Abstract
It has been hypothesized that transcriptional changes associated with lower mTORC1 activity in mice with reduced levels of Growth Hormone and Insulin-like Growth Factor 1 are responsible for the longer healthy lifespan of these mutant mice. Cell lines and tissues from these mice show alterations in the levels of many proteins that cannot be explained by corresponding changes in mRNAs. Such post-transcriptional modulation may be the result of preferential mRNA translation by the cap-independent translation of mRNA bearing the N6-methyl-adenosine (m6A) modification. The long-lived endocrine mutants – Snell dwarf, growth hormone receptor deletion, and pregnancy associated plasma protein-A knockout – all show increases in the N6-adenosine-methyltransferases (METTL3/14) that catalyze 6-methylation of adenosine (m6A) in the 5’ UTR region of select mRNAs. In addition, these mice have elevated levels of YTH domain containing protein 1 (YTHDF1), which recognizes m6A and promotes translation by a cap-independent mechanism. Consistently, multiple proteins that can be translated by the cap-independent mechanism are found to increase in these mice, including DNA repair and mitochondrial stress response proteins, without changes in corresponding mRNA levels. Lastly, a drug that augments cap-independent translation by inhibition of cap-dependent pathways (4EGI-1) was found to elevate levels of the same set of proteins and able to render cells resistant to several forms of in vitro stress. Augmented translation by cap-independent pathways facilitated by m6A modifications may contribute to the stress resistance and increased healthy longevity of mice with diminished GH and IGF-1 signals.
Keywords: Aging, Growth Hormone, IGF1, gene expression, protein translation
Introduction
Genetic or pharmacological inhibition of the mechanistic target of rapamycin (mTOR) induces lifespan extension in lower organisms (McCormick et al., 2011, Johnson et al., 2013, Wang and Miller, 2012b, Wang and Miller, 2012a), as well as mice treated with rapamycin (Zhou and Huang, 2010) or calorie restriction (Cummings and Lamming, 2016). Mutants with lower growth hormone and insulin like growth factor 1 (GH/IGF1) signaling, including Snell dwarf (DW), GH receptor deletion (GHR−/−) and pregnancy associated plasma protein A-1 deficient mice (PKO mice) all show a reduction in mTORC1 activity (Dominick et al., 2015, Dominick et al., 2017). Lower mTOR function diminishes cap-dependent mRNA translation; despite this, slow-aging endocrine mutant mice show increased mitochondrial function and elevated levels of proteins involved in cellular stress resistance (Dominick et al., 2017, Pickering et al., 2015, Gesing et al., 2011a, Fang et al., 2013). These include elevation of Direct DNA repair, mediated by O-6-methylguanidine-DNA methyltransferase (MGMT) and N-myc downstream regulated gene-1 (NDRG1) (Meira et al., 2014), and of proteins involved in mitochondrial biogenesis and stress resistance, such as peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1) (Gesing et al., 2011b) and mitochondrial transcription factor A (TFAM), which also increase in calorie restriction (Picca and Lezza, 2015).We have reported that elevated MGMT and NDRG1 proteins in the DW, GHR−/− and PKO mice are not the result of increases in their mRNA transcript levels, suggesting regulation at the translational level (Dominick et al., 2017). It is not yet clear how these proteins levels are increased in the absence of transcription changes, and in the face of the general inhibition of translation caused by lower mTORC1 activity. We have suggested that changes in the composition of the mammalian homolog of the yeast carbon catabolite repression 4 complex (CNOT complex), a posttranscriptional regulator of mRNA stability and translation (Miller and Reese, 2012), could play a role in selective translation of specific subsets of mRNAs (Dominick et al., 2017). However, new mechanistic insights are needed to account for the specificity of a selective upregulation in translation of this proteins.
Translation of a select group of proteins can be enhanced, during inhibition of cap-dependent translation, via a cap-independent translation (CIT) mechanism (Lacerda et al., 2017). Methylation at position 6 of adenosine (m6A) at the 5-prime end of certain mRNA sequences has been shown to increase the expression of HSP70 by CIT when cap-dependent translation is inhibited (Zhou et al., 2015, Wang et al., 2015). In this pathway, the nuclear N6-adenosine-methyltransferases 3 and 14 (METTL3 and METTL14) add m6A modifications to specific sets of mRNAs, while the alpha-ketoglutarate-dependent dioxygenase homolog 5 (ALKBH5) and the fat mass and obesity-associated protein (FTO) remove them. The balance between these opposing process determines the level of m6A-modified mRNAs. In the cytosol, m6A-mRNAs are recognized and bound by the YTH domain containing family of proteins (YTHDF). YTHDF1 can control preferential translation of m6A-tagged mRNAs by direct association to the translation machinery, bypassing the canonical cap-dependent interaction (Shi et al., 2017, Wang et al., 2015). In addition, overexpressing YTHDF2 can enhance translation of some proteins, including heat shock proteins (Zhou et al., 2015). Furthermore, YTHDF2 can interact with the CNOT complex to regulate mRNA stability and, in some cases, translation (Du et al., 2016). In addition, rapamycin (a drug that can extend mouse lifespan) seems to enhance the preferential translation of m6A-containing mRNAs (Bodi et al., 2015). All these observations, including lower mTOR activity in slow aging mouse models, suggests a mechanistic connection between m6A modification and preferential translation controlled by METTL3/14, the YTHDF proteins and the CNOT complex that may be involved in lifespan extension.
We hypothesized that mutations that extend lifespan by lowering signaling through GH and/or IGF-1 would lead to elevation of CIT. However, other mechanisms that modulate protein levels (without change in transcription) could also be involved; including increases in protein stability. To explore these hypothesis, we evaluated levels of the proteins involved in the m6A pathway (METTL3/14, ALKBH5, FTO and YTHDF1/2) and evaluated levels of proteins whose mRNAs can be translated by CIT, including MGMT, NDRG1, TFAM, PGC1 and the mitochondrial Lon Peptidase 1 (LONP1) (Wei et al., 2015, Gesing et al., 2011a, Westbrook et al., 2009). The analysis of fibroblasts and tissues samples from DW, GHR−/−, and PKO mice would test if upregulation of these proteins would reflect enhanced CIT or altered protein stability. Furthermore, we tested whether these slow-aging mice had changes in METTL3, YTHDF1 and YTHDF2 that controls the CIT translation of m6A tag proteins. Lastly, we tested whether 4EGI-1, a drug that inhibits cap-dependent translation (Salvi et al., 2016), enhances the levels of CIT relevant proteins.
Materials and Methods
Reagents and animal models
DW mice were produced as previously described (Dominick et al., 2015). Breeding stock for GHR−/− mice was provided by Andrzej Bartke (Southern Illinois University, IL) and John Kopchick (Edison Biotechnology Institute of Ohio University, OH). Breeding stock for PKO mice were provided by Cheryl Conover (Mayo Clinic, Rochester, MN). Genetically heterogenous mice of the UM-HET3 stock were produced as described (Harrison et al., 2009). All mice were used at ages from 5–6 months. All experiments were approved by the University of Michigan Institutional Animal Care and Use Committee. Sentinel animals were examined quarterly for evidence of viral infection; all tests were negative. Unless indicated in text, all mice were fed ad lib.
Chemical reagents (including 4EGI-1, thapsigargin and tunicamycin) were purchased from Millipore-Sigma (St Louis, MO). Primary mouse fibroblasts from DW, GHR−/− and UM-HET3 tail mice were obtain and cultured as described (Murakami et al., 2003). Clones corresponding to human YTHDF1-Flag and YTHDF2-Flag were purchased from Origene (Rockville. MD. USA; catalogs: RC207185 and RC200038). The ShRNA for human METTL3 was obtained from Millipore-Sigma (St Louis, MO. USA; catalog: SHCLNG-NM_019852) and Human SiRNA METTL3 from Invitrogen (Carlsbad, CA. USA; catalog: HSS125548). Transfections were performed with Lipofectamine 3000 (Thermofisher Scientific. Waltham, MA. USA) while SiRNA transfection was performed with Lipofectamine RNAiMAX from Invitrogen (Carlsbad, CA. USA). qRT-PCR probes were obtained from Integrated DNA technologies (Coralville, Iowa. USA). Primary antibodies for western blots were purchased from different sources as indicated in Supplemental Table 1. Peroxidase conjugated secondary antibodies were purchased from Jackson Immunoresearch (West Grove, PA. USA).
RNA isolation and cDNA synthesis
Total RNA from fibroblast and tissue samples of mice was isolated using the Trizol Plus RNA purification kit (Cat 1218355, Thermo Fisher Scientific, Waltham, MA. USA) according to the manufacturer’s instruction. RNA was further purified using the Qiagen RNeasy mini RNA cleanup protocol (Qiagen, Valencia, CA, USA). The concentration of total RNA was measured using the Nanodrop-One spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Quality control was estimated by 260/230 ratio and 260/280 ratio using Nanodrop-One; each individual sample showed a 1.88 ratio or higher. RNA integrity was also checked by evaluation of the 28/18S RNA using an Agilent 2100 Bioanalyzer (data not shown). Total RNA (1.0 μg) was reverse transcribed to cDNA using iScript cDNA reverse transcription kits (Bio-Rad, Hercules, CA. USA) according to the manufacturer’s instructions.
Quantitative real-time PCR
For each individual gene (human or mouse), three sets of primers were designed using the IDT QRT-PCR probe design website (Integrated DNA Technology, Coralville, Iowa, USA) based on their corresponding exon regions. Primers were designed with annealing temperature of approxiamately 61.5 °C. The specific probes shown in Supplemental Figure 2, used for the qRT-PCR analysis, were selected based on the presence of a single peak in the melt curve and a single amplicon band using agarose gel electrophoresis. The higher sensitivity probes were selected based on the lower CT values using a standard cDNA obtained from HEK (human) and mouse fibroblasts (UMHet fibroblast). In all cases, the selected probes shown in Supplemental Figure 2 show similar sensitivity to probes reported in the literature. The specific qRT-PCR was performed using Fast Start Universal SYBR Green Master Mix (Applied Biosystems, Foster City, CA. USA). Reactions were performed using an Applied Biosystems 7500 Real-Time RT-PCR System. RT-PCR was performed using quantitative PCR systems (Applied Biosystems® 7500 Real-Time PCR Systems; ThermoFisher Scientific; Waltham, MA, USA) with corresponding primers (see Supplementary Table 2). Actin and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were simultaneously assayed as loading controls. In all cases, the internal mRNA controls for Actin and GAPDH show no significant differences between samples from each experimental group (less than 1 CT value of differences between samples, data not shown); therefore the cycle time (CT) data are shown without normalization, to avoid introducing artifacts related to altered levels of Actin or GAPDH. Furthermore, two-way Anova analysis revealed no significant sex or interaction effects in any of the mRNA levels, and therefore males and females from each group were pooled for analysis. Data for each gene was analyzed by comparing control (or untreated) with respect to corresponding genetic mice model (or treatment) using Prism statistical analysis software, with the Student’s t-test at a significance level of p≤0.05.
In vitro cycloheximide treatments, METTL3 knock down, 4EGI-1 treatments and stress assays.
For the cycloheximide experiments, mouse fibroblasts from DW and GHR−/−, and their respective littermate controls, were incubated with 25 µg/ml of cycloheximide for varying intervals (as indicated in text); then cell lysates were obtained and western blots against specific proteins performed as described in western blots section of methods. The 25 µg/ml cycloheximide dose was selected after each cell line was tested with concentrations between 1 µg/ml and 100 µg/ml. Doses of 50 to 100 µg/ml led to significant cell death by apoptosis, while doses below 10 µg/ml showed little effect on protein degradation after 24 hours of treatment. The lack of antibodies suitable for immunoprecipitation precludes direct measures of protein turnover by radiolabel pulse methods; therefore, we measured protein levels by immunoblotting at different cycloheximide intervals (which blocks protein synthesis) to measure the rate of degradation over a period of 24 hours. To calculate protein degradation (protein half-life) we use the methods previously described (Salmon et al., 2005) with modifications.
For transfection experiments, human HEK cells or mouse fibroblasts were left untreated (Lipofectamine plus ShRNA controls) or treated with shRNA METTL3 (or siRNA METTL3) for 48 hours followed by treatment with 4EGI-1 at 50 µM or untreated control for an additional 48 hours. The dose of 50 µM for 4EGI-1 treatments was selected based on previously described results in cell culture (Chen et al., 2012). In addition, a dose curve from 5 to 150 µM was performed, and revealed that doses above 50 µM range showed no higher upregulation of the proteins analyzed in our experimental settings (data not shown). Cell lysates were obtained and adjusted to equivalent actin levels using western blots and levels of different target protein targets were analyzed by immunoblotting as described below.
Stress assays were performed as described (Salmon et al., 2005) with modifications: DMEM medium with 10% FCS was used for experiments in which cells were exposed to 4EGI-1 for 48 hours followed by the different stressors for 24-hour. The stressors MMS, thapsigargin, or tunicamycin were used over a range of concentrations selected based on preliminary dose titrations.
In vivo 4EGI-1 treatments, tissue processing and western blot analysis.
For in vivo 4EGI-1 assays, UM-HET3 mice were injected intraperitoneally with 75 mg/kg body weight of 4EGI-1 in 15 µl of DMSO each day for 5 consecutive days as previously described (Chen et al., 2012). Control mice received 15 µl of DMSO each day for 5 consecutive days. Then mice were fasted for 18 hours, and their tissues analyzed for proteins by Western blots as described below or for mRNA levels using qRT-PCR.
Tissues or tissue culture cells from mutant and littermate mice were extracted in RIPA buffer (Fisher Scientific, Pittsburgh, PA, USA) supplemented with Complete Protease Inhibitor Cocktail as previously described (Dominick et al., 2017). The protein extracts were separated by SDS/PAGE on a 4–15% running gel, and then transferred to Immobilon-P Transfer Membrane (Millipore, Billerica, MA, USA) for immune blot analyses. Membranes were blocked in Tris buffered saline containing 0.05% Tween20 (TBS-T) and 5% bovine serum albumin (BSA). All samples were normalized to the same amount of actin (determine by western blots) follow by western blots with the primary antibodies (see supplementary Table 1) using TBS-T 5% BSA for one hour at room temperature. Membranes were then washed with TBS-T and incubated with peroxidase conjugated secondary antibody for 1 hour. Membranes were then evaluated using an ECL Chemiluminescent Substrate (Fisher Scientific, Pittsburgh, PA, USA). Light intensity was measured by GE Image Quant LAS-4000 (General Electric, Pittsburgh, PA. USA), while General Electric, and quantification was performed using Image-Quant software.
For in vivo 4EGI-1 assays, the UM-HET3 mice were IP injected with 75 mg/Kg body weight of 4EGI-1 in 15 µl of DMSO each day for 5 consecutive days as been previously described (Chen et al., 2012). Control mice also received 15 µl of DMSO/day for 5 days. Then mice were fasted for 18 hours, then euthanized, and their tissues analyzed by Western blots or qRT-PCR as described above.
Statistical analysis.
Unless indicated otherwise, results are presented as mean ± standard error of the mean (SEM). To compare specific effects between groups, an unpaired t-test was used with p ≤ 0.05 as the criterion for significance. Statistical analysis and graph presentation were performed using GraphPad Prism software. Two-way Anova analysis reveals no statistical differences between males and females in any of the data sets, and therefore males and females from a single experimental group were pooled for analysis.
Results
Fibroblasts from DW and GHR−/− mice have higher levels of proteins regulating m6A-mediated cap-independent translation (“CIT”).
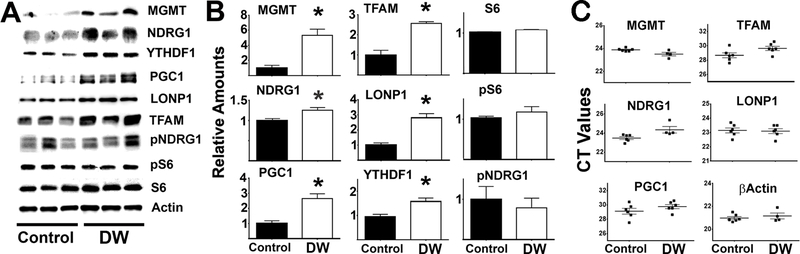
To test if preferential translation (Dominick et al., 2017) might be involved in the upregulation of proteins in fibroblasts of slow-aging mouse models (DW, GHR−/−) levels of YTHDF1 were evaluated, because YTHDF1 contributes to CIT translation (Zhou et al., 2015, Wang et al., 2015). In addition, levels of several proteins whose mRNA contain m6A modifications, and which were therefore likely to be targets of CIT, were also evaluated: MGMT, NDRG1, PGC1, LONP1 and TFAM (Linder et al., 2015, Dominissini et al., 2012). Figure 1 shows a typical western blot comparing DW and control fibroblasts. No changes in ribosomal S6, pS6 (an indicator of mTORC1 function) or pNDRG1 (an indicator of mTORC2 function) were found between DW and controls, suggesting no differences in mTOR signaling. In contrast, protein levels of YTHDF1 and CIT targets (MGMT, NDRG1, PGC1, TFAM and LONP1) were higher in DW fibroblasts (Figure 1A), consistent with the hypothesis that these mice might have higher CIT. Data from a total of 6 DW and 6 control mice (half males), confirmed that there were significantly higher levels of YTHDF1 and CIT target proteins with no changes in mTOR signaling (Figure 1B). For unknown reasons, YTHDF2 protein was not detected in fibroblasts from DW or GHR−/− mice using available antibodies. To test if changes in the level of these proteins reflects changes in transcription, mRNA levels were measured in parallel experiments. There were no significant differences between DW and control mice in respective levels of each mRNA (Figure 1C) ; the increase in protein without an increase in mRNA suggests that the abundance of these proteins is being controlled by a post-transcriptional mechanism.
Figure 1. Fibroblasts from DW mice show upregulation of proteins levels associated with m6A cap-independent translation without changes in their corresponding mRNA levels.
A) Representative western blots from 3 mice per group comparing the expression of proteins involved in the m6A and mTOR pathways between control and DW fibroblasts; extracts were normalized to actin. B) Bar graphs represent the mean SEM of protein levels from fibroblasts obtained from 6 mice per group. Error bars are S.E.M. All values are shown relative to mean levels of control mice. The (*) indicates statistical significance (p<0.05) relative to controls. C) No significant differences were found in the mean CT value (horizontal bar) of the corresponding mRNA levels between control and DW fibroblasts from 6 mice per group. Each symbol represents a different mouse.
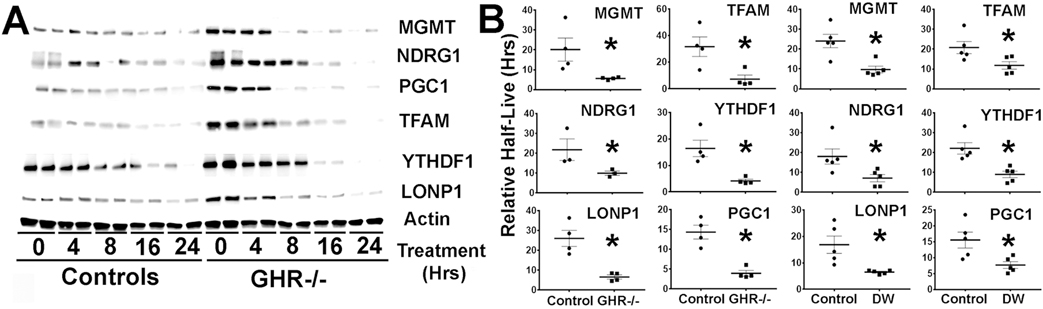
Increased protein levels without increases in the corresponding mRNAs might be due to increases in protein stability. To test this hypothesis, fibroblasts from DW and GHR−/− mice were treated with cycloheximide, and the rate of degradation over the next 24 hours was evaluated by immunoblotting. Figure 2A shows a typical result for GHR−/− fibroblasts; identical results were seen for cells from DW mice (not shown). As expected, levels of each tested protein decline after cycloheximide addition. Figure 2B shows summary statistics for half-life for each protein for both GHR−/− and DW cells. Half-life was significantly shorter for all six proteins in both DW and GHR−/− cells respect to their controls. The basis for the higher rate of degradation is unknown. A decrease in the half-lives of these proteins, despite their elevated steady state levels in fibroblasts of DW GHR−/− mice, suggests that these proteins have a higher production rate, likely to reflecting elevated CIT translation. Figure 2 shows that in the GHR−/− fibroblasts, there was upregulation of both YTHDF1 and CIT targets proteins (Figure 1; see t = 0 data for Figure 2) suggesting that both DW and GHR−/− cells have augmented CIT.
Figure 2. Fibroblasts from GHR−/− and DW mice show upregulation of protein degradation compared to control mice.
A) Representative western blots comparing degradation of MGMT, NDRG1, TFAM, LONP1, YTHDF1 and PGC1 proteins between control and GHR−/− fibroblasts at different time points during cycloheximide treatments. Extracts were normalized to actin. B) Bar graphs represent the mean ± S.E.M. of the half-life for each protein relative to actin from fibroblasts of 4 GHR−/− mice per group (2 females) and 5 DW mice per group (2 females). Each symbol represents a different mouse, and (*) indicates statistical significance (p<0.05) relative to controls.
DW, GHR−/− and PKO mice show upregulation of the m6A-CIT pathway in liver tissue.
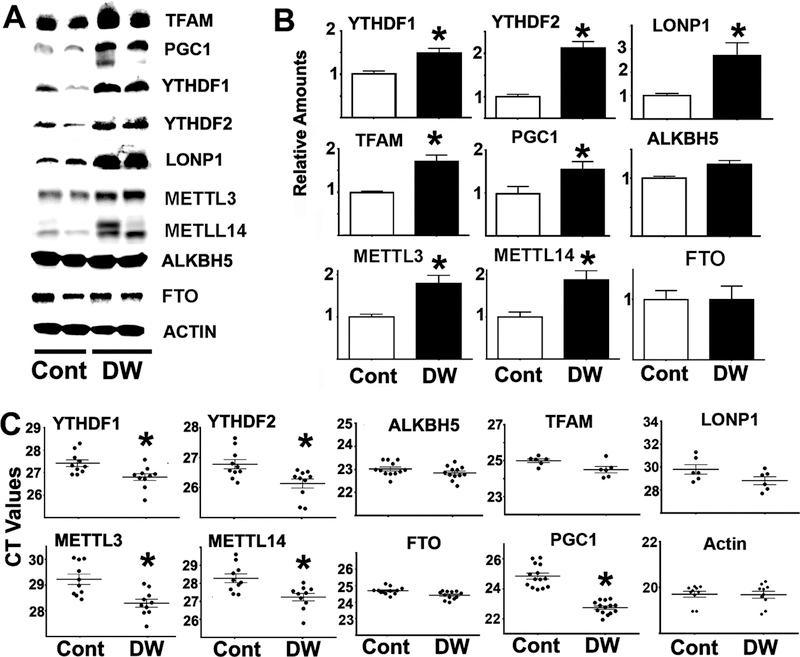
The activity of the m6A-CIT-pathway was evaluated in liver of DW mice and controls by measuring levels of METTL3, METTL14, ALKBH5, FTO, YTHDF1 and YTHDF2 proteins, as well as five CIT target protein (MGMT, NDRG1, PGC1, LONP1, and TFAM). The levels of METTL3, METTL14, YTHDF1 and YTHDF2 protein were all higher in DW liver than in controls (Figure 3A–B). Furthermore, consistent with the fibroblast results, liver of DW mice had elevated levels of CIT target proteins (TFAM, LONP1 and PGC1; Figure 3A–B) suggesting higher CIT. In contrast, we found no changes in levels of ALKBH5 or FTO (Figure 3A–B). DW livers showed increases in mRNA levels for METTL3, METTL14, YTHDF1 and YTHDF2 with respect to controls, but no changes in mRNA for ALKBH5 or FTO. Consistent with the fibroblast data, there were no changes in mRNA for the CIT targets (MGMT, NDRG1, TFAM and LONP-1). Our finding that, in DW liver, there are increased levels of CIT target proteins, METTL3/14 and YTHDF1/2, is consistent with the hypothesis that there is a systemic increase in m6A-dependent CIT in DW mice.
Figure 3. Liver from DW mice shows upregulation of several proteins involved in, or regulated by, the m6A pathway.
A) Representative western blots comparing protein levels from the m6A pathway as well as TFAM, LONP1 and PGC1 between control and DW livers. All extracts were normalized to actin. B) Bars represent the mean ± S.E.M. from at least 12 mice per group. All values are shown relative to mean levels of control mice. C) CT values from mRNA purified from at least 10 mice per group. LONP1 mRNA was performed with 6 mice per group in an independent experiment. Each dot represents a different mouse. The (*) indicates statistical significance (p<0.05) relative to controls.
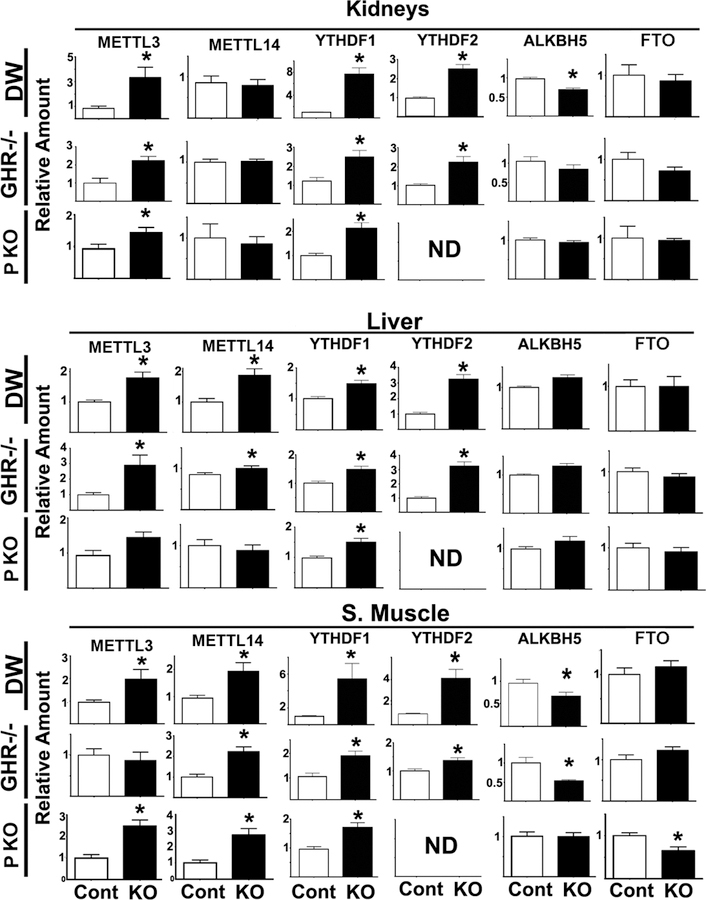
To broaden our survey, the levels of m6A pathway proteins were measured in kidney, skeletal muscle and liver from DW, GHR−/−, and PAPPA-KO (PKO) mice (Figure 4). METTL3 was significantly higher in all tissues from DW mice, and in kidney and liver of GHR−/− mice. PKO mice also show elevation of METTL3 in all three tissues, though the elevation in liver was not statistically significant. Two-way Anova analysis showed no significant effects of sex or interactions (using p≤0.05 threshold) for each of the tissues and proteins, and thus data were combined across sex for subsequent analyses. However, there was a trend, which did not reach statistical significance, for higher levels of METTL3 protein in some tissue of bot female control and DW mice.
Figure 4. METTL3/14, YTHDF1/2, ALKBH5 and FTO protein levels in different tissues of DW, GHR−/− and PKO mice.
Bars represent the mean ± S.E.M.SEM from 6 independent experiments totaling at least 20 mice per group (half males). All values are shown relative to mean levels of control mice. The (*) indicates statistical significance (p<0.05) relative to controls. ND = not detected.
Interestingly, all these long-lived mice show elevation of YTHDF1 in all tissues tested. Similarly, YTHDF2 was upregulated in all three tissues from DW and GHR−/− mice. We were not able to detect YTHDF2 in tissue samples from either PKO mice or littermate controls; the reason is unknown. The data on METTL14 are less consistent, with elevation seen in skeletal muscle of all three kinds of mice, and in liver of DW and GHR−/− mice. As in the case of METTL3, METTL14 protein tended to be higher in females than in males in some tissues, but the effect did not reach statistical significance. Levels of ALKBH5 and FTO in long-lived mice were either lower than or comparable to those in controls; lower levels, when present, could potentially contribute to elevation of CIT. Thus, the data suggest upregulation of the molecules that insert or read m6A signals (METTL3 or METTL14 and YTHDF1) and thus promote CIT, with some variation among tissues in these slow-aging mice.
Overexpression of YTHDF1 or YTHDF2 enhances translation of proteins that are upregulated in several slow-aging mouse models.
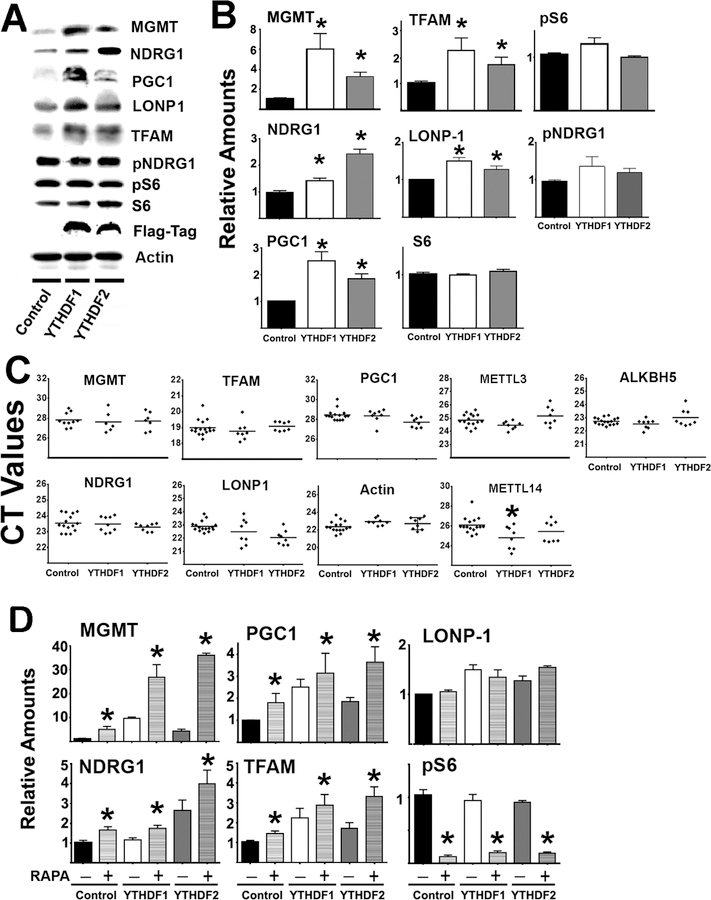
To test whether these m6A readers can regulate the expression of the relevant CIT target proteins, human YTHDF1-Flag tag or YTHDF2-Flag tag constructs were overexpressed in HEK cells. Lysates prepared 48 hours after transfection showed the expression of exogenous YTHDF1 or YTHDF2 containing the Flag tag (Figure 5A). As expected, S6, pS6, and pNDRG1 were not altered by the transfections of any of the constructs, implying that overexpression of YTHDF1 or YTHDF2 does not affect mTOR activity (mTORC1 and mTORC2) or downstream signaling. However, overexpressing either YTHDF1 or YTHDF2 led to upregulation of CIT targets (MGMT, NDRG1, PGC1, LONP1, and TFAM proteins, see Figure 5A–B), consistent with the results in primary fibroblasts (Figure 1) and in liver samples from DW mice (Figure 3). Importantly, there were no alterations in the corresponding mRNAs for these CIT target proteins after transfection, suggesting that protein upregulation was due to increased translation (Figure 5C). We noted a small, but significant, increase in mRNA for METTL14 (though not METTL3) after YTHDF1 transfection, suggesting a possible positive feedback effect that could further augment m6A modification.
Figure 5. Overexpression of YTHDF proteins enhances protein levels for several genes found to be upregulated in slow aging mice.
A) Representative western blot of target protein levels in HEK cells transfected with control vector, or with YTHDF1 or YTHDF2 vectors. B) Bars represent the mean ± S.E.M.SEM from 8 independent transfections. All values were normalized to 1 for the respective controls. C) CT values of qRT-PCR performed using mRNA purified from at least 8 independent transfections per group. The (*) indicates statistical significance (p<0.05) relative to controls. D) The bar represents the mean ± S.E.M. from at least 8 independent transfections, showing treatment with or without rapamycin (Rapa+ or Rapa-) in enhancing the effects of YTHDF on protein expression. (*) indicates statistical significance (p<0.05) relative to rapamycin untreated cells (Rapa -) from same group.
Because inhibition of cap-dependent translation might promote CIT, we tested whether rapamycin treatment (which inhibits mTORC1 and cap-dependent translation) would enhance these effects. Rapamycin treatment did indeed have an additional increase in levels of CIT targets (MGMT, NDRG1, TFAM, and PGC1; Figure 5D), suggesting that blockage of cap-dependent translation might enhance CIT.
Inhibition of cap-dependent translation with 4EGI-1 upregulates CIT through the m6A pathway.
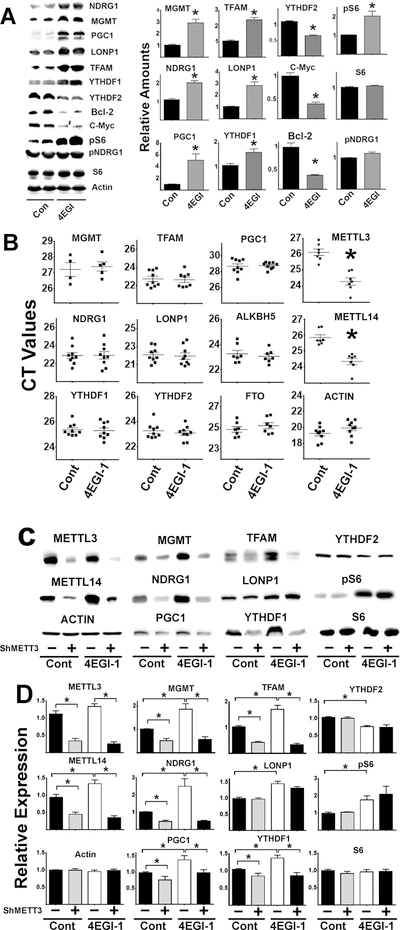
Lower mTORC1 activity might, in principle, lead to upregulation of this set of proteins by mechanisms independent of m6A-CIT (Saxton and Sabatini, 2017). To test this hypothesis, we evaluated the effects of 4EGI-1 on the m6A-CIT pathway and on CIT target proteins. 4EGI-1 can disrupt eIF4E/eIF4G interaction and thus inhibit cap-dependent translation without effects on CIT (Papadopoulos et al., 2014, Chen et al., 2012). Thus, 4EGI-1 exposure can test the idea that reduction in cap-dependent translation might enhance CIT independently of mTOR signaling. In addition, we used ShRNA METTL-3 to test if reduction in METTL-3 dependent m6A would control the expression of the CIT target proteins.
Results are shown in Figure 6. As expected, 4EGI-1 led to reduced expression of Bcl-2 and c-Myc protein, through inhibition of cap-dependent translation (Mazzoletti et al., 2011, Wu et al., 2016). But more important, as predicted by our models, HEK cells exposed to 4EGI-1 have elevated levels of CIT target proteins (MGMT, NDRG1, PGC1, TFAM and LONP1), consistent with CIT induction. 4EGI-1 also increases levels of YTHDF1, suggesting a second route to increased CIT. Unexpectedly, 4EGI-1 also enhanced phosphorylation of S6 (without changes in S6 protein levels, Figure 6A). The mechanism is unknown, but we note that 4EGI-1 elevation of pS6 is not seen in untransformed mouse fibroblasts (see below); compensatory increased TORC1 activity may be a property or HEK cells not seen in untransformed cell strains. 4EGI-1 treatments did not induce changes in mRNA for any of the CIT target proteins (Figure 6B), confirming that increased protein levels do not reflect corresponding changes in transcription. Interestingly, 4EGI-1 treatment lead to increases in METTL3 and METTL14 transcripts (corresponding to a lower CT values for both enzymes after 4EGI-1 treatments). This observation suggests a possible positive feed-back loop that enhances the m6A pathway (see Figure 6B) when cap-dependent translation is reduced. In contrast, 4EGI-1 treatment did not affect levels of ALKBH5 and FTO.
Figure 6. Effects of 4EGI-1 and downregulation of the METTL3/14 in HEK CIT proteins.
A) Representative western blots from two mice per group showing the effects of 4EGI-1 treatments on CIT protein expression; bars represent the mean ± S.E.M.SEM from, at least, 4 independent experiments. All values were normalized to the respective controls. B) CT values of qRT-PCR performed in mRNA purified from at least four 4EGI-1/controls treatments per group for MGMT and additional, at least, six 4EGI-1 treatments/controls for the other mRNAs tested. C) Representative western blot analysis of downregulation of METTL3/14 by shRNAMETTL3 followed by 4EGI-1 treatments. D) Graph bars represent the mean ± S.E.M. SEM from at least 4 independent experiments. All values were normalized to the respective controls. The (*) indicates statistical significance (p<0.05) relative to controls.
Using a shMETTL3 knockdown protocol would test causal links between the m6A pathway and changes in the levels of CIT target proteins. As shown in Figure 6C, lowering METTL3/14 enzymes by ShMETTL3 produces significant declines in CIT target proteins (MGMT, NDRG1, PGC1 and TFAM) and YTHDF1 proteins, suggesting that the upregulated proteins are indeed modulated by the m6A pathway via CIT. There were no effects on S6, YTHDF2 and unexpectedly LONP1 proteins levels. Interestingly, LONP1 does not seem to be affected by METLL-3 downregulation, but was upregulated by 4EGI-1, hinting that this protein may be regulated by other non-m6A CIT mechanisms (Lacerda et al., 2017).
To further test whether 4EGI-1 upregulation of these CIT proteins was m6A dependent, cells transfected or not transfected with ShMETTL3 were treated with 4EGI-1, and CIT targets then evaluated (Figure 6D). Treatment with 4EGI-1 alone led to a significant increase in CIT targets (MGMT, NDRG1, PGC1, TFAM, and LONP1). In contrast, cells treated with ShMETTL3 did not show increases in the levels of CIT targets (NDRG1, MGMT, PGC1, TFAM) or YTHDF1 protein after 4EGI-1 treatment. ShMETTL3 did not affect levels of LONP1, consistent with the data using SiMETTL3. As expected, we found no effects of either ShMETTL3 or 4EGI-1 on several control proteins (actin and S6) suggesting that the increases in many protein levels by 4EGI-1 is mediated by the m6A pathway rather than a more general effect on protein translation.
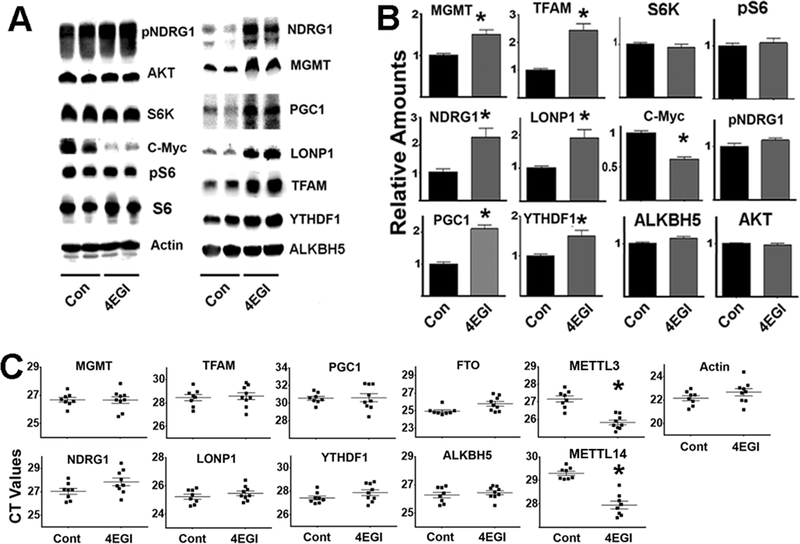
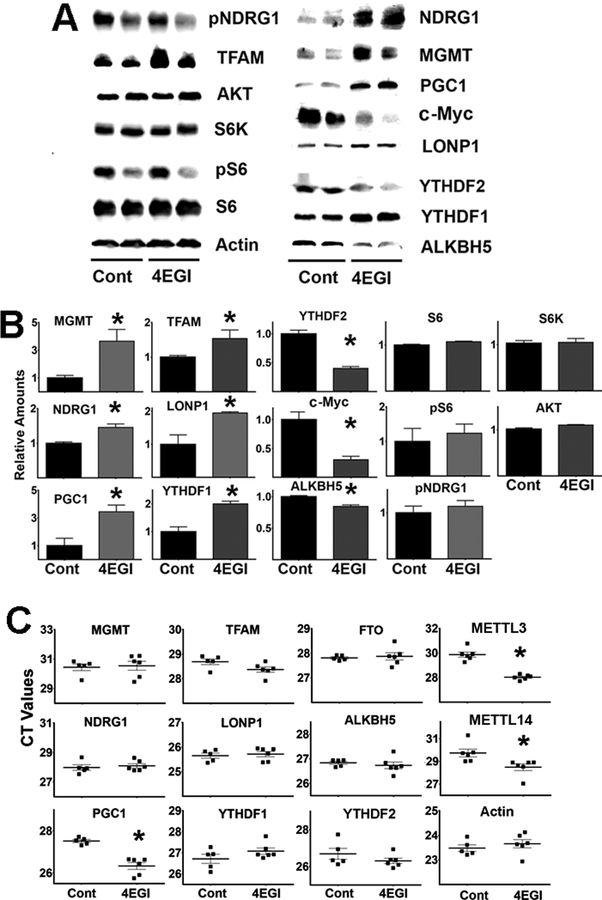
To test our model using non-transformed cells, primary fibroblasts freshly isolated from UM-HET3 mice were treated (or untreated) with 4EGI-1. The data are shown in Figure 7A–B. 4EGI-1 treatment led to downregulation of the c-Myc protein, reflecting inhibition of cap-dependent translation (Yi et al., 2014). Level of protein involved in mTORC1 and mTORC2 signaling pathway were not altered by 4EGI-1, including levels of AKT, S6K, S6, pS6 or pNDRG1. The results suggest that, under these conditions, 4EGI-1 does not alter mTORC1 or mTORC2 activity (Figure 7B). However, 4EGI-1 did enhance expression of all five tested CIT target proteins (MGMT, NDRG1, PGC1, TFAM, and LONP1), and increased levels of YTHDF1 protein without altering ALKBH5. These results are similar to those seen in HEK cells (Figure 6). As expected, 4EGI-1 did not alter any of the mRNA transcript levels of the CIT target proteins (Figure 7C). Interestingly, 4EGI-1 treatment did increase levels of mRNA transcripts for METTL3 and METTL14 (declines in the CT values for both mRNAs, Figure 7C), as previously noted in HEK cells, supporting models of a causal link between decline in cap-dependent translation and increases in the m6A-dependent pathway.
Figure 7. 4EGI-1 treatments can upregulate the expression of proteins associated with the m6A pathway in mouse fibroblasts.
A) Representative western blot analysis of 4EGI-1 treatments in UM-HET3 mouse fibroblast cells. B) Bars represent the mean ± S.E.M. from fibroblasts of 6 mice per group. C) CT values of qRT-PCR performed using at least 8 control and 8 4EGI-1 fibroblast preparations per group. The (*) indicates statistical significance (p<0.05) relative to controls.
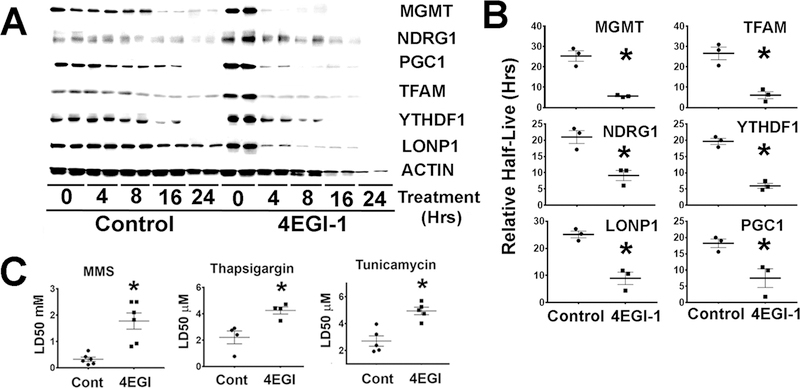
To test the alternative hypothesis that 4EGI-1 could reflect increases in stability (half-life) of CIT proteins, UM-HET3 tail fibroblasts were treated with 4EGI-1 and then incubated with cycloheximide to evaluate the rate of protein degradation as described in Figure 2. Interestingly, 4EGI-1 seemed to enhance protein degradation (Figure 5A) when compared to untreated controls. Analysis in multiple experiments suggested significant reduction in half-life of these proteins by 4EGI-1 treatments (Figure 5B). The results suggested that increases in the level of CIT target proteins by 4EGI-1 are not the result of enhanced protein stability.
Because CIT is involved in stress resistance (Lacerda et al., 2017), it is possible that 4EGI-1 pre-treatment might enhance cellular resistance to several forms of stress. To test this hypothesis, primary mouse fibroblasts were treated with 4EGI-1 as described in Figure 7, followed by 24-hour exposure to varying doses of MMS, thapsigargin, or tunicamycin; then the LD50 (i.e. the dose that leads to 50% cell death) were calculated. 4EGI-1 pre-treatment enhanced resistance to all three compounds (Figure 8C). The results suggest that 4EGI-1 can increase cellular resistance to some forms of injury, by a CIT mechanism.
Figure 8. 4EGI-1 upregulates protein degradation and enhances stress resistance in mouse fibroblasts.
A) Representative western blots comparing protein degradation rates of MGMT, NDRG1, TFAM, LONP1, YTHDF1 and PGC1 in UM-HET3 fibroblasts, comparing untreated (controls) or 4EGI-1 treated cells at different time points during cycloheximide treatments. B) Mean ± S.E.M. of the half-life relative to actin for each protein from at least 3 independent fibroblast lines for each genotype of mice. Each symbol represents cells from a different mouse, while (*) indicates statistical significance (p<0.05) compared to controls. C) Effects of 4EGI-1 on stress resistance: LD50 (50% lethal dose) values for MMS and tunicamycin treatments in at least 4 experiments from at least 4 different mouse fibroblast donors. The (*) indicates statistical significance (p<0.05) relative to controls.
Acute 4EGI-1 treatment of mice induces cap-independent translation in vivo
To evaluate 4EGI-1 effects in vivo, 4EGI-1 was injected daily into UMHET3 mice for five consecutive days. Tissues harvested and analyzed for the levels of m6A pathway and CIT target proteins. The results are shown in Figure 9A–B. As predicted from the work on cultured cells, 4EGI-1 upregulated CIT targets (MGMT, NDRG1, PCG1, TFAM, LONP1) and YTHDF1 protein in liver without altering mTOR downstream pathways. In addition, lower levels of ALKBH5 were found (as in liver of DW mice, Figure 3), which could promote CIT indirectly by increasing the levels of m6A-modified transcripts. The mRNA transcripts of the corresponding CIT targets were not altered by 4EGI-1 treatment, ruling out transcriptional regulation (Figure 9C). Unexpectedly, 4EGI-1 can downregulated YTHDF2 (Figure 9B), as previously noted in HEK cells (Figure 6). This effect was opposite in direction to the increase seen in slow-aging mice (see Figure 3 and 4). In the other hand, the METTL3 and METTL14 transcripts were also upregulated by 4EGI-1 treatment in vivo (see lower CT values in Figure 9C). The effects of 4EGI-1 in vivo suggest that suppression of cap-dependent translation, for example by rapamycin, could lead to upregulation of the m6A pathway and increases in the CIT activity.
Figure 9. 4EGI-1 treatments can upregulate the expression of proteins associated with the m6A pathway in mouse liver.
A) Representative western blot analysis of 4EGI-1 treatments in liver. B) Bar graph with the mean ± S.E.M. from at least four control mice and nine 4EGI-1 treated mice per group. All values were normalized to the respective controls. The (*) indicates statistical significance (p<0.05) relative to controls. C) CT values of qRT-PCR performed in mRNA purified from samples shown in panel B.
Discussion
At least six mutations that diminish GH and/or IGF1 action extend healthy lifespan in mice, retarding cancer and many other age-related diseases (Junnila et al., 2013). Pathways connecting lower GH/IGF1 signaling to health preservation have been postulated to involve improved insulin sensitivity and glucose tolerance, lower visceral fat (Anisimov and Bartke, 2013), and increased cellular stress resistance (Page et al., 2009). Current models suggest that lower mTOR activity plays in important role in these effects in long-lived Ames dwarf (Sharp and Bartke, 2005), DW, GHR−/− and PAPPA-KO mice (Dominick et al., 2015). Discussions of the role of lower mTOR as a mediator of disease resistance and slow aging have plausibly assumed that transcriptional changes are responsible for the phenotypes and extended lifespan of these mice. In contrast, the data presented here suggest an alternative and complementary mechanism, based on differential translation, that may contribute to the health benefits and lifespan effects of these endocrine mutants, and perhaps also drugs and diets that delay age-related changes in mice (Steffen and Dillin, 2016). Our current data and previous work (Dominick et al., 2017, Pickering et al., 2015) have demonstrated that several mouse models with low GH/IGF1 signaling show upregulation of proteins involved in metabolic and stress response, without corresponding changes in transcription. Our new data suggest a second set of changes in these mice: a switch from cap-dependent to cap-independent translation (CIT) leading to preferential translation of a specific group of m6A-tagged mRNAs. The corresponding proteins play known roles in mitochondrial biogenesis, prevention of mitochondrial oxidation damage, DNA repair, and glucose homeostasis, and our data show that CIT augmentation renders cells resistant to multiple forms of lethal injury. Thus m6A- translational regulation, mediated by m6A-triggered CIT, could represent a key, shared mechanism for longevity extension in both endocrine mutants and rapamycin-treated mice. Multiple tissues of these mice (DW, GHR−/− and PKO) show increases in the METTL3/14 enzymes or declines in the opposing pathway (ALKBH5/FTO) that regulate levels of m6A (Figure 3 and 4). We also note increases in proteins for which YTHDF1 (Figure 5) would lead to enhanced CIT targets (MGMT, NDRG1, TFAM and PGC) because their mRNA contain 5’ sequences susceptible to m6A modification (Meyer and Jaffrey, 2014, Dominissini et al., 2012). Consistent with our model, reduction of cap-dependent translation enhances the effect of YTHDF1 overexpression (Figure 6 and 7), suggesting that rapamycin, which suppresses cap-dependent translation (via mTOR reduction), can upregulate CIT. The turnover data (Fig 2) shows that the increases in the protein levels of these CIT targets are not the results of increases in proteins stability; to the contrary fibroblasts from DW and GHR−/− mice show enhanced protein degradation, through some unknown mechanism, and the same phenotype is seen in normal cells after 4EGI-1 treatment (Fig 8). These results are consistent with previous data showing enhanced protein degradation rates in fibroblasts from DW mice (Wang and Miller, 2012b, Wang and Miller, 2012a). Furthermore, 4EGI-1 treatments can enhance stress resistance in mouse fibroblasts (Fig 8) suggesting that enhancing CIT renders cells resistance to several forms of injury. These results are also consistent with previous data showing primary fibroblasts from DW mice are more resistant to several stressors (Miller, 2009, Murakami et al., 2003), including methyl-methane sulfonate (MMS), a DNA alkylating agent whose effects are repaired by MGMT/NDRG1.
The list of proteins known to be affected by this selective translation shift includes proteins that enhance stress resistance, metabolic and mitochondrial pathways, and could promote tissue repair and protect against age-related declines in tissue health. Further expansion of the list of proteins modulated by CIT is likely to provide useful information on how these pathways affect cell biology and age-related pathophysiology. The responses to downregulation METTL3/14 by ShMETTL3 experiments suggest that MGMT, NDRG1, TFAM, PGC1 are mechanistically modulated by the m6A pathway (Figures 6C–D). These results suggest that augmentation of the m6A pathway by endocrine hormonal changes or drug effects may be responsible for their in vivo upregulation. Our data suggest that these effects do not require mTOR-induced transcriptional changes some of key pathways that could lead to lifespan extension, since 4EGI-1 treatment does not alter mTORC1 or mTORC2 signaling in vitro in fibroblasts or in vivo (Fig. 7 and 9). 4EGI-1 also leads to increases in YTHDF1 and (unexpectedly) declines in YTHDF2 (Figure 6A and 9), implying regulatory circuits that may alter the balance of factors controlling m6A-mediated translation in response to declines in cap-dependent translation found in many models of lifespan extension. On the other hand, our data on LONP1 suggest that other mechanisms, perhaps including other forms of CIT (Lacerda et al., 2017), could contribute to these non-transcriptional control pathways.
Inhibition of cap-dependent translation by 4EGI-1 (Figure 6 and 9), and rapamycin (Gera et al., 2004), leads to a decline in c-Myc protein. Reduction of c-Myc has been shown to lead to lifespan extension in flies and in mice through an unknown mechanism (Hofmann et al., 2015, Greer et al., 2013). c-Myc is known to promote cap-dependent translation (Schmidt, 2004), suggesting a model in which c-Myc downregulation might lead to declines in cap-dependent translation, and relative or absolute increases in CIT. In addition, enhanced GH and IGF1 signaling are known to increase expression of c-Myc (Murphy et al., 1987). These data suggest a model where diminution of GH or IGF1 signals lead to lower c-Myc expression, in addition to lowering mTOR, and thereby promotes CIT with beneficial effects. Furthermore, overexpression of eukaryotic translation initiation factor binding protein (4E-BP1) produces an effect similar to 4EGI-1 treatment by inhibiting eIF4E-eIF4G interaction, thus blocking cap-dependent translation (Zid et al., 2009, Teleman et al., 2005). Upregulation of 4E-BP1 has also been suggested as a mechanism for dietary restriction mediated lifespan extension in Drosophila (Zid et al., 2009) and has beneficial effects in aged mice (Tsai et al., 2016, Tsai et al., 2015). GH and IGF1 can lead to enhanced 4E-BP1 dissociation from the translation complex and enhanced translation (Bush et al., 2003, Schiaffino and Mammucari, 2011). Considering the CIT-inducing effect of 4EGI-1, it is conceivable that 4E-BP1 upregulation might indirectly induce CIT. Consistent with this idea, increased 4E-BP1 leads to higher expression of nuclear encoded mitochondrial genes required for oxidative respiration (Zid et al., 2009). Assessment of changes in CIT and analysis of mRNA sequences of upregulated genes in these models would provide further insight.
There are no current well tested methods that allow evaluation of m6A modification levels in specific 5’ regions of specific low abundance mRNAs from tissues, including those proteins tested here. However, we do find increases in transcripts and proteins from the m6A “writers”, METTL3 and METTL14, in vivo (Figure 3 and 4) suggesting upregulation of m6A pathways at several control points. Our work implies strongly that post-transcriptional pathways, including those dependent on cap-independent translation of selected mRNAs, play a role in lifespan extension in rapamycin-treated mice, in endocrine mutants with diminished GH and/or IGF1 action, and perhaps in calorie restricted mice.
Supplementary Material
Acknowledgments:
We wish to thank Roxann Alonzo, Ilkim Erturk, Lori Roberts, Natalie Perry, William Kohler, Melissa Han, and Logan Beckmann for their technical assistance.
Funding: This work was supported by grants from the National Institutes of Health AG022303, and AG024824 and by the Paul F. Glenn Center for Biology of Aging Research at the University of Michigan.
Abbreviations:
- mTOR
mechanistic target of rapamycin
- mTORC1
mechanistic target of rapamycin complex 1
- mTORC2
mechanistic target of rapamycin complex 2
- DW
Snell dwarf mice
- GHR−/−
grow hormone receptor gene disrupted mice
- PKO
pregnancy associated plasma protein A-1 deficient mice
- MGMT
O-6-methylguanidine-DNA methyltransferase
- NDRG1
N-myc downstream regulated gene-1
- PGC1
peroxisome proliferator-activated receptor gamma coactivator 1-alpha
- LONP1
mitochondrial ATP-dependent Lon protease homolog 1
- TFAM
mitochondrial transcriptional factor A.
- YTHDF1
YT domain containing family of proteins-1
- YTHDF2
YTH domain containing family of proteins-2
- METTL3
nuclear N6-adenosine-methyltransferase 3
- METTL14
nuclear N6-adenosine-methyltransferase 14
- ALKBH5
alpha-ketoglutarate-dependent dioxygenase homolog 5
Footnotes
Competing interests: The authors have declared that no competing interests exist.
References
- ANISIMOV VN & BARTKE A 2013. The key role of growth hormone-insulin-IGF-1 signaling in aging and cancer. Crit Rev Oncol Hematol, 87, 201–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BODI Z, BOTTLEY A, ARCHER N, MAY ST & FRAY RG 2015. Yeast m6A Methylated mRNAs Are Enriched on Translating Ribosomes during Meiosis, and under Rapamycin Treatment. PLoS One, 10, e0132090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSH JA, KIMBALL SR, O’CONNOR PM, SURYAWAN A, ORELLANA RA, NGUYEN HV, JEFFERSON LS & DAVIS TA 2003. Translational control of protein synthesis in muscle and liver of growth hormone-treated pigs. Endocrinology, 144, 1273–83. [DOI] [PubMed] [Google Scholar]
- CHEN L, AKTAS BH, WANG Y, HE X, SAHOO R, ZHANG N, DENOYELLE S, KABHA E, YANG H, FREEDMAN RY, et al. 2012. Tumor suppression by small molecule inhibitors of translation initiation. Oncotarget, 3, 869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CUMMINGS NE & LAMMING DW 2016. Regulation of metabolic health and aging by nutrient-sensitive signaling pathways. Mol Cell Endocrinol [DOI] [PMC free article] [PubMed]
- DOMINICK G, BERRYMAN DE, LIST EO, KOPCHICK JJ, LI X, MILLER RA & GARCIA GG 2015. Regulation of mTOR activity in Snell dwarf and GH receptor gene-disrupted mice. Endocrinology, 156, 565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOMINICK G, BOWMAN J, LI X, MILLER RA & GARCIA GG 2017. mTOR regulates the expression of DNA damage response enzymes in long-lived Snell dwarf, GHRKO, and PAPPA-KO mice. Aging Cell, 16, 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOMINISSINI D, MOSHITCH-MOSHKOVITZ S, SCHWARTZ S, SALMON-DIVON M, UNGAR L, OSENBERG S, CESARKAS K, JACOB-HIRSCH J, AMARIGLIO N, KUPIEC M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature, 485, 201–6. [DOI] [PubMed] [Google Scholar]
- DU H, ZHAO Y, HE J, ZHANG Y, XI H, LIU M, MA J & WU L 2016. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun, 7, 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANG Y, WESTBROOK R, HILL C, BOPARAI RK, ARUM O, SPONG A, WANG F, JAVORS MA, CHEN J, SUN LY et al. 2013. Duration of rapamycin treatment has differential effects on metabolism in mice. Cell Metab, 17, 456–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERA JF, MELLINGHOFF IK, SHI Y, RETTIG MB, TRAN C, HSU JH, SAWYERS CL & LICHTENSTEIN AK 2004. AKT activity determines sensitivity to mammalian target of rapamycin (mTOR) inhibitors by regulating cyclin D1 and c-myc expression. J Biol Chem, 279, 2737–46. [DOI] [PubMed] [Google Scholar]
- GESING A, BARTKE A, WANG F, KARBOWNIK-LEWINSKA M & MASTERNAK MM 2011a. Key regulators of mitochondrial biogenesis are increased in kidneys of growth hormone receptor knockout (GHRKO) mice. Cell Biochem Funct, 29, 459–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GESING A, MASTERNAK MM, WANG F, JOSEPH AM, LEEUWENBURGH C, WESTBROOK R, LEWINSKI A, KARBOWNIK-LEWINSKA M & BARTKE A 2011b. Expression of key regulators of mitochondrial biogenesis in growth hormone receptor knockout (GHRKO) mice is enhanced but is not further improved by other potential life-extending interventions. J Gerontol A Biol Sci Med Sci, 66, 1062–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREER C, LEE M, WESTERHOF M, MILHOLLAND B, SPOKONY R, VIJG J & SECOMBE J 2013. Myc-dependent genome instability and lifespan in Drosophila. PLoS One, 8, e74641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARRISON DE, STRONG R, SHARP ZD, NELSON JF, ASTLE CM, FLURKEY K, NADON NL, WILKINSON JE, FRENKEL K, CARTER CS et al. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature, 460, 392–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOFMANN JW, ZHAO X, DE CECCO M, PETERSON AL, PAGLIAROLI L, MANIVANNAN J, HUBBARD GB, IKENO Y, ZHANG Y, FENG B, et al. 2015. Reduced expression of MYC increases longevity and enhances healthspan. Cell, 160, 477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHNSON SC, RABINOVITCH PS & KAEBERLEIN M 2013. mTOR is a key modulator of ageing and age-related disease. Nature, 493, 338–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JUNNILA RK, LIST EO, BERRYMAN DE, MURREY JW & KOPCHICK JJ 2013. The GH/IGF-1 axis in ageing and longevity. Nat Rev Endocrinol, 9, 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LACERDA R, MENEZES J & ROMAO L 2017. More than just scanning: the importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol Life Sci, 74, 1659–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDER B, GROZHIK AV, OLARERIN-GEORGE AO, MEYDAN C, MASON CE & JAFFREY SR 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods, 12, 767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAZZOLETTI M, BORTOLIN F, BRUNELLI L, PASTORELLI R, DI GIANDOMENICO S, ERBA E, UBEZIO P & BROGGINI M 2011. Combination of PI3K/mTOR inhibitors: antitumor activity and molecular correlates. Cancer Res, 71, 4573–84. [DOI] [PubMed] [Google Scholar]
- MCCORMICK MA, TSAI SY & KENNEDY BK 2011. TOR and ageing: a complex pathway for a complex process. Philos Trans R Soc Lond B Biol Sci, 366, 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEIRA LB, CALVO JA, SHAH D, KLAPACZ J, MOROSKI-ERKUL CA, BRONSON RT & SAMSON LD 2014. Repair of endogenous DNA base lesions modulate lifespan in mice. DNA Repair (Amst), 21, 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MEYER KD & JAFFREY SR 2014. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol, 15, 313–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER JE & REESE JC 2012. Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol, 47, 315–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLER RA 2009. Cell stress and aging: new emphasis on multiplex resistance mechanisms. J Gerontol A Biol Sci Med Sci, 64, 179–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURAKAMI S, SALMON A & MILLER RA 2003. Multiplex stress resistance in cells from long-lived dwarf mice. FASEB J, 17, 1565–6. [DOI] [PubMed] [Google Scholar]
- MURPHY LJ, BELL GI & FRIESEN HG 1987. Growth hormone stimulates sequential induction of c-myc and insulin-like growth factor I expression in vivo. Endocrinology, 120, 1806–12. [DOI] [PubMed] [Google Scholar]
- PAGE MM, SALMON AB, LEISER SF, ROBB EL, BROWN MF, MILLER RA & STUART JA 2009. Mechanisms of stress resistance in Snell dwarf mouse fibroblasts: enhanced antioxidant and DNA base excision repair capacity, but no differences in mitochondrial metabolism. Free Radic Biol Med, 46, 1109–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAPADOPOULOS E, JENNI S, KABHA E, TAKROURI KJ, YI T, SALVI N, LUNA RE, GAVATHIOTIS E, MAHALINGAM P, ARTHANARI H, et al. 2014. Structure of the eukaryotic translation initiation factor eIF4E in complex with 4EGI-1 reveals an allosteric mechanism for dissociating eIF4G. Proc Natl Acad Sci U S A, 111, E3187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PICCA A & LEZZA AM 2015. Regulation of mitochondrial biogenesis through TFAM-mitochondrial DNA interactions: Useful insights from aging and calorie restriction studies. Mitochondrion, 25, 67–75. [DOI] [PubMed] [Google Scholar]
- PICKERING AM, LEHR M & MILLER RA 2015. Lifespan of mice and primates correlates with immunoproteasome expression. J Clin Invest, 125, 2059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SALMON AB, MURAKAMI S, BARTKE A, KOPCHICK J, YASUMURA K & MILLER RA 2005. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab, 289, E23–9. [DOI] [PubMed] [Google Scholar]
- SALVI N, PAPADOPOULOS E, BLACKLEDGE M & WAGNER G 2016. The Role of Dynamics and Allostery in the Inhibition of the eIF4E/eIF4G Translation Initiation Factor Complex. Angew Chem Int Ed Engl, 55, 7176–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAXTON RA & SABATINI DM 2017. mTOR Signaling in Growth, Metabolism, and Disease. Cell, 168, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHIAFFINO S & MAMMUCARI C 2011. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet Muscle, 1, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHMIDT EV 2004. The role of c-myc in regulation of translation initiation. Oncogene, 23, 3217–21. [DOI] [PubMed] [Google Scholar]
- SHARP ZD & BARTKE A 2005. Evidence for down-regulation of phosphoinositide 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR)-dependent translation regulatory signaling pathways in Ames dwarf mice. J Gerontol A Biol Sci Med Sci, 60, 293–300. [DOI] [PubMed] [Google Scholar]
- SHI H, WANG X, LU Z, ZHAO BS, MA H, HSU PJ, LIU C & HE C 2017. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res, 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STEFFEN KK & DILLIN A 2016. A Ribosomal Perspective on Proteostasis and Aging. Cell Metab, 23, 1004–12. [DOI] [PubMed] [Google Scholar]
- TELEMAN AA, CHEN YW & COHEN SM 2005. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev, 19, 1844–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI S, SITZMANN JM, DASTIDAR SG, RODRIGUEZ AA, VU SL, MCDONALD CE, ACADEMIA EC, O’LEARY MN, ASHE TD, LA SPADA, et al. 2015. Muscle-specific 4E-BP1 signaling activation improves metabolic parameters during aging and obesity. J Clin Invest, 125, 2952–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TSAI SY, RODRIGUEZ AA, DASTIDAR SG, DEL GRECO E, CARR KL, SITZMANN JM, ACADEMIA EC, VIRAY CM, MARTINEZ LL, KAPLOWITZ BS, et al. 2016. Increased 4E-BP1 Expression Protects against Diet-Induced Obesity and Insulin Resistance in Male Mice. Cell Rep, 16, 1903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG M & MILLER RA 2012a. Augmented autophagy pathways and MTOR modulation in fibroblasts from long-lived mutant mice. Autophagy, 8, 1273–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG M & MILLER RA 2012b. Fibroblasts from long-lived mutant mice exhibit increased autophagy and lower TOR activity after nutrient deprivation or oxidative stress. Aging Cell, 11, 668–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG X, ZHAO BS, ROUNDTREE IA, LU Z, HAN D, MA H, WENG X, CHEN K, SHI H & HE C 2015. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell, 161, 1388–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEI Y, ZHANG YJ, CAI Y & XU MH 2015. The role of mitochondria in mTOR-regulated longevity. Biol Rev Camb Philos Soc, 90, 167–81. [DOI] [PubMed] [Google Scholar]
- WESTBROOK R, BONKOWSKI MS, STRADER AD & BARTKE A 2009. Alterations in oxygen consumption, respiratory quotient, and heat production in long-lived GHRKO and Ames dwarf mice, and short-lived bGH transgenic mice. J Gerontol A Biol Sci Med Sci, 64, 443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU M, ZHANG C, LI XJ, LIU Q & WANGGOU S 2016. Anti-Cancer Effect of Cap-Translation Inhibitor 4EGI-1 in Human Glioma U87 Cells: Involvement of Mitochondrial Dysfunction and ER Stress. Cell Physiol Biochem, 40, 1013–1028. [DOI] [PubMed] [Google Scholar]
- YI T, KABHA E, PAPADOPOULOS E & WAGNER G 2014. 4EGI-1 targets breast cancer stem cells by selective inhibition of translation that persists in CSC maintenance, proliferation and metastasis. Oncotarget, 5, 6028–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU H & HUANG S 2010. The complexes of mammalian target of rapamycin. Current protein & peptide science, 11, 409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU J, WAN J, GAO X, ZHANG X, JAFFREY SR & QIAN SB 2015. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature, 526, 591–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZID BM, ROGERS AN, KATEWA SD, VARGAS MA, KOLIPINSKI MC, LU TA, BENZER S & KAPAHI P 2009. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell, 139, 149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.