4 (6.3) | ||
Values are n (%). A significantly increasing frequency of positive expression of HMGB1 and BRG1 was detected in prostate cancer specimens compared to BPH tissues (P=0.045, χ2-test)
Table 2.
Correlation between HMGB1 and BRG1 expression with reference to clinicopathological characteristics was analyzed in prostate cancer (n=64)
|
Clinicopathological characteristics |
N |
HMGB1 |
p valueb |
N |
BRG1 |
p valueb |
||
|---|---|---|---|---|---|---|---|---|
| Positive (%) | Negative (%) | Positive (%) | Negative (%) | |||||
| Age(years) | ||||||||
| ≤ 69a | 29 | 28 (96.6) | 1 (3.4) | 0.123 | 30 | 29 (96.7) | 1 (3.3) | 0.353 |
| > 69 | 33 | 28 (84.8) | 5 (15.2) | 34 | 31 (91.2) | 3 (8.8) | ||
| pT status | ||||||||
| T1-T2 | 29 | 27 (93.1) | 2 (6.9) | 1.0 | 42 | 40 (95.2) | 2 (4.8) | 0.603 |
| T3-T4 | 27 | 26 (96.3) | 1 (3.7) | 22 | 20 (90.9) | 2 (9.1) | ||
| Clinical stage | ||||||||
| I- II | 15 | 10 (66.7) | 5 (33.3) | 0.002 | 16 | 13 (81.3) | 3 (18.8) | 0.045 |
| III- IV | 47 | 46 (97.9) | 1 (2.1) | 48 | 47 (97.9) | 1 (2.1) | ||
| Gleason Score | ||||||||
| < 7 | 7 | 4 (57.1) | 3 (42.9) | 0.01 | 8 | 5 (62.5) | 3 (37.5) | 0.006 |
| ≥ 7 | 51 | 49 (96.1) | 2 (3.9) | 52 | 51 (98.1) | 1 (1.9) | ||
| N - Regional lymph nodes | ||||||||
| N0 | 52 | 47 (90.4) | 5 (9.6) | 0.97 | 54 | 51 (94.4) | 3 (5.6) | 0.62 |
| N1 | 10 | 9 (90.0) | 1 (10.0) | 10 | 9 (90.0) | 1 (10.0) | ||
| M - Distant metastasis | ||||||||
| M0 | 53 | 48 (90.6) | 5 (9.4) | 0.88 | 55 | 51(92.7) | 4 (7.3) | 0.26 |
| M1 | 9 | 8 (88.9) | 1 (11.1) | 9 | 9 (100) | 0 (0.0) | ||
HMGB1 and BRG1 expression was determined by IHC; a: mean age. b: p-value is from χ2-test -test.
Discussion
Our previous study revealed a positive correlation between HMGB1 and RAGE expression in a cohort of patients with primary prostate cancer 14 suggesting that HMGB1 is implicated in the development and/or progression of PCa. To elucidate the pivotal role of increased expression of HMGB1 in PCa, we performed loss-of-function and gain-of-function experiments. Our findings demonstrated that shRNA-mediated silencing of HMGB1 expression in PC-3 and LNCaP cells significantly decreased cell proliferation, migration, and invasion and inhibited entry into the S-phase of the cell cycle. In the in vivo studies using a murine model, HMGB1 silencing resulted in the suppression of tumorigenesis and lung metastasis. On the contrary, ectopic expression of HMGB1 augmented the aggressive behavior of PCa cells. These observations underscored the key role of HMGB1 in the proliferation and metastasis of PCa.
HMGB1 is a chromatin-binding protein involved in DNA replication and DNA repair processes 25. Aberrant overexpression of HMGB1 has been shown in a variety of cancers and is closely associated with tumorigenesis 6, 10. Previously, we have shown upregulation and co-expression of RAGE and HMGB1 in PCa, which suggested a cooperative role of both proteins in the progression of PCa 14. Also, HMGB1 may regulate AR either by acting as a co-activator of AR or indirectly associating with RAGE signaling in prostate oncogenesis 26. This implied that HMGB1 may be involved in the tumorigenesis of PCa via multiple pathways. We, therefore, analyzed multiple key cell cycle proteins associated with the G1 to S phase transition by immunoblotting. There was a concomitant inhibition of C-myc, CDK4, cyclin D1, and cyclin E with the knockdown of HMGB1, whereas the expression of cyclin-dependent kinase inhibitors p27 and p21 was increased. HMGB1 has also been shown to be involved in cell proliferation and oncogenesis by activating the PI3K/Akt pathway 12, 27. However, it was unclear whether HMGB1 induced prostate carcinoma through PI3K/Akt pathway. We, therefore, analyzed the key proteins in this pathway and found that silencing of HMGB1 resulted in remarkably decreased phosphorylated Akt levels, whereas elevated expression of HMGB1 induced the phosphorylation of Akt. These results suggested that HMGB1 played a critical role in PCa progression.
HMGB1 has been implicated in the metastasis of different human malignancies including lung adenocarcinoma 28, triple-negative breast cancer 29, and osteosarcoma 30. Moreover, employing transgenic adenocarcinoma mouse prostate (TRAMP) model, He et al reported that HMGB1 promotes invasive carcinoma 31. In the present study, we have shown that HMGB1 is involved in the metastasis of PCa by regulating EMT in PCa cells. Tumor metastasis is attributed to many different mechanisms in various cancers; one of these mechanisms, EMT, is generally recognized to mediate the process of cancer cell invasion and metastasis 32. Our results demonstrated that knockdown of HMGB1 resulted in elevated the expression of E-cadherin and reduced expression of vimentin which are considered characteristic features of EMT. However, the molecular mechanisms underlying HMGB1-mediated EMT in PCa remain unclear.
In the present study, using LC-MS/MS analysis, we defined BRG1 from the candidate HMGB1- interacting proteins. BRG1 encodes an ATPase subunit of the SWI/SNF chromatin- remodeling complex, which regulates transcriptional activity by remodeling the chromatin structure and is involved in signal transduction, gene transcription, and protein stability 33, 34. BRG1 is highly conserved and located on chromosome 19p13.2, an area usually amplified n various tumors, including melanoma 35 and gastric 36 and PCa 37. Previous studies have shown that amplification of BRG1 was highly correlated with metastatic phenotype and malignant progression 38. Notably, stabilization of BRG1 suppressed E-cadherin expression in gastric cancer cells subsequently promoting metastasis 36. Furthermore, BRG1 has been reported as a corepressor of ZEB1 to regulate E-cadherin transcription and was required for the induction of EMT by ZEB1 24. It has been reported that activation of both Akt and other signaling pathways may induce EMT contributing to tumor metastasis 39. A recent study described that BRG1 acted as a prognostic indicator and a potential therapeutic target for PCa 40. Increased BRG1 expression in PTEN-deficient PCa cells led to chromatin remodeling into configurations that drove a pro-tumorigenic transcriptome, steering cells to become further addicted to BRG1 23.
Our data indicated that HMGB1 acted as an interacting partner of BRG1. By increasing BRG1 protein expression, HMGB1 promoted the growth and invasion of PCa and mediated EMT. The correlation between HMGB1 and BRG1 was confirmed by the following observations. First, in PCa cell lines as well as in clinical specimens, the expression of BRG1 significantly correlated with HMGB1 levels. Second, the introduction of HMGB1 dramatically increased the expression of BRG1 in PCa cells. Third, deletion of BRG1 partly counteracted the aggressive phenotype mediated by HMGB1. And finally, ectopic expression of BRG1 rescued the attenuation of cellular tumorigenesis and EMT by the knockdown of HMGB1 indicating that BRG1 was required for HMGB1-induced malignant progression in PCa. These results strongly demonstrate that BRG1 is a downstream effector of HMGB1.
Accumulating evidences have revealed that multiple cellular signaling pathways such as Wnt/β‐catenin, mitogen‐activated protein kinase (MAPK), TGF‐β/Smads were implicated in the progression of EMT 41-43. Evidence from recent literatures and our previous studies imply that PI3K/Akt signaling pathway palys a pivotal role in the EMT process of PCa 44, 45. It is has been widely reported that PI3K/AKT signaling pathway regulates EMT by promoting the phosphorylation of downstream target proteins (Bad, Caspase9, NF‐κB, GSK‐3β, mTOR, p21Cip1, p27 Kip1, etc.) to further mediates tumor proliferation, invasion and metastases 46-48. Furthermore, activation of p‐Akt could increase the expression of integrin‐linked kinase as well as induce key transcription factors, such as Snail, Slug, and Twist, which ultimately promotes the process of EMT 49-51. Both HMGB1 and BRG1 have been shown to be closely related with the PI3K/AKT pathway 12, 22. However, whether the activation of Akt contributes to the cellular metastasis function of PCa induced by HMGB1-BRG1 axis remains uncertain. In the present study, we observed that HMGB1 could activate PI3K/AKT signaling pathway by regulating the protein phosphorylation level and that HMGB1-induced activation of EMT occurs partly through phosphorylation of Akt. Furthermore, HMGB1 and BRG1 were required for increased cell migration and invasion of PCa cells by Akt. Meanwhile, blocking Akt activation with LY294002 largely increased the expression of E-cadherin, while showed no significant effect in vimentin inferred that p‐Akt protein levels fall below a certain threshold resulting in upregulation E-cadherin level and finally alleviated cell migration competency in PCa cells during EMT process.
No changes of vimentin protein expression implied E-cadherin was not interrelated to vimentin in PCa cells, which was consistent with previous reports 52, 53. Notably, loss of E-cadherin was reckoned as the crucial step to initiate EMT that sustained PCa metastasis 54. Based upon these observations, we propose that HMGB1 may strengthen BRG1 function and activate the Akt signaling pathway to promote EMT. In the future, it would be important to elucidate the mechanisms underlying the HMGB1-mediated upregulation of BRG1 as well as the molecular interactions of HMGB1 and BRG1 implicated in EMT in PCa cells.
Conclusions
In summary, our data indicated that HMGB1 plays a vital role in tumorigenesis and metastasis of PCa process. HMGB1 promotes PCa development via activation of the Akt signaling pathway and facilitates metastasis through modulating BRG1-mediated EMT (Figure 7E). HMGB1 may serve as a molecular marker to monitor the progression of PCa.
Supplementary Material
Supplementary figures and table legends.
Supplementary table 1.
Supplementary table 2.
Acknowledgments
This work was supported by the Medical Science and Technology Foundation of Guangdong Province (No. 2013B051000050, No. 2014A020212538, and No. 2016A020215175), the Natural Science Foundation of Guangdong Province (No. 2016A030313583), the Medical Scientific Research Foundation of Guangdong Province (No. A2016555), the Outstanding Youths Development Scheme of Nanfang Hospital, Southern Medical University (No. 2015J005), and the Science and Technology Planning Project of Guangzhou (No. 201704020070).
Author Contributions
L. D. J. and Z. S. C. designed research experiments; L. D. J., S. X. L., Z. H. B. and H. B. performed experiments; Y. Y. Z., W. C., C. H. and S. F. P. were responsible for data curation and formal analysis; Z. S. C. executed the funding acquisition and supervised the procedure of the experiments. L. D. J. and H. B. prepared and write the original draft. All authors have reviewed and approved the final version of the manuscript.
Abbreviations
- HMGB1
high mobility group box 1
- PCa
prostate cancer
- BRG1
Brahma-related gene 1
- EMT
epithelial-mesenchymal transition
- FBS
fetal bovine serum
- PCR
polymerase chain reaction
- qRT-PCR
Quantitative reverse transcription polymerase chain reaction
- RIPA
Radio-Immunoprecipitation Assay
- BCA
bicinchonininic acid
- SDS
dodecyl sulfate, sodium salt
- TBS-T
triethanolamine buffered saline solution-Tween
- ECL
enhanced chemiluminescence
- PBS
Phosphate Buffered Saline
- EDTA
Ethylene Diamine Tetraacetic Acid
- NP40
NobleRyder 40
- 2 × SDS-PAGE
2 × dodecyl sulfate, sodium salt-Polyacrylamide gel electrophoresis
- DAPI
4′, 6-Diamidino-2-phenylindole
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Shao N, Wang Y, Jiang WY, Qiao D, Zhang SG, Wu Y. et al. Immunotherapy and endothelin receptor antagonists for treatment of castration-resistant prostate cancer. Int J Cancer. 2013;133:1743–50. doi: 10.1002/ijc.28162. [DOI] [PubMed] [Google Scholar]
- 3.Raval AD, Thakker D, Negi H, Vyas A, Kaur H, Salkini MW. Association between statins and clinical outcomes among men with prostate cancer: a systematic review and meta-analysis. Prostate Cancer Prostatic Dis. 2016;19(2):151–62. doi: 10.1038/pcan.2015.58. [DOI] [PubMed] [Google Scholar]
- 4.Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L. et al. HMGB1 in health and disease. Mol Aspects Med. 2014;40:1–116. doi: 10.1016/j.mam.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishiguro H, Nakaigawa N, Miyoshi Y, Fujinami K, Kubota Y, Uemura H. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate. 2005;64:92–100. doi: 10.1002/pros.20219. [DOI] [PubMed] [Google Scholar]
- 6.Huang CYA-Ohoo, Chiang SF, Chen WT, Ke TW, Chen TW, You YS. et al. HMGB1 promotes ERK-mediated mitochondrial Drp1 phosphorylation for chemoresistance through RAGE in colorectal cancer. Cell Death Dis. 2018;9(10):1004. doi: 10.1038/s41419-018-1019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladoire S, Enot D, Senovilla L, Ghiringhelli F, Poirier-Colame V, Chaba K. et al. The presence of LC3B puncta and HMGB1 expression in malignant cells correlate with the immune infiltrate in breast cancer. Autophagy. 2016;12:864–75. doi: 10.1080/15548627.2016.1154244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yusein-Myashkova S, Stoykov I, Gospodinov A, Ugrinova I, Pasheva E. The repair capacity of lung cancer cell lines A549 and H1299 depends on HMGB1 expression level and the p53 status. J Biochem. 2016;160:37–47. doi: 10.1093/jb/mvw012. [DOI] [PubMed] [Google Scholar]
- 9.Morale MG, da Silva Abjaude W, Silva AM, Villa LL, Boccardo E. HPV-transformed cells exhibit altered HMGB1-TLR4/MyD88-SARM1 signaling axis. Sci Rep. 2018;8:3476. doi: 10.1038/s41598-018-21416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, Zhu S, Fan XG, Wang H, Lotze MT, Zeh HJ 3rd. et al. High mobility group protein B1 controls liver cancer initiation through yes-associated protein -dependent aerobic glycolysis. Hepatology. 2018;67:1823–41. doi: 10.1002/hep.29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang R, Zhang Q, Zeh HJ 3rd, Lotze MT, Tang D. HMGB1 in cancer: good, bad, or both? Clin Cancer Res. 2013;19:4046–57. doi: 10.1158/1078-0432.CCR-13-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan C, Wang Y, Qiu MK, Wang SQ, Liu YB, Quan ZW. et al. Knockdown of Hmgb1 Inhibits Cell Proliferation and Induces Apoptosis in Hemangioma Via Downregulation of Akt Pathway. J Biol Regul Homeost Agents. 2017;31:41–9. [PubMed] [Google Scholar]
- 13.Zhang J, Shao S, Han D, Xu Y, Jiao D, Wu J. et al. High mobility group box 1 promotes the epithelial-to-mesenchymal transition in prostate cancer PC3 cells via the RAGE/NF-kappaB signaling pathway. Int J Oncol. 2018;53:659–71. doi: 10.3892/ijo.2018.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao CB, Bao JM, Lu YJ, Zhao T, Zhou XH, Zheng DY. et al. Co-expression of RAGE and HMGB1 is associated with cancer progression and poor patient outcome of prostate cancer. Am J Cancer Res. 2014;4:369–77. [PMC free article] [PubMed] [Google Scholar]
- 15.Romero OA, Setien F, John S, Gimenez-Xavier P, Gomez-Lopez G, Pisano D. et al. The tumour suppressor and chromatin-remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO Mol Med. 2012;4:603–16. doi: 10.1002/emmm.201200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lv D, Wu H, Xing R, Shu F, Lei B, Lei C. et al. HnRNP-L mediates bladder cancer progression by inhibiting apoptotic signaling and enhancing MAPK signaling pathways. Oncotarget. 2017;8(8):13586–99. doi: 10.18632/oncotarget.14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi J, Li F, Yao X, Mou T, Xu Z, Han Z. et al. The HER4-YAP1 axis promotes trastuzumab resistance in HER2-positive gastric cancer by inducing epithelial and mesenchymal transition. Oncogene. 2018;37:3022–38. doi: 10.1038/s41388-018-0204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu ZY, Liu YP, Xie LY, Wang XY, Yang F, Chen SY. et al. AKAP-9 promotes colorectal cancer development by regulating Cdc42 interacting protein 4. Biochim Biophys Acta. 2016;1862:1172–81. doi: 10.1016/j.bbadis.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baidoo EEK, Teixeira Benites V. Mass Spectrometry-Based Microbial Metabolomics: Techniques, Analysis, and Applications. Methods Mol Biol. 2019;1859:11–69. doi: 10.1007/978-1-4939-8757-3_2. [DOI] [PubMed] [Google Scholar]
- 20.Nocito A, Bubendorf L, Tinner EM, Suess K, Wagner U, Forster T. et al. Microarrays of bladder cancer tissue are highly representative of proliferation index and histological grade. J Pathol. 2001;194:349–57. doi: 10.1002/1096-9896(200107)194:3<349::AID-PATH887>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Yi J, Tao L, Huang G, Chu X, Song H. et al. Wnt signaling induces radioresistance through upregulating HMGB1 in esophageal squamous cell carcinoma. Cell Death Dis. 2018;9:433. doi: 10.1038/s41419-018-0466-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muthuswami R, Bailey L, Rakesh R, Imbalzano AN, Nickerson JA, Hockensmith JW. BRG1 is a prognostic indicator and a potential therapeutic target for prostate cancer. J Cell Physiol; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Y, Li N, Dong B, Guo W, Wei H, Chen Q. et al. Chromatin remodeling ATPase BRG1 and PTEN are synthetic lethal in prostate cancer. J Clin Invest. 2019;129:759–73. doi: 10.1172/JCI123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Tillo E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A. et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29:3490–500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 25.Anggayasti WL, Mancera RL, Bottomley S, Helmerhorst E. The self-association of HMGB1 and its possible role in the binding to DNA and cell membrane receptors. FEBS Lett. 2017;591:282–94. doi: 10.1002/1873-3468.12545. [DOI] [PubMed] [Google Scholar]
- 26.Gnanasekar M, Kalyanasundaram R, Zheng G, Chen A, Bosland MC, Kajdacsy-Balla A. HMGB1: A Promising Therapeutic Target for Prostate Cancer. Prostate cancer. 2013;2013:157103. doi: 10.1155/2013/157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu L, Zhang DH, Xu Y, Bai G, Lv Y, Liang J. miR-505 enhances doxorubicin-induced cytotoxicity in hepatocellular carcinoma through repressing the Akt pathway by directly targeting HMGB1. Biomed Pharmacother. 2018;104:613–21. doi: 10.1016/j.biopha.2018.05.087. [DOI] [PubMed] [Google Scholar]
- 28.Hu WW, Liu PY, Yang YC, Chen PC, Su CM, Chao CC. et al. Association of HMGB1 Gene Polymorphisms with Lung Cancer Susceptibility and Clinical Aspects. Int J Med Sci. 2017;14:1197–202. doi: 10.7150/ijms.20933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang BF, Tzeng HE, Chen PC, Wang CQ, Su CM, Wang Y. et al. HMGB1 genetic polymorphisms are biomarkers for the development and progression of breast cancer. Int J Med Sci. 2018;15:580–6. doi: 10.7150/ijms.23462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu K, Huang J, Ni JD, Song DY, Ding ML, Wang JJ. et al. MALAT1 promotes osteosarcoma development by regulation of HMGB1 via miR-142-3p and miR-129-5p. Cell Cycle. 2017;16:578–87. doi: 10.1080/15384101.2017.1288324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He Y, Zha J, Wang Y, Liu W, Yang X, Yu P. Tissue damage-associated "danger signals" influence T-cell responses that promote the progression of preneoplasia to cancer. Cancer Res. 2013;73:629–39. doi: 10.1158/0008-5472.CAN-12-2704. [DOI] [PubMed] [Google Scholar]
- 32.Choupani J, Derakhshan SM, Bayat S, Alivand MR, Khaniani MS. Narrower insight to SIRT1 role in cancer: A potential therapeutic target to control epithelial-mesenchymal transition in cancer cells. J Cell Physiol. 2018;233:4443–57. doi: 10.1002/jcp.26302. [DOI] [PubMed] [Google Scholar]
- 33.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Haswell JR, Roberts CW. Molecular pathways: SWI/SNF (BAF) complexes are frequently mutated in cancer-mechanisms and potential therapeutic insights. Clin Cancer Res. 2014;20:21–7. doi: 10.1158/1078-0432.CCR-13-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laurette P, Strub T, Koludrovic D, Keime C, Le Gras S, Seberg H, Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. Elife; 2015. p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang LY, Zhao JJ, Chen H, Wan LX, Inuzuka H, Guo JP, SCFFBW7-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat Commun; 2018. p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun A, Tawfik O, Gayed B, Thrasher JB, Hoestje S, Li CY. et al. Aberrant expression of SWI/SNF catalytic subunits BRGI/BRM is associated with tumor development and increased invasiveness in prostate cancers. Prostate. 2007;67:203–13. doi: 10.1002/pros.20521. [DOI] [PubMed] [Google Scholar]
- 38.Pyo JS, Son BK, Oh D, Kim EK. BRG1 is correlated with poor prognosis in colorectal cancer. Hum Pathol. 2018;73:66–73. doi: 10.1016/j.humpath.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Cao ZQ, Wang XX, Lu L, Xu JW, Li XB, Zhang GR. et al. beta-Sitosterol and Gemcitabine Exhibit Synergistic Anti-pancreatic Cancer Activity by Modulating Apoptosis and Inhibiting Epithelial-Mesenchymal Transition by Deactivating Akt/GSK-3beta Signaling. Front Pharmacol. 2018;9:1525. doi: 10.3389/fphar.2018.01525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muthuswami R, Bailey L, Rakesh R, Imbalzano AN, Nickerson JA, Hockensmith JW. BRG1 is a prognostic indicator and a potential therapeutic target for prostate cancer. J Cell Physiol; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Wen Q, Luo J, Chu S, Chen L, Xu L. et al. Suppression Of beta-catenin Nuclear Translocation By CGP57380 Decelerates Poor Progression And Potentiates Radiation-Induced Apoptosis in Nasopharyngeal Carcinoma. Theranostics. 2017;7:2134–49. doi: 10.7150/thno.17665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ciaramella V, Sasso FC, Di Liello R, Corte CMD, Barra G, Viscardi G. et al. Activity and molecular targets of pioglitazone via blockade of proliferation, invasiveness and bioenergetics in human NSCLC. J Exp Clin Cancer Res. 2019;38:178. doi: 10.1186/s13046-019-1176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li GY, Wang W, Sun JY, Xin B, Zhang X, Wang T. et al. Long non-coding RNAs AC026904.1 and UCA1: a "one-two punch" for TGF-beta-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics. 2018;8:2846–61. doi: 10.7150/thno.23463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang B, Lv DJ, Wang C, Shu FP, Gong ZC, Xie T. et al. Suppressed epithelial-mesenchymal transition and cancer stem cell properties mediate the anti-cancer effects of ethyl pyruvate via regulation of the AKT/nuclear factor-kappaB pathway in prostate cancer cells. Oncol Lett. 2018;16:2271–8. doi: 10.3892/ol.2018.8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torrealba N, Vera R, Fraile B, Martinez-Onsurbe P, Paniagua R, Royuela M. TGF-beta/PI3K/AKT/mTOR/NF-kB pathway. Clinicopathological features in prostate cancer. Aging Male; 2019. pp. 1–11. [DOI] [PubMed] [Google Scholar]
- 46.Xiong XY, Bai L, Bai SJ, Wang YK, Ji T. Uric acid induced epithelial-mesenchymal transition of renal tubular cells through PI3K/p-Akt signaling pathway. J Cell Physiol; 2019. [DOI] [PubMed] [Google Scholar]
- 47.Wei R, Xiao Y, Song Y, Yuan H, Luo J, Xu W. FAT4 regulates the EMT and autophagy in colorectal cancer cells in part via the PI3K-AKT signaling axis. J Exp Clin Cancer Res. 2019;38:112. doi: 10.1186/s13046-019-1043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abate-Shen C, Banach-Petrosky WA, Sun X, Economides KD, Desai N, Gregg JP. et al. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 2003;63:3886–90. [PubMed] [Google Scholar]
- 49.Chin YR, Toker A. Akt isoform-specific signaling in breast cancer: uncovering an anti-migratory role for palladin. Cell Adh Migr. 2011;5:211–4. doi: 10.4161/cam.5.3.15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan Y, Li X, Duan J, Yuan L, Fan S, Fan J. et al. Enoxaparin sensitizes human non-small-cell lung carcinomas to gefitinib by inhibiting DOCK1 expression, vimentin phosphorylation, and Akt activation. Mol Pharmacol. 2015;87:378–90. doi: 10.1124/mol.114.094425. [DOI] [PubMed] [Google Scholar]
- 51.Zhao GX, Xu YY, Weng SQ, Zhang S, Chen Y, Shen XZ, CAPS1 promotes colorectal cancer metastasis via Snail mediated epithelial mesenchymal transformation. Oncogene; 2019. [DOI] [PubMed] [Google Scholar]
- 52.Putzke AP, Ventura AP, Bailey AM, Akture C, Opoku-Ansah J, Celiktas M. et al. Metastatic progression of prostate cancer and e-cadherin regulation by zeb1 and SRC family kinases. Am J Pathol. 2011;179:400–10. doi: 10.1016/j.ajpath.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang HY, Chen SY, Wu CH, Lu CC, Yen GC. Glycyrrhizin Attenuates the Process of Epithelial-to-Mesenchymal Transition by Modulating HMGB1 Initiated Novel Signaling Pathway in Prostate Cancer Cells. J Agric Food Chem. 2019;67:3323–32. doi: 10.1021/acs.jafc.9b00251. [DOI] [PubMed] [Google Scholar]
- 54.Lo UG, Lee CF, Lee MS, Hsieh JT. The Role and Mechanism of Epithelial-to-Mesenchymal Transition in Prostate Cancer Progression. Int J Mol Sci; 2017. p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and table legends.
Supplementary table 1.
Supplementary table 2.