Abstract
Two manganese peroxidases (MnPs), MnP1 and MnP2, and a laccase, Lac1, were purified from Trametes polyzona KU-RNW027. Both MnPs showed high stability in organic solvents which triggered their activities. Metal ions activated both MnPs at certain concentrations. The two MnPs and Lac1, played important roles in dye degradation and pharmaceutical products deactivation in a redox mediator-free system. They completely degraded Remazol brilliant blue (25 mg/L) in 10–30 min and showed high degradation activities to Remazol navy blue and Remazol brilliant yellow, while Lac1 could remove 75% of Remazol red. These three purified enzymes effectively deactivated tetracycline, doxycycline, amoxicillin, and ciprofloxacin. Optimal reaction conditions were 50 °C and pH 4.5. The two MnPs were activated by organic solvents and metal ions, indicating the efficacy of using T. polyzona KU-RNW027 for bioremediation of aromatic compounds in environments polluted with organic solvents and metal ions with no need for redox mediator supplements.
Keywords: Manganese peroxidase, Trametes polyzona, dye decolorization, pharmaceutical product degradation, redox mediator-free system
1. Introduction
Persistent contaminants include wide groups of chemicals as synthetic dyes, pharmaceutical products, polycyclic aromatic hydrocarbons, and pesticides. They remain biologically active at trace concentrations, and adversely impact on both aquatic environments and human health [1]. Synthetic dyes are highly toxic, mutagenic, and carcinogenic substances [2]. The World Health Organization (WHO) classifies azo dyes as carcinogens in categories I and II. Exposure to a non-lethal dose of pharmaceutical products, especially antibiotics, induces adaptation of drug resistant genes in pathogenic bacteria which causes serious public health issues [3].
Ligninolytic enzymes including manganese peroxidase (MnP), laccase, and lignin peroxidase (LiP), in particular from white rot fungi, have oxidizing abilities which make them attractive candidates for an eco-friendly approach to bioremediation of persistent phenolic and non-phenolic contaminants. [4–8]. Manganese peroxidase (EC 1.11.1.13, Mn(II): hydrogen-peroxide oxidoreductase) is a member of class II fungal secretion heme peroxidase in subfamily A.2. It catalyzes H2O2-dependent oxidation of Mn2+ to Mn3+ and forms complexes with dicarboxylic acids, oxalic acid or malonic acid, etc. Chelated Mn3+ subsequently oxidizes varieties of phenolic and non-phenolic compounds. MnPs, of Trametes versicolor, Bjerkandera adusta and Cerrena unicolor were proved stable in conditions with either Mn2+, Cu2+ , and Co2+ and also activated their enzyme activity [7,9,10]. Conversely, some MnPs were reported to be unstable with metal ions [11–13]. MnPs of T. versicolor and B. adusta were completely inhibited by 20 mM Hg2+ and 5 mM Ag+, respectively [7,9]. However, Mn2+, Ni2+, Li+, K+, and Ca2+ were not harmful to MnP of Trametes sp. [12]. Laccase (EC 1.10.3.2, benzenediol: dioxygen oxidoreductase) belongs to a multicopper oxidase family. The enzyme catalyzes oxidation of various phenolic compounds coupled with reducing oxygen to water. Laccases are widely distributed in fungi, insects, plants, and bacteria [14–18]. Many laccases have been reported from genus Trametes including T. versicolor [9], T. trogii [19], T. hirsuta [20], T. polyzona [21], T. maxima [22] and Trametes sp. [23]. These ligninolytic enzymes carry out many important functions involved in lignin synthesis and degradation of plant cell walls as well as morphogenesis of fungal fruiting body formation, pathogenicity, and stress responses [24–27]. These functions and applications of ligninolytic enzymes excite interest in studying and understanding enzyme structure, biochemical characteristics, and genes.
The white rot fungus Trametes polyzona KU-RNW027 has recently demonstrated high potential in decolorizing various synthetic dyes [28]. Here, purification and characterization of ligninolytic enzymes from T. polyzona KU-RNW027 gave two MnPs and one laccase which were proven to play important roles in dye degradation and pharmaceutical products deactivation. Both MnPs were remarkably stable in various organic solvents and metal ions which activated their activities. Results offered new insight into MnPs with novel properties for bioremediation.
2. Materials and methods
2.1. Strains and culture condition
T. polyzona KU-RNW027 was maintained on potato dextrose agar (PDA) and kept in 20% glycerol at −20 °C for long-term preservation. Cultivation of the fungus was carried out in Kirk’s liquid medium [29] supplemented with 25 mg/L of Remazol brilliant red F3B gran with shaking at 130 rpm for 5 days under room temperature. Culture supernatants were used as a source of enzymes.
2.2. Enzyme purification
Culture supernatant of T. polyzona KU-RNW027 was concentrated by an Amicon ultrafiltration system using a 30 kDa molecular weight cut off Millipore membrane at 4 °C. Concentrated enzyme was applied onto a Toyopearl® DEAE 650 M anion exchange chromatography column with 50 mM Tris-HCl (pH 7.5) as an elution buffer containing 0–1 M NaCl with an elution rate of 0.33 mL/min. Fractions of each MnP and laccase activities were collected separately and further subjected to a Toyopearl® HW-55 gel filtration chromatography column with 50 mM phosphate elution buffer (pH 7.0) at 0.33 mL/min.
It was noted that numbers of fraction collected would depend on the profiles of protein, activity, and heme. Non-denaturing polyacrylamide gel electrophoresis was used at the final step for laccase. Quantification of protein followed Lowry-Folin [30] or Bradford [31]. Bovine serum albumin (BSA) was used as the standard. Enzyme purification and molecular mass, as well as enzyme subunit, were determined using SDS-PAGE [32]. Molecular weight markers were obtained from Thermo Scientific (Waltham, MA). Protein bands were visualized with Coomassie brilliant blue R-250. After non-denaturing SDS-PAGE, the zymogram was visualized using a staining buffer consisted of 1 mM of 2,6-dimethoxyphenol (2,6-DMP), 1 mM of Mn2+, and 0.1 mM H2O2 in 50 mM malonate buffer, pH 4.5.
2.3. Enzyme assays
MnP and laccase assays followed previously described methods [6,33]. MnP and laccase activities were determined by monitoring oxidation of 2,6-DMP at 469 nm. One unit (U) of either MnP or laccase was defined as 1 µmol of 2,6-DMP oxidized per min. Control reaction with a denatured enzyme was carried out in parallel.
2.4. Kinetic measurements
Kinetic constants, Michaelis-Menten constant (K m) and maximum reaction velocity (V max) values, of the purified enzymes were determined to obtain their initial activities with various substrates. Results were computed by line interceptions on X and Y axes of Lineweaver-Burk plots. Each reaction was performed at optimal pH and temperature, varying concentrations of each substrate at 0.125–2.5 mM. Five substrates as Mn2+, H2O2, 2,6-DMP, 2,2'-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) (ABTS), and guaiacol, were used for MnPs. ABTS, 2,6-DMP, and guaiacol were used for laccase.
2.5. Effect of temperature on enzyme activity and stability
Temperatures were varied from 20 to 90 °C to determine optimal values for each enzyme activity. The reaction was carried out at pH 4.5. Thermal stability was estimated by pre-incubating the purified enzyme at 20–90 °C for 1 h. Residual activities were determined at pH 4.5 and 30 °C.
2.6. Effect of pH on enzyme activity and stability
Reactions were carried out at pH 2–10 at 30 °C to determine optimal pH while enzyme stabilities were estimated by pre-incubating each purified enzyme in buffers of pH 2–10 for 1 h. Residual activities were determined at pH 4.5 and 30 °C. Buffer systems (each 50 mM) were KCl-HCl for pH 2.0–2.5, glycine-HCl for pH 2.5–3.5, sodium malonate for pH 3.0–4.5, sodium citrate-phosphate for pH 4.0–5.5, sodium phosphate for pH 5.0–8.0, Tris-HCl for pH 7.5–9.0, and glycine-NaOH for pH 8.5–10.0.
2.7. Effect of metal ions
Metal ions as Na+, K+, NH4 +, Ca2+, Mg2+, Cu2+, Mn2+, Fe2+, Co2+, Zn2+, Ag+, and Hg2+ were added to each reaction mixture at final concentrations of 1, 5, and 10 mM. Reaction mixtures were incubated at 4 °C for 1 h, and then 2,6-DMP was added. Remaining activities were measured. The reaction without metal ions as the control was expressed as 100%. All experiments were performed in triplicate.
2.8. Effect of organic solvents and inhibitors
Organic solvents and inhibitors including methanol, ethanol, propanol, isopropanol, butanol, acetone, NaN3, ethylenediaminetetraacetic acid (EDTA), nitrilotriacetic acid (NTTA), Tween 20, and Tween 80 at various concentrations were added to each reaction mixture and incubated at 4 °C for 1 h. Remaining activities were measured. The reaction without organic solvents or inhibitors as the control was expressed as 100%. All experiments were performed in triplicate.
2.9. Substrate specificity
Substrate specificities were determined with ABTS, 2,6-DMP, guaiacol, catechol, hydroquinone, resorcinol, caffeic acid, syringic acid, 4-hydroxy cinnamic acid, veratryl alcohol, phenol, and tyrosine at final concentration of 1 mM. All experiments were performed in triplicate.
2.10. LC-MS/MS peptide fingerprinting of two MnPs
Purified MnP proteins were excised from the SDS-PAGE gel for trypsin in-gel digestion. Digested peptides were analyzed by LC-MS/MS at Proteomics Service Center, Faculty of Medical Technology, Mahidol University, Thailand. LC-MS/MS data were analyzed using the MASCOT search program (www.matrixscience.com) against the National Center for Biotechnology Information (NCBI) database.
2.11. Enzyme capability in dye decolorization
One anthraquinone dye, Remazol brilliant blue R (RBBR, λ max 596) and 5 azo dyes as Remazol golden yellow RGB gran (RGY, λ max 410), Remazol brilliant yellow 4GL gran (RBY, λ max 405), Remazol red RGB gran (RR, λ max 520), Remazol brilliant red F3B gran (RBR, λ max 540), and Remazol navy blue RGB 150% gran (RNB, λ max 610) were used at final concentration 25 mg/L. The reaction mixture contained 1 U/mL of each purified enzyme in 50 mM malonate buffer (pH 4.5). Final concentration of 0.1 mM H2O2 and 1 mM Mn2+ were added to develop MnP activity. The reaction was shaken at 100 rpm for 7 days at 30 °C. Control with denatured enzyme was performed in parallel. All experiments were conducted in triplicate. Decolorization capability (%) was calculated following Equation (1).
| (1) |
where, Cc and Ct are dye concentrations of control and test experiment, respectively. Standard curves were plotted between absorbance at λ max of each dye and dye concentration from 0 to 60 mg/L.
2.12. Enzyme capability for antibiotics deactivation
Tetracycline, doxycycline, amoxicillin or ciprofloxacin at 25 mg/L were added to the reaction containing 1 U/mL of each purified enzyme in 50 mM malonate buffer, pH 4.5 at 30 °C. For MnP activity, 0.1 mM H2O2, and 1 mM Mn2+ were added and the reaction mixture was shaken at 100 rpm for 7 days. Enzyme capability in deactivating each antibiotic was evaluated using agar well diffusion assay on trypticase soy agar (TSA) with Staphylococcus aureus as the test organism. The bacterial suspension was adjusted to McFarland No. 0.5 turbidity and swabbed on the TSA. Fifty microliters of each sample were loaded into 5 mm diameter wells. Plates were incubated at 37 °C for 24 h. The control with denatured enzymes was included in parallel. All experiments were performed in triplicate. Antibiotic deactivation capability (%) was calculated following Equation (2).
| (2) |
where, Sc and St are sizes of the inhibition zones of the control and test experiment, respectively.
3. Results and discussion
3.1. Purification of MnP and laccase from T. polyzona KU-RNW027
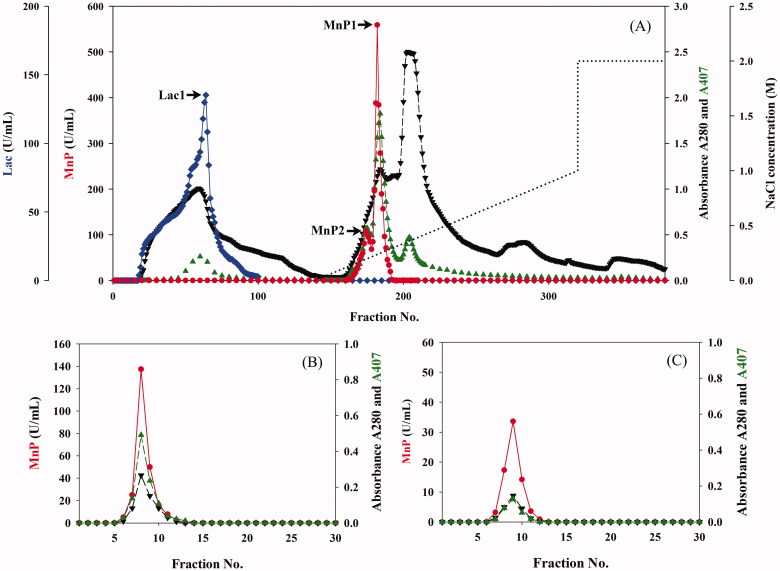
Two MnPs and a laccase of T. polyzona KU-RNW027 were successfully purified through 4 steps of ultrafiltration concentration, DEAE-Toyopearl® ion exchange, Toyopearl® 55HW gel filtration chromatography, and polyacrylamide gel electrophoresis as the main ligninolytic enzyme proteins. The three ligninolytic enzymes were partial purification on DEAE-toyopearl® as shown in Figure 1(A). Protein with laccase activity was eluted in void volume. Almost all of the fractions with laccase activity was collected and further loaded on Toyopearl® 55HW gel filtration column chromatography with 3 bed volume of 50 mM phosphate buffer, pH 7.0 as a elution buffer at a flow rate of 0.33 mL/min. Protein fraction with laccase activity were further purified on non-denaturing polyacrylamide gel electrophoresis. A protein band with laccase activity was carefully cut and eluted from gel with 50 mM malonate buffer, pH 4.5. The protein was proved to be pure and gave a single band on SDS-PAGE with laccase activity on zymogram (Figure 2(A,B)).
Figure 1.
Protein, activities and heme profile of three ligninolytic enzymes of Trametes polyzona KU-RNW027 on Toyopearl® DEAE 650M anion exchange chromatography (A) and the profiles of MnP1 (B) and MnP2 (C) on Toyopearl® HW-55 gel filtration chromatography. MnP activity ( ), laccase activity (
), laccase activity ( ), protein at 280 nm (
), protein at 280 nm ( ) and heme at 407 nm (
) and heme at 407 nm ( ) and NaCl gradient (
) and NaCl gradient ( ).
).
Figure 2.
Ligninolytic enzymes produced by Trametes polyzona KU-RNW027. SDS-PAGE of the enzymes staining by Coomassie brilliant blue R-250 (A) and Zymogram (B) Lane M: molecular weight markers, lane 1: Lac1, lane 2: MnP1, and lane 3: MnP2.
Protein with MnP activity was adsorbed with DEAE ion exchange chromatography and eluted with gradient 50 mM Tris-HCl, pH 7.5 containing 0–1 mM NaCl. With 9 times of the elution buffer to the column bed volume, two peaks of MnP activity and heme was partially purified from the other contaminated protein. Only one fraction at the top peak of each MnP activity was separately collected. Each fraction was further purified on to Toyopearl® 55HW gel filtration column chromatography and eluted with 3 bed volume of 50 mM phosphate buffer, pH 7.0. Protein, MnP activity and heme profiles on gel filtration showed single peaks at the same position (Figure 1(B,C)). On SDS-PAGE, bands of the 2 MnPs showed a single band with different Rf that indicated the molecular weight differences (Figure 2). Collections of laccase and the two MnPs were repeatedly done for characterization. Two MnP isozymes, named MnP1 and MnP2, were 29 and 13-folds purified with high specific activities of 484.56 and 207.18 U/mg, respectively (Table 1). A protein with laccase activity, named Lac1, was 15-fold with specific activities of 291.04 U/mg. Molecular masses of MnP1, MnP2, and Lac1 on SDS-PAGE were 44, 42, and 71 kDa, respectively (Figure 2(A)). A single activity band of each purified enzyme on zymogram is shown as Figure 2(B). Though, molecular mass of MnP1 and MnP2 were very close to each other, zymogram of the two MnPs showed clearly separation of their activities at different Rf. All three enzymes were monomeric proteins. Lac1 was different from plant laccase as hexameric proteins with high molecular weight ∼260 kDa [17]. Lac1 molecular mass was similar to that of T. polyzona WR710-1 [21]. Spectral analysis showed that purified MnP1 and MnP2 contained two peaks of amino acid, at 280 nm, and heme, at 407 nm. A single peak at 280 nm was observed in the spectrum of purified Lac1. Data indicated that MnP1 and MnP2 but not Lac1 contained heme as cofactor [34]. These purified enzymes were used for biochemical characterization.
Table 1.
Purification of MnPs (A) and laccase (B) from Trametes polyzona KU-RNW027.
| Purification step | Total activity (U) | Total protein (mg) | Specific activity | Recovery (%) | Purification fold | |
|---|---|---|---|---|---|---|
| (A) | ||||||
| Crude enzyme | 47610.00 | 2880.00 | 16.53 | 100.00 | 1.00 | |
| Ultrafiltration | 51447.90 | 2408.70 | 21.36 | 108.06 | 1.29 | |
| DEAE-toyopearl | MnP1 | 26271.95 | 231.84 | 113.32 | 55.18 | 6.85 |
| MnP2 | 8140.86 | 54.81 | 148.53 | 17.10 | 8.98 | |
| Gel-filtration | MnP1 | 2543.94 | 5.25 | 484.56 | 5.34 | 29.31 |
| MnP2 | 763.56 | 3.69 | 207.18 | 1.60 | 12.53 | |
| (B) | ||||||
| Crude enzyme | 56790.00 | 2880.00 | 19.72 | 100.00 | 1.00 | |
| Ultrafiltration | 42900.90 | 2408.70 | 17.81 | 75.54 | 0.90 | |
| DEAE-toyopearl | 17201.89 | 138.70 | 124.02 | 30.29 | 6.29 | |
| Gel-filtration | 1337.39 | 7.68 | 174.08 | 2.35 | 8.83 | |
| Electrophoresis | Lac1 | 849.68 | 2.92 | 291.04 | 1.50 | 14.76 |
3.2. Kinetics of the purified enzymes
Kinetics values, K m and V max, of the purified MnPs and laccase were determined with various substrates (Table 2). Lowest K m values of MnP1 and MnP2 were 0.005 and 0.036 mM, respectively for H2O2. Results indicated that both MnPs from T. polyzona KU-RNW027 had the highest affinity toward H2O2 unlike the MnP of Echinodontium taxodii 2538 that showed higher affinity for Mn2+ than H2O2 [13]. The K m value of Lac1 showed the highest affinity toward ABTS at 0.29 mM, comparable to the K m value of a laccase from T. polyzona WR701-1 that also had higher affinity for ABTS (0.15 mM) than 2,6-DMP (0.503 mM) [21]. By contrast, a bacterial laccase of Klebsiella pneumoniae showed higher affinity for 2,6-DMP than ABTS [35].
Table 2.
Kinetic parameters of purified ligninolytic enzymes produced by Trametes polyzona KU-RNW027.
| Substrate | Λ (nm) |
Ε (cm-1·mM-1) |
Km (mM) |
Vmax (mM·min-1) |
||||
|---|---|---|---|---|---|---|---|---|
| MnP1 | MnP2 | Lac1 | MnP1 | MnP2 | Lac1 | |||
| Mn2+ | 240 | 6.5 | 0.150 | 0.330 | ND | 769.23 | 833.33 | ND |
| H2O2 | 240 | 6.5 | 0.005 | 0.036 | ND | 666.67 | 909.09 | ND |
| 2,6-DMP | 469 | 49.6 | 0.006 | 0.340 | 0.320 | 125.00 | 169.49 | 294.12 |
| ABTS | 420 | 36.0 | 0.110 | 0.290 | 0.290 | 384.61 | 416.67 | 1428.52 |
| Guaiacol | 465 | 12.1 | 0.310 | 1.720 | 0.440 | 114.94 | 344.82 | 30.30 |
ND: not determined.
3.3. Effect of temperature
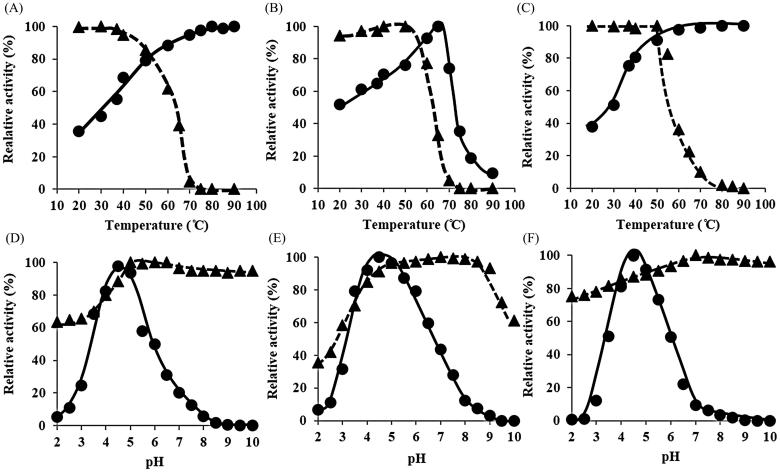
MnP1 exhibited higher activity with increasing reaction temperature up to 90 °C but was not stable at high temperature (Figure 3(A)). MnP1 activity remained 100% after 1 h-incubation only at temperatures not higher than 37 °C. Incubation at 70 °C almost completely negated MnP1 activity. On the other hand, MnP2 showed optimal temperature at 70 °C (Figure 3(B)). The enzyme rapidly lost activity at temperatures higher than 50 °C. Activity of MnP2 was stable at temperatures not higher than 50 °C. At 60 °C, the enzyme retained 80% of the original activity but was rapidly denatured with increasing temperature within 1 h. Lac1 activity increased progressively as temperature increased from 20 to 50 °C and reached 100% at 60–90 °C (Figure 3(C)). As with MnPs, Lac1 was stable only up to 50 °C. No activity was exhibited at 80 °C. In brief, MnP1, MnP2, and Lac1 of T. polyzona KU-RNW027 showed high optimal temperature at over 50 °C but were stable up to 40 or 50 °C. As with Lac1, KUlac3 of Ganoderma sp. KU-Alk4 exhibited activity at high temperature (90 °C) but was unstable [14]. Each MnP of B. adusta CX-9 and Trametes sp. 48424 was optimally active at 70 °C but stable at lower temperatures of 60 and 50 °C, respectively [7,12].
Figure 3.
Effect of temperature and pH on the activities of 3 purified ligninolytic enzymes produced by Trametes polyzona KU-RNW027 was shown as optimal temperature ( ) and thermal stability (
) and thermal stability ( ) of MnP1 (A), MnP2 (B) and Lac1 (C) Optimal pH (
) of MnP1 (A), MnP2 (B) and Lac1 (C) Optimal pH ( ) and pH stability (
) and pH stability ( ) of MnP1 (D), MnP2 (E) and Lac1 (F). Values are averages of three independent experiments with maximal mean deviation of ± 5%.
) of MnP1 (D), MnP2 (E) and Lac1 (F). Values are averages of three independent experiments with maximal mean deviation of ± 5%.
3.4. Effect of pH
MnP1, MnP2, and Lac1 had the same optimal pH at 4.5 (Figure 3(D–F)), similar to ligninolytic enzymes of Trametes sp. 48424 and T. polyzona WR710-1 [12,21]. Activities of ligninolytic enzymes of B. adusta and Ganoderma sp. had optimal pH at 3.0–3.5 [7,14], while those of Trichoderma harzianum were at pH 6.0 [15]. By contrast, MnP of Schizophyllum sp. F17 had optimal activity at neutral pH (pH 6.8) [11]. All of the three purified enzymes of T. polyzona KU-RNW027 showed stability over a wide range of pH from 5.0 to alkaline pH (Figure 3(D–F)). MnP1 had more promising pH stability with activity after incubation at pH 5.0–10.0 for 1 h still at 100%. MnP1 activity in low pH, 4.0–5.0 and 2.0–3.0 still retained 80 and 60% of initial activity, respectively, while MnP2 activity decreased more at low pH range. On the other hand, at pH 10.0, MnP2 activity was retained 60%. Lac1 activity was 100% stable at pH 7.0–10.0 and then slightly decreased at pH value ranging below 7.0 (Figure 3(F)). However, at pH 2.0, its activity still remained more than 70%. All ligninolytic enzymes of T. polyzona KU-RNW027 were 100% stable in natural and alkaline pH. Both MnPs were 100% stable at pH 5.0. Stabilities of the three enzymes at pH lower than 5.0 were still 40-75%. Due to optimum pH at low pH of 4.5 and wide range pH stability of the three purified enzymes, it showed promising for bioremediation of polluted lignin derivatives, pharmaceutical products, pesticide, herbicide, and insecticide in the acidic soil areas with pH 3.5–5.0. The low pH soil and water have been found in the provinces north of Bangkok, Thailand, which is agricultural and agro-industrial area.
3.5. Effect of metal ions
Effects of metal ions on the activity of each purified enzyme are shown in Table 3. MnP1 was activated by almost all of the metal ions at 10 mM concentration except for K+ that slightly inhibited its activity. Notably MnP1 was activated by Ag+ and Hg2+ at concentrations as high as 10 mM. In most cases, MnP and laccase were fully inhibited by Hg2+ and Ag+ [7,22]. MnP2 was activated by, 10 mM Hg2+ and also by Mn2+, Na+, and K+ but not Ag+, Fe2+, Cu2+, and Zn2+. In case of Lac1, almost all metal ions except Ag+ and Mn2+ inhibited its activity. Ag+ at low concentration (1 mM) highly activated Lac1. Higher concentrations of Mn2+ increased activity of Lac1. Fe2+ and Co2+ were the most potent inhibitors leading to reduction of Lac1 activity. Results were similar to laccase from T. maxima [22].
Table 3.
Effect of metal ions on activity of purified ligninolytic enzymes produced by Trametes polyzona KU-RNW027.
| Relative activity (%) |
||||
|---|---|---|---|---|
| Reagent | Final concentration | MnP1 | MnP2 | Lac1 |
| Metal ion (mM) | ||||
| Na+ | 1 | 101.3 ± 0.2 | 120.5 ± 0.7 | 95.4 ± 1.1 |
| 5 | 102.9 ± 0.5 | 127.5 ± 1.0 | 86.8 ± 0.3 | |
| 10 | 108.3 ± 1.2 | 143.0 ± 2.3 | 65.5 ± 0.5 | |
| K+ | 1 | 96.2 ± 0.3 | 149.5 ± 1.5 | 99.0 ± 0.6 |
| 5 | 91.8 ± 0.6 | 135.5 ± 2.0 | 95.0 ± 0.1 | |
| 10 | 90.1 ± 0.5 | 124.0 ± 0.5 | 89.5 ± 0.2 | |
| NH4+ | 1 | 131.0 ± 0.2 | 93.00 ± 0.9 | 100.4 ± 0.3 |
| 5 | 136.5 ± 0.7 | 92.7 ± 0.3 | 100.0 ± 0.5 | |
| 10 | 133.0 ± 0.5 | 89.5 ± 0.7 | 100.8 ± 1.0 | |
| Ca2+ | 1 | 138.0 ± 0.4 | 95.5 ± 1.1 | 93.3 ± 0.6 |
| 5 | 134.5 ± 0.7 | 95.2 ± 0.3 | 89.4 ± 1.0 | |
| 10 | 126.5 ± 1.0 | 91.1 ± 0.5 | 82.3 ± 0.3 | |
| Mg2+ | 1 | 140.0 ± 0.5 | 97.1 ± 0.5 | 99.2 ± 0.7 |
| 5 | 141.5 ± 1.4 | 97.1 ± 0.8 | 92.9 ± 0.9 | |
| 10 | 137 ± 0.3 | 94.6 ± 0.9 | 91.7 ± 1.2 | |
| Cu2+ | 1 | 146.5 ± 0.2 | 102.2 ± 0.7 | 100.4 ± 0.2 |
| 5 | 135.0 ± 0.5 | 93.3 ± 0.4 | 100.8 ± 0.6 | |
| 10 | 90.5 ± 0.1 | 77.4 ± 0.5 | 100.0 ± 0.5 | |
| Mn2+ | 1 | 151.5 ± 0.5 | 105.7 ± 0.7 | 99.6 ± 0.6 |
| 5 | 160.5 ± 0.9 | 108.9 ± 0.4 | 102.7 ± 0.9 | |
| 10 | 159.5 ± 0.2 | 115.0 ± 0.5 | 109.5 ± 1.1 | |
| Fe2+ | 0.01 | 141.0 ± 0.3 | 103.5 ± 1.2 | 99.6 ± 0.5 |
| 0.02 | ND | ND | 65.8 ± 0.3 | |
| 0.03 | ND | ND | 12.2 ± 0.5 | |
| 0.04 | ND | ND | 2.0 ± 0.2 | |
| 0.05 | 151.0 ± 0.5 | 96.5 ± 0.2 | 0.0 ± 0.0 | |
| 0.1 | 132.5 ± 0.7 | 70.7 ± 0.5 | 0.0 ± 0.0 | |
| 0.5 | 53.0 ± 1.1 | 33.4 ± 0.2 | 0.0 ± 0.0 | |
| 1 | 0.0 ± 0.1 | 2.2 ± 0.3 | 0.0 ± 0.0 | |
| 5 | ND | 0 ± 0.0 | 0.0 ± 0.0 | |
| 10 | ND | ND | ND | |
| Co2+ | 0.01 | ND | ND | 99.2 ± 0.7 |
| 0.05 | ND | ND | 106.3 ± 0.8 | |
| 0.1 | ND | ND | 103.1 ± 0.5 | |
| 0.5 | ND | ND | 102.4 ± 1.1 | |
| 0.75 | ND | ND | 39.8 ± 0.9 | |
| 1 | 153.5 ± 0.7 | 100.3 ± 0.8 | 0.0 ± 0.0 | |
| 5 | 139.0 ± 1.5 | 100.0 ± 0.5 | ND | |
| 10 | 108.0 ± 0.2 | 99.4 ± 0.7 | ND | |
| Zn2+ | 1 | 132.0 ± 0.3 | 100.6 ± 0.4 | 100.4 ± 0.8 |
| 5 | 120.5 ± 0.5 | 89.5 ± 0.5 | 109.1 ± 1.0 | |
| 10 | 114.0 ± 0.3 | 85.7 ± 0.2 | 99.6 ± 0.3 | |
| Ag+ | 1 | 161.5 ± 1.2 | 107.6 ± 0.8 | 172.1 ± 0.5 |
| 5 | 134.0 ± 0.5 | 83.8 ± 0.6 | 134.6 ± 1.3 | |
| 10 | 101.0 ± 0.7 | 60.2 ± 0.5 | 119.3 ± 0.7 | |
| Hg2+ | 1 | 100.5 ± 0.5 | 117.8 ± 0.4 | 100.4 ± 0.5 |
| 5 | 130.5 ± 1.0 | 115.0 ± 0.5 | 59.5 ± 0.1 | |
| 10 | 211.0 ± 0.9 | 102.2 ± 1.5 | 28.4 ± 0.1 | |
ND: not determined. Activation of the enzyme activities were indicated in bold.
3.6. Effect of organic solvents
In the presence of various organic solvents, MnP1 and MnP2, were activated, while Lac1 was mostly inhibited (Table 4). Unlike MnPs of Trametes sp. 48424 and Echinodontium taxodii 2538, their activities reduced in low water content systems with organic solvents including methanol, ethanol, and isopropanol [12,13]. MnP2 was more stable to organic solvents than MnP1. The former could tolerate to 20–50% concentration of all organic solvents. Butanol was the best solvent and activated MnP2 2-fold at 50% concentration. MnP2 was also activated by methanol, ethanol, propanol, isopropanol, and acetone, though not as high as butanol. MnP1 showed less stability to organic solvents, low concentrations (20–30%) almost all organic solvents activated its activity. Butanol was also the best organic solvent to trigger MnP1 activity 1.5-fold at 50% concentration. Lac1 could not tolerate low water content systems containing organic solvents. After incubation for 1 h in the organic solvent phase, Lac1 activity was mostly inhibited by all solvents tested, except 10–30% butanol. Under these conditions, its activity was similar to that in the aqueous phase. However, 40–50% butanol reduced Lac1 activity by only 2–4%.
Table 4.
Effect of organic solvent and inhibitor on activity of purified ligninolytic enzymes produced by Trametes polyzona KU-RNW027.
| Final concentration | Relative activity (%) |
|||
|---|---|---|---|---|
| MnP1 | MnP2 | Lac1 | ||
| Organic solvent (%) | ||||
| Methanol | 10 | 126.8 ± 0.2 | 161.0 ± 0.9 | 68.2 ± 0.8 |
| 20 | 112.1 ± 0.5 | 176.5 ± 1.2 | 52.6 ± 1.0 | |
| 30 | 84.7 ± 0.3 | 153.0 ± 0.5 | 37.0 ± 0.4 | |
| 40 | 32.1 ± 0.9 | 112.0 ± 0.3 | 29.2 ± 0.3 | |
| 50 | 22.1 ± 0.1 | 81.5 ± 0.5 | 13.0 ± 0.5 | |
| Ethanol | 10 | 127.4 ± 0.2 | 169.5 ± 0.8 | 71.4 ± 0.1 |
| 20 | 132.1 ± 0.3 | 188.5 ± 1.0 | 44.8 ± 0.5 | |
| 30 | 132.6 ± 0.5 | 176.0 ± 0.7 | 32.5 ± 1.0 | |
| 40 | 77.4 ± 1.1 | 117.5 ± 0.4 | 18.2 ± 0.1 | |
| 50 | 37.9 ± 0.4 | 85.0 ± 0.9 | 13.6 ± 0.4 | |
| Propanol | 10 | 159.4 ± 1.0 | 170.0 ± 1.1 | 76.0 ± 0.3 |
| 20 | 141.0 ± 0.3 | 104.5 ± 0.5 | 60.4 ± 0.9 | |
| 30 | 130.0 ± 0.5 | 61.0 ± 0.7 | 44.2 ± 1.0 | |
| 40 | 100.4 ± 0.4 | 49.5 ± 0.5 | 39.0 ± 0.3 | |
| 50 | 89.7 ± 0.9 | 48.0 ± 0.4 | 33.8 ± 0.5 | |
| Isopropanol | 10 | 119.5 ± 0.9 | 170.5 ± 0.1 | 65.6 ± 0.4 |
| 20 | 107.4 ± 0.5 | 131.0 ± 0.5 | 44.2 ± 0.2 | |
| 30 | 85.3 ± 0.3 | 57.0 ± 0.9 | 24.0 ± 0.7 | |
| 40 | 56.3 ± 0.2 | 43.0 ± 0.1 | 14.9 ± 0.1 | |
| 50 | 16.3 ± 1.0 | 39.5 ± 0.5 | 9.7 ± 0.5 | |
| Butanol | 10 | 151.1 ± 0.2 | 212.5 ± 0.8 | 105.2 ± 0.7 |
| 20 | 151.6 ± 0.5 | 226.0 ± 2.0 | 100.0 ± 0.8 | |
| 30 | 151.1 ± 0.3 | 215.5 ± 0.5 | 100.0 ± 0.3 | |
| 40 | 153.2 ± 0.5 | 206.5 ± 0.1 | 98.0 ± 0.5 | |
| 50 | 155.8 ± 0.3 | 201.5 ± 0.5 | 96.1 ± 0.3 | |
| Acetone | 10 | 130.5 ± 0.8 | 171.5 ± 0.4 | 99.4 ± 0.6 |
| 20 | 105.3 ± 1.2 | 131.5 ± 0.5 | 31.8 ± 0.5 | |
| 30 | 97.9 ± 0.3 | 120.0 ± 0.5 | 17.5 ± 0.8 | |
| 40 | 84.7 ± 0.5 | 97.5 ± 0.7 | 9.2 ± 0.1 | |
| 50 | 74.2 ± 0.1 | 74.0 ± 0.3 | 6.4 ± 0.3 | |
| Inhibitor (mM) | ||||
| NaN3 | 1 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Chelating agent (mM) | ||||
| EDTA | 1 | 76.1 ± 0.9 | 90.5 ± 0.7 | 100.0 ± 0.3 |
| 2 | 41.1 ± 0.1 | 81.0 ± 0.5 | 96.1 ± 0.7 | |
| 5 | 4.5 ± 0.5 | 73.0 ± 0.1 | 87.2 ± 1.0 | |
| 10 | 1.2 ± 0.1 | 45.0 ± 0.2 | 37.5 ± 0.4 | |
| 20 | 0.0 ± 0.0 | 6.0 ± 0.4 | 9.21 ± 0.2 | |
| 50 | ND | 0.0 ± 0.0 | 0.0 ± 0.0 | |
| NTTA | 1 | 95.2 ± 0.2 | 100.5 ± 0.5 | 100.0 ± 0.1 |
| 2 | 74.8 ± 0.5 | 100.0 ± 0.3 | 94.7 ± 0.5 | |
| 5 | 62.1 ± 0.3 | 100.0 ± 0.2 | 89.5 ± 0.3 | |
| 10 | 52.2 ± 0.8 | 96.0 ± 0.7 | 65.8 ± 0.8 | |
| 20 | 26.8 ± 0.4 | 61.0 ± 0.3 | 52.0 ± 0.2 | |
| 50 | 6.1 ± 0.2 | 9.0 ± 0.5 | 35.2 ± 0.6 | |
| Nonionic surfactant (%) | ||||
| Tween 20 | 1 | 107.0 ± 1.0 | 76.8 ± 0.4 | 93.8 ± 0.1 |
| 2 | 106.0 ± 1.2 | 73.9 ± 0.5 | 86.5 ± 0.6 | |
| 5 | 103.5 ± 0.5 | 70.7±0.1 | 52.3 ± 0.5 | |
| Tween 80 | 1 | 100.5 ± 0.5 | 94.9 ± 0.3 | 69.7 ± 0.2 |
| 2 | 100.0 ± 0.1 | 83.8 ± 0.5 | 55.3 ± 0.9 | |
| 5 | 100.0 ± 0.3 | 66.0 ± 0.1 | 39.1 ± 0.2 | |
ND: not determined. Activation of the enzyme activities were indicated in bold.
This phenomenon of enzyme activation by organic solvents, in particular MnP1 and MnP2, occurred because organic solvents lowered the water activity of enzymatic reactions, which changed the equilibrium resulted enzyme reactions in reverse [36]. Equilibrium-controlled enzyme activation was reported in an α-amylase, a hydrolase from Bacillus licheniformis. With 20–40% methanol, ethanol, propanol, butanol, propadiol, DMSO, and 1,4-dioxane, the α-amylase reactions were equilibrium-controlled and its activity was elevated. For application, MnPs of T. polyzona KU-RNW027 were more advantageous when both exhibited higher activities in the solvent phase than in the aqueous phase. Hazardous aromatic substances such as pesticides, insecticides, and herbicides which contaminate the environment dissolve better in the solvent phase than in the aqueous phase. Thus, properties of these two MnPs are promising for aromatic compound degradation in environment.
3.7. Effect of inhibitors
NaN3 at 1 mM, completely inhibited activity of purified MnP1, MnP2, and Lac1 (Table 4) by blocking electron transfer [37]. This observation contrasted with a laccase of Bacillus sp. on which NaN3 had no effect [18]. Two chelating agents as EDTA and NTTA had inhibitory effects on MnP1 even at 1 mM. A small amount of EDTA also inhibited MnP2 but it was not affected by 5 mM NTTA. Lac1 was less sensitive to 1 mM of both chelating agents. However, higher concentrations inhibited Lac1. EDTA and NTTA caused the chelation of metal ions from catalytic sites [9]. On the other hand, MnP1 was not inhibited by the two nonionic surfactants, Tween 20, and Tween 80 with activation in the condition containing 1–5% Tween 20. By contrast, both surfactants at 1–5% drastically inhibited MnP2 and Lac1 with increasing surfactant concentrations. Tween 20 inhibited enzyme activities less efficiently than Tween 80.
3.8. Substrate specificity
Substrate specificity of the purified enzymes toward 12 kinds of phenolic and non-phenolic substrates showed that all purified enzymes gave the highest substrate specificity with ABTS (Table 5). MnP1, MnP2, and Lac1 showed 51, 64, and 55% relative activity with 2,6-DMP, respectively. MnP1 had 30 and 25% activity with guaiacol and phenol, respectively. Less than 25% activity was observed with the others including catechol and no activity with veratryl alcohol, tyrosine, 4-hydroxy cinnamic acid, and hydroquinone. MnP2 had almost 43% activity with caffeic acid, 21% relative activity with guaiacol and low activity (1–16% relative activity) with syringic acid, catechol, phenol, resorcinol, 4-hydroxy cinnamic acid, and hydroquinone. MnP2 showed no activity with veratryl alcohol and tyrosine. Lac1 had very low activity toward the substrates except for ABTS and 2,6-DMP, with 12, 9 and 1% relative activity toward phenol, guaiacol, and catechol, respectively. Considering substrate structure including substitution on the phenolic ring, all three enzymes showed high preference for dimethoxyl-substituted phenol at the ortho-position, but not for substrates substituted at meta- and para-positions. By contrast, enzymes of Ganoderma sp. KU-Alk4 had strong preference for meta- and para-dimethoxy substituted phenol but not for ortho-diphenol [14]. Moreover, the three enzymes showed higher activity on guaiacol than catechol, implying that all enzymes preferred ortho-monomethoxy substituted phenol.
Table 5.
Substrate specificity of ligninolytic enzymes produced by Trametes polyzona KU-RNW027.
| Reagent | Wavelength (nm) |
Relative activity (%) |
||
|---|---|---|---|---|
| MnP1 | MnP2 | Lac1 | ||
| ABTS | 420 | 100.0 ± 0.0 | 100.0 ± 0.1 | 100.0 ± 0.2 |
| 2,6-DMP | 469 | 50.8 ± 0.5 | 63.5 ± 0.5 | 54.7 ± 0.8 |
| Guaiacol | 470 | 30.0 ± 0.7 | 20.8 ± 0.5 | 9.0 ± 0.3 |
| Catechol | 400 | 11.6 ± 1.2 | 10.4 ± 0.2 | 1.0 ± 0.2 |
| Hydroquinone | 400 | 0.0 ± 0.3 | 1.0 ± 0.1 | 0.0 ± 0.0 |
| Resorcinol | 400 | 2.4 ± 0.1 | 5.2 ± 0.4 | 4.3 ± 0.3 |
| Caffeic acid | 400 | 14.4 ± 0.4 | 42.7 ± 0.9 | 3.3 ± 0.5 |
| Syringic acid | 400 | 21.6 ± 1.1 | 15.6 ± 0.8 | 4.3 ± 0.8 |
| 4-hydroxy cinnamic acid | 400 | 0.0 ± 0.0 | 3.1 ± 0.4 | 0.0 ± 0.0 |
| Veratryl alcohol | 280 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Phenol | 270 | 25.2 ± 1.5 | 10.4 ± 0.6 | 11.9 ± 0.8 |
| Tyrosine | 280 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 |
3.9. Peptide fingerprinting of two MnPs
LC-MS/MS-based peptide fingerprinting confirmed that both MnP1 and MnP2 are fungal MnP. Results demonstrated their peptide coverages as 20.32 and 10.16% manganese peroxidase isozyme precursors of Lenzites gibbosa CB-1 (ACO92620.2) and T. versicolor (AAT90348.1), respectively.
3.10. Dye decolorization capability of purified MnP1, MnP2, and Lac1
MnP1, MnP2, and Lac1 of T. polyzona KU-RNW027 demonstrated efficiency in decolorizing all selected anthraquinone and azo dyes commonly used in industries, in particular, the textile industry (Table 6). Reactions for dye decolorization of the three purified enzymes were carried out in a redox mediator-free system. All enzymes showed the highest efficiency on RBBR, with complete decolorization of the anthraquinone dye by MnP1, MnP2, and Lac1 within 10, 10, and 30 min, respectively. A laccase of Trametes sp. F1635 was shown to have high efficiency to decolorize RBBR but only in a redox mediator system [38]. However, decolorization was 90% in 24 h in a reaction supplemented with violuric acid as a redox mediator. Peroxidases of B. adusta CX-9, LiP and MnP were reported to have strong efficiency on RBBR decolorization [7]. Its LiP played a role on dye degradation rather than MnP. Efficiency of the MnP of B. adusta CX-9 on RBBR decolorization was only 38% within 12 h, less than MnP of T. polyzona KU-RNW027. MnP of Irpex lacteus was reported to decolorize Reactive black 5, an anthraquinone dye 80% in 90 min [34]. The three enzymes of T. polyzona KU-RNW027 decolorized RNB at high efficiency in 7 days. MnP1 and MnP2 decolorized RNB at 92 and 83%, respectively, while complete decolorization of RNB was observed with Lac1. Lac1 was also effective for RR decolorization at 75% color elimination in 7 days. All enzymes decolorized 69% of RBY while less than 50% decolorization of RGY and RBR was detected in 7 days. Laccase of Trametes sp., Lac-01, had decolorizing ability on triarylmethane dyes, bromthymol blue and malachite green [23]. Laccase of Ganoderma sp. KU-Alk4 also showed high ability to decolorize Indigo carmine and 12 U/mL of the laccase completely degraded 25 mg/L of Indigoid dye in 24 h. Complete indigo carmine degradation was successfully demonstrated in a 5 L-airlift reactor for 14 cycles [6]. Addition of a redox mediator as violuric acid and acetosyringone was suggested to improve activity of ligninolytic enzymes in dye decolorization [38]. However, addition of a redox mediator in the reaction had disadvantages for practical use since the process resulted in higher environmental toxic release and increased cost [39]. All ligninolytic enzymes of T. polyzona KU-RNW027 showed promise for practical dye decolorization and the reactions could be handled without a redox mediator. Thus, these enzymes are promising catalysts for treatment of wastewater containing synthetic dyes.
Table 6.
Decolorization of 25 mg/L synthetic dyes by 1 U/mL of purified ligninolytic enzymes from Trametes polyzona KU-RNW027.
| Dye | Color index name (Chemical formula) | Chemical class | λmax (nm) | Chemical structure | Decolorization (%) |
||
|---|---|---|---|---|---|---|---|
| MnP1 | MnP2 | Lac1 | |||||
| RBBR | Reactive Blue 19 (C22H16N2Na2O11S3) | Anthraquinone | 595 |  |
100.0 ± 0.0 a | 100.0 ± 0.0 a | 100.0 ± 0.0 b |
| RNB | Reactive Blue 120 (C20H16N3Na3O15S4) | Azo | 610 |  |
92.0 ± 3.5 | 83.3 ± 1.2 | 100.0 ± 0.0 |
| RBY | Reactive Yellow 160 (C25H22ClN9Na2O12S3) | Azo | 405 |  |
73.1 ± 2.3 | 69.2 ± 1.5 | 71.15 ± 0.5 |
| RGY | Reactive Orange 107 (C25H22ClN9Na2O12S3) | Azo | 410 |  |
45.8 ± 3.2 | 41.7 ± 1.2 | 33.3 ± 0.6 |
| RR | Reactive Red 198 (C27H18ClN7Na4O16S5) | Azo | 530 | 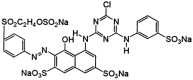 |
49.8 ± 2.8 | 41.7 ± 0.5 | 75.0 ± 2.1 |
| RBR | Reactive Red 180 (C29H19N3Na4O17S5) | Azo | 540 |  |
33.3 ± 0.6 | 41.7 ± 1.7 | 45.7 ± 3.3 |
Decolorization was completed in 10 min.
Decolorization was completed in 30 min.
3.11. Deactivation of pharmaceutical products by purified enzymes
Degradation of pharmaceuticals and personal care products were reviewed for both whole cell and ligninolytic enzymes of white rot fungi [40]. All the purified ligninolytic enzymes of T. polyzona KU-RNW027 were proved to deactivate selected pharmaceutical products in chemical classes of tetracycline, β-lactam, and quinolone (Table 7). MnP2 had the highest capability in deactivating the four antibiotics. The three enzymes showed the highest capabilities toward tetracycline and doxycycline as antibiotics in the class tetracycline. MnP1, MnP2, and Lac1 at 1 U/mL completely deactivated both antibiotics at a dose of 25 mg/L in 1, 1, and 3 days, respectively. MnP2 showed 100% deactivation ability against amoxicillin, a β-lactam antibiotics while MnP1 and Lac1 showed only 45 and 25%, respectively. Deactivation ability of MnP2 against ciprofloxacin, a quinolone, was moderated with deactivation ability at 73%. Notably the reaction was carried out with no redox mediator. 1-Hydroxybenzotriazole as a redox mediator improved T. versicolor IFO-6482 laccase activity for tetracycline degradation [41], while veratryl alcohol enhanced LiP activity of Phanerochaete chrysosporium in degrading tetracycline and oxytetracycline [42]. For implementation in the environment, enzymatic reactions that do not require addition of redox mediators are more promising.
Table 7.
Deactivation of 25 mg/L pharmaceutical products by 1 U/mL of purified ligninolytic enzymes from Trametes polyzona KU-RNW027.
| Pharmaceutical product | Chemical class | Chemical structure | Deactivation (%) |
||
|---|---|---|---|---|---|
| MnP1 | MnP2 | Lac1 | |||
| Tetracycline | Tetracyclines | 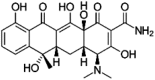 |
100 ± 0.0 a | 100 ± 0.0 a | 100 ± 0.0 b |
| Doxycycline | Tetracyclines |  |
100 ± 0.0 a | 100 ± 0.0 a | 100 ± 0.0 b |
| Amoxicillin | β-lactams | 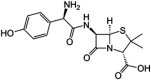 |
45 ± 2.5 | 100 ± 0.0 c | 25 ± 3.3 |
| Ciprofloxacin | Quinolones |  |
20 ± 1.3 | 73.3 ± 0.5 | 6.7 ± 1.5 |
Complete inactivation of pharmaceutical product observed within 1 day.
Complete inactivation of pharmaceutical product observed within 3 days.
Complete inactivation of pharmaceutical product observed within 5 days.
4. Conclusions
Ligninolytic enzymes of T. polyzona KU-RNW027 that played roles in synthetic dye degradations and pharmaceutical product deactivations were determined as MnP1, MnP2, and Lac1. Two MnPs had novel properties of high stability and activation ability by various organic solvents and metal ions. These three purified enzymes degraded an anthraquinone dye and 5 azo dyes. They also deactivated antibiotics of tetracycline, β-lactam, and quinolone classes with preferences in oxidizing dimethoxyl substituted phenol at the ortho-position. All reactions were carried out under a redox mediator-free system. Properties, especially regarding organic solvents and metal ions, offered advantages for implementation of T. polyzona KU-RNW027 or its enzymes as environmentally effective in the biodegradation and bioremediation of aromatic compounds in environments polluted with organic solvents and metal ions without any redox mediator supplement.
Funding Statement
This work was supported by a grant from the Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program [grant number PHD/0214/2553]
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1. Alharbi OML, Basheer A, Khattab RA, et al. . Health and environmental effects of persistent organic pollutants. J Mol Liq. 2018;263:442–453. [Google Scholar]
- 2. Leo L, Loong C, Ho XL, et al. . Occurrence of azo food dyes and their effects on cellular inflammatory responses. Nutrition. 2018;46:36–40. [DOI] [PubMed] [Google Scholar]
- 3. Thai PK, Ky LX, Binh VN, et al. . Occurrence of antibiotic residues and antibiotic-resistant bacteria in effluents of pharmaceutical manufacturers and other sources around Hanoi, Vietnam. Sci Total Environ. 2018;645:393–400. [DOI] [PubMed] [Google Scholar]
- 4. Teerapatsakul C, Pothiratana C, Chitradon L, et al. . Biodegradation of polycyclic aromatic hydrocarbons by a thermotolerant white rot fungus Trametes polyzona RYNF13. J Gen Appl Microbiol. 2016;62:303–312. [DOI] [PubMed] [Google Scholar]
- 5. Bilal M, Asgher M, Parra-Saldivar R, et al. . Immobilized ligninolytic enzymes: an innovative and environmental responsive technology to tackle dye-based industrial pollutants - A review. Sci Total Environ. 2017;576:646–659. [DOI] [PubMed] [Google Scholar]
- 6. Teerapatsakul C, Roberto P, Keshavarz T, et al. . Repeated batch for dye degradation in an airlift bioreactor by laccase entrapped in copper alginate. Int Biodeterior Biodegrad. 2017;120:52–57. [Google Scholar]
- 7. Bouacem K, Rekik H, Jaouadi NZ, et al. . Purification and characterization of two novel peroxidases from the dye-decolorizing fungus Bjerkandera adusta strain CX-9. Int J Biol Macromol. 2018;106:636–646. [DOI] [PubMed] [Google Scholar]
- 8. Mir-Tutusaus JA, Baccar R, Caminal G, et al. . Can white-rot fungi be a real wastewater treatment alternative for organic micropollutants removal? A review. Water Res. 2018;138:137–151. [DOI] [PubMed] [Google Scholar]
- 9. Asgher M, Ramzan M, Bilal M. Purification and characterization of manganese peroxidases from native and mutant Trametes versicolor IBL-04. Chinese J Catal. 2016;37:561–570. [Google Scholar]
- 10. Zhang H, Zhang J, Zhang X, et al. . Purification and characterization of a novel manganese peroxidase from white-rot fungus Cerrena unicolor BBP6 and its application in dye decolorization and denim bleaching. Process Biochem. 2018;66:222–229. [Google Scholar]
- 11. Xiaobin C, Rong J, Pingsheng L, et al. . Purification of a new manganese peroxidase of the white-rot fungus Schizophyllum sp. F17, and decolorization of azo dyes by the enzyme. Enzyme Microb Technol. 2007;41:258–264. [Google Scholar]
- 12. Zhang H, Zhang S, He F, et al. . Characterization of a manganese peroxidase from white-rot fungus Trametes sp.48424 with strong ability of degrading different types ofdyes and polycyclic aromatic hydrocarbons. J Hazard Mater. 2016;320:265–277. [DOI] [PubMed] [Google Scholar]
- 13. Kong W, Chen H, Lyu S, et al. . Characterization of a novel manganese peroxidase from white-rotfungus Echinodontium taxodii 2538, and its use for the degradation of lignin-related compounds. Process Biochem. 2016;51:1776–1783. [Google Scholar]
- 14. Teerapatsakul C, Parra R, Bucke C, et al. . Novel laccases of Ganoderma sp. KU-Alk4, regulated by different glucose concentration in alkaline media. World J Microbiol Biotechnol. 2007;23:1519–1527. [Google Scholar]
- 15. Bagewadi ZK, Mulla SI, Ninnekar HZ. Purification and immobilization of laccase from Trichoderma harzianum strain HZN10 and its application in dye decolorization. J Genet Eng Biotechnol. 2017;15:139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lang M, Kanost MR, Gorman MJ. Multicopper oxidase-3 is a laccase associated with the peritrophic matrix of Anopheles gambiae . PLoS One. 2012;7:e33985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jaiswal N, Pandey VP, Dwivedi UN. Purification of a thermostable alkaline laccase from papaya (Carica papaya) using affinity chromatography. Int J Biol Macromol. 2015;72:326–332. [DOI] [PubMed] [Google Scholar]
- 18. Siroosi M, Amoozegar MA, Khajeh K. Purification and characterization of an alkaline chloride-tolerant laccase from a halotolerant bacterium, Bacillus sp. strain WT. J Mol Catal B Enzym. 2016;134:89–97. [Google Scholar]
- 19. Campos PA, Levin LN, Wirth SA. Heterologous production, characterization and dye decolorization ability of a novel thermostable laccase isoenzyme from Trametes trogii BAFC 463. Process Biochem. 2016;51:895–903. [Google Scholar]
- 20. Patil PD, Yadav GD. Application of microwave assisted three phase partitioning method for purification of laccase from Trametes hirsuta . Process Biochem. 2018;65:220–227. [Google Scholar]
- 21. Chairin T, Nitheranont T, Watanabe A, et al. . Purification and characterization of the extracellular laccase produced by Trametes polyzona WR710–1 under solid-state fermentation. J Basic Microbiol. 2014;54:35–43. [DOI] [PubMed] [Google Scholar]
- 22. Suman SK, Khatri M, Dhawaria M, et al. . Potential of Trametes maxima IIPLC-32 derived laccase for the detoxification of phenolic inhibitors in lignocellulosic biomass prehydrolysate. Int Biodeterior Biodegradation. 2018;133:1–8. [Google Scholar]
- 23. Ling Z, Wang S, Zhu M, et al. . An extracellular laccase with potent dye decolorizing ability from white rot fungus Trametes sp. LAC-01. Int J Biol Macromol. 2015;81:785–793. [DOI] [PubMed] [Google Scholar]
- 24. Teerapatsakul C, Chitradon L. Physiological regulation of an alkaline-resistant laccase produced by Perenniporia tephropora and efficiency in biotreatment of pulp mill effluent. Mycobiology. 2016;44:260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fang W, Fernandes EKK, Roberts DW, et al. . A laccase exclusively expressed by Metarhizium anisopliae during isotropic growth is involved in pigmentation, tolerance to abiotic stresses and virulence. Fungal Genet Biol. 2010;47:602–607. [DOI] [PubMed] [Google Scholar]
- 26. Ujor VC, Monti M, Peiris DG, et al. . The mycelial response of the white-rot fungus, Schizophyllum commune to the biocontrol agent, Trichoderma viride . Fungal Biol. 2012;116:332–341. [DOI] [PubMed] [Google Scholar]
- 27. Zhang J, Chen H, Chen M, et al. . Cloning and functional analysis of a laccase gene during fruiting body formation in Hypsizygus marmoreus . Microbiol Res. 2015;179:54–63. [DOI] [PubMed] [Google Scholar]
- 28. Lueangjaroenkit P, Teerapatsakul C, Chitradon L. Morphological characteristic regulation of ligninolytic enzyme produced by Trametes polyzona . Mycobiology. 2018;46:396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirk TK, Tien M, Kersten PJ, et al. . Lignin peroxidase from fungi: Phanerochaete chrysosporium . Methods Enzymol. 1990;188:159–171. [Google Scholar]
- 30. Lowry OH, Rosebrough NJ, Farr AL, et al. . Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 31. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. [DOI] [PubMed] [Google Scholar]
- 32. Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. [DOI] [PubMed] [Google Scholar]
- 33. Kondo R, Harazono K, Sakai K. Bleaching of hardwood kraft pulp with manganese peroxidase secreted from Phanerochaete sordida YK-624. Appl Environ Microbiol. 1994;60:4359–4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen W, Zheng L, Jia R, et al. . Cloning and expression of a new manganese peroxidase from Irpex lacteus F17 and its application in decolorization of reactive black 5. Process Biochem. 2015;50:1748–1759. [Google Scholar]
- 35. Liu Y, Huang L, Guo W, et al. . Cloning, expression, and characterization of a thermostable and pH-stable laccase from Klebsiella pneumoniae and its application to dye decolorization. Process Biochem. 2017;53:125–134. [Google Scholar]
- 36. Chitradon L, Mahakhan P, Bucke C. Oligosaccharide synthesis by reversed catalysis using a-amylase from Bacillus licheniformis . J Mol Catal B Enzym. 2000;10:273–280. [Google Scholar]
- 37. Zhou X, Qu Y, Kim BH, et al. . Effects of azide on electron transport of exoelectrogens in air-cathode microbial fuel cells. Bioresour Technol. 2014;169:265–270. [DOI] [PubMed] [Google Scholar]
- 38. Wang S, Chen Q, Zhu M, et al. . An extracellular yellow laccase from white rot fungus Trametes sp. F1635 and its mediator systems for dye decolorization. Biochimie. 2018;148:46–54. [DOI] [PubMed] [Google Scholar]
- 39. Moldes D, Sanroman MA. Amelioration of the ability to decolorize dyes by laccase: relationship between redox mediators and laccase isoenzymes in Trametes versicolor . World J Microbiol Biotechnol. 2006;22:1197–1204. [Google Scholar]
- 40. Asif MB, Hai FI, Singh L, et al. . Degradation of pharmaceuticals and personal care products by white-rot fungi–a critical review. Curr Pollution Rep. 2017;3:88–103. [Google Scholar]
- 41. Suda T, Hata T, Kawai S, et al. . Treatment of tetracycline antibiotics by laccase in the presence of 1-hydroxybenzotriazole. Bioresour Technol. 2012;103:498–501. [DOI] [PubMed] [Google Scholar]
- 42. Wen X, Jia Y, Li J. Degradation of tetracycline and oxytetracycline by crude lignin peroxidase prepared from Phanerochaete chrysosporium – A white rot fungus. Chemosphere. 2009;75:1003–1007. [DOI] [PubMed] [Google Scholar]