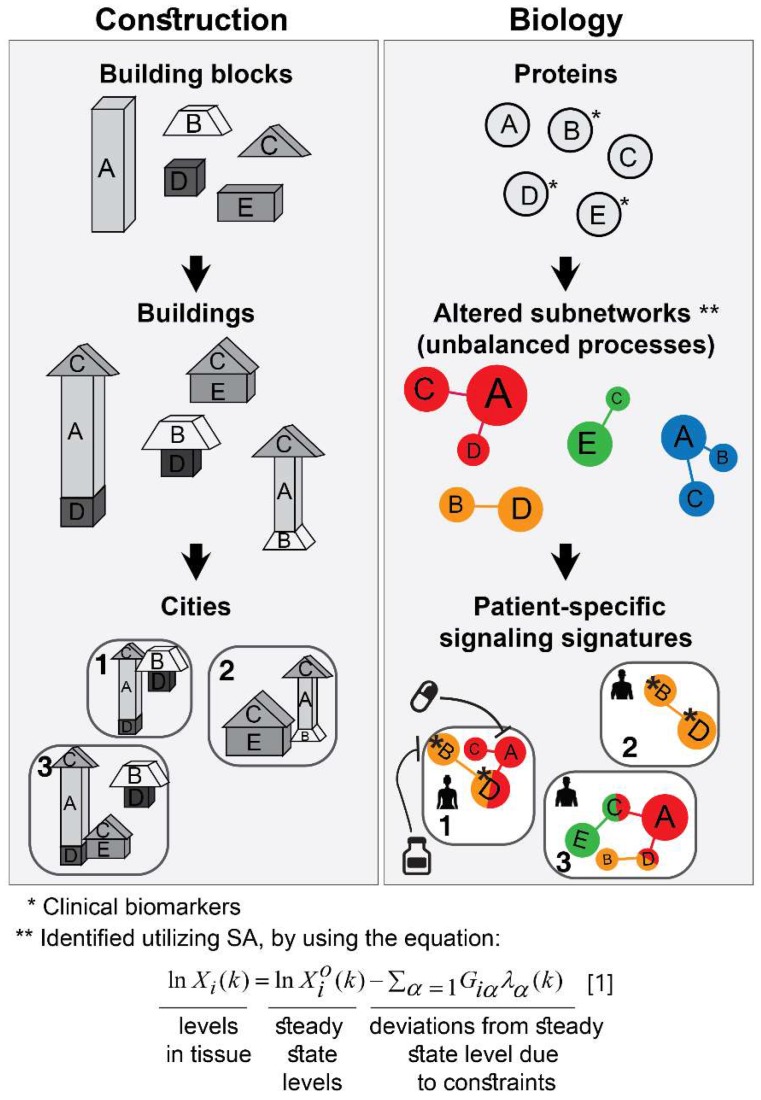
Figure 1.
Overview of our approach. In construction, a collection of building blocks can be used to assemble different types of buildings (left). Specific subsets of buildings can exist in different cities (left). Similarly, in biology, altered protein subnetworks, or unbalanced molecular processes (buildings) are made up of a collection of proteins (blocks) (right). Each biological sample (city) can contain a unique subset of unbalanced processes that maintain its current state (right). This state is different from the steady state, since proteins participate in different unbalanced processes reflecting the genomic and environmental constraints on the tumor. Each protein can be influenced by several processes simultaneously (e.g. protein D in patient 1, right panel). In the figure, the proteins are represented by circles, such that the diameter of the circle denotes its relative weight (=importance), G, in the process (analogous to size of blocks in each building), e.g. protein A is important in the red process, less important in the blue process, and does not participate in the green and orange processes (right). The size of the entire process in each sample (or size of building in each city) denotes its sample-specific amplitude (λ). For example, the red unbalanced process is relatively important in patient 3 and insignificant in patient 2 (right). The common procedure today is to test biomarkers in cancer tissue (proteins B, D, E here). If B regulates D, the conventional approach would suggest that inhibition of B should inhibit D. While this inhibition may be effective in patient 2, it is expected to be inefficient in patient 1, because protein D participates in red and orange processes simultaneously in this patient. Refer to Supplementary Information for complete details regarding the mathematical algorithm.