Abstract
Background
Host-parasite protein interactions (HPPI) are those interactions occurring between a parasite and its host. Host-parasite protein interaction enhances the understanding of how parasite can infect its host. The interaction plays an important role in initiating infections, although it is not all host-parasite interactions that result in infection. Identifying the protein-protein interactions (PPIs) that allow a parasite to infect its host has a lot do in discovering possible drug targets. Such PPIs, when altered, would prevent the host from being infected by the parasite and in some cases, result in the parasite inability to complete specific stages of its life cycle and invariably lead to the death of such parasite. It therefore becomes important to understand the workings of host-parasite interactions which are the major causes of most infectious diseases.
Objective
Many studies have been conducted in literature to predict HPPI, mostly using computational methods with few experimental methods. Computational method has proved to be faster and more efficient in manipulating and analyzing real life data. This study looks at various computational methods used in literature for host-parasite/inter-species protein-protein interaction predictions with the hope of getting a better insight into computational methods used and identify whether machine learning approaches have been extensively used for the same purpose.
Methods
The various methods involved in host-parasite protein interactions were reviewed with their individual strengths. Tabulations of studies that carried out host-parasite/inter-species protein interaction predictions were performed, analyzing their predictive methods, filters used, potential protein-protein interactions discovered in those studies and various validation measurements used as the case may be. The commonly used measurement indexes for such studies were highlighted displaying the various formulas. Finally, future prospects of studies specific to human-plasmodium falciparum PPI predictions were proposed.
Result
We discovered that quite a few studies reviewed implemented machine learning approach for HPPI predictions when compared with methods such as sequence homology search and protein structure and domain-motif. The key challenge well noted in HPPI predictions is getting relevant information.
Conclusion
This review presents useful knowledge and future directions on the subject matter.
Keywords: Host-parasite protein interactions (HPPI), machine learning, Plasmodium falciparum parasite, human host, computational methods, Inter-species protein interaction predictions
1. INTRODUCTION
Recently, infectious diseases became a major health concern to the world as a result of several millions of sicknesses and deaths recorded every year, despite clinical efforts and development. Several research works tried to identify host-parasite interactions from different perspectives to get a thorough understanding of parasite and defensive means to curb parasitic infections. In host-parasite interactions, proteins are involved, and these proteins control all biological systems in a cell such as molecular functions and biological processes. Proteins interrelate with other proteins to generate a protein interaction network. Therefore, interactions between proteins become vital keys in various biological activities.
Host-parasite protein interactions play critical roles in host and parasite infection and therefore identifying the protein-protein interactions (PPIs) that allow parasites infect the host would be of great assistance in discovering potential drug targets [1]. Unfortunately, there is a limitation in the presently available knowledge on genes and proteins implicated in such interactions because only a few works have been experimentally proven as protein interactions between host and pathogen in several host-parasite systems. Computational method of identifying host-parasite protein interactions became popular because of this limitation in experimental method. The computational approaches are divided basically into methods based on; sequence homology, protein structure, domain and motif and machine learning [2].
2. Methods of host-parasite protein interactioN predictions used in literature
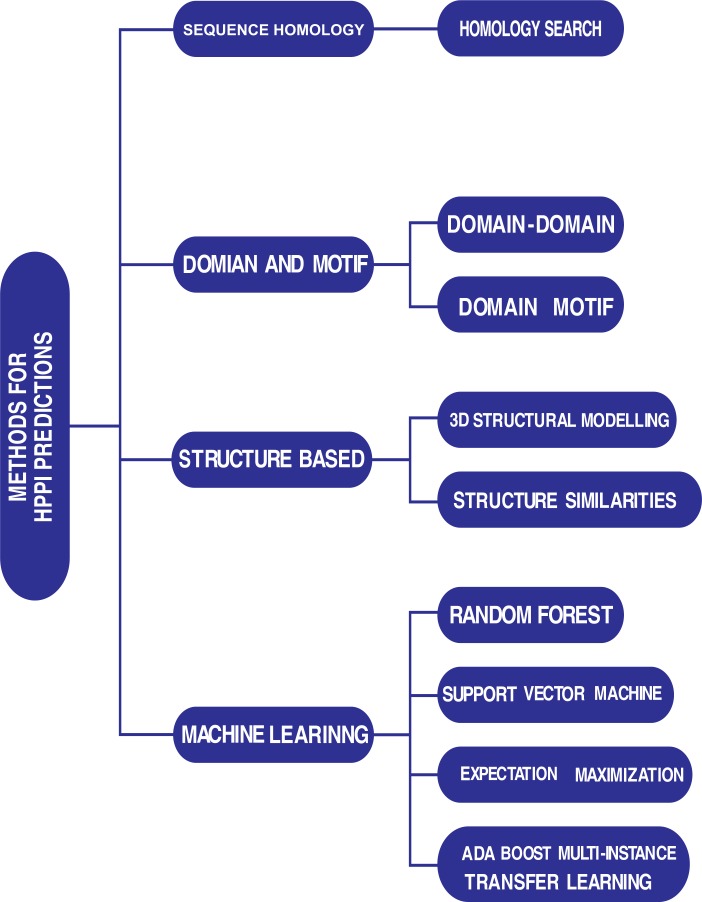
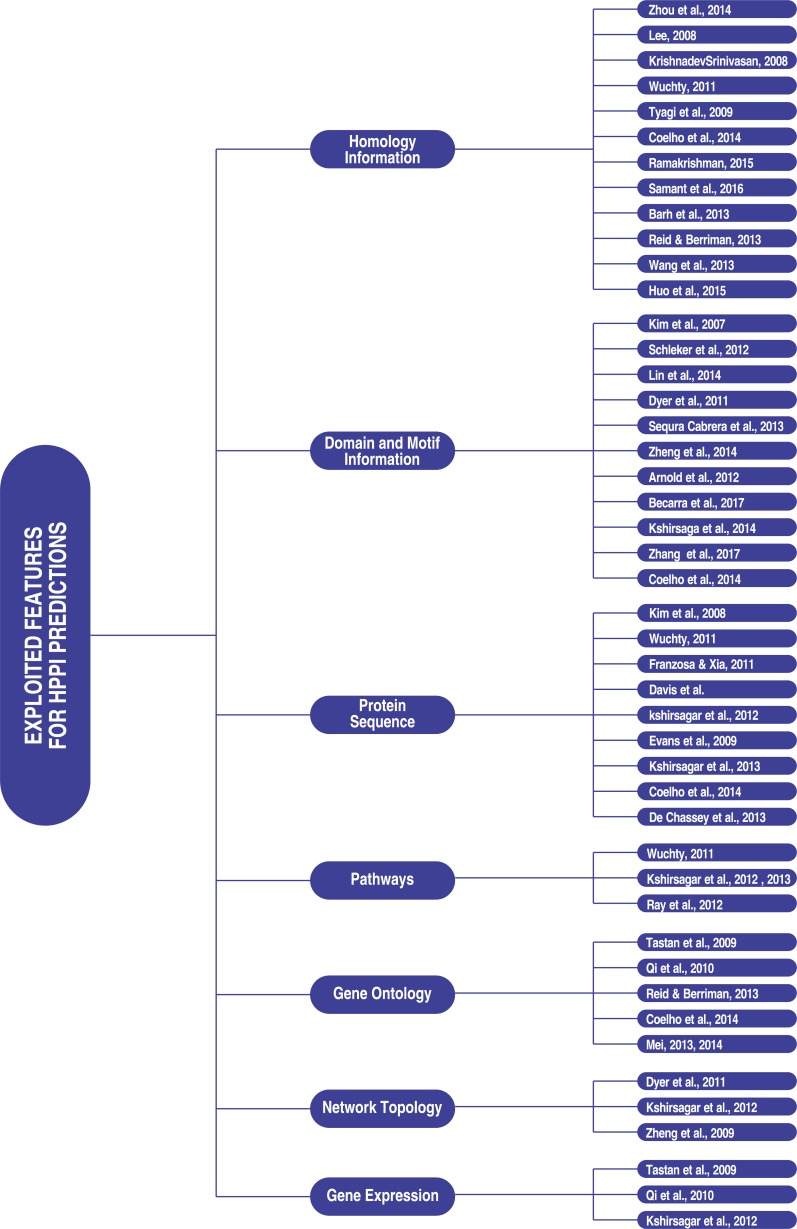
There are several computational approaches used for predictions, but the popularly used ones in inter-species/host-parasite protein interaction predictions are presented here. They are sequence homology search, domain-motif, structure-based and machine learning approaches. Fig. (1) presents a graphical view of such methods. Fig. (2) is a graphical view of features exploited by previous studies in host-parasite/inter-species protein-protein interaction predict-tions.
Fig. (1).
Methods used in Literature to predict Host-parasite protein interactions.
Fig. (2).
Exploited features for HPPI and Inter-species prediction from literature.
2.1. Sequence Homology-Based Method
The homology-based method is a traditional means of intra-specie protein-protein interactions prediction. This method has been embraced in the prediction of inter-species host-parasite protein-protein interactions. The idea behind sequence homology method is that the interaction between two proteins in a particular species will probably be conserved in related species [3]. The conserved interactions are called Interologs. What this means is that pairs of homologous protein originate from similar family pairs of interacting proteins which will likely take the structure and function as well as interactions of the family proteins. The operation of homology method for identification of inter-species protein-protein interaction involves first getting the template PPI pair (x y), and finding the homolog x' in the host and the homolog y' in the parasite and then infer that (x', y') interact [3]. Some of studies utilized this method as a method of host-parasite protein interactions prediction. Such studies include; [4-13]. The limitation of this method of protein interaction prediction is that inferences cannot be made about interactions between specie-specific families of genes.
2.2. Domain and Motif Interaction-Based Method
Domains are the core determinants of the structure and function of proteins which perform specific function in facilitating proteins interaction with other molecules [14]. Majority of PPIs are facilitated by domain-motif interaction by binding domains in protein to short linear motifs in interacting partners. These interactions are mostly implicated in major cellular processes, requiring their tight regulation [15]. A number of studies have used this method as a building block for predicting protein-protein interactions in single species [16-18]. Study [1] happens to be the first to explore this method for inter-species protein interaction prediction, although, the number of interactions predicted was few and their biological significance has not been assessed. A procedure that predicts interacting protein between parasite and host from the integration of protein domain profiles and interacting protein between proteins from the same organism was used by [1].
Here, Bayesian statistics were used to find the probability of proteins interactions for every pair of functional domains found in the protein pair. This procedure was applied in the host-parasite system by identifying domains in the individual host and parasite protein having at least one domain. Then, the probability of the interaction was computed for each pair of host and parasite with at least one domain. Other studies [19-22] also used domain and motif interaction based method for HPPI.
2.3. Structure-Based and Structural Modeling Approaches
In the structure-based method of prediction, two proteins with similar structures to a known proteins interaction pairs are likely interacting in a structurally similar way. Quite a number of studies already employed the structural information to predict the similarities between query proteins such as host-parasite protein interaction and template PPIs to infer that interaction exists in host-parasite protein pairs that match some template PPIs [23]. Doolittle and colleague [24] constructed a map interaction of HIV-1 and human. The same Doolittle and colleague in another study [25] applied similar approach for building a network of interactions to Dengue virus and its host. A study by [26], presumes interactions between proteins that are structurally homologous using Human and Influenza A NS1. Protein structure-based protocols were used by [27] to discover potential protein interactions in P. falciparum and host erythrocytes. For structural modeling, study [28] conducted HPPI comparative modeling of 3D structures and applied the techniques to 10 pathogens including Mycobacterium, apicomplexa, kinetoplastida and Plasmodium falciparum which are responsible for neglected human diseases.
2.4. Machine Learning
Machine learning approach is a robust method of HPPI predictions, although this method has not been used extensively in HPPI prediction.
Random Forest (RF) is a classifier algorithm made up of decision trees. Individual tree in the training phase is built by random feature vector sampled from a dataset independently. A little part of the variables is selected at random and individual classification tree is raised for every node in a tree. In order to group a new object, the input vector is set up for each of the trees in the forest. Based on the largest vote, a class is allocated to the object. Random forest is a practical classifier when large datasets with a large number of features are concerned because there will be no need to feature select or feature delete. The Random forest can also classify features based on importance and can also be used to recover missing data. Nevertheless, specific databases with noisy data may overfit. Random forest and Decision trees are extensively employed in bioinformatics and computational biology for classifying biological data [29] especially for PPI prediction [30]. The RF approach was used by study [31] for feature evaluation in order to accurately predict protein-protein interactions from negative dataset. In predicting cytokine-receptor interactions, Random forest classifier method was used by [32]. The studies that implemented Random forest classifier approach to predict host-parasite/inter-species PPIs that this work is interested in are studies [19, 33]. Study [19] employed RF method for HPPI predictions by integrating thirty-five features within eight groups. The same random forest classifier was used by [33] to assess the quality of conserved interactions for predictions of putative PPIs between human host and malaria parasite.
Support vector machine (SVM) is another classifier approach used in HPPI predictions. SVM classifier is reinforced by margin optimization. This margin for an object is connected to the certainty of its classification. Objects with correct classification will have large margins and objects with classification unclear will probably have small margins. Training SVM can be achieved using a labeled dataset with each data labeled to indicate that it belongs to a class or two. Support vector machine is a very powerful classification method for arbitrary complex problems. Studies [34-38] employed this approach. Here, fixed feature vector length that connotes relative frequency of conserved amino acids in the protein sequence was used.
The other machine learning approaches employed in Literature include; Expectation maximization algorithm by [37]. Here, the study predicted, HPPI in human RBCs and merozoite membrane proteins from estimates of domain-domain interaction probabilities. Also, study [38], proposed Multi-instance transfer learning method called AdaBoost which was used to re-construct the proteome-wide Salmonella and human protein-protein interaction networks. The training dataset was improved using homolog knowledge transfer in the form of independent homolog instances. AdaBoost instance re-weighting was employed to offset the noise from homolog instances. Fig. 1 is a graphical display of computational methods used in literature for such predictions.
2.5. Strength of Methods Reviewed
Homology-based method: The simplicity and the seeming biological basis of homology-based method is a major advantage. Prediction using this method only requires template PPIs and protein sequences data. The method is also scalable with application to several host-parasite systems. Studies [4, 6, 7], used homology-based approach alone for prediction while [33] combined homology-based method with other methods to predict host-parasite protein-protein interaction.
Domain-domain/motif: Protein domain is a key in protein structure prediction. In order to also determine protein structure, annotate functions, mutagenesis analysis, and protein engineering, protein domain prediction is important. Ability to predict domains from sequence information increases identification of tertiary structure; improve annotation of protein function, assist the determination of structure and give direction to engineering and mutagenesis of protein. The identification of domains within a protein sequence also serves as a foundation for other methods.
Structure-based prediction approach: This method of prediction is quite efficient when the structure of the target protein has not been resolved experimentally. The similarity between structure leads to an identification of homologs.
Machine learning is a group of computational methods used to identify complicated patterns in a given dataset and make decisions on previously unseen data.
3. tabulations of hppi predictions in human host & plasmodium falciparum and other inter-species protein interaction predictions
This part is divided into two sections. Section 3.1 presents specific studies conducted on human host and Plasmodium falciparum parasite PPI predictions. It looks at the method of prediction by each study, the predictive method, filters used, potential PPIs identified and measurements used. Table 1 is a tabular presentation of section 3.1.
Table 1.
Cross section of studies on human host and Plasmodium falciparum PPIs predictions
| Host-Parasite | Predictive Method | Potential PPIs Identified | Filter | Measurements | Ref |
|---|---|---|---|---|---|
| Human-Plasmodium falciparum | Combined interaction probability of domains Bayesian statistics for assessment |
A total of 516 PPIs between human and Pf were predicted. Important PPIs predicted are PfEMP1s and MSP1s, Q8IAS3, plasminogen (Q5TEH4) and pfEMP1, Q8IAL6 and Q8I339. They all interact with human blood coagulation proteins which may play a role in disrupting human blood coagulation pathways. Q8IHZ5, a known subtilisin-like protease, interacts with a number of blood coagulation proteins, which suggests that it may be involved in the degradation of blood platelets. Also, hypothetical Plasmodium protein Q8IKP8 interacts with the predicted partners of Q8IHZ5. | Gene ontology terms | Area under the Curve (AUC), Sensitivity and Precision |
[1] |
| Human-Plasmodium falciparum | Interlogs inferred from ortholog information | Interactions between putative HSP40 homologs of P. falciparum and the H. sapiens TNF receptor associated factor family was revealed here, suggesting a role for these interactions in the interference of the human immune response to P. falciparum. Calmodulin (PF14_0323), interacts with 50 human proteins. Among the 50 human proteins interacting with PF14_0323, thirteen (13) of them interact with human calmodulin (CALM3). This suggests that P. falciparum calmoduin shares some of the targets of human calmodulin, and may hijack these PPIs for its purpose. PF14_0359 and the TNF receptor associated factor family (TRAF1, TRAF2 and TRAF6) are predicted also to interact. |
Gene ontology annotations and Presence/ absence of translocational signals. |
Sensitivity | [4] |
| Human-Plasmodium falciparum | Homology detection method using template PPI databases, DIP, and iPfam | Remarkable interactions are: Plasmepsins and host cytoskeletal proteins, interaction between TRAP and ICAMs | Database of Interacting Proteins (DIP) sequences | [5] | |
| Human-Plasmodium falciparum | Interolog | The study observed that most of the highly interacting proteins were involved in structural assembly of the pathogen such as actin, tubulin, and histone. 𝛼-tubulin was finalized as an important protein involved in the infection process. | Cellular location, Gene ontology, and Functional role. | [13] | |
| Human-Plasmodium falciparum | Homology-based approach | A total of 208 physicochemically viable interactions were predicted. The key interacting proteins are: SAR1 and the host ADP-ribosylation factor-binding protein GGA3 (Q9NZ52), Host calcium-activated potassium channel protein 4, KCNN4 (UniProt ID: O15554), and conserved parasitic protein of unknown function, PF3D7_1463900. | Intrachain heterodomain interactions from iPfam, Intra host and intra pathogen interactions and Expression profile of parasite proteins from PlasmoDB. | [27] | |
| Human-Plasmodium falciparum | Comparative Modelling | The key prediction from this study relating to P. falciparum and human are; P. falciparum thrombospondin-related adhesive protein (TRAP, SSP2, PF13_0201) interact with human Toll-like receptor 4 (TLR4, ENSP00000346893), based on a template structure of Glycoprotein IBa bound to Von Willenbrand factor (PDB1M10). TRAP, animmunogenic protein used as a component of several vaccine candidates, interacts with TLR4. |
Biological context and Network-level Information |
[28] | |
| Host-Parasite | Predictive Method | Potential PPIs Identified | Filter | Measurements | Ref |
| Human-Plasmodium falciparum | Sequence Orthology/ Homology and Random forest |
The discovery here is that parasite proteins predominantly target central proteins to take control of a human host cell. Several prominent pathways of signaling and regulation proteins were predicted to interact with parasite chaperones. | Expression data and molecular properties | Area under the curve (AUC) | [33] |
| Human-Plasmodium falciparum | Estimation maximization | A network consisting of 205 PPIs between parasite and human membrane proteins were predicted. A further prediction shows that SNARE proteins of parasites and APP of humans may function in the invasion of RBCs by parasites. | Gene expression data | Area under the curve (AUC) | [37] |
| Human-Plasmodium falciparum | Mining of combined HPPI data | The analysis in the study revealed; apolipoproteins and temperature/Hsp expression on PfEMP1 presentation, the essence of MSP-1 in platelet activation, role of parasite proteins in TGF-β regulation and the contribution of albumin in astrocyte dysfunction. | Gene Ontology, Tissue-specific annotation |
[39] |
The second section, 3.2 reviews other studies on host/parasite PPI predictions and inter-species PPIs, presenting the type of host, pathogen or species used in the study. The methods of prediction employed the predictive method, filters used, potential PPIs identified and measurements used as in section 1. Both sections are presented in the form of a table. Table 2 is a tabular representation of section 3.2.
Table 2.
Cross section of other studies on host-parasite/inter-species protein-protein interaction predictions.
| Host- Pathogen | Predictive Method | Potential PPIs Identified | Filter | Measurements | Ref. | |
|---|---|---|---|---|---|---|
| Mycobacterium Tuberculosis – Homo Sapien | Homology detection approach based on sequence motif | A total of 118 pairs of HPIs were obtained from 43 Mycobacterium tuberculosis proteins and 48 Homo sapiens proteins were predicted and stored in the PATH database | Domain-Domain Interactions (DDIs), and Functional annotations of protein and publicly available experimental results for further filter | F1 Score | [7] | |
| Salmonella-Human | Sequence and interacting domain similarity approach | This study predicted 29 out of 59 gold standard PPIs used. With Domain-based prediction feature, nine (9) of the gold standard interactions were predicted. These nine interactions are also part of the set of 29 PPIs formerly predicted. | Domain-based prediction feature | [8] | ||
| Candida albicans-Zebrafish | Ortholog-based PPIs and Multivariate linear dynamic model of regulatory responses |
This study developed a computational framework. Some of the predictions between Candida albicans-Zebrafish were done. An important discovery here is that redox status is critical during the battle between the host and pathogen, which could determine the outcome of infection. | Sequence-targeted probes derived from the individual genome | [11] | ||
| Mycobacterium Tuberculosis H37Rv –Human | Stringent homology-based approach | An interesting discovery made aside from PPI predictions include host proteins and pathogen proteins that partake in the host-pathogen PPIs which tend to be hubs in their own intra-species PPI network. Again, host and pathogen proteins that are involved in host-pathogen PPIs might have a lengthier primary sequence, more domains, more hydrophilic and others. |
PATRIC database | [12] | ||
| HIV 1- Human protein | Supervised learning using Random Forest Classifier | A key prediction from this study is HIV-1 protein tat and human vitamin D receptor (VDR) Tat is a regulatory protein of HIV-1. The interaction has also been validated experimentally. | Eukaryotic Linear Motif (ELM) database | ROC-AUC, Precision-Recall | [19] | |
| Host- Pathogen | Predictive Method | Potential PPIs Identified | Filter | Measurements | Ref. | |
| Human- microbial Oral | Ensemble methodology for prediction naïve Bayes classifier for training and validation |
The study revealed important pathways involved in the onset of infectious oral diseases, and also potential drug-targets and biomarkers. Also, the first computational model of the Human-Microbial oral interactome was constructed. | PPI pairs from the five databases | Area under the ROC-AUC, F1, score, Accuracy, Precision-Recall | [22] | |
| HIV virus-Human | Structural similarity | A total of 502 interactions involving 137 human proteins were predicted. Three interactions consistent with two other studies predicted by this study are; gp41 and LCK, gp41 and PLK1, IN and XPO1. Twenty-two (22) of true positives predictions out of 265 predictions were made. |
RNAi functional data and shared Gene Ontology cellular component annotation for further filter | [24] | ||
| Dengue virus-Human and Insect hosts | Structural similarity | They predicted 2,073 interactions among viral and human proteins and found 7 out of 19 true positives. The study revealed the possibility that some of the protein interactions which enable DENV to manipulate the cellular pathways of two hosts are conserved between the species. |
Functional information from recent literature and Gene Ontology cellular component annotation (Subcellular co-localiza tion) |
[25] | ||
| Influenza A NS1–Human | Method based on structural homologous proteins interactions | The study predicted that out of 41 human proteins of influenza–human PIN, twelve (12) have been identified to be host factors for influenza virus replication. When Influenza–human interactome were combined with predicted and literature data, forty-seven (47) of 364 human proteins were identified to be host factors directly controlling viral replication |
Predicted and literature data | [26] | ||
| Human-Human papillomaviruses (HPV) and hepatitis C virus (HCV) | Support vector machine | This study predicted interactions between viruses and human proteins. The comparative analysis of HCV and HPV viral interaction networks gave 11 common human proteins that are targeted by both viruses. The SVM model revealed an average accuracy of 81.6% to predict human-HCV proteins, and accuracy of 83.3% to predict Human-HPV proteins. |
BLAST and Gene Ontology | Sensitivity, specificity accuracy | [34] | |
| Human- Yersinia Pestis, Francisella Tularensis, Salmonella and Bacillus anthracis |
Multitask learning approach | The study carried out a host-pathogen protein-protein interaction (PPI) prediction involving a fixed host and pathogens with various bacterial species. A set of interologs were predicted to exist between the four datasets. |
BLAST | Precision-Recall and F1 score | [35] | |
| HIV-1 and Human | Ensemble Transfer Learning method and Support Vector Machine for classification | The study deployed a model that is robust against data unavailability with less demanding data constraint. Analysis of overlapped predictions between the model in this study and the other existing models were carried out and the model was applied to novel host- pathogen PPIs identification. |
Gene Ontology | ROC-AUC, F1 Score, Precision-Recall | [36] | |
| Host- Pathogen | Predictive Method | Potential PPIs Identified | Filter | Measurements | Ref. | |
| Human T-cell leukemia viruses (HTLV) retroviruses-Human | Multi-instance Ada boost transfer learning method | The study used homology knowledge (GO) in the form of auxiliary homolog instance to address the problem of scarcity and unavailability of data. The study concluded that the method is effective in enriching information abundance evidenced by the HTLV-human PPI networks predicted. |
AdaBoost instance reweighting | ROC-AUC, Precision recall Curve, Specificity, Sensitivity and F1 score | [38] | |
| Xanthomonas oryzae pathovar oryzae (Xoo) oryzae-Rice | XooNET uses Structural Interactome MAP (PSIMAP), Protein interactions Experimental Interactome MAP (PEIMAP) and Domain-Domain interactions from iPfam | This study discovered 15 annotated AvrBs3 homologues in Xoo; Xoo1125, a hypothetical protein has over 60 interaction partners including the Avr proteins, responsible for the loss of pathogenicity when transposon insertion eukaryotic linear motifs |
Psi-Blast and hmm pfam for domain assignment | [40] | ||
| HIV1-Human | Method of domain-motif based on Multiple sequence alignments | The study predicted 109 true positives HPPIs from a total of 4,523 predictions. A total of 56 of the 133 Eukaryotic Linear Motif (ELM) resource were conserved on some HIV-1 protein. The essential discovery here is that ELMs that are conserved may appear frequently on human proteins. ELM LIG_PDZ_3 occurred on 90% of human proteins while some other ELMs such as LIG_EH1_1, occurred on quite a few human proteins. |
Conserved Eukaryotic Linear Motifs (ELMs) in Protein's multiple alignments | [41] | ||
| Dengue virus-Human | Domain and motif based method | A total of 79 human proteins (out of 1654) were identified to have interactions with viral proteins deposited in the VirHostNet database. The Functional enrichment analysis of the remaining 1,574 human proteins revealed 1,224 proteins that share biological processes annotations with the 79 identified human proteins targeted by the virus. |
Human domain set was used to filter the 3DID database in order to obtain motif-domain interactions involving only domains in the human proteome |
[42] | ||
| Plasmodium berghei-Mouse | Correlated gene expression profiles | The first network of mouse/mosquito malaria host-parasite interactions was predicted in the study. Several host genes involved in malaria infections were discovered. Specific ones include chromatin remodeling which is important for malaria interacting with its host to control gene expression timing. Also, genes involved in vesicle transport to the Golgi are important in host–parasite interactions for both Plasmodium and mouse especially to export proteins to the host cell surface. |
Yeast two-hybrid interologues and CSS interactions |
[43] | ||
| Hepatitis C virus (HCV)-Human | Method based on Domain-domain interactome. Topological and functional analysis of the network. |
Domain-centric perspective was used to construct a global landscape of virus-host interactions. The study identified that viruses use unique domains to interact the same host partners with fundamental functions and it also employed conserved DDIs occurring in host interactomes to mediate the interspecies interaction. |
Integrated domain-domain interaction (IDDI) database | [44] | ||
| Host- Pathogen | Predictive Method | Potential PPIs Identified | Filter | Measurements | Ref. | |
| Mycobacterium tuberculosis - Homo sapiens | Interolog method and domain-domain interactions to filter HPPIs | The study predicted 118 pairs of HPIs. A biological interaction network between M. tuberculosis and Homo sapiens was then constructed using the predicted inter- and intra-species interactions based on the 118 pairs of HPIs. Finally, a web accessible database named PATH was built. |
Protein sequences and Functional annotations of protein and publicly available experimental results | [45] | ||
| Francisella-human | Comparative genomics and Literature | The study identified 222 unique PPIs between 18 Francisella tularensis proteins and 183 human proteins. Twelve (12) Human-F. tularensis interactions were chosen for re-testing. There was a confirmation of four interactions in this assay. They are FTT0482c with WD repeat-containing protein 48 (WDR48), FTT1538c with 78-kDa glucose-regulated protein (HSPA5), FTT1538c with WDR48, and FTT1597 with AP-3 complex subunit mu-1 (AP3M1). |
Proteome-scale yeast two-hybrid (Y2H) | [46] | ||
|
Human-Staphylococcus aureus and Human-Neisseria. Meningitides |
Genome-wide protein microarray analysis | The study revealed interactions between the S. aureus immune evasion protein FLIPr (formyl-peptide receptor like-1 inhibitory protein) and the human complement component C1q, as key players of the offense-defense fighting; and of the interaction between meningococcal NadA and human LOX-1 (low-density oxidized lipoprotein receptor), an endothelial receptor | Human recombinant proteins from the GNF library | [47] | ||
| Grass carp-grass carp reovirus (GCRV) | Structural motif-domain interactions | A systems-based framework for the understanding of the GCRV infectome and diseasome was provided by the study. JAM-A protein was predicted to interact with GCRV Sigma1-like protein motifs, sharing similar binding mode compared with orthoreovirus. |
RNA-seq data from previous work | [48] | ||
| Human- human immunodeficiency virus 1 (HIV-1). | Short linear motifs | A method that predicts virus-host SLiM mediated PPIs and rank ranks candidate interactions were developed. The study discovered that the majority of conserved linear motifs in the HIV-1 virus are located in disordered regions. |
NIAID HIV-1-human interactions and the set of ELM mediated HIV-1-human interactions. | [49] | ||
| Human- Mycobacterium tuberculosis | Pairwise structure similarities | Secreted proteins of the STPK, ESX-1, and PE/PPE family in M. tuberculosis targeted human proteins involved in immune response and phagocytosis. M. tuberculosis also targeted host factors known to regulate HIV replication |
Cellular localization information | [50] | ||
3.1. Tabulation of Studies on Human Host and Plasmodium falciparum PPI Predictions
Table 1 is a tabular cross section of results from literature of host-parasite protein-protein interaction predictions specifically for human host and Plasmodium falciparum parasite. The table highlights the methods used and also described the work done.
3.2. Tabulation of Studies on Other Host-Parasite/Inter-Species Protein Interaction Predictions
Table 2 presents other studies on inter-species/host-parasite protein interaction predictions in literature, the methods of prediction employed, the filters used, potential Protein-protein Interactions identified and measurements used.
4. some MEASUREMENTS INDEXES used in hppi predictions and future prospects
4.1. Some Evaluation Measurements Indexes used in HPPI Predictions
Common measurements indexes identified in literature for host-parasite protein-protein interactions include; Sensitivity as used by [1, 4, 34] and Specificity by [34], Accuracy by [22, 34], AUC (area under the roc curve) by [1, 22, 33, 36-38]; Precision-Recall by [1, 22, 36, 37] and F1score by [7, 22, 34].
The formula for calculating the evaluation indexes for Sensitivity, Specificity, Accuracy, ROC-AUC and Precision-Recall are;
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
4.2. Future Prospects
From the above review, it is eminent that HPPI predictions will uncover the required knowledge in gaining understanding and discovering the crucial interactions that might be responsible for diseases. Such computational studies will shed more light on taking experimental work further and also assist in therapeutic development. It will further enhance deeper insight into the disease under study and open rooms of opportunities to explore new therapy development through the important protein interactions and proteins predicted. The need to explore machine learning methods is proposed here to ensure efficient results.
The future prospects in studies carried out on HPPI for human, and Plasmodium falciparum include; the need to include reliability assessment of protein-protein interactions identified through high-throughput screens in study [1]. For study [4], multiple methods could be combined to improve sensitivity and identify drug targets for drug discovery and design. The extent to which the method by [5] can predict HPPI should be investigated further, through a comprehensive experimental database for PPIs between P. falciparum and human. There is the need to identify molecular strategies that allow the parasite, Plasmodium falciparum to have power over the host in study [33]. This will expand the knowledge of the parasite unique re-modelling processes of the host cell and give potential leads to disease mediation.
Furthermore, in study [36], more studies to understand the significant relationship between the SNPs and parasite invasion are required. There is also the need to experimentally confirm the identified interactions. For study [27], experimental attempts are required to understand possible means of pathogenesis of red blood cells protein-protein interactions predicted in the study. Finally, an in vivo testing and validation of inhibitors identified in study [13] is required to understand the properties as antimalarial drugs
CONCLUSION
Host-parasite interaction prediction is gaining more ground in the recent years because the knowledge from it can give a better understanding of how parasite infects its host, which protein interactions are important for such infection to take place and also identifying drug targets. Computational methods will therefore, play an essential part in creating pathways for experimental HPPI validation by identifying and presenting important interactions that could then be taken further experimentally. In this review, we have been able to identify up to date studies conducted in host-parasite protein interactions with a table showing the host-pathogen involved in the interaction, the methods such studies used in conducting their studies, filters employed and measurements as the case may be. The future prospects of the studies were also mentioned with a specific focus on human-Plasmodium falciparum predictions. This kind of review will help researchers who want to work in this area to know what has been done so far and thus give direction to further studies.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
This work is partially supported by the Federal Polytechnic, Ilaro Staff Development Programme and NIH-H3ABioNet grant U24HG006941.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Dyer M.D., Murali T.M., Sobral B.W. Computational prediction of host-pathogen protein-protein interactions. Bioinformatics. 2007;23:i159–i66. doi: 10.1093/bioinformatics/btm208. [DOI] [PubMed] [Google Scholar]
- 2.Zhou H., Jin J., Wong L. Progress in Computational studies of host-pathogen interactions. J. Bioinform. Comput. Biol. 2013;11(2):123001. doi: 10.1142/S0219720012300018. [DOI] [PubMed] [Google Scholar]
- 3.Itzhaki Z., Akiva E., Margalit H. Preferential use of protein domain pairs as interaction mediators: order and transitivity. Bioinformatics. 2010;26(20):2564–2570. doi: 10.1093/bioinformatics/btq495. [DOI] [PubMed] [Google Scholar]
- 4.Lee S.A., Chan C.H., Tsai C.H., et al. Ortholog-based protein-protein interaction prediction and its application to inter-species interactions. BMC Bioinformatics. 2008;9(Suppl. 12):S11. doi: 10.1186/1471-2105-9-S12-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krishnadev O., Srinivasan N. A data integration approach to predict host-pathogen protein-protein interactions: application to recognize protein interactions between human and a malarial parasite. In Silico Biol. 2008;8(34):235–250. [PubMed] [Google Scholar]
- 6.Tyagi N., Krishnadev O., Srinivasan N. Prediction of protein-protein interactions between Helicobacterpylori and a human host. Mol. Biosyst. 2009;5:1630–1635. doi: 10.1039/b906543c. [DOI] [PubMed] [Google Scholar]
- 7.Krishnadev O., Srinivasan N. Prediction of protein-protein interactions between human host and a pathogen and its application to three pathogenic bacteria. Int. J. Biol. Macromol. 2011;48:613–619. doi: 10.1016/j.ijbiomac.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 8.Schleker S., Garcia-Garcia J., Klein-Seetharaman J., Oliva B. Prediction and comparison of Salmonella-human and Salmonella-Arabidopsis interactomes. Chem. Biodivers. 2012;9:991–1018. doi: 10.1002/cbdv.201100392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z.G., He F., Zhang Z., Peng Y.L. Prediction of protein-protein interactions between Ralstonia solanacearum and Arabidopsisthaliana. Amino Acids. 2012;42:2363–2371. doi: 10.1007/s00726-011-0978-z. [DOI] [PubMed] [Google Scholar]
- 10.Barh D., Gupta K., Jain N., et al. Conserved host-pathogen PPIs Globally conserved inter-species bacterial PPIs based conserved host-pathogen interactome derived novel target in C. pseudotuberculosis, C. diphtheriae, M. tuberculosis, C. ulcerans, Y.pestis, and E.coli targeted by Piper betel compounds. Integr. Biol. 2013;5:495–509. doi: 10.1039/c2ib20206a. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y.C., Lin C., Chuang M.T., Hsieh W.P., Lan C.Y., Chuang Y.J. Inter species protein-protein interaction network construction for characterization of host-pathogen interactions: a Candida albicans-zebra fish interaction study. BMC Syst. Biol. 2013;7(79):1–11. doi: 10.1186/1752-0509-7-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H., Gao S., Nguyen N., Fan M., Jin J. Stringent homology-based prediction of H. sapiens-M. tuberculosis H37Rvprotein-protein interactions. Biol. Direct. 2014;9:1–30. doi: 10.1186/1745-6150-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramakrishnan G., Srinivasan N., Padmapriya P., Natarajan V. Homology-based prediction of potential protein-protein interaction between Human erythrocytes and Plasmodium falciparum. Bioinform. Biol. Insights. 2015;9:195–206. doi: 10.4137/BBI.S31880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ideker T., Sharan R. Protein networks in disease. Genome Res. 2008;18(4):644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akiva E., Friedlander G., Itzhaki Z., Margalit H.A. Dynamic view of Domain-Motif Interactions. PLOS Comput. Biol. 2012;8(1):e1002341. doi: 10.1371/journal.pcbi.1002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojcik J., Schächter V. Protein-protein interaction map inference using interacting domain profile pairs. Bioinformatics. 2001;17:S296–S305. doi: 10.1093/bioinformatics/17.suppl_1.s296. [DOI] [PubMed] [Google Scholar]
- 17.Pagel P., Wong P., Frishman D. A domain interaction map based on phylogenetic profiling. J. Mol. Biol. 2002;344:1331–1346. doi: 10.1016/j.jmb.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Han D.S., Kim H.S., Jang W.H., Lee S.D., Suh J.K. PreSPI: a domain combination based prediction system for protein-protein interaction. Nucleic Acids Res. 2014;32(21):6312–6320. doi: 10.1093/nar/gkh972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tastan O., Qi Y., Carbonell J.G., Klein-Seetharaman J. Prediction of interactions between HIV-1 and human proteins by information integration. Pac. Symp. Biocomput. 2009;14:516–527. [PMC free article] [PubMed] [Google Scholar]
- 20.Qi Y., Tastan O., Carbonell J.G., Klein-Seetharaman J., Weston J. Semi-supervised multi-task learning for predicting interactions between HIV-1 and human proteins. Bioinformatics. 2010;26:i645–i52. doi: 10.1093/bioinformatics/btq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kshirsagar M., Carbonell J., Klein-Seetharaman J. Techniques to cope with missing data in host-pathogen protein interaction prediction. Bioinformatics. 2012;28:i466–i72. doi: 10.1093/bioinformatics/bts375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coelho E.D., Arrais J.P., Matos M., et al. Computational prediction of human-microbial oral interactome. BMC Syst. Biol. 2014;8(24):1–12. doi: 10.1186/1752-0509-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao V.S., Srinivas K., Sajiini G.N., Kumar S. Protein-protein interaction detection: Methods and Analysis. Int. J. Proteomics. 2014;2014:147648. doi: 10.1155/2014/147648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doolittle J.M., Gomez S.M. Structural similarity-based predictions of protein interactions between HIV-1 and Homo sapiens. Virol. J. 2010;7:82–97. doi: 10.1186/1743-422X-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doolittle J.M., Gomez S.M. Mapping protein interactions between Dengue virus and its human and insect hosts. PLoS Negl. Trop. Dis. 2011;5(2):e954–e69. doi: 10.1371/journal.pntd.0000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Chassey B., Meyniel-Schicklin L., Aublin-Gex A., Navrati V., Chantier T., André P. Structure homology and interaction redundancy for discovering virus-host protein interactions. EMBO Rep. 2013;14:938–944. doi: 10.1038/embor.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramakrishnan G., Srinivasan N., Padmapriya P., Natarajan V. Homology-based prediction of potential protein-protein interactions between human erythrocytes and plasmodium falciparum. Bioinform. Biol. Insights. 2015;9:195–206. doi: 10.4137/BBI.S31880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis F.P., Barkan D.T., Eswar N., McKerrow J.H., Sali A. Host pathogen protein interactions predicted by comparative modeling. Protein Sci. 2007;16:2585–2596. doi: 10.1110/ps.073228407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Wang M., Zhang H. The use of classification trees for bioinformatics. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2011;1(1):55–63. doi: 10.1002/widm.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X.W., Liu M. Prediction of protein–protein interactions using random decision forest framework. Bioinformatics. 2005;21(24):4394–4400. doi: 10.1093/bioinformatics/bti721. [DOI] [PubMed] [Google Scholar]
- 31.Zeng J., Li D., Wu Y., Zou Q., Liu X. An empirical study of features fusion techniques for protein-protein interaction prediction. Curr. Bioinform. 2016;11:4–12. [Google Scholar]
- 32.Wei L., Zou Q., Liao M., Lu H., Zhao Y. A novel machine learning method for cytoskine-receptor interaction prediction. Comb. Chem. High Throughput Screen. 2016;19(2):144–152. doi: 10.2174/1386207319666151110122621. [DOI] [PubMed] [Google Scholar]
- 33.Wuchty S. Computational prediction of host-parasite protein interactions between P. falciparum and H. sapiens. PLoS One. 2011;6(11):e26960. doi: 10.1371/journal.pone.0026960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui G., Fang C., Han K. Prediction of protein-protein interactions between viruses and human by an SVM model. BMC Bioinformatics. 2013;13(7):S5–S15. doi: 10.1186/1471-2105-13-S7-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kshirsagar M., Carbonell J., Klein-Seetharaman J. Multitask learning for host-pathogen protein interactions. Bioinformatics. 2013;29:i217–i26. doi: 10.1093/bioinformatics/btt245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mei S. Probability weighted ensemble transfer learning for predicting interactions between HIV-1 and human proteins. PLoS One. 2013;8:e79606. doi: 10.1371/journal.pone.0079606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Huang Y., Liang J., et al. Computational prediction of protein interactions related to the invasion of erythrocytes by malarial parasites. BMC Bioinformatics. 2014;15(1):393. doi: 10.1186/s12859-014-0393-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mei S. Computational reconstruction of proteome-wide protein interaction networks between HTLVretro viruses and Homo sapiens. BMC Bioinformatics. 2014;15:245. doi: 10.1186/1471-2105-15-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao A., Kumar M.K., Joseph T., Bulusu G. Celebral malaria: insight from host-parasite protein-protein interactions. Malar. J. 2010;9(155):1–7. doi: 10.1186/1475-2875-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J.G., Park D., Kim B.C., et al. Predicting the interactome of Xanthomonasoryzae pathovar oryzae for tarrget selection and DB service. BMC Bioinformatics. 2008;9(41):1–7. doi: 10.1186/1471-2105-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evans P., Dampier W., Ungar L., Tozeren A. Prediction of HIV-1 virus-host protein interactions using virus and host sequence motifs. BMC Med. Genomics. 2009;2:27. doi: 10.1186/1755-8794-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segura-Cabrera A., García-Pérez C.A., Guo X., Rodríguez-Pérez M.A. A viral-human interactome based on structural motif-domain interactions captures the human infectome. PLoS One. 2013;8(8):e71526. doi: 10.1371/journal.pone.0071526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid A.J., Berriman M. Genes involved in host-parasite interactions can be revealed by their correlated expression. Nucleic Acids Res. 2013;41(3):1508–1518. doi: 10.1093/nar/gks1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng L.L., Li C., Ping J., Zhou Y., Li Y., Hao P. The domain landscape of virus-host interactomes. BioMed Res. Int. 2014;2014:867235. doi: 10.1155/2014/867235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huo T., Liu G., Yang C., Lin J., Rao Z. Prediction of host-pathogen interactions between Mycobacterium tuberculosis and Homo sapiens using sequence motifs. BMC Bioinformatics. 2015;16:100. doi: 10.1186/s12859-015-0535-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wallqvist A., Memisevic V., Zavaljevski N., et al. Using host-pathogen protein interactions to identify and characterize Francisella tularensis virulence factors. BMC Genomics. 2015;16:1106. doi: 10.1186/s12864-015-2351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scietti L., Sampieri K. Exploring host-pathogen interactions through genome wide protein microarray analysis. Sci. Rep. 2016;6:27996. doi: 10.1038/srep27996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang A., He L., Wang Y. Prediction of GCRV virus-host protein interactome based on structural motif-domain interactions. BMC Bioinformatics. 2017;18(1):145. doi: 10.1186/s12859-017-1500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Becerra A., Bucheli V.A., Moreno P.A. Prediction of virus-host protein-protein interactions mediated by short linear motifs. BMC Bioinformatics. 2017;18:163. doi: 10.1186/s12859-017-1570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui T., Li W., Liu L., Huang Q., He Z.G. Uncovering new pathogen-host protein-protein interactions by pairwise structure similarity. PLoS One. 2016;11(1):e147612. doi: 10.1371/journal.pone.0147612. [DOI] [PMC free article] [PubMed] [Google Scholar]