Abstract
Talaromyces marneffei is the only dimorphic species in its genus and causes a fatal systemic mycosis named talaromycosis. Our previous study indicated that knockdown of AcuD gene (encodes isocitrate lyase of glyoxylate bypass) of T. marneffei by RNA interference approach attenuated the virulence of T. marneffei, while the virulence of the AcuD knockout strains was not studied. In this study, T. marneffei-zebrafish infection model was successfully established through hindbrain microinjection with different amounts of T. marneffei yeast cells. After co-incubated at 28°C, the increasing T. marneffei inoculum doses result in greater larval mortality; and hyphae generation might be one virulence factor involved in T. marneffei-zebrafish infection. Moreover, the results demonstrated that the virulence of the ΔAcuD was significantly attenuated in this Zebrafish infection model.
Keywords: Talaromycosis, AcuD, virulence, zebrafish
1. Introduction
Talaromycosis (formerly called penicilliosis) caused by Talaromyces marneffei (T. marneffei) is a serious invasive fungal infection [1]. The disease is endemic in southeastern Asia, including southern China, Thailand, the Philippines, and Vietnam. The disease occurs in immuno-compromised patients, especially those with AIDS. Recently, with the increasing number of HIV infections, the incidence of talaromycosis has increased, and this disease has become an important diagnostic indicator of AIDS [2]. Talaromycosis has high morbidity and mortality, making T. marneiffei a clinically significant pathogenic fungi. Therefore, understanding the pathogenic mechanism of T. marneffei is essential for the prevention and treatment of this disease.
T. marneffei is the only known thermally dimorphic species in genus Penicillium, with a mycelial phase at 25 °C and the yeast phase, which is the pathogenic state in the human body or culture at 37 °C. A study by Cánovas and Andrianopoulos (2006) indicated that the AcuD gene, which encodes the glyoxylate bypass key enzyme isocitrate lyase, is highly expressed in the yeast phase of this fungus [3]. At nearly the same time, our studies showed that the AcuD gene is highly expressed in yeast phase cells in a systematic screening of differentially expressed proteins between yeast and mycelial phases, and also in an infection model with macrophages during the infection process [4,5]. By investigating the expression of AcuD after co-culturing of T. marneffei conidia with murine macrophages, Thirach et al. [6] observed similar results. These results collectively suggest a link between the glyoxylate bypass and pathogenicity in T. marneffei.
The glyoxylate cycle is a bypass of the tricarboxylic acid cycle that allows two-carbon (C2) compounds to be used as carbon sources in gluconeogenesis, in which the unique enzymes are isocitrate lyase (ICL) and malate synthase (MS) [7]. The role of the glyoxylate cycle and its enzymes in microbial virulence has been reported [8]. Recent studies of AcuD (ICL-encoding gene)-deleted mutants (ΔAcuD) in many fungal pathogens showed that a functional role for the glyoxylate cycle in fungal virulence is widespread, but not universal. In the human-related pathogens Aspergillus fumigatus and Cryptococcus neoformans, no differences in survival were observed between a ΔAcuD and a wild-type in the animal infection model (Table 1) [9–16]. Cánovas and Andrianopoulos [3] constructed the AcuD stable knockout mutant (ΔAcuD) and the complement mutant (ΔAcuD+) for understanding the developmental regulation factors of the glyoxylate cycle in T. marneffei, while they did not test the virulence of these mutants. In our previous study by Sun et al. [10], the AcuD gene of T. marneffei was knocked down using an RNA interference approach, and the data in a murine-infection model indicated that the RNAi-mediated silencing of AcuD expression could attenuate virulence of T. marneffei (Table 1). As our previous research showed that siRNA could not generate continuous effects to the target gene due to adaption of the host cell, we constructed ΔAcuD and ΔAcuD+ successfully based on the protocol kindly provided by Andrianopoulos with some optimization [17,18].
Table 1.
Virulence phenotypes of AcuD gene mutant during interactions with host organisms.
| Fungal pathogen | Mutant | Host model | Virulence | Reference |
|---|---|---|---|---|
| Human-related pathogenic fungi | ||||
| Candida glabrata | ΔAcuD | Murine | Less virulent | [9] |
| Talaromyces marneffei |
AcuDacuD-RNAi AcuDacuD-RNAqb |
Murine | Less virulent | [10] |
| Aspergillus fumigatus | ΔAcuD | Murine | Virulent | [11] |
| Cryptococcus neoformans | ΔAcuD | Rabbit Murine |
Virulent | [12] |
| Candida albicans | ΔAcuDa | Murine | Less virulent | [13] |
| Plant-related pathogenic fungi | ||||
| Colletotrichum lagenarium | ΔAcuD | Cucumber | Less virulent | [14] |
| Magnaporthe grisea | ΔAcuD | Rice | Less virulent | [15] |
| Leptosphaeria maculans | ΔAcuD | Cotyledons | Less virulent | [16] |
ΔAcuD stand for AcuD knockout mutant.
Various names have been used in the literature for the gene encoding ICL in different organisms; for clarity, the designation AcuD is used here.
AcuD gene was knocked down by RNAi-mediated silencing.
While traditional mammalian models such as murine and rabbits have the advantage of relatively close similarity to humans, operational complexity and economic factors limited their use. We had successfully established Caenorhabditis elegans (C. elegans) and Galleria mellonella infection models for T. marneffei virulence studies [19,20]. However, because huge racial differences exist between invertebrates and humans, these two models also have limited value for mechanistic studies of T. marneffei-host interactions. Therefore, development of a new host model is necessary. The zebrafish, which is a transitional model between lower animals and mammals, is a desirable host model for studying medically important microbial infections because it has evolved closer to humans than Drosophila and C. elegans, is easier to manipulate and more economical than murine [21].
In the present work, we established a T. marneffei-zebrafish infection model to investigate the virulence of T. marneffei, and found that hyphae growth of T. marneffei might be a crucial virulence factor involved in zebrafish killing. Also, we tested the virulence of ΔAcuD of T. marneffei in this model, and found that AcuD gene deletion decreased the virulence of T. marneffei.
2. Materials and methods
2.1. Ethics statement
Animal experiments in this study were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and received permission from the Ethics Committee on the Care and Use of Laboratory Animals of Sun Yat-sen University, Guangzhou, China.
2.2. Zebrafish care and maintenance
Zebrafish wild-type AB line were mated, staged, and raised according to the standard protocols (http://zfin/org) at the animal facility of Sun Yat-sen University. Eggs were collected following natural spawning and treated with N-phenylthiourea starting at 12 hours post-fertilization (hpf) to prevent pigmentation. A total of 150 eggs/dish were kept in E3 (5 mM sodium chloride, 0.174 mM potassium chloride, 0.33 mM calcium chloride, 0.332 mM magnesium sulfate, and 2 mM HEPES in Nanopure water, pH 7) at 28 °C with 0.3 mg/l methylene blue (Sigma-Aldrich, St. Louis, MO) for the first 24 hours to prevent microbial growth. Debris and unfertilized eggs were removed twice daily until hatching.
For the infection experiments, larvae were switched to E3 without methylene blue (E3-MB) at 24 hpf and manually dechorionated between 48 and 52 hpf. Prior to microinjection or imaging, the larvae were anesthetized in E3-MB containing 200 µg/ml of 3-aminobenzoic acid (Tricaine, Sigma-Aldrich, St. Louis, MO) and euthanized with an anesthetic overdose with physical assurance.
2.3. Strains, growth conditions and inocula preparation
The wild-type strain of T. marneffei SUMS0594 was a clinical isolate stored in Sun Yat-sen Memorial Hospital, Guangzhou, China. SPM4 strain (Genotype: niaD1pyrG1), studied by Borneman et al. [22] was kindly gifted by Prof. Cao CW from Guangxi Medical University. The ΔAcuD and ΔAcuD+ were constructed and deposited in our lab.
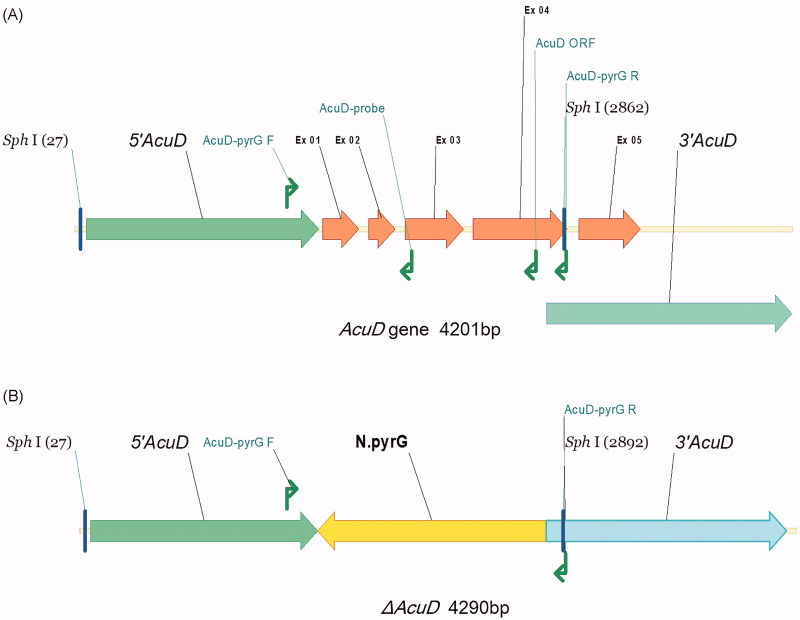
Genomic DNA of T. marneffei was extracted from frozen mycelia as described previously [10]. PCR was performed using two pairs of specific primers (AcuD-pyrG F and AcuD-pyrG R, AcuD-pyrG F and AcuD-ORF) to confirm that the AcuD gene was the targeted disruption (Figure 1, Table 2). Deletion of AcuD was further confirmed by southern blot analysis with a 751-bp PCR product, generated by primers AcuD-pyrG F and AcuD-Probe. The genomic DNA was digested with Sph I, and southern blot was performed as previously reported [23].
Figure 1.
Schematic diagram of the gene structure in AcuD gene of the (A) T. marneffei wild-Type and (B) ΔAcuD Strains. 5′AcuD, 3′AcuD: the homologous sequence regions at either ends of the AcuD structural gene, Ex: exon, F: forward primer, R: reverse primer, ORF: open reading frame, N. pyrG: pyrG cassette of Emericella nidulans.
Table 2.
Primers used in the article.
| Primer name | Sequence (5′–3′) |
|---|---|
| AcuD-pyrG F | GCCCTTGGTTTTCGGTTATGT |
| AcuD-pyrG R | GGGGAGTGGCTTACTTTTGCT |
| AcuD-ORF | GTTCGGGGCGCATCCCAGTTC |
| AcuD-Probe | TTTCGGTCGTGGAAGAGTTGG |
All the T. marneffei strains were grown on potato dextrose agar (PDA) at 25 °C for 7 days for the collection of conidia. Yeast cells were collected from liquid cultures of T. marneffei by rotating conidia for 48 h in BHI medium at 37 °C. The cultures were harvested with sterile water supplemented with 0.01% tween and gently rubbed with an L-style spreader, passed through two layers of sterile Miracloth into a sterile 50 ml screw-cap tube, and made up to a volume of 50 ml. Following centrifugation at 900×g for 10 min, the yeast cell pellets were thoroughly resuspended in 50 ml of sterile phosphate-buffered saline (PBS), pelleted again, and made up to a volume of approximately 5 ml in sterile PBS before passing once more through a double layer of sterile Miracloth. Then, the yeast cells were enumerated with a hemocytometer and brought to final concentrations of 1 × 107 CFU/ml, 5 × 107 CFU/ml, and 1 × 108 CFU/ml in PBS prior microinjecting into zebrafish larvae. For heat inactivation, the fungal cells were incubated at 65 °C for 30 min with gentle agitation. All fungal cell suspensions were prepared on the day of microinjection. The suspensions were diluted 1000 times, and 100 µl of which was applied to three PDA plates per group and incubated at 37 °C. To confirm the viability of fungal cells and verify the accuracy of the count, colony form units (CFUs) of the yeast cells were counted 24 hours later.
2.4. Zebrafish killing assay
In preparation of hindbrain ventricle infection, yeast cells stocked in PBS at different amounts (PBS control, 1 × 107 CFU/ml, 5 × 107 CFU/ml, 1 × 108 CFU/ml and heat-inactivated control) were mixed with sterile phenol red (final concentration 0.1%) for clear visualization of injection success. The microinjection was conducted under a stereoscope (PLI-90A; Harvard apparatus, Holliston, MA) using fine needles.
Embryos were manually dechorionated at 48–52 hpf. Afterwards, juveniles were anesthetized in a prepared solution of 200 µg/ml of ethyl 3-aminobenzoate. The anesthetized embryos (48–52 hpf) were lined up on a flat 1% agarose injecting plate. The position of the fish and the tip of the needle were adjusted such that the embryos were positioned with their dorsal sides toward the needle tip. The needle carefully punctured into the otic vesicle of the fish, and 3 nanoliter (nl) infection volumes were injected as described previously [24]. Under these conditions, individual larvae in the 1 × 107 CFU/ml, 5 × 107 CFU/ml, or 1 × 108 CFU/ml groups received an average 30, 150, or 300 yeast cells, respectively. The CFUs/injection was monitored via injection into 0.1 ml of sterile PBS on PDA plates. Each group contained 100 larvae, and each experiment was performed in triplicate. The delivered yeast dosage was determined by immediate CFU enumeration on a group of randomly selected injected embryos to ensure proper inoculation. For fungal viability assays, primary homogenates of infected zebrafish embryos were prepared by adding 500 µl of sterile PBS per embryo. Serial 10-fold dilutions of the primary homogenates were plated in triplicate on PDA with 1% ampicillin at 25 °C for 3 to 4 days before the colonies were counted.
2.5. Microscopic observations
Infected larvae were individually sorted into single wells of a 96-well plate and monitored every 6 hours starting at 6 hours post-infection (hpi). Morphological changes of larvae were observed under the same microscope. The discoloration and cessation of the heart beat and blood flow in the larvae was considered as the standard of death. Hyphae growth in the zebrafish as well as its relationship with death was also recorded. The observations were terminated at 96 hpi. Infection and mortality data were collected from three independent experiments. All dead zebrafish larvae were placed on PDA medium with 1% ampicillin for 7 to 8 days at 25 °C to monitor contamination of other fungi.
2.6. Statistical analyses
Survival data from at least three independent experimental replicates were pooled and analyzed using log-rank and Wilcoxon tests. The survival distributions for the different experimental conditions were displayed in a graphical format using Kaplan–Meier plots. All p values were two sided, and a p < .05 was used to define statistical significance. The data analyses were conducted using GraphPad Prism version 6 and the STATA 6 statistical softwares.
3. Results
3.1. The verification of ΔAcuD knockout mutant
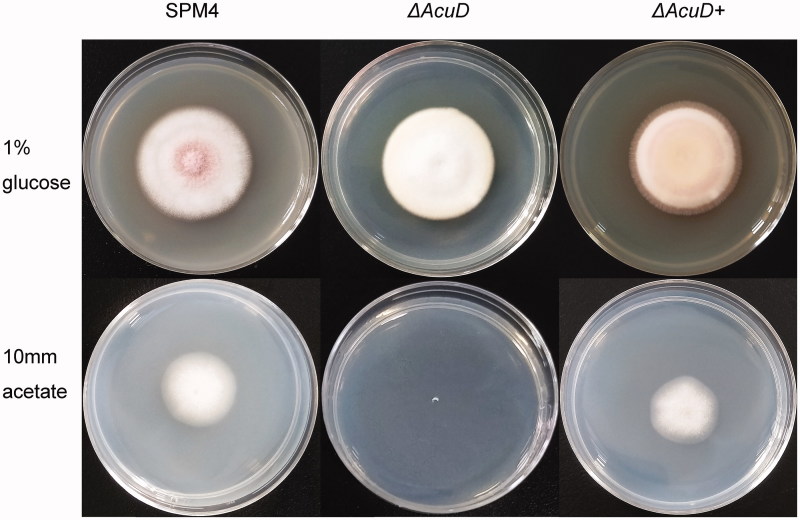
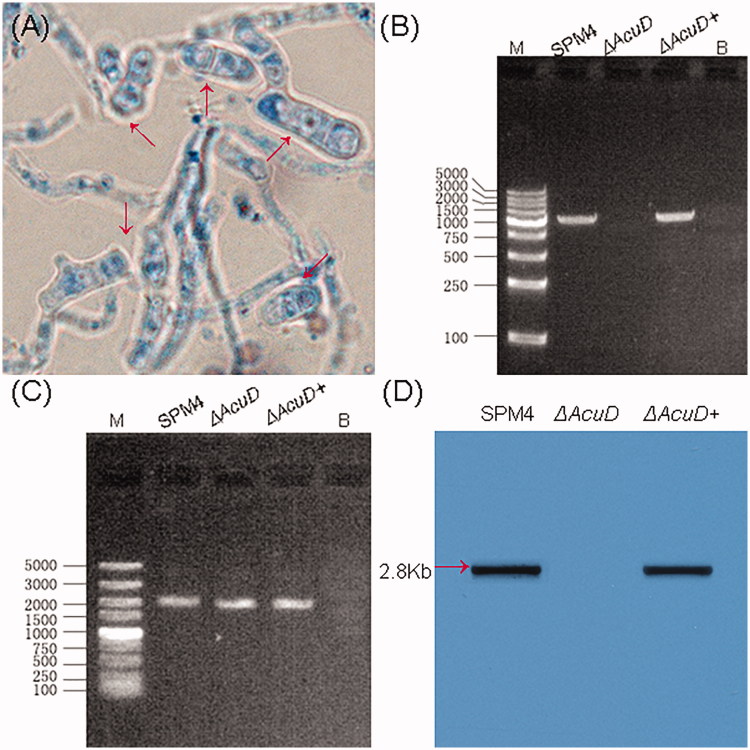
The ΔAcuD were constructed by stably replacing the AcuD structural gene with the screening tag pyrG gene via homologous recombination from the parental strain SPM4 (Figure 1). The phenotypic tests show that the ΔAcuD mutants were unable to grow on medium with acetate as a sole carbon source. But no growth difference between ΔAcuD mutants and SPM4 was observed on the glucose medium except the former’s lack of red pigment. Reintroduction of AcuD gene into ΔAcuD mutants restores the ability to assimilate acetate just as previously reported (Figure 2) [3]. The yeast-like cells were round or oval in shape with a sausage appearance or a septum and split breeding (Figure 3(A)).
Figure 2.
Colony morphology of the T. marneffei strain SPM4, ΔAcuD and ΔAcuD+ incubated in the media containing 1% glucose or 10 mm acetate as the sole carbon source, 25 °C, 7 days. 1% glucose: 1% glucose + YNB (Yeast Nitrogen Base without Amino Acids); 10 mm Acetate: 10 mm acetate + YNB.
Figure 3.
(A) T. marneffei yeast phase cells grown in BHI liquid broth at 37 °C for 48 h (lactophenol cotton blue stain 100×), red arrows indicate T. marneffei yeast-like cells. (B, C) PCR using gene-specific primers for the verification of AcuD knockout mutant. (B) Size of bands: SPM4 1479 bp, ΔAcuD, ΔAcuD+ 1479 bp; (C) Size of bands: SPM4 1658 bp, ΔAcuD 1688 bp, ΔAcuD+ 1658 bp. M: DL5000, B:Blank control. (D) Southern blot analysis: Southern blot of genomic DNA from the SPM4, ΔAcuD, ΔAcuD + T. marneffei strains.
PCR analyses of ΔAcuD, SPM4, ΔAcuD+ revealed that ΔAcuD mutant lacked AcuD gene and obtained pyrG cassette (Figure 3(B,C)). Southern blot analysis showed that AcuD encoding gene was not detected in the ΔAcuD mutant, conversely, AcuD encoding gene were detected in the SPM4 and ΔAcuD+ strain (Figure 3(D)). The sequenced results revealed that ΔAcuD was targeted and knocked out by the pyrG gene at that locus (unpublished data).
3.2. Assays of zebrafish killing by T. marneffei
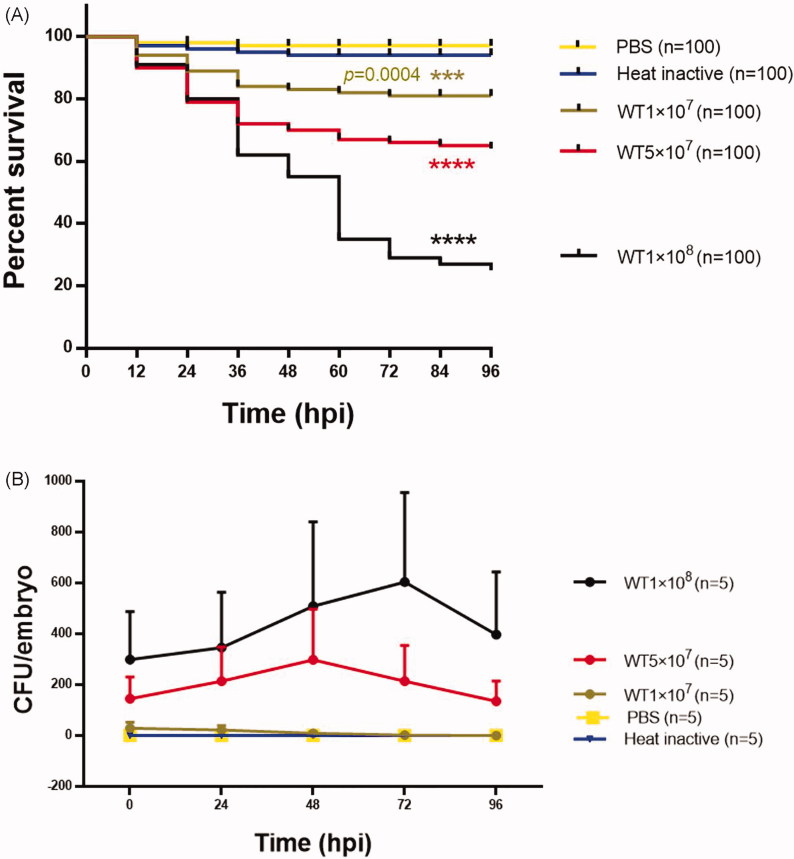
To determine whether the zebrafish was a suitable infection model for studying T. marneffei virulence mechanisms, the hindbrain ventricle microinjection method was used. First, we used T. marneffei wild-type strain SUMS0594 to infect zebrafish larvae in different amounts of yeast cells at 1 × 107 CFU/ml, 5 × 107 CFU/ml, and 1 × 108 CFU/ml respectively. As shown in Figure 4(A), inoculation with 1 × 108 CFU/ml dose of the SUMS0594 resulted in rapid killing of the larvae. The survival rates were significantly dependent on the amounts of injected T. marneffei yeast cells.
Figure 4.
Assays of zebrafish killing by the wild-type strain of T. marneffei SUMS0594. (A) The survival rates of zebrafish larvae inoculated with different amounts of yeast cells and co-incubated at 28 °C (n = 100). (B) T. marneffei CFU time-course counts of different groups, mean ± SD, n = 5 larvae/group/experiment. Different colors indicate five independent experiments. WT1 × 107, WT5 × 107, WT1 × 108 stands for the inoculated amounts of yeast cells were 1 × 107CFU/ml, 5 × 107CFU/ml, 1 × 108 CFU/ml, respectively; Heat inactive: heat-inactivated yeast cells of SUMS0594 at the amount of 1 × 108 CFU/ml; PBS: sterile phosphate-buffered saline. ****indicates a p value< .0001.
There was no significant difference in the survival rates between the control groups which received heat-inactivated yeast cells and the sham injection with PBS (p = .3149 > .05, Gehan–Breslow–Wilcoxon test). However, the survival rates significantly differed between each experimental group and the control groups. Meanwhile, significant differences were found in the survival rates between the groups that received different amounts of yeast cells (1 × 107 CFU/ml vs 5 × 107 CFU/ml: **p = .0097 < .05; 5 × 107 CFU/ml vs 1 × 108 CFU/ml: ***p = .0002 < .001; 1 × 107 CFU/ml vs 1 × 108 CFU/ml: ****p < .0001, Gehan–Breslow–Wilcoxon test). In particular, the group that received the greatest amount of inocula (1 × 108 CFU/ml) exhibited obvious decrease in the larval survival rates to 80%, 55%, and 29% at 24 hpi, 48 hpi, and 72 hpi respectively (Figure 4(A)). Therefore, we propose that the hindbrain ventricle microinjection method is available for establishing T. marneffei-zebrafish infection model. T. marneffei CFU time-course counts indicated the doses of injected yeast in 5 × 107 CFU/ml or 1 × 108 CFU/ml could maintain a relatively stable fungal load. There was a slight rise from the initial infection time to 72 hpi, especially during 24 hpi to 48 hpi. After 72 hpi, the CFU counts declined in both the groups (Figure 4(B)). This is consistent with the above survival rate results. To facilitate observation, the recommended initial amount for injection of yeast cells of T. marneffei is 5 × 107 CFU/ml, and the optimal amount is 1 × 108 CFU/ml.
3.3. Hyphae generation is involved in T. marneffei virulence
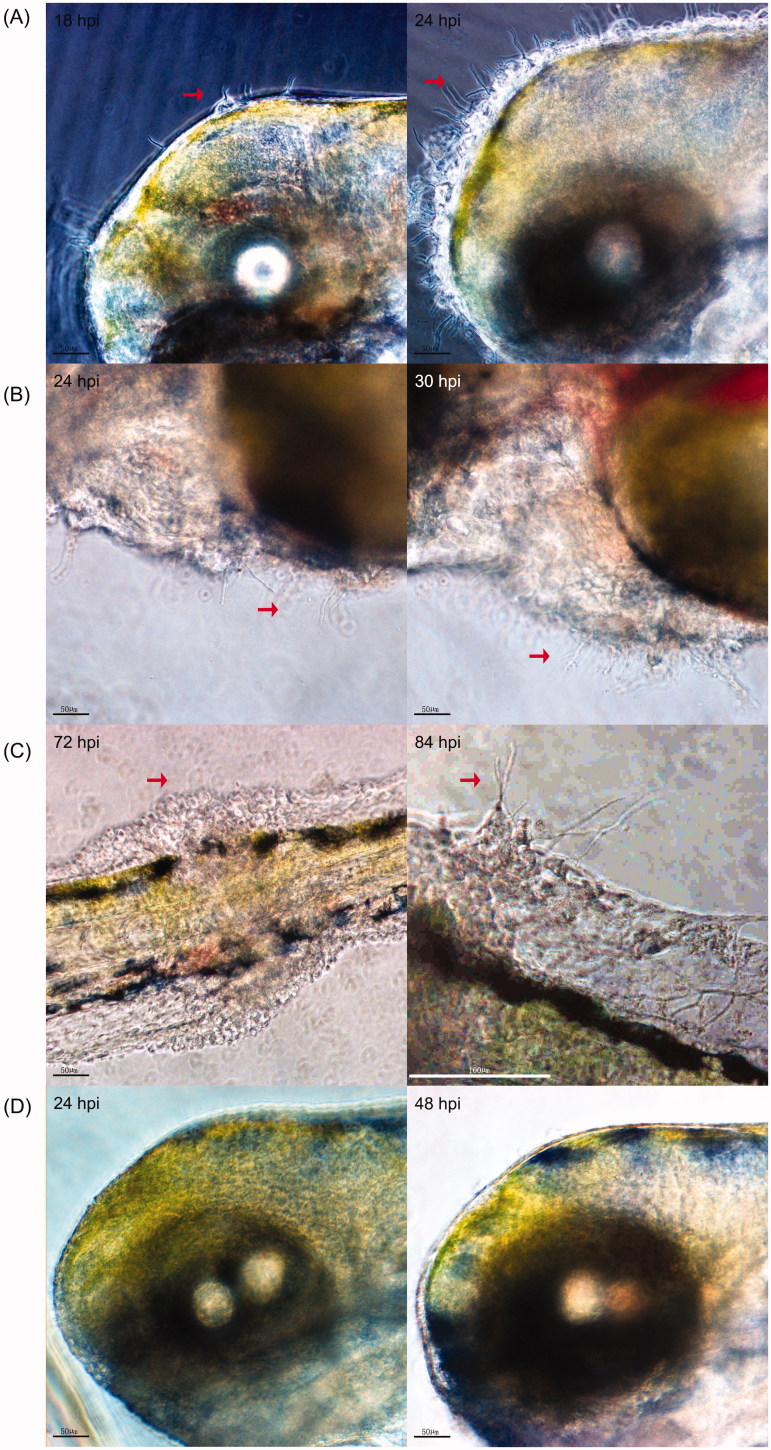
The morphology of zebrafish larvae after T. marneffei inoculation was observed at different times. The growing hyphae could penetrate the tissue of zebrafish larvae (Figure 5(A–C)) when the larvae were inoculated with 1 × 108 CFU/ml yeast. At 18 hpi, the hyphae could be seen extruding out of the larvae brain and some granular aggregation could be seen surrounding the hyphae in the injected location. This happened to about 69% (59/86) of living larvae and the amount of larvae affected rapidly increased. At 30 hpi, the aggregated growth of hyphae around the heart region could be seen in about 29% (19/66) of the living larvae. At 72 hpi, only 30 larvae remained alive, of which four had a granuloma formation surrounding the tail. At 84 hpi, the hyphae penetrated through the granuloma. The 96-hour observation reminded us that systemic infection of zebrafish could be established by the local injection through hindbrain ventricle. All of the infected zebrafish larvae died within 24 hours after their tissues were penetrated by hyphae (Figure 5(A–C)). However, growing hyphae could barely be detected in the 26 survived larvae. In the PBS control groups, no hyphae penetration or granuloma formation was observed (Figure 5(D)), which indicated that hyphae generation was involved in the virulence of T. marneffei interaction with zebrafish.
Figure 5.
In vivo visualization of T. marneffei in the infected zebrafish larvae. (A) Hyphae generation of T. marneffei in the hindbrain cavity, (B) heart region and tail (C right) of zebrafish larvae from the SUMS0594 1 × 108 CFU/ml infection group. C left shows the granuloma formation surrounding the tail before hyphal penetration. A/B/C were three different zebrafish larvae. (D) No granular bulge and mycelial growth can be observed on the zebrafish surface in the PBS group. The observation time was marked in the upper left corner of the image. The red arrows indicate the granuloma formation and the extruding fungal hyphae.
3.4. Knockout of AcuD gene attenuated virulence of T. marneffei
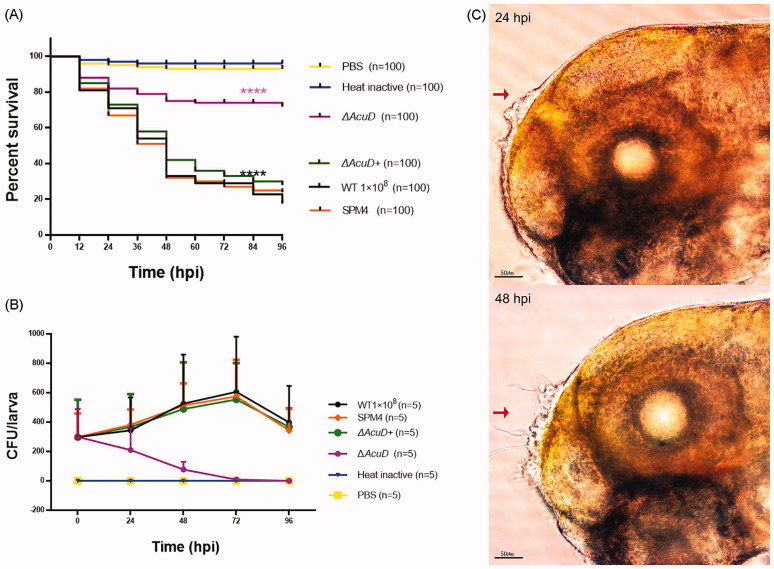
To evaluate the effect of the AcuD gene on the virulence of T. marneffei in zebrafish, the wild-type strain SUMS0594, SPM4, ΔAcuD, and ΔAcuD+ were used. According to previous experimental results, the amount of 1 × 108 CFU/ml yeast cells of T. marneffei were used to infect zebrafish larvae. As shown in Figure 6(A), there is no significant difference between the survival rates of the groups infected with SPM4 and wild-type (p = .9377, Gehan–Breslow–Wilcoxon test). 76% (76/100) of the zebrafish larvae died within 96 hpi in the SPM4 strain-infected groups compared with 28% (28/100) in the ΔAcuD groups (p < .0001). Complementation of AcuD gene (ΔAcuD+) restored the virulence and 72% (72/100) of the zebrafish larvae died within 96 hpi in the ΔAcuD+ strain-infected groups (SPM4 versus ΔAcuD+, p = .2555 > .05). These results indicated that the virulence of ΔAcuD was significantly attenuated. The decline in ΔAcuD CFU numbers over time confirmed these results (Figure 6(B)). The hyphae generation was much fewer and could only be seen at the injection location, especially during 24–48 hpi (Figure 6(C)) unlike the wild-type and SPM4 groups.
Figure 6.
Knockout of AcuD attenuated virulence of T. marneffei. (A) The survival rates of zebrafish larvae inoculated with different T. marneffei strains and co-incubated at 28 °C (n = 100); (B) T. marneffei CFU time-course counts of different groups, mean ± SD, n = 5 larvae/group/experiment. Different colors indicate six independent experiments. WT1 × 108, SPM4, ΔAcuD, ΔAcuD+ stands for inoculated at an amount of 1 × 108 CFU/ml yeast cells of wild-type strain SUMS0594, SPM4, AcuD knockout mutant, AcuD complement mutant respectively; Heat inactive: heat-inactivated yeast cells of SUMS0594 at an amount of 1 × 108 CFU/ml; PBS: sterile phosphate-buffered saline. ****indicates a p value < .0001. (C) A large number of granular bulges can be seen at the injection site of the zebrafish at 24 hpi (left), few mycelia grow out through the granular bulge at 48 hpi. Zebrafish larvae from the ΔAcuD group.
4. Discussion
Zebrafish, also known as bluefish or flower fish, is a derived member of genus Danio from the family Cyprinidae [25]. As an emerging model organism in recent years, the zebrafish has many advantages. The zebrafish grows fast, is genetically tractable, and is more evolutionarily close to humans than invertebrate model organisms (i.e., nematodes, Drosophila etc.). Furthermore, zebrafish larvae are transparent and amenable to prolonged in vivo imaging and small molecule screening [26]. Therefore, the zebrafish model is widely used in scientific researches such as developmental genetics, environmental toxicology, and human diseases [27–29]. In recent years, the zebrafish model is increasingly being used for studying the pathogenesis of microorganisms, including several clinically relevant fungal pathogens, such as Aspergillus fumigatus, Candida albicans, and Cryptococcus neoformans [24,30,31]. Recently, Ellett et al. [32] established a reproducible systemic invasive filamentous form of talaromycosis in zebrafish larvae at 28 °C, and studied the different roles of neutrophils and macrophages in T. marneffei infection.
Under natural conditions, pathogenic micro-organisms can infect zebrafish through the gastrointestinal tract, spasms or a damaged epidermis [33]. In laboratory experiments, zebrafish can be infected with different inoculation approaches at different growth stages according to different experimental design [21]. During the embryonic and juvenile stages, the following inoculation methods are most commonly used: (1) bloodstream infections, such as intravenous inoculations, are performed via either the tail vein or the duct of Couvier; (2) yolk sac microinjection; (3) hindbrain ventricle microinjection, directly or via otic vesicle; (4) embryos immersion infection; and (5) spinal cord tissues and axial muscle injections. In our pilot study, yolk sac and hindbrain ventricle microinjection routes were attempted to establish a disseminated infection in transparent zebrafish larvae. The attempts at infection of the yolk sac had resulted in universal lethality within 24 h, which was similar to the study reported by Brothers et al. [30]. In contrast, our initial inoculation of zebrafish larvae through the hindbrain microinjection yielded a stable and reproducible infection. In this study, we used the wild-type strain to assess the virulence of T. marneffei in zebrafish larvae and observed that T. marneffei was lethal to the larvae. The death of infected larvae had a positive correlation with the increasing amounts of T. marneffei, which is similar to the study on C. albicans while not consistent with C. neoformans [30,34]. Meanwhile, we found that T. marneffei disseminated throughout the fish, fungal hyphae were even present at the tail of fish (Figure 1(A)), which is consistent with the observations in C. albicans [30]. Thus, the hindbrain ventricle microinjection inoculation method is available to establish the T. marneffei-zebrafish systemic infection model.
We also compared the virulence of the wild-type and ΔAcuD of T.marneffei using the zebrafish infection model. The results showed that the virulence of the ΔAcuD was significantly attenuated, which was consistent with the results reported in the literature [10]. These findings indicate that the AcuD gene, which encodes the key enzyme ICL of the glyoxylate cycle is relevant to the virulence of T.marneffei and also indicate that zebrafish can be a viable infectious model organism for assessing the virulence of different T.marneffei mutants.
Funding Statement
This study was supported by a grant from the National Natural Science Foundation of China NSFC (http://www.nsfc.gov.cn/), grant number 81171545.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Samson RA, Yilmaz N, Houbraken J. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 2011;70:159–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Le T, Wolbers M, Chi NH, et al. Epidemiology, seasonality, and predictors of outcome of AIDS-associated Penicillium marneffei infection in Ho Chi Minh City, Viet Nam. Clin Infect Dis. 2011;52:945–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canovas D, Andrianopoulos A. Developmental regulation of the glyoxylate cycle in the human pathogen Penicillium marneffei. Mol Microbiol. 2006;62:1725–1738. [DOI] [PubMed] [Google Scholar]
- 4.Xi L, Xu X, Liu W, et al. Differentially expressed proteins of pathogenic Penicillium marneffei in yeast and mycelial phases. J Med Microbiol. 2007;56:298–304. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Xi LY, Liu HF, et al. Differential expression of isocitrate lyase in P. marneffei phagocytized by nonstimulated and stimulated murine macrophages. Nan Fang Yi Ke Da Xue Xue Bao. 2007;27:631–633. [PubMed] [Google Scholar]
- 6.Thirach S, Cooper CJ, Vanittanakom N. Molecular analysis of the Penicillium marneffei glyceraldehyde-3-phosphate dehydrogenase-encoding gene (gpdA) and differential expression of gpdA and the isocitrate lyase-encoding gene (acuD) upon internalization by murine macrophages. J Med Microbiol. 2008;57:1322–1328. [DOI] [PubMed] [Google Scholar]
- 7.Dolan SK, Welch M. The glyoxylate shunt, 60 years on. Annu Rev Microbiol. 2018;72:309–330. [DOI] [PubMed] [Google Scholar]
- 8.Dunn MF, Ramírez-Trujillo JA, Hernández-Lucas I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology (Reading, Engl). 2009;155:3166–3175. [DOI] [PubMed] [Google Scholar]
- 9.Chew SY, Ho KL, Cheah YK, et al. Glyoxylate cycle gene ICL1 is essential for the metabolic flexibility and virulence of Candida glabrata. Sci Rep. 2019;9:2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, Li X, Feng P, et al. RNAi-mediated silencing of fungal acuD gene attenuates the virulence of Penicillium marneffei. Med Mycol. 2014;52:167–178. [DOI] [PubMed] [Google Scholar]
- 11.Schobel F, Ibrahim-Granet O, Ave P, et al. Aspergillus fumigatus does not require fatty acid metabolism via isocitrate lyase for development of invasive aspergillosis. Infect Immun. 2007;75:1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rude TH, Toffaletti DL, Cox GM, et al. Relationship of the glyoxylate pathway to the pathogenesis of Cryptococcus neoformans. Infect Immun. 2002;70:5684–5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412:83–86. [DOI] [PubMed] [Google Scholar]
- 14.Asakura M, Okuno T, Takano Y. Multiple contributions of peroxisomal metabolic function to fungal pathogenicity in Colletotrichum lagenarium. Appl Environ Microbiol. 2006;72:6345–6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ZY, Thornton CR, Kershaw MJ, et al. The glyoxylate cycle is required for temporal regulation of virulence by the plant pathogenic fungus Magnaporthe grisea. Mol Microbiol. 2003;47:1601–1612. [DOI] [PubMed] [Google Scholar]
- 16.Idnurm A, Howlett BJ. Isocitrate lyase is essential for pathogenicity of the fungus Leptosphaeria maculans to canola (Brassica napus). Eukaryot Cell. 2002;1:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borneman AR, Hynes MJ, Andrianopoulos A. The abaA homologue of Penicillium marneffei participates in two developmental programmes: conidiation and dimorphic growth. Mol Microbiol. 2000;38:1034–1047. [DOI] [PubMed] [Google Scholar]
- 18.Chen C, Feng J, Chen Z, et al. Optimization of the protoplast-mediated transformation system of Penicillium marneffei. Chin J Mycol. 2016;3:140–144. [Google Scholar]
- 19.Huang X, Li D, Xi L, et al. Caenorhabditis elegans: a simple nematode infection model for Penicillium marneffei. PLoS One. 2014;9:e108764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Li D, Xi L, et al. Galleria mellonella larvae as an infection model for Penicillium marneffei. Mycopathologia. 2015;180:159–164. [DOI] [PubMed] [Google Scholar]
- 21.Rosowski EE, Knox BP, Archambault LS, et al. The Zebrafish as a model host for invasive fungal infections. J Fungi (Basel). 2018;4:136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borneman AR, Hynes MJ, Andrianopoulos A. An STE12 homolog from the asexual, dimorphic fungus Penicillium marneffei complements the defect in sexual development of an Aspergillus nidulans steA mutant. Genetics. 2001;157:1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao X, Feng J, Li Y, et al. Agrobacterium tumefaciens-mediated transformation: an efficient tool for targeted gene disruption in Talaromyces marneffei. World J Microbiol Biotechnol. 2017;33:183. [DOI] [PubMed] [Google Scholar]
- 24.Knox BP, Deng Q, Rood M, et al. Distinct innate immune phagocyte responses to Aspergillus fumigatus conidia and hyphae in zebrafish larvae. Eukaryot Cell. 2014;13:1266–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauta PR, Nayak B, Das S. Immune system and immune responses in fish and their role in comparative immunity study: a model for higher organisms. Immunol Lett. 2012;148:23–33. [DOI] [PubMed] [Google Scholar]
- 26.Howe DG, Bradford YM, Eagle A, et al. The Zebrafish Model Organism Database: new support for human disease models, mutation details, gene expression phenotypes and searching. Nucleic Acids Res. 2017;45:D758–D768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raldua D, Pina B. In vivo zebrafish assays for analyzing drug toxicity. Expert Opin Drug Metab Toxicol. 2014;10:685–697. [DOI] [PubMed] [Google Scholar]
- 28.Tavares B, Santos LS. The importance of Zebrafish in biomedical research. Acta Med Port. 2013;26:583–592. [PubMed] [Google Scholar]
- 29.Simmich J, Staykov E, Scott E. Zebrafish as an appealing model for optogenetic studies. Prog Brain Res. 2012;196:145–162. [DOI] [PubMed] [Google Scholar]
- 30.Brothers KM, Newman ZR, Wheeler RT. Live imaging of disseminated candidiasis in zebrafish reveals role of phagocyte oxidase in limiting filamentous growth. Eukaryot Cell. 2011;10:932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenor JL, Oehlers SH, Yang JL, et al. Live imaging of host-parasite interactions in a zebrafish infection model reveals cryptococcal determinants of virulence and central nervous system invasion. Mbio. 2015;6:e1415–e1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellett F, Pazhakh V, Pase L, et al. Macrophages protect Talaromyces marneffei conidia from myeloperoxidase-dependent neutrophil fungicidal activity during infection establishment in vivo. PLoS Pathog. 2018;14:e1007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Sar AM, Appelmelk BJ, Vandenbroucke-Grauls CM, et al. A star with stripes: zebrafish as an infection model. Trends Microbiol. 2004;12:451–457. [DOI] [PubMed] [Google Scholar]
- 34.Bojarczuk A, Miller KA, Hotham R, et al. Cryptococcus neoformans intracellular proliferation and capsule size determines early macrophage control of infection. Sci Rep. 2016;6:21489. [DOI] [PMC free article] [PubMed] [Google Scholar]