ABSTRACT
Aspergillus
species contain pathogenic and opportunistic fungal pathogens which have the potential to cause mycosis (invasive aspergillosis) in humans. The existing antifungal drugs have limitation largely due to the development of drug-resistant isolates. To gain insight into the mechanism of action and antifungal drug resistance in Aspergillus species including biofilm formation, we have reviewed protein data of Aspergillus species during interaction with antifungals drugs (polynes, azoles and echinocandin) and phytochemicals (artemisinin, coumarin and quercetin). Our analyses provided a list of Aspergillus proteins (72 proteins) that were abundant during interaction with different antifungal agents. On the other hand, there are 26 proteins, expression level of which is affected by more than two antifungal agents, suggesting the more general response to the stress induced by the antifungal agents. Our analysis showed enzymes from cell wall remodelling, oxidative stress response and energy metabolism are the responsible factors for providing resistance against antifungal drugs in Aspergillus species and could be explored further in clinical isolates. Also, these findings have clinical importance since the effect of drug targeting different proteins can be potentiated by combination therapy. We have also discussed the opportunities ahead to study the functional role of proteins from environmental and clinical isolates of Aspergillus during its interaction with the antifungal drugs.
Abbreviations
IPA: invasive pulmonary aspergillosis; IA: invasive aspergillosis; AmB: Amphotericin B; CAS: Caspofungin; VRC: Voriconazole; ITC: Itraconazole; POS: Posaconazole; ART: Artemisinin; QRT: Quercetin; CMR: Coumarin; MIC: minimal inhibitory concentration
KEYWORDS: Antifungal agents, proteomic, azole resistance, phytochemicals, Hsp70
Introduction
Aspergilli contain approximately 340 species that perform range of biological functions in the environment and about 40 of them are known to cause health problems (Samson et al. 2014; Thakur et al. 2015). Aspergillus fumigatus, A. terreus, A. flavus, A. niger and A. oryzae are among the most studied Aspergilli due to their medical, agricultural and industrial importance (Krijgsheld et al. 2013; Sugui et al. 2015). Aspergillus oryzae is widely used for traditional food fermentations in EastAsia, and Aspergillus niger is used to produce various enzymes (e.g. amylases and pectinases) and organic acids (Powell et al. 2013). Aspergillosis is a severe clinical problem caused by Aspergillus species, especially in immunocompromised patients, hence Aspergilli have emerged as important opportunistic fungal pathogens (Chowdhary et al. 2014). Also, a large number of patients have been diagnosed with chronic pulmonary aspergillosis worldwide following the treatment of pulmonary tuberculosis (Denning 2011). Aspergillus conidia are present in air, soil, food-products, indoor environment and plant debris. Conidia being smaller in size (2–5 µm) (Mousavi et al. 2016), are the main source for Aspergilli conidia distribution into the environment (Latgé 1999; Zmeili and Soubani 2007). Minuscule size of conidia enhances its existence in the air for a longer duration because of which it is inhaled by human beings and if these conidia are not cleared by phagocytic cells, may germinate into hyphae in respiratory mucosa (Zmeili and Soubani 2007). Conidia showed metabolically less active and possess prolonged viability in adverse conditions (Lamarre et al. 2008). Aspergillus causes invasive aspergillosis (IA) to extreme complications. Huge rise in drug-resistant isolates of Aspergillus species possess the additional threat to human beings (Hagiwara et al. 2016; Sanglard 2016). Currently, three class of antifungal drugs are commonly used for the treatment against Aspergillus mediated infections; polyenes, triazoles and echinocandin. These compounds target the cell wall either by disrupting ergosterol biosynthesis or β-1, 3-glucan or by targeting the ergosterol directly (Groll and Kolve 2004). We now have understood on how few of the existing antifungal drugs work. However, transcriptional factors and signalling cascades that are involved in providing antifungal drug resistance in Aspergillus isolates are partly known. Thus, we have reviewed proteins/enzymes that showed regulated expression during the interaction with antifungal agents in Aspergillus species.
Present treatment strategies for IA
Treatment options have evolved which includes three major classes of antifungal viz. triazoles, echinocandin and polyenes. The triazoles include Itraconazole (ITC), VRC and POS. Isavuconazole is newly introduced azole against Aspergillus and approved against IA (Miceli and Kauffman 2015). Echinocandins include Caspofungin (CAS), Anidulafungin (AND) and micafungin (Supplementary file-1A). Treatment of IA using Amphotericin (AmB), possesses ergosterol binding tendency, forming fungicidal sterol sponge and disables membrane functions (Anderson et al. 2014). The major disadvantage of AmB is toxicity to humans. It has been observed that antifungal compounds showed adverse effects in children suffering from aspergillosis such as 3–5 mg/kg dosage of AmB per day for aspergillosis treatment causes infusion-related infection, hypokalemia and nephrotoxicity (Canadian Paediatric Society ID, UD ICA 2010). However, the liposomal AmB have shown the minimal toxic effect in IA patients and have prolonged persistence against azole-resistant Aspergillus species (Seyedmousavi et al. 2013). AmB is not recommended for aspergillosis caused by A. terreus and also AmB-resistant Aspergillus species, hence the combination therapy with a synergistic response is considered as alternative approach (Dannaoui et al. 2004; Elefanti et al. 2013). Antifungal combinational studies against A. fumigatus using in vivo or in vitro, and clinical methods have shown effective results (Ben-Ami et al. 2011; Stergiopoulou et al. 2011).
Triazoles are used as the more preferred choice of antifungals in clinical practices. Inhibition of ergosterol biosynthesis leads to the disruption of the structural unit of the cell membrane of fungi (Sanglard and Odds 2002). ITC, one of the triazoles, was the first drug introduced in azole class for aspergillosis patients in 1997 and other azoles evolved later. According to current status, azole resistance in A. fumigatus has emerged globally, thereby threatening the azole therapy against aspergillosis (Verweij et al. 2016). Van der Linden et al. have provided the mortality rate data in azole-resistant strains infected aspergillosis patients. Azoles resistant Aspergillus isolates were categorised on the basis of MIC values; ITC > 2 μg/ml, VRC > 2 μg/ml and posaconazole (> 0.5 μg/ml). These data were collected as per the guidelines mentioned in the CLSI reference method (van der Linden et al. 2011). Recent treatment strategies for azole-resistant as well as susceptible isolates of A. fumigatus are focused on the combinational drug therapy which includes azole (VRC) and echinocandin. Echinocandin inhibits cell wall biosynthesis by blocking the catalytic subunit of β-glucan synthase (Arendrup 2014).
According to a report by Ming Zhang 2014, A. flavus and A. niger mediated IPA was effectively treated by a combined effect of CAS and VRC. However, A. fumigatus mediated IPA treatment showed less efficacy in response to the combination of echinocandin and triazole (Krishnan-Natesan et al. 2012; Zhang et al. 2014). In a clinical report, 27.5% mortality rates were observed in monotherapy, whereas for combined (VRC and AND) therapy it was 19.3% (Marr et al. 2015). Denning, D.W. et al., reported that AmB- and azole-resistant Aspergillus isolates, a more frequently occurring clinical isolates, however, azoles are used as first-line therapy hence, these therapeutic options for IA need revision due to emergence of drug resistance Aspergillus isolates and intervention of novel therapeutic strategies to overcome this issue (Denning and Bowyer 2013). Isavuconazole is a recently approved drug (2015) for treatment of aspergillosis and mucormycosis, however as per the recent U.S. guidelines voriconazole has been recommended as a first-line therapy for aspergillosis (Misch and Safdar 2016).
The antifungal response is less effective when A. fumigatus forms a biofilm. Higher MIC of antifungal drug is required to destroy the biofilm structures, which has been one of the reason for drug resistance in A. fumigatus against the polyene, azole, and echinocandin (Mowat et al. 2008; Seidler et al. 2008). It has been hypothesised that extracellular matrix in biofilm confers drug resistance via absorbing antifungal molecules, thus disallowing their diffusion to the site of fungal cells. This has been supported by the formation of extracellular matrix that sequesters antifungal drugs and reduces drug susceptibility in C. albicans (Nett et al. 2010). Activation of multidrug resistance protein that pumps out antifungal drugs has been reported in biofilm structure. Thus, the role of efflux-pump in azole resistance has been observed (Rajendran et al. 2011) that possibly could be one of the reasons for treatment failure in aspergillosis cases.
Drug toxicity and pesticides exposure to humans are the major issues worldwide (Dorner 2004; Damalas and Eleftherohorinos 2011). Phytochemicals have acquired a lot of significance as a new candidate over the present drug discovery methods (Butler 2004) and are discussed in the later part of the review.
Epidemiological pattern of drug resistance in Aspergillus species
Olga Rivero-Menendez recommended the need of antifungal susceptibility assay on the Aspergillus isolates derived from the clinical samples for the treatment of aspergillosis cases (Rivero-Menendez et al. 2016). In clinical practices, long-term use of azole drugs for aspergillosis is the major reason for the emergence of azole resistance (Hagiwara et al. 2016). Another route for the increase in azole resistance in Aspergillus is the extensive application of fungicides in the agriculture (Snelders et al. 2009; Chowdhary et al. 2013). Thus, investigations need to focus on to screen environmental samples that are resistant to azoles. Restricted use of azoles in agricultural, i.e. rotation of antifungal products, change in dosage and application periods could be the best possible strategy to reduce the burden of azole in the environment (Chowdhary et al. 2013; Berger et al. 2017). Additionally, some of the Aspergillus species has intrinsic resistant to certain antifungal and other species are susceptible to a certain class of drug but may become resistance due to the prolonged incomplete dosages of antifungal drugs. Recently, Rivero-Menendez et al. briefly showed the dominance and occurrence of azole drug resistance in Aspergillus species and reported the highest number of azole resistance isolates in European countries (Rivero-Menendez et al. 2016).
The occurrence of azoles resistant isolates of A. fumigatus varies from 6.6 to 28% worldwide. In the UK, it is 2.1–20%. In the Netherlands, Germany and France resistance level has up to 10–12% in clinical and environmental isolates. While in other continents (Asia, Africa, America and Australia) resistance level is also around 10% (Rivero-Menendez et al. 2016). Recently, 32.4% of A. fumigatus isolates in clinical samples were observed in India and out of the 1.75% were azole-resistant. Thus, lower occurrence of resistant isolates in India was observed in comparison to European countries that is probably the limited application of azole fungicides in Asia (Chowdhary et al. 2015; Directorate-General. ECHCP 2016).).
In general, resistant isolates are reported when MIC values are above the epidemiologic cut-off values based on these EUCAST defined breakpoints Aspergillus spp. (susceptible or resistant) for azoles (POS > 0.25 µg/ml, ISA > 1 µg/ml, ITC > 2 µg/ml and VRC > 2 µg/ml) (mEUCAST 2016). The Clinical and Laboratory Standards Institute has also defined MIC cut-off for various azoles viz. POS > 0.5 µg/ml, VRC > 1 µg/ml and ITC > 1 µg/ml (Espinel-Ingroff et al. 2010).
In addition, drug resistance in Aspergillus spp. is also mediated by the development of biofilms that provided temporary antifungal drug resistance and protects the pathogen in the hostile environment (Seidler et al. 2008; Villena et al. 2009; Bruns et al. 2010; Kaur and Singh 2014; Paul et al. 2017). In earlier studies, Mowat et al. showed the formation of biofilm structures in A. fumigatus cultures which are observed to be resistant to antifungal drugs (Mowat et al. 2007). Beauvais et al. also reported the extracellular matrix on the colony surface of A. fumigatus (Beauvais et al. 2007). Extracellular matrix helps hyphae to hold together to form biofilm structure, which permits reduced drug susceptibility. Also, major changes in metabolic activities have been observed during biofilm formation which might be associated with virulence (Muszkieta et al. 2013). Biofilm-associated infections have very high mortality rate and difficult to cure with existing drug therapies, thus further studies are needed to understand the role of biofilm in drug resistance.
Aspergillus terreus is another major cause of aspergillosis, reported in University and Hospital of Innsbruck in Austria and medical centres in Houston, Texas (Lass-Florl et al. 2007; Blum et al. 2008). As per the recent reports in India, 6.6% of A. terreus isolates were found among the aspergillosis cases in a referral Chest Hospital in Delhi (Chowdhary et al. 2015). In another study from India, it has been observed that only 8% of A. terreus isolates were susceptible to AmB with MICs (0.5–1 mg/L) and showed no particular genotypic pattern (Kathuria et al. 2015). Previously, it has been evident from previous reports that A. terreus isolates are naturally resistant to AmB (Steinbach et al. 2004a; Lass-Florl et al. 2007). Also, A. terreus has shown high in vitro and in vivo MICs for AmB which confirms the AmB resistance (Graybill et al. 2004; Steinbach et al. 2004b; Lass-Florl et al. 2005). Recently, failure of azole drug treatment against A. terreus has also been observed in Danish clinical samples by Arendrup et al. In their study (Arendrup et al. 2012), they have reported the development of ITC resistance in A. terreus which may be associated with M217I Cyp51A mutation.
A. terreus isolates (approximately 5%) showed resistance against posaconazole in in-vitro studies. High percentage (10%) posaconazole resistance A. terreus s.s. isolates were isolated from Austria, Germany and the UK (Zoran et al. 2018). Hence, lack of AmB response and azole (VRC) resistance has made A. terreus an infectious threat in immunocompromised patients (Pastor and Guarro 2014). As per another report from Alcazar-Fuoli, Laura et al. A. niger rarely showed varying MICs to ITC and the isolates which showed higher MIC for ITC also had higher MIC values to VRC and Isavuconazole compared with A. fumigatus MIC values (Alcazar-Fuoli et al. 2009). Higher MICs for other Aspergillus species such as A. awamori and A. niger have been observed in comparison to A. tubingensis (Szigeti et al. 2012). Additionally, biofilm formation has been reported in A. niger (Villena et al. 2009; Paul et al. 2017) probably accounting for high MIC against the drug.
Aspergillus flavus is mostly prevalent in arid climates and can tolerate extreme conditions and frequently occur in Africa, the Middle East and Southeast Asian countries (Krishnan et al. 2009). A. flavus is known to produce aflatoxins (potent carcinogen). This fungus contaminates various crops leads to economic losses in agriculture. Consumption of aflatoxin-contaminated foods or feeds causes severe illness in animal and humans such as aflatoxicosis, liver necrosis/liver cancer (Tiwari. 2018). Prevalence of A. flavus in India is about 45.4% and about 2.5% are resistant to VRC (Chowdhary et al. 2015; Sharma et al. 2018). In recent reports, some of the clinical isolates of A. flavus showed resistance to VRC and the high MICs linked to being T788G and Y319H alterations in the cyp51C gene (Liu et al. 2012; Paul and Rudramurthy 2015). Also, Aspergillus alliaceus (genomic similarity with A. flavus) showed high MICs value to AmB and echinocandins, which varies for different azoles (Balajee et al. 2007). From the above data, we could hypothesise that there is a large increase in the antifungal drug resistance from environmental and clinical Aspergillus isolates. Also, most of the antifungal agents are also used in crop protection and to preserve materials from fungal decay (van der Linden et al. 2015). Thus, it leads to the emergence of acquired resistance in this fungal species and there is a need for systematic surveillance programmes worldwide to reduce the use of antifungals in the environment.
Proteomic approach to characterise the antifungal response in Aspergilli
Proteomic analysis has been applied to elucidate the resistant mechanism in resistant vs. susceptible strains and also in the identification of potential biomarkers (Vermeulen et al. 2018). Various research groups focused on the characterisation of Aspergillus spp. at development stages (Asif et al. 2006; Suh et al. 2012; Tiwari et al. 2016; Thakur and Shankar 2017). Comparative proteome analysis of resting conidia to mycelia provided biochemical and cellular pathway during the morphotypes of Aspergilli (Asif et al. 2006; Vodisch et al. 2009; Teutschbein et al. 2010; Tiwari et al. 2016; Thakur and Shankar 2017; Shankar et al. 2018). To date, proteomic-based analysis on how Aspergilli adapt to host condition has conceded information of Aspergilli infection mechanisms (Suh et al. 2012; Kubitschek-Barreira et al. 2013). To get a comprehensive picture of the response of drugs and phytochemical on Aspergillus spp., we have found limited reports in Aspergilli on proteome response under antifungal agents in case of drugs AmB, CAS and azoles (ITC and VRC) (Gautam et al. 2008, 2016; Cagas et al. 2011a; Amarsaikhan et al. 2017) and some of the phytochemicals like artemisinin (ART), coumarin-derivative (CMR) and quercetin (QRT) (Gautam et al. 2011; Singh et al. 2012; Tiwari and Shankar 2018b) which have been described. In the remaining part of the review, we discussed the major proteins and pathways involved during the exposure of drugs and phytochemicals.
Proteome analysis in response to antifungal drugs
AmB, a class of polyene, acts via primarily binding to cell membrane ergosterol, thereby disrupting membrane function as well as ROS accumulation (Valiante et al. 2015). Gautam et al. (2008) studied the response of AmB in A. fumigatus and observed down-regulation of translation machinery and energy metabolism. Their study showed the abundance of 48 proteins using MALDI, 44 proteins were highly expressed and 4 of them showed less abundance. Additionally, ergosterol biosynthesis protein Erg13 (AFUA_3G10660) was up-regulated under AmB exposure (Gautam et al. 2008) Also, increased abundance of Hem13 (AFUA_1G07480), a heme biosynthetic protein on AmB exposure reflects the need of more heme-molecules; it acts as cofactor and prerequisite for the ergosterol biosynthesis. Most of the iron enzyme lost their activities on iron-deficiency (Shakoury-Elizeh et al. 2010). Induction of oxidative stress responses involves up-regulation of proteins such as catalase, manganese superoxide dismutase and Prx1/LsfA upon AmB treatment. Which further provide evidence that AmB damages cell due to oxidative stress (Gautam et al. 2008). ITC targets ergosterol leading to the accumulation of sterols that is toxic to the cells (Valiante et al. 2015). Gautam et al. (2016) also studied proteome of A. fumigatus under ITC stress that resulted in the differential abundance of 54 proteins. It has been observed that 12 proteins with the increased level of expression and 42 proteins with decrease in abundance (Gautam et al. 2016). The increased level of oxidative stress proteins like catalase, Cat1 were observed similar to AmB exposure, most abundant proteins in response to more than two antifungal agents are summarised in Table 1. Another study by Gautam et al. (2016) demonstrated the synergy of ITC with the ART, indicating a positive effect of this combination (Gautam et al. 2016). On the other hand, Cagas, Jain et al. studied the proteomic response of A. fumigatus against CAS using iTRAQ, at 24 and 48h and provided updated protein data set. Previously, they attempted to profile proteins at different morphotypes in A. fumigatus that study they provide differential protein expression patterns at various developmental stages in A. fumigatus (Cagas et al. 2011b) .Cagas et al., has observed 58 ribosomal proteins that are differentially expressed in their study, suggesting a shift in ribosomal programming in the cell. Also, in comparative study, ribosomal proteins at 24h post drug exposure of the susceptible strain was observed and only 4 out 19 proteins showed increased abundance in the resistant strain. Their results speculated a ribosomal reshuffling response to the CAS (Cagas et al. 2011a, 2011b). Gautam et al. (2008) observed that most of the ribosomal proteins in response to AmB were down-regulated in microarray data (Gautam et al. 2008). Interestingly, earlier reports suggested that mitochondrial hypoxia response domain protein (AFUA_1G12250) would be most promising biomarker which was down-regulated up to 16-fold at both 24 and 48h in susceptible strain and was relatively unaltered in the resistant strain under CAS but not observed during exposure of VRC and AmB to A. fumigatus, suggested as a biomarker specific to CAS (Gautam et al. 2008; Cagas et al. 2011a; Amarsaikhan et al. 2017) . From the above data, it is reflected that each drug molecule affects proteome of Aspergilli with a common target pathway. Some drugs may have different targets but broadly affects the common pathways which ultimately lead to cell cycle arrest. On the other hand, there is limited available protein data on biofilms in Aspergillus spp. We have observed that sets of protein are required for biofilm formation in A. fumigatus. Protein associated with the biofilms formation showed abundance in translational regulatory proteins (Muszkieta et al. 2013). Whereas, in the case of A. niger intracellular protein analysis of biofilm showed the 19% overexpressed and 32% differentially expressed protein spots when compared free-living submerged cultures using 2D-PAGE and MS-TOF analysis (Villena et al. 2009). Results suggested that proteins are involved in cell adhesion, which allow biofilm development and surface adhesion fermentation. Thus, from the discussed data available in Table 1 suggested that antifungal drugs mediated proteome response in Aspergilli majorly involves proteins that were found in managing the energy oxidative stress, alteration of cell wall biosynthesis and ribosomal reprogramming. Also, induction of bypass energy metabolism pathways is evident upon exposure to all the antifungal drugs Figure 1. These pathways and proteins might be involved in a resistant mechanism and may also be explored as new drug targets in drug resistance Aspergilli.
Table 1.
Most abundant proteins in-response to more than two antifungal agents in Aspergillus species through proteomic studies.
| Antifungal
drugs |
Phytochemicals |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| S. no. | Name of the protein | Name of organisms | AmB (24h) (Gautam et al. 2008) | CAS (24h & 8h)(Cagas et al. 2011a) | VRC (4h) (Amarsaikhan et al. 2017) |
ITC (24) (Gautam et al. 2016) | ART (3h) (Gautam et al. 2011) |
QRT (7h) (Tiwari and Shankar 2018b) | CMR (16h) (Singh et al. 2012) |
| 1 | Mitochondrial Hsp70 chaperone (Ssc70), putative | Aspergillus fumigatus, Aspergillus flavus | + | − | + | + | − | + | + |
| 2 | Enolase | Aspergillus fumigatus | + | − | + | + | − | − | + |
| 3 | Ubiquinol-cytochrome C reductase complex core protein 2) | Aspergillus fumigatus | − | + | + | − | + | − | + |
| 4 | Glutamate/Leucine/Phenylalanine/Valine dehydrogenase, putative | Aspergillus fumigatus | + | − | + | + | − | − | + |
| 5 | Glutamate carboxypeptidase, putative | Aspergillus fumigatus, Aspergillus flavus | − | − | + | + | − | + | + |
| 6 | Allergen Asp F3 | Aspergillus fumigatus | − | + | + | + | − | − | + |
| 7 | Thioredoxin (Thioredoxin TrxA | Aspergillus fumigatus | + | − | + | + | − | − | + |
| 8 | Phosphoglycerate kinase | Aspergillus fumigatus | + | + | − | + | − | − | + |
| 9 | NAD-dependent formate dehydrogenase | Aspergillus fumigatus | + | + | − | + | − | − | + |
| 10 | Mycelial catalase Cat1 | Aspergillus fumigatus, Aspergillus flavus | + | + | − | + | + | − | |
| 11 | Fumarate hydratase | Aspergillus fumigatus | + | − | + | + | − | − | − |
| 12 | Cobalamin-independent methionine synthase MetH | Aspergillus fumigatus | − | + | + | − | − | − | + |
| 13 | NADH-ubiquinone oxidoreductase 213 kDa subunit | Aspergillus fumigatus | − | − | + | − | + | − | + |
| 14 | Translation elongation factor EF-2 subunit, putative | Aspergillus fumigatus | + | − | + | − | − | − | + |
| 15 | Antigenic mitochondrial protein HSP60, putative | Aspergillus fumigatus | − | + | + | − | − | − | + |
| 16 | Aminopeptidase P, putative | Aspergillus fumigatus, Aspergillus flavus | − | − | + | − | − | + | + |
| 17 | Autophagic serine protease Alp2 | Aspergillus fumigatus | − | − | + | + | − | − | + |
| 18 | Proteasome regulatory particle subunit (RpnL), putative | Aspergillus fumigatus | + | − | + | − | − | − | + |
| 19 | Conidial hydrophobin RodB | Aspergillus fumigatus | + | − | + | − | + | − | − |
| 20 | 1,3-beta-Glucanosyltransferase Bgt1 | Aspergillus fumigatus, Aspergillus flavus | − | − | + | − | + | + | |
| 21 | Cobalamin-independent methionine synthase MetH/D | Aspergillus fumigatus | − | + | + | − | − | − | + |
| 22 | Integral membrane protein | Aspergillus fumigatus, Aspergillus flavus | + | + | − | − | − | + | − |
| 23 | Antioxidant protein LsfA | Aspergillus fumigatus | + | + | − | − | − | − | + |
| 24 | Malate dehydrogenase, NAD-dependent | Aspergillus fumigatus | + | − | − | + | − | − | + |
| 25 | ATP synthase proteolipid P2, putative | Aspergillus fumigatus | + | + | − | − | + | − | − |
| 26 | Aldehyde reductase | Aspergillus fumigatus | + | − | − | + | − | − | + |
Time-points for drug treatment has been mentioned against the antifungal agents, “(+) represents the presence of the protein”, “(−) represents the absence of the protein”
Figure 1.
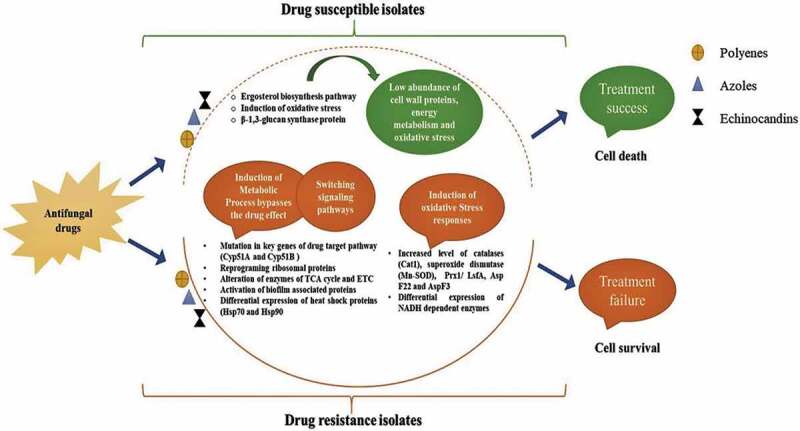
Probable determinants involved in the drug-resistance mechanism of Aspergillus species. Different colours of boxes represent antifungal drugs. In susceptible isolates, antifungal drugs targets cell wall, and generates oxidative stress. In resistance isolates, antifungal drugs showed the increased level of proteins from cell stress pathways and alternative metabolic pathways.
Proteome analysis in response to phytochemicals
Natural plant products with antifungal properties may offer a safe and effective alternative treatment strategy against aspergillosis. As of now the antimicrobial activity of these compounds, even studied comprehensively, still, none of them is available for practice at the clinical level. Screening of phytochemicals such as ART and CMR as anti-Aspergillus agents have been carried out (Gautam et al. 2011; Singh et al. 2012). Recently, we explored the anti-Aspergillus property of QRT along with other phytochemicals, QRT showed the strongest activity with MIC50 at 36 µg/ml in A. parasiticus and 113 µg/ml in toxigenic A. flavus (Tiwari et al. 2017). In addition, proteome response under QRT exposure showed the abundance of proteins of oxidative stress, cell wall proteins and membrane transport activity (influx and efflux proteins). Also under QRT stress switching of the signalling from MAPK to cAMP/PKA in A. flavus has been observed (Tiwari and Shankar 2018b). The similar kind of trend in signalling pathway was observed in Saccharomyces cerevisiae and Candida albicans, a remarkable decrease in PKC signalling was noted. In case of CAS drug treatment which involves drug resistance through calcineurin function, the activation of downstream MAPK pathway was also noted (LaFayette et al. 2010). In addition, HPLC analysis of A. flavus conidia grown for 48 h time point under influence of QRT showed a significant decrease in production of AFB1 (up to 51%) (Tiwari and Shankar 2018b). Gautam et al. (2008) investigated the exposure of ART to A. fumigatus using MALDI-ToF/ToF that leads to the differential abundance of 85 proteins; 29-increased and 56-decreased. Decreased expression of proteins included conidial hydrophobin (RodB), thaumatin domain protein (PhiA), galactomannan protein indicating remodelling of the cell wall, similar to AmB exposure (Gautam et al. 2008). Also, down-regulation of two mitochondrial genes, NADH dehydrogenase and NADH-ubiquinone oxidoreductase in microarray data and absence in protein dataset under ART exposure, reflects ART targets NADH dehydrogenase of A. fumigatus (Gautam et al. 2011). It was supported by the earlier study that deletion of NADH gene in yeast confers more resistance to ART, whereas overexpression of the NADH gene leads to more susceptibility in the yeast (Li et al. 2005). However, most of the other genes related to oxidative phosphorylation pathway showed up-regulation under ART exposure. As, it has been reported by Gautam et al. (2011) that ART may disrupt the membrane potential. Up-regulated transcripts may allow to re-established membrane potential by over-expressing these genes belonging to this pathway (Gautam et al. 2011). Importantly, the oxidative phosphorylation pathway was not affected significantly in C. albicans, S. cerevisiae and A. fumigatus (Zhang et al. 2002; Agarwal et al. 2003; Liu et al. 2005; Da Silva Ferreira et al. 2006; Yu et al. 2007; Gautam et al. 2008) upon standard antifungal drug treatment.
Subsequently, Singh et al., provided the proteome profile in A. fumigatus under exposure to synthetic coumarin-derivatives (SCD-1) and demonstrated the antifungal mechanism. Previously, Singh et al. showed SCD-1 a potent inhibitor of pathogenic Aspergilli at MIC90 of 15.62 μg/ml. In their study, differential abundance of 143 proteins was observed; 96-up and 30-down. Also, 4 proteins in control alone and 13 proteins in SCD-1 treated sample were reported. Proteins, involved in riboflavin biosynthesis fall under the category of decreased abundance, have been suggested as a novel target of SCD-1 (Singh et al. 2012). The proteomic analysis of antifungal treated A. fumigatus and A. flavus indicated that the different targets with similar or different altered pathways. Whereas there is no such proteomic data on A. terreus and A. niger in response to antifungal drugs. Researcher attempted to profile proteomic data on various morphotypes and at various stress conditions in A. terreus and A. niger (Sorensen et al. 2009; Lu et al. 2010; Thakur and Shankar 2017) and recently it has been reviewed (Shankar et al. 2018) on underlying mechanism to exit conidial dormancy in Aspergillus species.
Most abundant proteins under the exposure of both antifungal drug and phytochemicals
Proteins involved in cell stress viz. Aspf3 and enolase (glycolytic enzyme) which were frequently seen under the exposure of all drugs (AmB, VRC, CAS, ITC) and CMR (Gautam et al. 2008, 2016; Singh et al. 2012; Amarsaikhan et al. 2017). iTRAQ analysis was performed to check the effect of CAS on the expression level of Aspf3 at 48 h time point in A. fumigatus susceptible strain showed an increase of 3.5-fold and the resistant strain showed a decrease of 1.5-fold (Cagas et al. 2011a). Aspf3 encodes, a thioredoxin peroxidase, and an increase in the expression has been reported in response to hydrogen peroxide oxidative stress (Lessing et al. 2007; Cagas et al. 2011a) which clearly indicate that CAS mediated oxidative injury can easily cope up in resistant strains. Differential expression of a variety of antioxidant enzymes and enzymes of carbohydrate metabolism indicate the sensitivity of these metabolic pathways to antifungals. In case of oxidative stress mycelial catalase (Cat1) and aldehyde reductase was reported which induces oxidative stress, revealed that drugs mediate damage of fungal cell membrane resulting in oxidative stress conditions via ROS activation. Whereas, antifungal agents induce the proteins of regulation of ROS homeostasis significantly (Cowen and Lindquist 2005; Blum et al. 2013; Jukic et al. 2017) validate the existing reports on the role of oxidative stress response in drug resistance studies.
Enolase (Aspf 22) has been reported an allergen which stimulates a strong IFNγ immune response in humans and its homolog in C. albicans showed partial protection as a vaccine candidate (Denikus et al. 2005; Chaudhary et al. 2010). Presence of enolase (involved in energy metabolism) in hyphae of Aspergilli suggests that this enzyme may facilitate tissue invasion in the host (Denikus et al. 2005; Moloney et al. 2016; Shankar et al. 2018). In A. fumigatus, enolase was abundantly expressed under the influence of antifungal drug suggesting energy metabolism is vital to overcoming the drug stress. We also observed heat shock protein Hsp70 was overexpressed during antifungal drug treatment. The role of other heat shock proteins in response to an antifungal drug has been recently discussed (Cowen and Lindquist 2005; Blatzer et al. 2015; Tiwari et al. 2015; Tiwari and Shankar 2018a). Various enzymes of the glycolytic pathway, TCA cycle and electron transport chain were abundant under antifungal agent exposure (Table 1). Similarly, the abundance of TCA cycle proteins has also been observed in the proteome of biofilm of A. fumigatus that may contribute towards the persistence of the organism inside the host (Muszkieta et al. 2013). These results reflect a shift in energy metabolism to glyoxylate cycle under antifungal and biofilm conditions to combat the shortage of energy. Thus, up-regulation of TCA cycle proteins under antifungal stress may suggest their role in resistance mechanism in Aspergilli. Under the exposure of CAS and AmB, ribosomal proteins were found to be highly expressed, which indicates drug-mediated ribosomal reshuffling (Gautam et al. 2008; Cagas et al. 2011a). It reflects the requirement of more protein synthesis under antifungal stress to overcome the inhibitory effects of antifungal agents. Proteasome regulatory protein, RpnL (Gautam et al. 2008; Singh et al. 2012; Amarsaikhan et al. 2017) was also observed under the influence of antifungal agents (AmB, VRC and CMR).
The proteins/enzymes which are commonly targeted by antifungals are of great interest to increase the efficacy of treatment by using it in combination therapy. After reviewing the existing literature, we have compiled a dataset with similar proteins under the antifungal stress in different Aspergillus species (Supplementary file-1B). Out of 72 most abundant proteins, 26 proteins belonging to different metabolic pathways showed higher abundance underexposure of more than two antifungal agents (Table 1) whereas nine proteins are specific to one antifungal agent and remaining showed differential presence in antifungal agents. Most of these antifungal agents targeted the metabolism of the cell wall, ergosterol and oxidative stress proteins.
Insight into the drug-resistance mechanism in Aspergillus species
Antifungal drugs used for the treatment of various forms of aspergillosis face challenges due to the development of resistance in Aspergillus. Also, the prolonged use of the antifungal drug is one of the major causes of acquired drug resistance. It also depends on the type of Aspergillus species, antifungal drugs as well as on the geographical location. Though, the data on drug-resistance genes and mutations in the genome are available, but therapeutic choices are limited that make it difficult to control the invasive secondary fungal infections.
The major categories of mechanism of drug resistance include (1) Alterations in drug targets, due to mutations in target which reduces binding of drug to the target, (2) loss in drug efficacy, due to increase in drug efflux, overexpression of drug targets and sequester of antifungal agents and (3) Metabolic bypass, involves the activation of compensatory mechanisms which nullifies the toxic effects exerted by antifungals (Sanglard 2016). More recently, different resistance patterns with new mechanisms were observed, including intrinsic resistance in Aspergillus spp. and the emergence of simultaneous resistance to more than one class of drugs. Further, to increase the efficacy of existing drugs (combination of drugs) and through targeted drug therapy could be the future. The occurrence of drug resistance in Aspergilli and known mechanism of resistance of standard antifungal drugs are summarised in (Supplementary file-1A).
Aspergillus fumigatus and A. terreus are most extensively studied in the clinical spectrum due to the presence of high level of resistant isolates in comparison to other Aspergillus species. The major targets of the azoles reported by Mellado, E et al., are cyp51A and cyp51B (Cyp51 proteins (Mellado et al. 2001). These two encoded by different genes sharing 63% of sequence identity. Azole-resistant strains of A. fumigatus contain point mutations or overexpress cyp51A to provide resistance. The cyp51A encodes 14-sterol-demethylase, ergosterol is one of the major constituents of fungal cell structure (Snelders et al. 2010). Triazoles interact with the active site of Cyp51A, thus, hinder the ergosterol biosynthesis. Fungal cell death occurs due to the alteration in membrane fluidity (Snelders et al. 2009; Chowdhary et al. 2014). The wide use of other azole fungicides in agricultures exhibited a similar molecular structure to medical triazoles causing the evolution of cross-resistance in clinical practice (Snelders et al. 2012). Hagiwara et al. recently reviewed mutation at different sites in the cyp51 genes or tandem repeats in promoter region (TR34/L98H and TR46/Y121F/T289A) in Aspergilli contributing to resistance against azoles (Hagiwara et al. 2016). The whole genome sequencing of azole-resistant and susceptible (clinical and environmental) strains of A. fumigatus (from India, Netherlands and UK) have revealed that the environmental route is dominating with mutations in the cyp51A gene (TR34/L98H) in providing azole resistance (Abdolrasouli et al. 2015). On the other side, mutant study on biofilms of A. fumigatus, it has been observed that glycophosphatidylinositol-anchored cell wall protein (cspA) plays role in biofilm development, cell wall integrity and affects the drug-response (Fan et al. 2015). In addition, the substitution of S678P in Fks1p, the major subunit of glucan synthase, imparts resistance against echinocandin in Aspergillus fumigatus (Rocha et al. 2007). Thus, the data suggested that Alterations in drug targets is the most common strategy for resistance against antifungal drugs in Aspergilli.
A. terreus has been observed to be an intrinsic resistant to AmB in comparison to other Aspergilli, however, underlying molecular machinery is less unclear. The up-regulation of ergosterol biosynthesis genes (ERG5, ERG6 and ERG25) has been suggested to provide resistance against AmB (Walsh et al. 2003; Barker et al. 2004; Deak et al. 2009). Whereas, another study suggested that ergosterol content in A. terreus may have a little role in providing resistance against AmB. They further added AmB resistance Aspergillus strains absorb less amount of AmB drug, and also showed better protection management against oxidative damage due to the drug in A. terreus resistance isolates (Blum et al. 2013). According to a recent report in AmB resistance strains, SOD activity was more in comparison to susceptible isolates in Aspergilli (Jukic et al. 2017). Superoxide dismutase detoxifies superoxide anions, which are the precursor of ROS. Also, high production of gliotoxin (redox-active-metabolite) in biofilms of A. fumigatus was observed which confers resistance by enabling its growth and persistence in the host tissue (Bruns et al. 2010).
Another important key protein Hsp70 could be the regulator of AmB resistance, Blatzer et al. Use of inhibitors of Hsp70 and Hsp90, significantly increase the efficacy of AmB and azole drugs in resistant A. terreus isolates (Cowen and Lindquist 2005; Blatzer et al. 2015; Tiwari et al. 2015). However, the mechanisms by which heat shock proteins involved in drug resistance needs to be fully investigated. It primarily suggested that Metabolic bypass is the common strategy for the AmB resistance in A. terreus. Protein level analysis of host and pathogen under antifungal exposure may reveal the role of enzyme and protein which will be further investigated for their role in signalling and stress pathways. Thus, the advancement of existing technologies to fill the research gap is the challenges ahead.
Future aspects and conclusion
The mechanisms contributing to drugs-resistance include reducing drug-target interactions by increasing/decreasing expression of proteins involved in cell wall modulation, oxidative stress, heat shock proteins and energy metabolism. Also, other modes such as redox imbalance, ROS homeostasis and alteration in membrane fluidity contribute to the drug resistance. Another strategy of antifungal drug resistance includes biofilm formation, which allows adhesion of fungal cells on host surface. Similarly, efforts have been made to identify the specific protein in response to antifungal drugs that include overexpression of Cat1, Prx1/LsfA, enolase, thioredoxin peroxide (Aspf 3), Sod2. Fewer proteins enlisted are less abundant belonged to cytochrome C, RodA, PhiA. The major targets of the azoles are Cyp51A and Cyp51B. Hsp70 and Hsp90 contribute to protect cells during stress conditions. Proteins such as Sod2, Cat1, thioredoxin peroxide, Hsp70 and Hsp90 could be explored further as targets in resistant Aspergillus isolates. Following the facts that the level of certain protein was changed under drug exposure does not really mean its involvement in the drug-resistant mechanism, but provide opportunities that it can be tested by comparing proteome from drug-susceptible and resistant Aspergillus. Alterations in drug targets and metabolic bypass are common strategy for resistance against antifungal drugs in Aspergilli. Further, expression and mutation studies are required to understand the exact resistance pattern of these probable resistance determinants in Aspergilli. Synergistic drug combinations affecting different targets could provide an effective and alternate treatment strategy against drug-resistance fungal pathogens in immunocompromised patients. Furthermore, advances in the proteomic analysis of clinical vs. environmental isolates of Aspergillus may add detail insight into the drug-resistance mechanism.
Supplementary Material
Acknowledgments
We are thankful to Jaypee University of Information Technology for providing facilities and Ph.D. assistantship to SKS and ST.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary data
Supplementary data for this article can be accessed here.
References
- Abdolrasouli A, Rhodes J, Beale MA, Hagen F, Rogers TR, Chowdhary A, Meis JF, Armstrong-James D, Fisher MC.. 2015. Genomic context of azole resistance mutations in Aspergillus fumigatus determined using whole-genome sequencing. MBio. 6(3):e00536–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal AK, Rogers PD, Baerson SR, Jacob MR, Barker KS, Cleary JD, Walker LA, Nagle DG, Clark AM.. 2003. Genome-wide expression profiling of the response to polyene, pyrimidine, azole, and echinocandin antifungal agents in Saccharomyces cerevisiae. J Biol Chem. 278(37):34998–35015. [DOI] [PubMed] [Google Scholar]
- Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Cuenca-Estrella M, Rodriguez-Tudela JL. 2009. Species identification and antifungal susceptibility patterns of species belonging to Aspergillus section Nigri. Antimicrob Agents Chemother. 53(10):4514–4517. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarsaikhan N, Albrecht-Eckardt D, Sasse C, Braus GH, Ogel ZB, Kniemeyer O. 2017. Proteomic profiling of the antifungal drug response of Aspergillus fumigatus to voriconazole. Int J Med Microbiol. 307(7):398–408. [DOI] [PubMed] [Google Scholar]
- Anderson TM, Clay MC, Cioffi AG, Diaz KA, Hisao GS, Tuttle MD, Nieuwkoop AJ, Comellas G, Maryum N, Wang S, et al. 2014. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat Chem Biol. 10(5):400–406. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup MC. 2014. Update on antifungal resistance in Aspergillus and Candida. Clin Microbiol Infect. 20(Suppl 6):42–48. eng. [DOI] [PubMed] [Google Scholar]
- Arendrup MC, Jensen RH, Grif K, Skov M, Pressler T, Johansen HK, Lass-Florl C. 2012. In vivo emergence of Aspergillus terreus with reduced azole susceptibility and a Cyp51a M217I alteration. J Infect Dis. 206(6):981–985. eng. [DOI] [PubMed] [Google Scholar]
- Asif AR, Oellerich M, Amstrong VW, Riemenschneider B, Monod M, Reichard U. 2006. Proteome of conidial surface associated proteins of Aspergillus fumigatus reflecting potential vaccine candidates and allergens. J Proteome Res. 5(4):954–962. eng. [DOI] [PubMed] [Google Scholar]
- Balajee SA, Lindsley MD, Iqbal N, Ito J, Pappas PG, Brandt ME. 2007. Nonsporulating clinical isolate identified as Petromyces alliaceus (anamorph Aspergillus alliaceus) by morphological and sequence-based methods. J Clin Microbiol. 45(8):2701–2703. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker KS, Crisp S, Wiederhold N, Lewis RE, Bareither B, Eckstein J, Barbuch R, Bard M, Rogers PD. 2004. Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans. J Antimicrob Chemother. 54(2):376–385. [DOI] [PubMed] [Google Scholar]
- Beauvais A, Schmidt C, Guadagnini S, Roux P, Perret E, Henry C, Paris S, Mallet A, Prévost MC, Latgé JP. 2007. . An extracellular matrix glues together the aerial grown hyphae of Aspergillus fumigatus. Cell Microbiol. 9(6):1588–1600. [DOI] [PubMed] [Google Scholar]
- Ben-Ami R, Lewis RE, Kontoyiannis DP. 2011. In vitro interactions among echinocandins against Aspergillus fumigatus: lack of concordance among methods. Med Mycol. 49(3):285–288. [DOI] [PubMed] [Google Scholar]
- Berger S, El Chazli Y, Babu AF, Coste AT. 2017. Azole resistance in Aspergillus fumigatus: a consequence of antifungal use in agriculture? Front Microbiol. 8:1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatzer M, Blum G, Jukic E, Posch W, Gruber P, Nagl M, Binder U, Maurer E, Sarg B, Lindner H, et al. 2015. Blocking Hsp70 enhances the efficiency of amphotericin B treatment against resistant Aspergillus terreus strains. Antimicrob Agents Chemother. 59(7):3778–3788. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, Hörtnagl C, Jukic E, Erbeznik T, Pümpel T, Dietrich H, Nagl M, Speth C, Rambach G, Lass-Flörl C. 2013. New insight into Amphotericin B resistance in Aspergillus terreus. Antimicrob Agents Chemother. 57(4):1583–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, Perkhofer S, Grif K, Mayr A, Kropshofer G, Nachbaur D, Kafka-Ritsch R, Dierich MP, Lass-Florl C. 2008. A 1-year Aspergillus terreus surveillance study at the university hospital of innsbruck: molecular typing of environmental and clinical isolates. Clin Microbiol Infect. 14(12):1146–1151. eng. [DOI] [PubMed] [Google Scholar]
- Bruns S, Seidler M, Albrecht D, Salvenmoser S, Remme N, Hertweck C, Brakhage AA, Kniemeyer O, Muller FM. 2010. Functional genomic profiling of Aspergillus fumigatus biofilm reveals enhanced production of the mycotoxin gliotoxin. Proteomics. 10(17):3097–3107. eng. [DOI] [PubMed] [Google Scholar]
- Butler MS. 2004. The role of natural product chemistry in drug discovery. J Nat Prod. 67(12):2141–2153. [DOI] [PubMed] [Google Scholar]
- Cagas SE, Jain MR, Li H, Perlin DS. 2011a. Profiling the Aspergillus fumigatus proteome in response to caspofungin. Antimicrob Agents Chemother. 55(1):146–154. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagas SE, Jain MR, Li H, Perlin DS. 2011b. The proteomic signature of Aspergillus fumigatus during early development. Mol Cell Proteomics. 10(11):M111.010108 eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canadian Paediatric Society ID, UD ICA 2010. Antifungal agents for the treatment of systemic fungal infections in children. Paediatr Child Health. 15(9):603–608. [PMC free article] [PubMed] [Google Scholar]
- Chaudhary N, Staab JF, Marr KA. 2010. Healthy human T-cell responses to Aspergillus fumigatus Antigens. PLoS One. 5(2):e9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A, Kathuria S, Xu J, Meis JF. 2013. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 9(10):e1003633 eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A, Sharma C, Hagen F, Meis JF. 2014. Exploring azole antifungal drug resistance in Aspergillus fumigatus with special reference to resistance mechanisms [Research Support, Non-U S Gov’t Review]. Future Microbiol. 9(5):697–711. [DOI] [PubMed] [Google Scholar]
- Chowdhary A, Sharma C, Kathuria S, Hagen F, Meis JF. 2015. Prevalence and mechanism of triazole resistance in Aspergillus fumigatus in a referral chest hospital in Delhi, India and an update of the situation in Asia [Original Research]. Front Microbiol. 6(428). English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science (New York, NY). 309(5744):2185–2189. eng. [DOI] [PubMed] [Google Scholar]
- Da Silva Ferreira ME, Malavazi I, Savoldi M, Brakhage AA, Goldman MHS, Kim HS, Nierman WC, Goldman GH. 2006. Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Curr Genet. 50(1):32–44. [DOI] [PubMed] [Google Scholar]
- Damalas CA, Eleftherohorinos IG. 2011. Pesticide exposure, safety issues, and risk assessment indicators. Int J Environ Res Public Health. 8(5):1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannaoui E, Lortholary O, Dromer F. 2004. In vitro evaluation of double and triple combinations of antifungal drugs against Aspergillus fumigatus and Aspergillus terreus. Antimicrob Agents Chemother. 48(3):970–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deak E, Wilson SD, White E, Carr JH, Balajee SA. 2009. Aspergillus terreus accessory conidia are unique in surface architecture, cell wall composition and germination kinetics. PLoS One. 4(10):e7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denikus N, Orfaniotou F, Wulf G, Lehmann PF, Monod M, Reichard U. 2005. Fungal antigens expressed during invasive aspergillosis. Infect Immun. 73(8):4704–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D. 2011. Global burden of chronic pulmonary aspergillosis as a sequel to pulmonary tuberculosis. Bull World Health Organ. 89:864–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DW, Bowyer P. 2013. Editorial commentary: voriconazole resistance in Aspergillus fumigatus: should we be concerned? Clin Infect Dis. 57(4):521–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Directorate-General. ECHCP (accessed 2016 April25). Opinion on azole antimycotic resistance.
- Dorner JW. 2004. Biological control of aflatoxin contamination of crops. Toxin Rev. 23(2–3):425–450. [Google Scholar]
- Elefanti A, Mouton JW, Verweij PE, Tsakris A, Zerva L, Meletiadis J. 2013. Amphotericin B- and voriconazole-echinocandin combinations against Aspergillus spp.: effect of serum on inhibitory and fungicidal interactions. Antimicrob Agents Chemother. 57(10):4656–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel-Ingroff A, Diekema DJ, Fothergill A, Johnson E, Pelaez T, Pfaller MA, Rinaldi MG, Canton E, Turnidge J. 2010. Wild-type MIC distributions and epidemiological cutoff values for the triazoles and six Aspergillus spp. for the CLSI broth microdilution method (M38-A2 document). J Clin Microbiol. 48(9):3251–3257. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Li Z, Xu Z, Li H, Li L, Ning C, Ma L, Xie X, Wang G, Yu H. 2015. cspA influences biofilm formation and drug resistance in pathogenic fungus Aspergillus fumigatus. Biomed Res Int. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam P, Mushahary D, Hassan W, Upadhyay SK, Madan T, Sirdeshmukh R, Sundaram CS, Sarma PU. 2016. In-depth 2-DE reference map of Aspergillus fumigatus and its proteomic profiling on exposure to itraconazole. Med Mycol. 54(5):524–536. [DOI] [PubMed] [Google Scholar]
- Gautam P, Shankar J, Madan T, Sirdeshmukh R, Sundaram CS, Gade WN, Basir SF, Sarma PU. 2008. Proteomic and transcriptomic analysis of Aspergillus fumigatus on exposure to amphotericin B. Antimicrob Agents Chemother. 52(12):4220–4227. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam P, Upadhyay SK, Hassan W, Madan T, Sirdeshmukh R, Sundaram CS, Gade WN, Basir SF, Singh Y, Sarma PU. 2011. Transcriptomic and proteomic profile of Aspergillus fumigatus on exposure to artemisinin [journal article]. Mycopathologia. 172(5):331. [DOI] [PubMed] [Google Scholar]
- Graybill JR, Hernandez S, Bocanegra R, Najvar LK. 2004. Antifungal therapy of murine Aspergillus terreus infection. Antimicrob Agents Chemother. 48(10):3715–3719. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll AH, Kolve H. 2004. Antifungal agents: in vitro susceptibility testing, pharmacodynamics, and prospects for combination therapy. Eur J Clin Microbiol Infect Dis. 23(4):256–270. [DOI] [PubMed] [Google Scholar]
- Hagiwara D, Watanabe A, Kamei K, Goldman GH. 2016. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus fungi. Front Microbiol. 7:1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukic E, Blatzer M, Posch W, Steger M, Binder U, Lass-Florl C, Wilflingseder D. 2017. Oxidative stress response tips the balance in Aspergillus terreus amphotericin B resistance. Antimicrob Agents Chemother. 61(10):e00670-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathuria S, Sharma C, Singh PK, Agarwal P, Agarwal K, Hagen F, Meis JF, Chowdhary A. 2015. Molecular epidemiology and in-vitro antifungal susceptibility of Aspergillus terreus species complex isolates in Delhi, India: evidence of genetic diversity by amplified fragment length polymorphism and microsatellite typing. PLoS One. 10(3):e0118997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S, Singh S. 2014. Biofilm formation by Aspergillus fumigatus. Med Mycol. 52(1):2–9. [DOI] [PubMed] [Google Scholar]
- Krijgsheld P, Bleichrodt R, van Veluw GJ, Wang F, Müller WH, Dijksterhuis J, Wösten HAB. 2013. Development in Aspergillus. Stud Mycol. 74(1):1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan S, Manavathu EK, Chandrasekar PH. 2009. Aspergillus flavus: an emerging non-fumigatus Aspergillus species of significance. Mycoses. 52(3):206–222. eng. [DOI] [PubMed] [Google Scholar]
- Krishnan-Natesan S, Wu W, Chandrasekar PH. 2012. In vitro efficacy of the combination of voriconazole and anidulafungin against voriconazole-resistant cyp51A mutants of Aspergillus fumigatus. Diagn Microbiol Infect Dis. 73(2):135–137. [DOI] [PubMed] [Google Scholar]
- Kubitschek-Barreira PH, Curty N, Neves GW, Gil C, Lopes-Bezerra LM. 2013. Differential proteomic analysis of Aspergillus fumigatus morphotypes reveals putative drug targets. J Proteomics. 78:522–534. eng. [DOI] [PubMed] [Google Scholar]
- LaFayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, Gunatilaka AAL, Perfect JR, Cowen LE. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, Calcineurin, and Hsp90. PLoS Pathog. 6(8):e1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarre C, Sokol S, Debeaupuis JP, Henry C, Lacroix C, Glaser P, Coppee JY, Francois JM, Latge JP. 2008. Transcriptomic analysis of the exit from dormancy of Aspergillus fumigatus conidia. BMC Genomics. 9:417 eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass-Florl C, Grif K, Kontoyiannis DP. 2007. Molecular typing of Aspergillus terreus isolates collected in Houston, Texas, and Innsbruck, Austria: evidence of great genetic diversity. J Clin Microbiol. 45(8):2686–2690. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass-Florl C, Griff K, Mayr A, Petzer A, Gastl G, Bonatti H, Freund M, Kropshofer G, Dierich MP, Nachbaur D. 2005. Epidemiology and outcome of infections due to Aspergillus terreus: 10-year single centre experience. Br J Haematol. 131(2):201–207. eng. [DOI] [PubMed] [Google Scholar]
- Latgé J-P. 1999. Aspergillus fumigatus and Aspergillosis. Clin Microbiol Rev. 12(2):310–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessing F, Kniemeyer O, Wozniok I, Loeffler J, Kurzai O, Haertl A, Brakhage AA. 2007. The Aspergillus fumigatus transcriptional regulator AfYap1 represents the major regulator for defense against reactive oxygen intermediates but is dispensable for pathogenicity in an intranasal mouse infection model. Eukaryot Cell. 6(12):2290–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Mo W, Shen D, Sun L, Wang J, Lu S, Gitschier JM, Zhou B. 2005. Yeast model uncovers dual roles of mitochondria in the action of artemisinin. PLoS Genet. 1(3):e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Lee RE, Barker KS, Lee RE, Wei L, Homayouni R, Rogers PD. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob Agents Chemother. 49(6):2226–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Sun Y, Chen W, Liu W, Wan Z, Bu D, Li R. 2012. The T788G mutation in the cyp51C gene confers voriconazole resistance in Aspergillus flavus causing aspergillosis. Antimicrob Agents Chemother. 56(5):2598–2603. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Sun J, Nimtz M, Wissing J, Zeng A-P RU. 2010. The intra- and extracellular proteome of Aspergillus niger growing on defined medium with xylose or maltose as carbon substrate [journal article]. Microb Cell Fact. 9(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr KA, Schlamm HT, Herbrecht R, Rottinghaus ST, Bow EJ, Cornely OA, Heinz WJ, Jagannatha S, Koh LP, Kontoyiannis DP, et al. 2015. Combination antifungal therapy for invasive aspergillosis: a randomized trial [multicenter study randomized controlled trial research support, N I H, extramural research support, non-U S Gov’t]. Ann Intern Med. 162(2):81–89. [DOI] [PubMed] [Google Scholar]
- Mellado E, Diaz-Guerra TM, Cuenca-Estrella M, Rodriguez-Tudela JL. 2001. Identification of two different 14-alpha sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J Clin Microbiol. 39(7):2431–2438. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- mEUCAST 2016. European committee on antimicrobial susceptibility testing Antifungal breakpoint tables for interpretation of MICs v 8.0. accessed 2016 April25
- Miceli MH, Kauffman CA. 2015. Isavuconazole: a new broad-spectrum triazole antifungal agent. Clin Infect Dis. 61(10):1558–1565. eng. [DOI] [PubMed] [Google Scholar]
- Misch EA, Safdar N. 2016. Updated guidelines for the diagnosis and management of aspergillosis. J Thorac Dis. 8(12):E177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney NM, Owens RA, Doyle S. 2016. Proteomic analysis of Aspergillus fumigatus–clinical implications. Expert Rev Proteomics. 13(7):635–649. [DOI] [PubMed] [Google Scholar]
- Mousavi B, Hedayati MT, Hedayati N, Ilkit M, Syedmousavi S. 2016. Aspergillus species in indoor environments and their possible occupational and public health hazards. Med Mycol. 2(1):36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat E, Butcher J, Lang S, Williams C, Ramage G. 2007. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J Med Microbiol. 56(Pt 9):1205–1212. eng. [DOI] [PubMed] [Google Scholar]
- Mowat E, Lang S, Williams C, McCulloch E, Jones B, Ramage G. 2008. Phase-dependent antifungal activity against Aspergillus fumigatus developing multicellular filamentous biofilms. J Antimicrob Chemother. 62(6):1281–1284. [DOI] [PubMed] [Google Scholar]
- Muszkieta L, Beauvais A, Pähtz V, Gibbons JG, Anton Leberre V, Beau R, Shibuya K, Rokas A, Francois JM, Kniemeyer O, et al. 2013. Investigation of Aspergillus fumigatus biofilm formation by various “omics” approaches. Front Microbiol. 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett JE, Crawford K, Marchillo K, Andes DR. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob Agents Chemother. 54(8):3505–3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor FJ, Guarro J. 2014. Treatment of Aspergillus terreus infections: a clinical problem not yet resolved. Int J Antimicrob Agents. 44(4):281–289. eng. [DOI] [PubMed] [Google Scholar]
- Paul RA, Rudramurthy SM. 2015. A novel Y319H substitution in CYP51C associated with azole resistance in Aspergillus flavus. Antimicrob Agents chemother . 59(10):6615–6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Ludeña Y, Villena GK, Yu F, Sherman DH, Gutiérrez-Correa M. 2017. High-quality draft genome sequence of a biofilm forming lignocellulolytic Aspergillus niger strain ATCC 10864 [journal article]. Stand Genomic Sci. 12(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell KA, Renwick A, Peberdy JF, eds. 2013. The genus Aspergillus: from taxonomy and genetics to industrial application Vol. 69 USA: Springer Science & Business Media. [Google Scholar]
- Rajendran R, Mowat E, McCulloch E, Lappin DF, Jones B, Lang S, Majithiya JB, Warn P, Williams C, Ramage G. 2011. Azole resistance of Aspergillus fumigatus biofilms is partly associated with efflux pump activity. Antimicrob Agents Chemother. 55(5):2092–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero-Menendez O, Alastruey-Izquierdo A, Mellado E, Cuenca-Estrella M. 2016. Triazole resistance in Aspergillus spp.: a worldwide problem? J Fungi. 2(3):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EMF, Garcia-Effron G, Park S, Perlin DS. 2007. A Ser678Pro substitution in Fks1p confers resistance to echinocandin drugs in Aspergillus fumigatus. Antimicrob Agents Chemother. 51(11):4174–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CH, Perrone G, Seifert KA, Susca A, Tanney JB, et al. 2014. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 78:141–173. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D. 2016. Emerging threats in antifungal-resistant fungal pathogens. Front Med. 3:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Odds FC. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect Dis. 2(2):73–85. eng. [DOI] [PubMed] [Google Scholar]
- Seidler MJ, Salvenmoser S, Müller F-MC. 2008. Aspergillus fumigatus forms biofilms with reduced antifungal drug susceptibility on bronchial epithelial cells. Antimicrob Agents Chemother. 52(11):4130–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedmousavi S, Melchers WJG, Mouton JW, Verweij PE. 2013. Pharmacodynamics and dose-response relationships of liposomal amphotericin b against different azole-resistant Aspergillus fumigatus isolates in a murine model of disseminated aspergillosis. Antimicrob Agents Chemother. 57(4):1866–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoury-Elizeh M, Protchenko O, Berger A, Cox J, Gable K, Dunn TM, Prinz WA, Bard M, Philpott CC. 2010. Metabolic response to iron deficiency in Saccharomyces cerevisiae. J Biol Chem. 285(19):14823–14833. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar J, Tiwari S, Shishodia SK, Gangwar M, Hoda S, Thakur R, Vijayaraghavan P. 2018. Molecular insights into development and virulence determinants of Aspergilli: a proteomic perspective [Review]. Front Cell Infect Microbiol. 8(180):180 English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C, Kumar R, Kumar N, Masih A, Gupta D, Chowdhary A. 2018. Investigation of multiple resistance mechanisms in voriconazole-resistant Aspergillus flavus clinical isolates from a chest hospital surveillance in Delhi, India. Antimicrob Agents Chemother. 62:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Gupta S, Singh B, Sharma SK, Gupta VK, Sharma GL. 2012. Proteomic characterization of Aspergillus fumigatus treated with an antifungal coumarin for identification of novel target molecules of key pathways. J Proteome Res. 11(6):3259–3268. [DOI] [PubMed] [Google Scholar]
- Snelders E, Camps SM, Karawajczyk A, Schaftenaar G, Kema GH, van der Lee HA, Klaassen CH, Melchers WJ, Verweij PE. 2012. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS One. 7(3):e31801 eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E, Huis in ‘T Veld RA, Rijs AJ, Kema GH, Melchers WJ, Verweij PE. 2009. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl Environ Microbiol. 75(12):4053–4057. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E, Karawajczyk A, Schaftenaar G, Verweij PE, Melchers WJ. 2010. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob Agents Chemother. 54(6):2425–2430. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen LM, Lametsch R, Andersen MR, Nielsen PV, Frisvad JC. 2009. Proteome analysis of Aspergillus niger: lactate added in starch-containing medium can increase production of the mycotoxin fumonisin B2 by modifying acetyl-CoA metabolism. BMC Microbiol. 9:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach WJ, Benjamin DK Jr., Kontoyiannis DP, Perfect JR, Lutsar I, Marr KA, Lionakis MS, Torres HA, Jafri H, Walsh TJ. 2004a. Infections due to Aspergillus terreus: a multicenter retrospective analysis of 83 cases. Clin Infect Dis. 39(2):192–198. eng. [DOI] [PubMed] [Google Scholar]
- Steinbach WJ, Perfect JR, Schell WA, Walsh TJ, Benjamin DK Jr.. 2004b. In vitro analyses, animal models, and 60 clinical cases of invasive Aspergillus terreus infection. Antimicrob Agents Chemother. 48(9):3217–3225. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulou T, Meletiadis J, Sein T, Papaioannidou P, Walsh TJ, Roilides E. 2011. Synergistic interaction of the triple combination of amphotericin B, ciprofloxacin, and polymorphonuclear neutrophils against Aspergillus fumigatus. Antimicrob Agents Chemother. 55(12):5923–5929. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui JA, Kwon-Chung KJ, Juvvadi PR, Latgé J-P, Steinbach WJ. 2015. Aspergillus fumigatus and related species. Cold Spring Harb Perspect Med. 5(2):a019786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M-J, Fedorova ND, Cagas SE, Hastings S, Fleischmann RD, Peterson SN, Perlin DS, Nierman WC, Pieper R, Momany M. 2012. Development stage-specific proteomic profiling uncovers small, lineage specific proteins most abundant in the Aspergillus fumigatus conidial proteome. Proteome Sci. 10:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szigeti G, Kocsube S, Doczi I, Bereczki L, Vagvolgyi C, Varga J. 2012. Molecular identification and antifungal susceptibilities of black Aspergillus isolates from otomycosis cases in Hungary. Mycopathologia. 174(2):143–147. eng. [DOI] [PubMed] [Google Scholar]
- Teutschbein J, Albrecht D, Potsch M, Guthke R, Aimanianda V, Clavaud C, Latge JP, Brakhage AA, Kniemeyer O. 2010. Proteome profiling and functional classification of intracellular proteins from conidia of the human-pathogenic mold Aspergillus fumigatus. J Proteome Res. 9(7):3427–3442. eng. [DOI] [PubMed] [Google Scholar]
- Thakur R, Anand R, Tiwari S, Singh AP, Tiwary BN, Shankar J. 2015. Cytokines induce effector T-helper cells during invasive aspergillosis; what we have learned about T-helper cells? Front Microbiol. 6:429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur R, Shankar J. 2017. Proteome profile of Aspergillus terreus Conidia at germinating stage: identification of probable virulent factors and enzymes from mycotoxin pathways. Mycopathologia. 182(9–10):771–784. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Shankar J. 2018a. Hsp70 in fungi: evolution, function and vaccine candidate In: Asea AAA, Kaur P, editors. HSP70 in human diseases and disorders. Cham: Springer International Publishing; p. 381–400. [Google Scholar]
- Tiwari S, Gupta N, Malairaman U, Shankar J. 2017. Anti-aspergillus properties of phytochemicals against aflatoxin producing aspergillus flavus and Aspergillus parasiticus. Natl. Acad. Sci. Lett. 40:267. [Google Scholar]
- Tiwari S. 2018. Evaluation of proteins involved in germination of toxigenic Aspergillus flavus Conidia and studies on phytochemicals as anti-aflatoxigenic agents. PhD thesis, Jaypee University of Information technology; Solan; HP. http://hdl.handle.net/10603/221822 [Google Scholar]
- Tiwari S, Shankar J. 2018b. Integrated proteome and HPLC analysis revealed quercetin-mediated inhibition of aflatoxin B1 biosynthesis in Aspergillus flavus. 8(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Thakur R, Goel G, Shankar J. 2016. Nano-LC-Q-TOF analysis of proteome revealed germination of Aspergillus flavus Conidia is accompanied by MAPK signalling and cell wall modulation. Mycopathologia. 181(11–12):769–786. [DOI] [PubMed] [Google Scholar]
- Tiwari S, Thakur R, Shankar J. 2015. Role of heat-shock proteins in cellular function and in the biology of fungi. Biotechnol Res Int. 2015:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiante V, Macheleidt J, Föge M, Brakhage AA. 2015. The Aspergillus fumigatus cell wall integrity signaling pathway: drug target, compensatory pathways, and virulence. Front Microbiol. 6:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden JWM, Arendrup MC, Warris A, Lagrou K, Pelloux H, Hauser PM, Chryssanthou E, Mellado E, Kidd SE, Tortorano AM, et al. 2015. Prospective multicenter international surveillance of Azole resistance in Aspergillus fumigatus. Emerg Infect Dis. 21(6):1041–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden JWM, Snelders E, Kampinga GA, Rijnders BJA, Mattsson E, Debets-Ossenkopp YJ, Kuijper EJ, Van Tiel FH, Melchers WJG, Verweij PE. 2011. Clinical implications of Azole resistance in Aspergillus fumigatus, the Netherlands, 2007–2009. Emerg Infect Dis. 17(10):1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen E, Carpentier S, Kniemeyer O, Sillen M, Maertens J, Lagrou K. 2018. Proteomic differences between Azole-susceptible and -resistant Aspergillus fumigatus strains. Adv Microbiol. 08(01):23. [Google Scholar]
- Verweij PE, Chowdhary A, Melchers WJG, Meis JF. 2016. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal Azoles? Clin Infect Dis. 62(3):362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villena GK, Venkatesh L, Yamazaki A, Tsuyumu S, Gutiérrez-Correa M. 2009. Initial intracellular proteome profile of Aspergillus niger biofilms. Revista Peruana de Biología. 16:1. [Google Scholar]
- Vodisch M, Albrecht D, Lessing F, Schmidt AD, Winkler R, Guthke R, Brakhage AA, Kniemeyer O. 2009. Two-dimensional proteome reference maps for the human pathogenic filamentous fungus Aspergillus fumigatus. Proteomics. 9(5):1407–1415. eng. [DOI] [PubMed] [Google Scholar]
- Walsh TJ, Petraitis V, Petraitiene R, Field-Ridley A, Sutton D, Ghannoum M, Sein T, Schaufele R, Peter J, Bacher J. 2003. Experimental pulmonary aspergillosis due to Aspergillus terreus: pathogenesis and treatment of an emerging fungal pathogen resistant to amphotericin B. J Infect Dis. 188(2):305–319. [DOI] [PubMed] [Google Scholar]
- Yu L, Zhang W, Wang L, Yang J, Liu T, Peng J, Leng W, Chen L, Li R, Jin Q. 2007. Transcriptional profiles of the response to ketoconazole and amphotericin B in Trichophyton rubrum. Antimicrob Agents Chemother. 51(1):144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang Y, Zhou Y, An S, Zhou Y, Cheng J. 2002. Response of gene expression in Saccharomyces cerevisiae to amphotericin B and nystatin measured by microarrays. J Antimicrob Chemother. 49(6):905–915. [DOI] [PubMed] [Google Scholar]
- Zhang M, Su X, Sun W-K, Chen F, Xu X-Y SY. 2014. Efficacy of the combination of Voriconazole and Caspofungin in experimental Pulmonary Aspergillosis by different Aspergillus species. Mycopathologia. 177(1–2):11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmeili OS, Soubani AO. 2007. Pulmonary aspergillosis: a clinical update. Qjm. 100(6):317–334. eng. [DOI] [PubMed] [Google Scholar]
- Zoran T, Sartori B, Sappl L, Aigner M, Sánchez-Reus F, Rezusta A, Chowdhary A, Taj-Aldeen SJ, Arendrup MC, Oliveri S, et al. 2018. Azole-resistance in Aspergillus terreus and Related Species: an emerging problem or a rare phenomenon? [Original Research]. Front Microbiol. 9:516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


