Abstract
Background
This meta-analysis aimed to clarify the diagnostic role of plasma methylated SEPT9 (mSEPT9) in colorectal cancer (CRC) and examined its association with CRC.
Material/Methods
A systematic review was conducted prior to July 2018. Summary sensitivity, specificity, and positive and negative likelihood ratio (PLR/NLR) were calculated for the diagnostic value of mSEPT9 for CRC. The areas under the receiver operating curves (AUCs) were used to summarize the overall test performance.
Results
Twenty-two studies with 2271 CRC patients were enrolled. The summary sensitivity, specificity, PLR, NLR, DOR, and AUC of the overall analysis of mSEPT9 were 0.69, 0.92, 8.1, 0.34, 24, and 0.89, respectively. Subgroup and meta-regression analyses demonstrated that the diagnostic value was higher for the Epi proColon 2.0 assay, Asian ethnicity, and mSEPT9 test combined with fecal occult blood test (FOBT) or fecal immunochemical test (FIT) than for other test methods, white ethnicity, and mSEPT9 test alone. The rate of mSEPT9 positivity was higher in advanced CRC cases compared with early-stage CRC cases, and was higher in CRC cases than in adenoma cases. No significant difference in mSEPT9 positivity rate was found between left- and right-sided CRC.
Conclusions
Plasma mSEPT9 has a high diagnostic value for CRC, especially on the newly developed Epi proColon test 2.0 method. The diagnostic sensitivity is superior among Asians compared to whites, and the combination of mSEPT9 and FOBT/FIT has a better performance than mSEPT9 alone. Finally, the expression of mSEPT9 is associated with CRC stage but not with location.
MeSH Keywords: Colorectal Neoplasms, Meta-Analysis as Topic, O(6)-Methylguanine-DNA Methyltransferase
Background
Colorectal cancer (CRC) is the third most frequent malignancy worldwide, and its prognosis is significantly dependent on the staging at diagnosis [1]. Therefore, early diagnosis of CRC is crucial to improve patients’ outcomes. Currently, several noninvasive methods are available to screen for CRC, the most common of which are fecal occult blood test (FOBT) and fecal immunochemical test (FIT). However, the inadequate sensitivity and specificity of these methods limit their application in the detection of CRC [2]. Although colonoscopy is the criterion standard for CRC screening, with a high diagnostic value, it is an invasive method that requires bowel preparation and experienced operators [3]. To date, several other biomarkers have also used to screen for CRC, including CPNE3 [4], CNPY2 isoform 2 [5], and SATB2 [6]; however, few have sufficiently satisfactory performance for clinical use. Therefore, the search for more patient-friendly and less-invasive approaches with high sensitivity and specificity is imperative to improve CRC patients’ outcome.
Septin 9 is a member of the conserved Septin family of GTP-binding proteins [7]. Aberrant methylation of v2 transcript of SEPT9 gene has been observed in almost 100% of CRC tissues, leading to significantly decreased SEPT9 expression in CRC [8,9]. Recently, emerging evidence has demonstrated cell-free circulating methylated SEPT9 (mSEPT9) to be a promising biomarker for CRC detection. However, the reported sensitivity and specificity values of plasma mSEPT9 have been highly variable across studies, with the sensitivity ranging from 50.9% to 93.1%, and specificity from 62.2% to 93.8% [10–13]. Previously, Yan et al. [14] performed a meta-analysis to assess the diagnostic value of mSEPT9 for CRC; however, their study had several limitations, including a limited number of studies recruited, inclusion of only English articles, and the fact that they did not analyze the diagnostic value for different stages of CRC. In the present study, we conducted a comprehensive meta-analysis using all eligible published articles, and analyzed the diagnostic value of mSEPT9 for CRC, as well as its association with CRC.
Material and Methods
Search strategy
This meta-analysis was performed in accordance with the guidelines of the Preferred Reporting Items for Systematic Review and Meta-Analyses statement (PRISMA). Using the electronic databases PubMed, Cochrane Library, Web of Science, Google Scholar, and Chinese National Knowledge Infrastructure (CNKI), a comprehensive search was performed to identify eligible articles that were published before July 2018. The following search terms were used: “colorectal cancer” or “CRC,” “methylated SEPT9” or “mSEPT9,” and “diagnosis” or “diagnose.” Included articles were limited to human studies, but not limited by language. Relevant articles were also searched using the related articles function in PubMed. In addition, references within the identified articles were also searched manually. The study was approved by the Review Boards of the First Affiliated Hospital of Guangxi Medical University.
Inclusion and exclusion criteria
The inclusion criteria were as follows: (1) CRC was pathologically diagnosed and none of the patients received chemotherapy, radiotherapy, or surgical intervention before colonoscopy examination; (2) studies that analyzed the diagnostic value of plasma mSEPT9 for CRC using a clear test method; and (3) the sensitivity and specificity data of mSEPT9 for CRC were provided. The exclusion criteria were as follows: review articles, letters, case reports and studies on cell lines, non-plasma/serum samples, unknown detection methods, and animal experiments. When the same patient cohort was reported in several articles, the most recent study was selected.
Data extraction and quality assessment
Data extraction included the name of the first author, year of publication, study region (country), mean age of patients, cut-off of plasma mSEPT9, number of CRC and controls, sensitivity and specificity value of mSEPT9, and test method for plasma mSEPT9. The Quality Assessment of Diagnostic Accuracy Studies (QUADAS), an evidence-based quality assessment tool developed for systematic reviews of studies of diagnostic accuracy (maximum score of 14, in which a study with a score of over 9 is viewed as high quality), was applied to assess the quality of individual studies [15]. Two reviewers independently judged the eligibility of the studies. Disagreements between reviewers were resolved by consensus.
Statistical analysis
Summary measures, including sensitivity and specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), and diagnostic odds ratio (DOR) with the corresponding 95% CIs, were calculated for each study. A publication bias of diagnostic studies was assessed using Deek’s test [16]. Pooled odds ratios (ORs) with the corresponding 95% CIs were used to estimate the association between mSEPT9 positivity rate and different stages or sites of CRC, while the publication bias was estimated using Egger’s and Begg’s tests.
The Cochran Q test and inconsistency index (I2) were used to assess the threshold effect as an important component of the source of variation of the studies. An I2 <25% indicates mild heterogeneity, while I2 values from 25% to 50% indicate moderate heterogeneity, and I2 >50% indicates significant heterogeneity. A fixed-effects model (Mantel-Haenszel method) was used when there was mild heterogeneity; otherwise, a random-effects model (DerSimonian and Laird) was used. Meta-regression analyses were employed to compare the diagnostic value among different studies. All statistical tests in this meta-analysis were performed using the Stata 11.2 software (Stata Corp, College Station, TX) with 2-tailed p-values. A p-value <0.05 was considered statistically significant.
Results
Study selection process
The primary retrieval based on the search terms obtained 136 articles. By screening the titles or abstracts of the articles, 87 articles were excluded because they were either reviews, animal studies, case reports, or irrelevant to the current meta-analysis. Next, the 49 remaining articles were evaluated further by screening the full text. Then, 27 articles were excluded for the following reasons: 20 articles did not provide the sensitivity and specificity data for mSEPT9, and 7 studies used non-serum or non-plasma specimens. Finally, 22 studies [10–13,17–34] with 2271 subjects (1801 CRC patients and 470 controls) were included in this meta-analysis. A flow chart of the article selection process is shown in Figure 1.
Figure 1.

Flow diagram of the study selection process.
Characteristics of included studies
The characteristics of included studies are shown in Table 1. All included CRC patients were confirmed by pathological diagnosis. Tumor stages were defined according to the TNM staging system. Most of the studies did not provided the cut-off value. Testing methods for mSEPT9 included the Epi proColon test 1.0, Epi proColon test 2.0, and Reverse transcription-polymerase chain reaction (RT-PCR) assay. All studies were prospective in nature. Five studies provided data for FIT or FOBT in the diagnosis of CRC. Twelve studies analyzed the mSEPT9 positivity rate in different stages of CRC, and 6 studies compared the mSEPT9 positivity rate for left- and right-sided CRC. The QUADAS scores ranged between 12 and 14 across the studies, indicating that they were of high quality.
Table 1.
Characteristic of included studies.
| First author | Country/year | Mean age | Test method | Cut-off | Index test | Sensitivity | Specificity | QUADAS scores |
|---|---|---|---|---|---|---|---|---|
| Lofton-Day C | Germany/2008 | 56 | RT-PCR | NA | Septin9 | 69% | 86% | 11 |
| Grützmann R | USA/2008 | 59 | RT-PCR | 2/3rule | Septin9 | 58% | 90% | 11 |
| deVos T | Germany/2009 | 62.5 | Epi proColon test 1.0 | 2/3 rule | Septin9 | 57% | 98% | 12 |
| He Q | China/2010 | 58 | RT-PCR | NA | Septin9 | 75% | 96.47% | 10 |
| Tänzer M | USA/2010 | 67 | RT-PCR | 2/3 rule | Septin9 | 73% | 91% | 11 |
| Herbst A | Germany/2011 | 63 | RT-PCR | NA | Septin9 | 46.6% | 81.3% | 13 |
| Warren JD | USA/2011 | 62 | Epi proColon test 1.0 | NA | Septin9 | 90% | 88% | 13 |
| Tóth K | Germany/2012 | 67.8 | RT-PCR | 2/3 rule | Septin9 | 79.3% | 99% | 12 |
| FOBT | 68.2% | 29.4% | ||||||
| Lee HS | Korea/2013 | 65.76 | RT-PCR | NA | Septin9 | 36.6% | 90.6% | 12 |
| Church TR | USA/2014 | 60 | Epi proColon test 1.0 | 1/3 rule | Septin9 | 48.2% | 91.5% | 13 |
| Johnson DA | Germany/2014 | 66 | Epi proColon test 1.0 | NA | Septin9 | 73.3% | 81.5% | 13 |
| Kang Q | China/2014 | 61.2 | Epi proColon test 2.0 | NA | Septin9 | 75% | 98.1% | 13 |
| FIT | 53.8% | 93.8% | ||||||
| Potter NT | USA/2014 | 60 | Epi proColon test 1.0 | NA | Septin9 | 68% | 78% | 12 |
| Tóth K | Germany/2014 | 67.8 | Epi proColon test 2.0 | NA | Septin9 | 82.8% | 91.7% | 12 |
| Jin P | China/2015 | 60.9 | Epi proColon test 2.0 | 2/3 rule | Septin9 | 74.8% | 87.4% | 12 |
| FIT | 58% | 82.4% | ||||||
| Li SJ | China/2015 | 56.4 | Epi proColon test 2.0 | NA | Septin9 | 72.5% | 91.1% | 13 |
| Ding QQ | China/2015 | 71 | Epi proColon test 2.0 | NA | Septin9 | 73.2% | 95.6% | 13 |
| Ørntoft MB | Denmark/2015 | 59 | Epi proColon test 1.0 | 2/3 rule | Septin9 | 59% | 82% | 13 |
| Wu D | China/2016 | 52.5 | Epi proColon test 2.0 | NA | Septin9 | 76.6% | 95.9% | 12 |
| Fu B | China/2018 | 60 | Epi proColon test 2.0 | 2/3 rule | Septin9 | 61.22% | 93.7% | 13 |
| Li Y | China/2018 | 62 | Epi proColon test 2.0 | NA | Septin9 | 81.94% | 83.61% | 12 |
| FOBT | 52.78% | 81.97% | ||||||
| Xie L | China/2018 | 66.07 | RT-PCR | NA | Septin9 | 61.8% | 89.6% | 13 |
| FOBT | 61.4% | 70.3% |
NA – not available; QUADAS – Quality Assessment of Diagnostic Accuracy Studies; FIT – fecal immunochemical test; FOBT – fecal occult blood test.
Overall result for mSEPT9 in the diagnosis of CRC
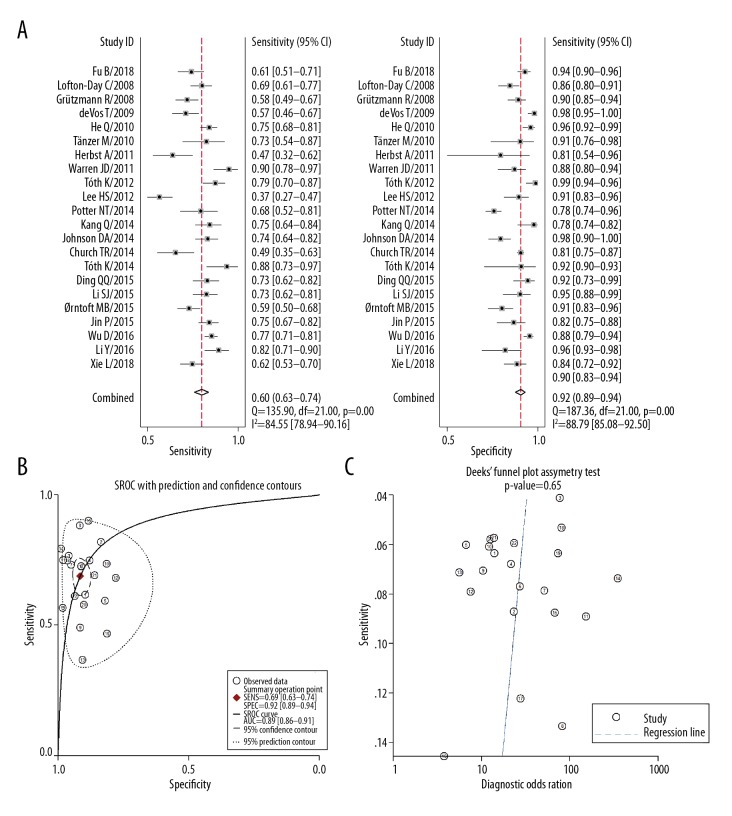
Twenty-two studies analyzed the diagnostic value of mSEPT9 for CRC. The pooled results demonstrated that the summary sensitivity, specificity, PLR, NLR, and DOR were 0.69 (range, 0.63–0.74), 0.92 (range, 0.89–0.94), 8.1 (range, 5.9–11.2), 0.34 (range, 0.28–0.40), and 24 (range, 16–37), respectively. There was significant heterogeneity in the summary sensitivity (I2=84.5%, p<0.001) and specificity (I2=88.8%, p<0.001) (Figure 2A). The summary area under the receiver operating curve was 0.89 (range, 0.86–0.91) (Figure 2B). No published bias was found across the studies using Deek’s test (p=0.653) (Figure 2C).
Figure 2.
Diagnostic value of plasma mSEPT9 in diagnosis for CRC. (A) Summary sensitivity and specificity plotted on forest graphs for plasma mSEPT9 in diagnosis for CRC. (B) SROC curve graph for plasma mSEPT9 in diagnosis of CRC. (C) Deek’s funnel plot asymmetry test of plasma mSEPT9 in diagnosis for CRC.
Subgroup analysis of different parameters in diagnosis of CRC
Using the meta-regression method, we found that mSEPT9 has a higher diagnostic sensitivity and specificity than FOBT, with p-values of 0.54 and 0.51, respectively. This suggests that mSEPT9 has a higher diagnostic value for CRC compared with FOBT. We also compared the diagnostic value of mSEPT9 for CRC located in different regions. We divided patients into Asian and white ethnicities, and found that the diagnostic sensitivity increased in patients of Asian ethnicity compared to those of white ethnicity (p=0.04), indicating that the diagnostic value of mSEPT9 varies by ethnicity.
However, we failed to demonstrate that the different controls (i.e., none-CRC diseases vs. healthy controls) of included studies affected the diagnostic value of mSEPT9 in CRC. Three studies presented the results of mSEPT9 or mSEPT9 combined with FOBT/FIT test for the diagnosis of CRC. When we compared their diagnostic values, we found that the sensitivity of mSEPT9 combined with FOBT was higher than that of mSEPT9 alone, although there was no significant difference (Table 2).
Table 2.
Different methods and regions in diagnosis of CRC.
| N | Sensitivity | P1 | Specificity | P2 | AUC | |
|---|---|---|---|---|---|---|
| Septin9 test | 22 | 0.69 (0.64–0.74) | 0.54 | 0.92 (0.89–0.94) | 0.01 | 0.88 |
| FIT/FOBT test | 5 | 0.59 (0.48–0.70) | 0.88 (0.81–0.96) | 0.64 | ||
| Asian ethnicity | 12 | 0.70 (0.63–0.76) | 0.04 | 0.91 (0.87–0.95) | 0.10 | 0.93 |
| White ethnicity | 15 | 0.65 (0.59–0.71) | 0.91 (0.88–0.95) | 0.87 | ||
| None-CRC diseases | 21 | 0.72 (0.58–0.83) | 0.72 | 0.90 (0.81–0.95) | 0.69 | 0.88 |
| Healthy controls | 6 | 0.68 (0.55–0.79) | 0.92 (0.87–0.95) | 0.91 | ||
| Septin9 test alone | 3 | 0.74 (0.675–0.789 | 0.01 | 0.79 (0.741–0.839) | 0.24 | 0.92 |
| Septin9+FOBT/FIT | 3 | 0.89 (0.839–0.923 | 0.75 (0.691–0.796) | 0.98 |
N – number; P1 – p-value of sensitivity comparison; P2 – p-value of specificity comparison; AUC – area under the curve.
Comparison of diagnostic value of different methods for CRC
In this meta-analysis, 3 methods were used to determine the diagnostic value for CRC: the Epi proColon test 1.0, the Epi proColon test 2.0, and RT-PCR. As shown in Table 3, the diagnostic sensitivity of the Epi proColon test 2.0 was higher than that of the Epi proColon test 1.0 and RT-PCR. The diagnostic sensitivity of the Epi proColon test 1.0 was also higher than that of RT-PCR method, suggesting that these newly developed methods have a higher diagnostic value.
Table 3.
Comparison of diagnostic value of different methods on CRC.
| N | Sensitivity | P1 | Specificity | P2 | AUC | |
|---|---|---|---|---|---|---|
| Epi proColon test 1.0 | 8 | 0.68 (0.59–0.76) | <0.01 | 0.91 (0.86–0.95) | 0.15 | 0.89 |
| Epi proColon test 2.0 | 10 | 0.74 (0.66–0.82) | 0.91 (0.87–0.95) | 0.92 | ||
| Epi proColon test 1.0 | 8 | 0.73 (0.68–0.77) | 0.04 | 0.91 (0.86–0.96) | 0.10 | 0.89 |
| RT-PCR | 9 | 0.60 (0.56–0.65) | 0.91 (0.86–0.95) | 0.66 | ||
| Epi proColon test 2.0 | 10 | 0.74 (0.66–0.82) | 0.02 | 0.92 (0.87–0.96) | 0.43 | 0.92 |
| RT-PCR | 9 | 0.60 (0.52–0.68) | 0.91 (0.86–0.96) | 0.66 |
N – number; P1 – p-value of sensitivity comparison; P2 – p-value of specificity comparison; AUC – area under the curve.
Comparison of the mSEPT9 positivity rate in different stages of CRC
Twelve studies provided data on mSEPT9 positivity rates in different stages of CRC. We divided the CRC patients into early-stage (stage I+II) and advanced-stage (stage III+IV), and compared the mSEPT9 positivity rates of different stages. As illustrated in Figure 3A, the mSEPT9 positivity rate was much higher in advanced-stage cases of CRC than in early-stage cases (OR=0.79, 95% CI=0.66–0.94, p=0.010). There was no significant heterogeneity among the studies (I2=0, p=0.999), and no published bias was found by Egger’s test (p=0.599) and Begg’s test (p=0.537) (Figure 3B).
Figure 3.
(A) Meta-analysis of plasma mSEPT9 positive rate in early or advanced stage of CRC. (B) Funnel plots of plasma mSEPT9-positive rate in early- or advanced-stage CRC.
Comparison of the mSEPT9 positivity rate in different sides of CRC
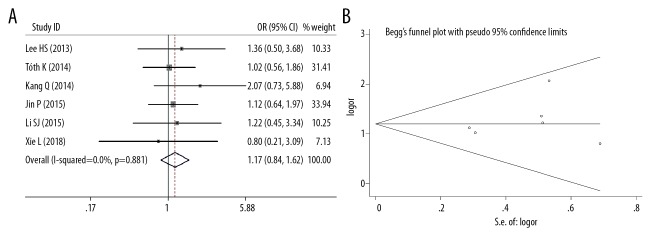
Six studies analyzed the mSEPT9 positivity rate in both left- and right-sided CRC. We found no significant difference in mSEPT9 positivity rates between left- and right-sided CRC (OR=1.17, 95% CI=0.84–1.62, p=0.352), and no significant heterogeneity among the studies (I2=0, p=0.881) (Figure 4A). Egger’s (p=0.470) and Begg’s tests (p=0.707) indicated limited publication bias across the studies (Figure 4B).
Figure 4.
(A) Meta-analysis of plasma mSEPT9-positive rate in left side or right side of CRC. (B) Funnel plots of plasma mSEPT9 positive rate in left side or right side of CRC.
Comparison of the mSEPT9 positivity rate in CRC and adenoma cases
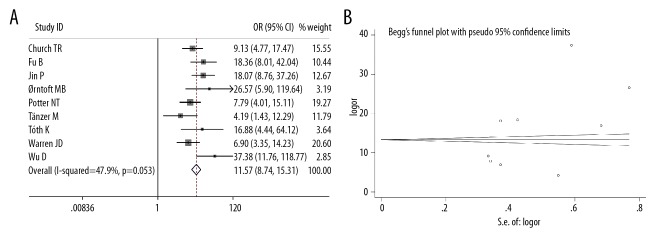
Nine studies included data of mSEPT9 positivity rates in CRC and adenoma cases. We found that the mSEPT9-positivity rate was significantly higher in CRC cases than in adenoma cases (OR=11.57, 95% CI=8.74–15.31, p<0.001); however, there was moderate heterogeneity among the studies (I2=0, p=47.9%) (Figure 5A). Egger’s (p=0.124) and Begg’s tests (p=0.251) indicated limited publication bias across the studies (Figure 5B).
Figure 5.
(A) Meta-analysis of plasma mSEPT9-positive rate in CRC and adenoma cases. (B) Funnel plots of plasma mSEPT9-positive rate in CRC and adenoma cases.
Discussion
In this meta-analysis, by pooling the data of a large number of studies, we found that plasma mSEPT9 has a higher specificity and a moderate sensitivity in the diagnosis of CRC. Furthermore, the diagnostic value was much higher than that of FOBT/FIT. After dividing the studies into subgroups, we observed that the more recently developed method, the Epi proColon 2.0 assay, has a much higher diagnostic value than the other methods. We also observed that the diagnostic value of mSEPT9 was higher in Asians than in whites, and that mSEPT9 combined with FOBT/FIT had higher diagnostic value than did the mSEPT9 test alone. Moreover, we estimated the association between mSEPT9 and stage as well as location of CRC, and found that the mSEPT9 positivity rate was remarkably increased in advanced stages compared with early stages of CRC, and was much higher in CRC cases compared with adenoma cases. However, no significant difference was observed for mSEPT9 positivity rate between left- and right-sided CRC.
A growing number of studies indicate that alteration of DNA methylation is one of the most common aberrant epigenetic modifications, which play essential roles in CRC initiation and progression [35,36]. In recent years, mSEPT9 has been used for early-stage CRC screening; however, its diagnostic value was variable among studies. In the present study, the sensitivity and specificity of mSEPT9 was 0.69 and 0.92, respectively, indicating that the sensitivity was moderate; however, after dividing the studies based on the test method and region, we found that the sensitivity increased. Currently, the plasma-based Epi proColon test is used to detect the expression of mSEPT9 in CRC, and the Epi proColon 2.0 assay was approved by the US FDA as the first blood-based CRC screening test. The sensitivity and specificity of Epi proColon 2.0 assay is much higher than the Epi proColon 1.0 assay and traditional RT-PCR method [37], perhaps because each assay used a different algorithm. It has been reported that the 2/3 algorithm provided the best balance between sensitivity and specificity, as it detected 3/4 of cancer patients with a less than 3% false-positive rate [38]. Of note, 2 studies [12, 23] using RT-PCR to detect mSEPT9 reported sensitivities of 36.6% and 46.6%, which were much lower than both the Epi proColon 1.0 and 2.0 assays. This suggests that RT-PCR might not be suitable as a screening tool for CRC.
The association between mSEPT9 and CRC clinicopathological parameters was another issue that we investigated; by pooling the available data, we found that the mSEPT9-positivity rate increased in later stages of CRC, suggesting that increased mSEPT9 positivity may represent high malignancy and possibly be associated with poor prognosis. As evidence has demonstrated that left- and right-sided CRC have different epidemiology, clinical presentation, pathology, genetic mutations, and survival rate [39], we further combined the available data; however, we were unable to show any difference in mSEPT9 positivity rates between the left- and right-sided CRC, indicating that tumor location does not influence the expression of plasma mSEPT9.
Previously, Yan et al. [14], Li et al. [40], and Nian et al. [41] reported the diagnostic value of mSEPT9 in colorectal cancer using meta-analyses, which included 14, 25, and 12 studies, respectively. Compared with previous meta-analyses, our study has several advantages. First, this meta-analysis included more studies, and we included English and Chinese studies, thus enlarging the sample size and reducing selection bias. Second, this meta-analysis compared more indexes compared with previous studies, including the comparison of Septin 9 test alone and a combination of Septin 9 test with other methods, Epi proColon test 1.0 and Epi proColon test 2.0, and Asians and whites. Third, we also examined the association between mSEPT9 and different stages and location of CRC by combing the available data. Our analysis verified that the mSEPT9-positivity rate was much higher in advanced-stage cases compared to early-stage cases of CRC, which was not previously reported. Fourth, the minimal heterogeneity across the studies in our analysis of the association between mSEPT9 and different stages and location of CRC enhances the reliability of the results. Fifth, the high quality of included studies and minimal publication bias indicate the robustness of the results. Taken together, this meta-analysis provided more information than previous meta-analyses, with more reliable results. However, further studies are required to verify that mSEPT9 could serve as a reliable biomarker to diagnose CRC.
The present study has several limitations. First, some included studies did not provide the cut-off value when analyzing the diagnostic value; thus, we could not exclude that different cut-off values could significantly influence the diagnostic value. Second, the controls of some included studies were variable, with some using non-CRC patients, some using healthy persons, and some using both. Although we have divided the included studies into non-CRC diseases and healthy controls, we could not further divide them into specific colorectal diseases; this might decrease the robustness of the resulting diagnostic value. Third, there was significant heterogeneity across the studies in terms of sensitivity and specificity, which might also undermine the reliability of our results. Fourth, this meta-analysis only selected English and Chinese articles; the exclusion of other languages might also induce selected bias. Fifth, the included studies did not account for the effects of risk factors for CRC, such as age, sex, smoking, diet, and genetic factors on their findings, which may undermine the robustness of their results. Thus, these results require cautious interpretation.
Conclusions
This meta-analysis demonstrated that plasma mSEPT9 has a high diagnostic value for CRC, especially with newer diagnostic methods. The diagnostic sensitivity of mSEPT9 is superior in Asians than in whites, while the combination of mSEPT9 with FOBT/FIT demonstrated better performance than mSEPT9 alone. The expression of mSEPT9 was also found to be associated with CRC stage, but not with the location. Further studies are required to verify that mSEPT9 could serve as a reliable biomarker to diagnose CRC.
Abbreviations
- CRC
colorectal cancer
- mSEPT9
methylated SEPT9
- PLR
likelihood ratio
- NLR
negative likelihood ratio
- DOR
diagnostic odds ratio
- OR
odds ratio
- FOBT
fecal occult blood test
- FIT
fecal immunochemical test
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. Cancer J Clin. 2017;67:177–93. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 2.Morikawa T, Kato J, Yamaji Y, et al. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129:422–28. doi: 10.1016/j.gastro.2005.05.056. [DOI] [PubMed] [Google Scholar]
- 3.Uraoka T, Hosoe N, Yahagi N. Colonoscopy: Is it as effective as an advanced diagnostic tool for colorectal cancer screening? Expert Rev Gastroenterol Hepatol. 2015;9:129–32. doi: 10.1586/17474124.2015.960397. [DOI] [PubMed] [Google Scholar]
- 4.Sun B, Li Y, Zhou Y, et al. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol. 2019;234(2):1416–25. doi: 10.1002/jcp.26936. [DOI] [PubMed] [Google Scholar]
- 5.Peng J, Ou Q, Pan Z, et al. Serum CNPY2 isoform 2 represents a novel biomarker for early detection of colorectal cancer. Aging (Albany NY) 2018;10(8):1921–31. doi: 10.18632/aging.101512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabir PD, Svanholm H, Christiansen JJ. SATB2 is a supplementary immunohistochemical marker to CDX2 in the diagnosis of colorectal carcinoma metastasis in an unknown primary. APMIS. 2018;126:494–500. doi: 10.1111/apm.12854. [DOI] [PubMed] [Google Scholar]
- 7.Hall PA, Russell SE. The pathobiology of the septin gene family. J Pathol. 2004;204:489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 8.Toth K, Galamb O, Spisak S, et al. The influence of methylated septin 9 gene on RNA and protein level in colorectal cancer. Pathol Oncol Res. 2011;17:503–9. doi: 10.1007/s12253-010-9338-7. [DOI] [PubMed] [Google Scholar]
- 9.Wasserkort R, Kalmar A, Valcz G, et al. Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer. 2013;13:398. doi: 10.1186/1471-2407-13-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.deVos T, Tetzner R, Model F, et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55:1337–46. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 11.Fu B, Yan P, Zhang S, et al. Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Markers. 2018;2018 doi: 10.1155/2018/6437104. 6437104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herbst A, Rahmig K, Stieber P, et al. Methylation of NEUROG1 in serum is a sensitive marker for the detection of early colorectal cancer. Am J Gastroenterol. 2011;106:1110–18. doi: 10.1038/ajg.2011.6. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DA, Barclay RL, Mergener K, et al. Plasma Septin9 versus fecal immunochemical testing for colorectal cancer screening: A prospective multicenter study. PLoS One. 2014;9:e98238. doi: 10.1371/journal.pone.0098238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan S, Liu Z, Yu S, Bao Y. Diagnostic value of methylated Septin9 for colorectal cancer screening: A meta-analysis. Med Sci Monit. 2016;22:3409–18. doi: 10.12659/MSM.900590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whiting P, Rutjes AW, Reitsma JB, et al. The development of QUADAS: A tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol. 2003;3:25. doi: 10.1186/1471-2288-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–93. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Church TR, Wandell M, Lofton-Day C, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–25. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding QQ, Zhang H, Xu HC, et al. [Value of methylated Septin 9 detection in screening colorectal cancer in elderly people]. Chinese Journal of Geriatrics. 2015;34:1348–50. [in Chinese] [Google Scholar]
- 19.Grutzmann R, Molnar B, Pilarsky C, et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3:e3759. doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Q, Chen HY, Bai EQ, et al. Development of a multiplex MethyLight assay for the detection of multigene methylation in human colorectal cancer. Cancer Genet Cytogenet. 2010;202:1–10. doi: 10.1016/j.cancergencyto.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 21.Jin P, Kang Q, Wang X, et al. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol. 2015;30:830–33. doi: 10.1111/jgh.12855. [DOI] [PubMed] [Google Scholar]
- 22.Kang Q, Jin P, Yang L, et al. [Significance of Septin9 gene methylation detection of plasma circulation DNA in colorectal cancer screening]. Zhonghua Yi Xue Za Zhi. 2014;94:3839–41. [in Chinese] [PubMed] [Google Scholar]
- 23.Lee HS, Hwang SM, Kim TS, et al. Circulating methylated septin 9 nucleic Acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl Oncol. 2013;6:290–96. doi: 10.1593/tlo.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li SJ, Liu YG, Wang J, et al. [Peripheral blood Septin9 gene methylation detection in colorectal cancer screening]. Chinese Journal of General Surgery. 2015;24:1756. [in Chinese] [Google Scholar]
- 25.Li Y, Hui LY, Wang YW. [Diagnostic evaluation of Septin9 methylated DNA detection in the screening of colorectal cancer]. Clinical Research and Practice. 2018;3:1. [in Chinese] [Google Scholar]
- 26.Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 2008;54:414–23. doi: 10.1373/clinchem.2007.095992. [DOI] [PubMed] [Google Scholar]
- 27.Orntoft MB, Nielsen HJ, Orntoft TF, Andersen CL. Performance of the colorectal cancer screening marker Sept9 is influenced by age, diabetes and arthritis: A nested case-control study. BMC Cancer. 2015;15:819. doi: 10.1186/s12885-015-1832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potter NT, Hurban P, White MN, et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 2014;60:1183–91. doi: 10.1373/clinchem.2013.221044. [DOI] [PubMed] [Google Scholar]
- 29.Tanzer M, Balluff B, Distler J, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010;5:e9061. doi: 10.1371/journal.pone.0009061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toth K, Sipos F, Kalmar A, et al. Detection of methylated SEPT9 in plasma is a reliable screening method for both left- and right-sided colon cancers. PLoS One. 2012;7:e46000. doi: 10.1371/journal.pone.0046000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toth K, Wasserkort R, Sipos F, et al. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PLoS One. 2014;9:e115415. doi: 10.1371/journal.pone.0115415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9:133. doi: 10.1186/1741-7015-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu D, Zhou G, Jin P, et al. Detection of colorectal cancer using a simplified SEPT9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn. 2016;18:535–45. doi: 10.1016/j.jmoldx.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 34.Xie L, Jiang X, Li Q, et al. Diagnostic value of methylated Septin9 for colorectal cancer detection. Front Oncol. 2018;8:247. doi: 10.3389/fonc.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behrouz Sharif S, Hashemzadeh S, Mousavi Ardehaie R, et al. Detection of aberrant methylated SEPT9 and NTRK3 genes in sporadic colorectal cancer patients as a potential diagnostic biomarker. Oncol Lett. 2016;12:5335–43. doi: 10.3892/ol.2016.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toiyama Y, Okugawa Y, Goel A. DNA methylation and microRNA biomarkers for noninvasive detection of gastric and colorectal cancer. Biochem Biophys Res Commun. 2014;455:43–57. doi: 10.1016/j.bbrc.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.deVos T, Molnar B. Screening for colorectal cancer based on the promoter methylation status of the Septin 9 gene in plasma cell-free DNA. J Clin Epigenet. 2017;3:1. [Google Scholar]
- 38.Song L, Li Y, Jia J, et al. Algorithm optimization in methylation detection with multiple RT-qPCR. PLoS One. 2016;11:e0163333. doi: 10.1371/journal.pone.0163333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stintzing S, Tejpar S, Gibbs P, et al. Understanding the role of primary tumour localisation in colorectal cancer treatment and outcomes. Eur J Cancer. 2017;84:69–80. doi: 10.1016/j.ejca.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, Gan A, Chen X, et al. Diagnostic performance of DNA hypermethylation markers in peripheral blood for the detection of colorectal cancer: A meta-analysis and systematic review. PLoS One. 2016;11:e0155095. doi: 10.1371/journal.pone.0155095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nian J, Sun X, Ming S, et al. Diagnostic accuracy of methylated SEPT9 for blood-based colorectal cancer detection: A systematic review and meta-analysis. Clin Transl Gastroenterol. 2017;8:e216. doi: 10.1038/ctg.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]