Abstract
Objective
Ovarian cancer (OC) is a common female disease with a poor prognosis. But the possible mechanism of OC tumor progression remains an active area of research. This study is intended to explore the effect of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) on proliferation and apoptosis of OC and its mechanism.
Materials and methods
MALAT1 and miR-503-5p expressions in human OC cell lines and normal human ovarian epithelial (HOSE) cell line were measured using qRT-PCR. OC cell line SKOV3 is divided into 4 groups: pcDNA3.1 group, pcDNA3.1-MALAT1 group, si-NC group, and si-MALAT1 group. MTT assay and 5-ethynyl-2ʹ-deoxyuridine (EdU) assay were applied for the detection of cell proliferation. Relationship of MALAT1 with miR-503-5p was verified using luciferase assay and RNA pull-down. The luciferase activity in cells was normalized to RNA concentrations determined by Bradford assays.
Results
MALAT1 expression in OC cells was elevated compared with HOSE cells. MTT assay and EdU assay supported that si-MALAT1 could inhibit cell proliferation in OC cells. Treatment of si-MALAT1 results in increased cell apoptosis rate in both SKOV3 cells and OVCAR3 cells. The expression of lncRNA-MALAT1 was negatively associated with the expression of miR-503-5p in OC cells, while luciferase assay and RNA pull-down together supported the direct binding of MALAT1 with miR-503-5p. Knockdown of MALAT1 was able to inhibit the activation of JAK2/STAT3 signal pathway, and MALAT1 overexpression was accompanied by activation of these factors.
Conclusion
lncRNA-MALAT1 can negatively target miR-503-5p expression to further promote proliferation and depress apoptosis of OC cells through the JAK2-STAT3 pathway.
Keywords: lncRNA-MALAT1, miR-503-5p, ovarian cancer, cell proliferation, cell apoptosis, JAK2/STAT3
Introduction
Ovarian cancer (OC) is a common cancer in females with 22,280 newly diagnosed cases and a mortality of 15,500 in the United States in 2012.1,2 The overall survival for OC patients is unsatisfactory with a 5-year survival rate of approximately 30%, which can be partially attributed to the difficulties in early diagnosis due to lack of early symptoms and effective biomarkers.3,4 Therefore, unfortunately, majority of OC patients are diagnosed at an advanced stage when the tumor has spread to other parts of the body.5 Therefore, identification of potential biomarkers and understanding of detailed pathology of OC is urgently needed for therapeutic benefits.
LncRNAs are implicated in various cell contexts mainly responsible for regulation at posttranscriptional level, organization of protein complexes and cell–cell cross talk.6 Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a lncRNA identified to regulate cell migration and proliferation in many carcinomas, such as hepatocellular carcinoma, colon adenocarcinoma, and esophageal squamous cell carcinoma.7–10 Current evidence supported the cooperation between lncRNAs and miRNAs in regulating both transcriptional and posttranscriptional gene expressions in tumorigenesis.11 Although much remains to be learned regarding the biological relationship between miRNAs and lncRNAs in tumor progression, the dysregulation of miRNAs has evidently proved to be associated with tumor progression and cell development. A previous study supported that expression of miR-503 was increased in follicular developmental stage resulting in the downregulation of genes related to granulosa cell proliferation and luteinization.12 In addition, downregulation of miR-503 was reported to associate with cisplatin resistance in OC cells.13 Therefore, those results were suggestive of the involvement of miR-503 in OC progressions and development. Previous evidence reported that the reaction of lncRNAs with miRNAs can disturb the stability of miRNAs.14 However, there has been no evidence supporting the interplay between MALAT1 and miR-503 in OC.
Several signaling pathways were known to associate with cell proliferation and apoptosis, among which MAPK signaling pathway, PI3K-AKT signaling pathway, and JAK2/STATS signal pathway attracted the most attention.15–17 JAK2/STAT3 pathway was activated in many tumors, including OC.18,19 However, whether MALAT1 mediates miR-503-5p in OC cells through JAK2/STAT3 pathway remains to be determined. In this regard, this study is performed to clarify the potential cross talk between MALAT1 and miR-503-5p, and to verify the implication of JAK2/STAT3 signaling pathway in proliferation and apoptosis of OC cells as well.
Materials and methods
Cell lines and cell culture
Cells were cultured in RPMI-1640 medium supplemented with 10% (v/v) fetal calf serum (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified chamber with 5% CO2 for 24 h. OC cells (CaOV3, SKOV3, OVCAR3, and OV90) and human ovarian epithelial cell line (HOSE) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA).
Cell transfection
Cell transfection was performed before cultured cells were grouped into four groups: pcDNA3.1 (empty vector pcDNA3.1), pcDNA3.1-MALAT1 (to induce MALAT1 overexpression), si-MALAT1 (to induce MALAT1 knockdown), and si-NC (control for MALAT1 knockdown).
The following sequence of si-RNA oligonucleotides (si-MALAT1) was used to knockdown MALAT1 expression: 5ʹ-CACAGGGAAAGCGAGUGGUUGGUA-3ʹ. The sequence of the noncoding control siRNA (si-NC) was 5ʹ-UUCUCCGAACGUGUCACGU-3ʹ. si-NC and si-MALAT1 were synthesized by Shanghai Sangon Biotechnology Co., Ltd. (Shanghai, China). pcDNA3.1 and pcDNA3.1-MALAT1 were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China).Cell transfection was performed by introducing 100 nM of miR-503-5p mimic, 100 nM of miR-503-5p inhibitor, mimics negative control (NC), inhibitor negative control, si-MALAT1, si-NC, pcDNA3.1, or pcDNA3.1-MALAT1 into cells for incubation at 37°C in a humidified chamber with 5% CO2 for correspondently 24, 48, and 72 hrs. Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fischer Scientific, Inc.) was used according to the manufacturer’s protocol. After transfection for 48 hrs, RT-qPCR was performed to assess the transfection efficiency of MALAT1 knockdown and overexpression.
RT-qPCR
TRIzol reagent (Invitrogen; Thermo Fisher Scientific) was used to extract total RNA and RNeasy Maxi kit (Qiagen GmbH) was applied according to kit protocols. Then, cDNAs were synthesized according to protocols of Prime-Script RT-PCR kit (Takara Biotechnology Co., Ltd., Dalian, China). PCR was conducted on an ABI PRISM 7700 Sequence Detection System (Applied Biosystems; Thermo Fisher Scientific, Inc.) using a SYBR Premix EX Taq II PCR kit (Takara Biotechnology Co., Ltd.) with a miRNA-specific 5ʹ primer (has-miR-503-5P; CTG CAG AAC TCT TCC CGC TGC) and the mRO 3ʹ primer supplied with the kit for all miRNAs. The sequences for MALAT1 primers were as follows: forward, 5ʹ-AAAGCAAGGTCTCCCCACAA-3ʹ, and reverse, 5ʹ-GGTCTGTGCTAGATCAAAAGGCA-3ʹ. GAPDH was used as an internal control for mRNAs with primers as follows: forward, 5ʹ-GAAGGTGAAGGTCGGAGTC-3ʹ and reverse, 5ʹ-GAAGATGGTGATGGGATTTC-3ʹ. U6 was used as an internal control for microRNAs whose primers were designed and synthesized by Sangon Biotech (Shanghai, China). Analysis of gene expressions was measured using the ΔΔCq method.20
MTT assay
Cell proliferation was determined using MTT assay after cell transfection. All procedures were performed strictly according to the manufacturer’s instructions. OC cells were inoculated into a 96-well plate with 5000 cells in each well and maintained for 24 hrs before 20 µL of 5 mg/mL MTT (Sigma-Aldrich, St Louis, MO, USA) was added for incubation. About 4 hrs later, the supernatant was abandoned and 200 µL of dimethyl sulfoxide was added. Absorbance was measured at 490 nm using an enzyme immunoassay analyzer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
5-Ethynyl-2ʹ-deoxyuridine (EdU)
Cell proliferation of OVCAR3 and SKOV3 cells was also verified using EdU Apollo in-vitro flow cytometry kit (RiboBio, Guangzhou, China) based on the protocols indicated in the instructions. About 48 hrs after cell treatment, cells were incubated with EdU solution (50 µmol/L) for 2 hrs. Then, cells were washed in PBS and fixed by 4% paraformaldehyde. After that, cells were stained with Apollo and then Hoechst (San Francisco, CA) for 30 mins before cell nucleus was observed. Cell proliferation was observed and photographed on MetaXpress software (Molecular Devices, Sunnyvale, CA, USA) using a high-power microscope.
Flow cytometry
OVCAR3 and SKOV3 cells (5×106 per well) were cultured in a 6-well plate at 37°C before 100 nM si-MALAT1 or si-NC was transfected within 24 hrs using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Flow cytometry was performed as previously described.21 At 48 hrs posttransfection, cells were digested with trypsin reagent (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany), followed by PBS wash and re-suspending in 100 μL of 1× binding buffer (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, 5 μL of propidium iodide and 5 μL of Annexin V-fluorescein isothiocyanate stain (Invitrogen; Thermo Fisher Scientific, Inc.) were added for staining. The apoptotic rate of cells was analyzed using flow cytometry (Beckman Coulter, Inc., Brea, CA, USA).
Luciferase assay
OVCAR3 cells were cotransfected with luciferase-reporter plasmids and miR-503-5p mimics or miR-503-5p inhibitor using Lipofectamine 2000 transfection reagent. Lysis buffer (25 mM of Tris phosphate, 1% Triton X-100, 1 mM of dithiothreitol, 2 mM of ethylenediaminetetraacetic acid, 10% glycerol, PH =7.8) was added at 48 hrs posttransfection. Then, cells were centrifuged at 14,000 rpm for 3 mins and then the supernatant was transferred to a 1.5-mL tube. Luciferase assays were performed using a luciferase assay kit (Promega). An enzyme-linked immunosorbent-assay plate reader (Bio-Rad Laboratories, Hercules, CA, USA) was applied to measure O-nitrophenol at a wavelength of 490 nm to evaluate β-galactosidase activity.
Pull-down assay
The biotinylated miR-503-5p probe, biotinylated lncRNA-MALAT1 probe, or scram probe was dissolved in 500 µL of washing/binding buffer (0.5 M NaCl, 20 mM Tris-HCL, PH 7.5, and 1 mM EDTA). The probes were incubated with streptavidin-coated magnetic beads (Sigma) at 25°C for 2 hrs to generate probe-coated magnetic beads. The pro-adipocyte lysates were incubated with probe-coated beads. After washing/binding with the wash buffer, the RNA complexes bound to the beads were eluted and extracted for Northern blot analysis or qPCR.
Northern blot analysis
The RNA complexes were collected and run on a 15% polyacrylamide-urea gel and then transferred to positively charged nylon membranes (Millipore) followed by cross-linking through UV irradiation. The membranes were subjected to hybridization with 100 pmol 3ʹ-digoxigenin (DIG)-labeled probes overnight at 43°C, and the detection was performed using a DIG luminescent detection kit (Sigma) according to the manufacturer’s instructions.
Western blot analysis
Protein was extracted from cell lysate based on the manufacturer’s instructions with the application of Radio Immunoprecipitation Assay Lysis Buffer (Beyotime, Shanghai, China). Western blot was performed strictly according to instructions described in a previous study.22 Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was applied to separate the protein samples before the protein was transferred to a polyvinylidene fluoride membrane. Then, 5% skim milk was added for incubation for 2 hrs at room temperature and then the blot was incubated in the specific primary antibodies for the factors and their phosphorylation forms in JAK2/STAT3 pathways phosphor-STAT3 (Tyr705; ab9145S, 1:1000; Cell Signaling Technology, Beverly, MA, USA), STAT3 (ab9139S, 1:1000; Cell Signaling Technology, Beverly, MA, USA), β-actin (ab4970S, 1:1000; Cell Signaling Technology, Beverly, MA, USA), phosphor-JAK2 (Tyr221; bs3206R, 1:1000; Bioss, Woburn, MA, USA), and JAK2 (bs-23003R, 1:1000; Bioss, Woburn, MA, USA), and apoptotic-related proteins p53 (ab2527S, 1:1000; Cell signaling technology), Bax (ab5023S, 1:1000; Cell signaling technology), Bcl-2 (ab15071S, 1:1000; Cell signaling technology), and Survivin (ab2803S, 1:1000; Cell signaling technology) at 4°C overnight. After that, the blot was washed in PBS before incubation with HRlabeledP- secondary antibodies (abcam) for 2 hrs at room temperature. ECL Plus Western Blot Substrate (Pierce, Carlsbad, CA) was added for color development and the grayscale of each band was calculated based on the grayscale of β-actin.
Statistical analysis
The data were exhibited in the form of mean ± standard error of three independent experiments. Data were processed based on SPSS 17.0 statistical software (SPSS, Chicago, IL, USA). Expressions of MALAT1 on different cell lines were compared using paired Student’s t-test. Differences among multiple groups were assessed by one-way ANOVA. P<0.05 was considered to have statistical significance.
Results
MALAT1 is increased in OC cells and si-RNA downregulates MALAT1 expression
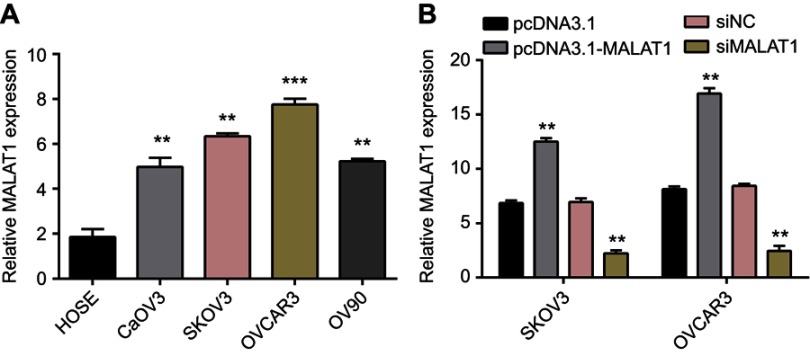
First of all, qRT-PCR was performed to determine MALAT1 expression in OC cell lines and HOSE cells. The expression of MALAT1 was increased in OC cell lines (CaOV3, SKOV3, OVCAR3, and OV90) in comparison to that in normal HOSE cells (all P<0.05; Figure 1A). Furthermore, pcDNA3.1a-MALAT1 was used to induce overexpression of MALAT1 and si-RNA was transfected to inhibit MALAT1 expression. Results suggested that pcDNA3.1a-MALAT1 treatment greatly promoted MALAT1 expression in SKOV3 and OVCAR3 cells, while MALAT1 expression was inhibited by si-MALAT1. Compared with pcDNA3.1a group, expressions of MALAT1 in pcDNA3.1a-MALAT1 group of SKOV3 and OVCAR3 cells were elevated by 1.67-fold and 1.79-fold, respectively (P<0.01, Figure 1B). The expressions of MALAT1 in si-MALAT1 were reduced by, respectively, 0.32-fold and 0.29-fold in SKOV3 and OVCAR3 cells, relative to si-NC samples (P<0.05; Figure 1C). The results showed that MALAT1 expression level is possibly related to OC cell viability.
Figure 1.
MALAT1 was upregulated in ovarian cancer cells relative to normal ovarian epithelial cells, and RNAi downregulated MALAT1 expression.
Notes: (A) The relative expression level of MALAT1 was detected by reverse transcription-quantitative polymerase chain reaction analysis in four ovarian cancer cell lines (CaOV3, SKOV3, OVCAR3, and OV90) and a normal ovarian epithelial cell line (HOSE). **P<0.01, ***P<0.001 vs HOSE. (B) The fold changes of MALAT1 expression in SKOV3 and OVSAR3 cells were analyzed 48 hrs after treating with pcDNA3.1, pcDNA3.1-MALAT1, si-MALAT1, or si-NC. Data are presented as the mean ± standard error of the mean. Experiments were performed in triplicate. **P<0.01 vs si-NC transfections.
Abbreviations: MALAT1, metastasis-associated lung adenocarcinoma transcript 1; si-MALAT1, MALAT1 small interfering RNA; si-NC, noncoding small interfering RNA.
MALAT1 promotes proliferation and inhibits apoptosis of OC cells
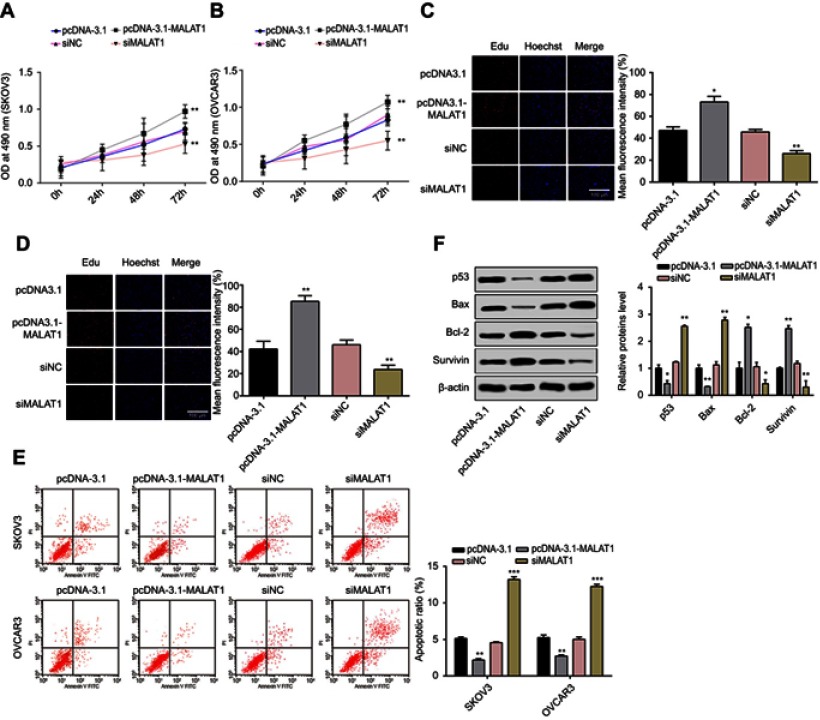
Whether MALAT1 can affect cell proliferation and cell apoptosis was assessed by MTT assay, Edu assay, and flow cytometry assay. MALAT1 overexpression was induced by pcDNA3.1-MALAT1 transfection and MALAT1 knockdown was induced by si-MALAT1 transfection. Overexpression of MALAT1 can increase cell proliferation at 24, 48, and 72 hrs after pcDNA3.1-MALAT1 transfection, and si-MALAT1 transfection can inhibit cell proliferation at 24, 48, and 72 hrs after transfection compared to si-NC transfected cells (all P<0.05, Figure 2A and B). EdU staining showed that the number of positive cells were substantially increased in SKOV3 and OVCAR3 cells after transfected with pcDNA3.1-MALAT, while transfection of si-MALAT1 could evidently suppress the number of positive cells (all P<0.05, Figure 2C and D). Flow cytometry assay found that the apoptotic rate of SKOV3 cells treated with pcDNA3.1-MALAT1 or pcDNA3.1 was 2.17% and 5.13%, respectively (P<0.01), while those in OVCAR3 cells, respectively, were 2.72% and 5.25% (P<0.01; Figure 2E). The effect of si-MALAT1 on SKOV3 and OVCAR3 cell apoptosis showed that the apoptotic rates of SKOV3 cells and OVCAR3 cells treated with si-MALAT1 were significantly higher than those treated with si-NC (all P<0.01; si-MALAT1 vs si-NC in SKOV3: 13.21% vs 4.56%; si-MALAT1 vs si-NC in OVCAR3: 12.26% vs 5.02%) (Figure 2E). Further, the effect of MALAT1 overexpression or knockdown on apoptotic-related proteins was evaluated. The differential p53, Bax, Bcl-2, and Survivin protein levels upon MALAT1 knockdown or overexpression were examined using Western blot assays, suggesting that the proapoptotic proteins p53 and Bax levels were upregulated and the antiapoptotic proteins Bcl-2 and Survivin levels were downregulated in si-MALAT1 group, compared to si-NC group. Meanwhile, the expression patterns of pcDNA3.1-MALAT1 treatment group were reversed (Figure 2F).
Figure 2.
MALAT1 promoted cell proliferation and inhibited cell apoptosis in SKOV3 and OVCAR3 cells.
Notes: The cell growths of SKOV3 (A) and OVCAR3 (B) cells were monitored using MTT assays in response to si-MALAT1 or pcDNA3.1-MALAT1 induced MALAT1 knockdown or overexpression, respectively. Effects of MALAT1 knockdown or overexpression on cell proliferations of SKOV3 (C) and OVCAR3 (D) were further determined by Edu staining (200×). (E) Apoptosis of SKOV3 and OVCAR3 cells transfected with si-MALAT1, si-NC, pcDNA3.1-MALAT1, or pcDNA3.1 was analyzed by flow cytometry. The percentage displayed on the histograms indicates apoptotic rate. Comparison of apoptotic rates of cells transfected with si-MALAT1, si-NC, pcDNA3.1-MALAT1, or pcDNA3.1. (F) Proteins were detected by Western blot. β-Actin was used as internal control to ensure equal loadings. Graphs are represented as the mean density of p53, Bax, Bcl-2, and Survivin bands normalized against the mean density of β-actin band from three independent experiments (presented as relative density of individual protein). Data are presented as the mean ± standard error of the mean (n=3). *P<0.05, **P<0.01, ***P<0.001 vs si-NC, or pcDNA3.1.
Abbreviations: MALAT1, metastasis-associated lung adenocarcinoma transcript 1; si-MALAT1, MALAT1 small interfering RNA; si-NC, noncoding small interfering RNA.
miR-503-5p directly binds MALAT1 and serves as a negative regulator
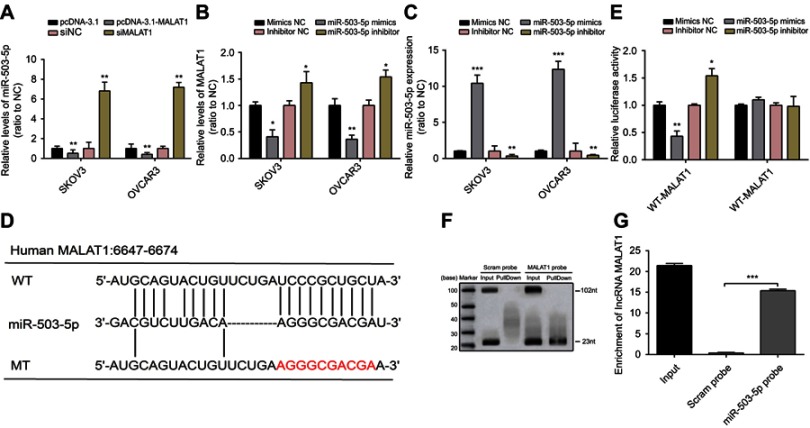
To investigate the mechanism by which MALAT1 regulates OC cell proliferation and apoptosis, a lot of studies have reported the correlation between lncRNAs and microRNAs.23,24 In our study, miR-503-5p expression was upregulated after MALAT1 was suppressed by si-MALAT1, while down-regulated when SKOV3 and OVCAR3 cells were transfected with pcDNA3.1-MALAT1 plasmids (Figure 3A). Then, mimics NC/miR-503-5p mimics or inhibitor NC/miR-503-5p inhibitor was transfected into SKOV3 and OVCAR3 cells to achieve miR-503-5p inhibition or overexpression, and RT-PCR was used to verify the efficiency of transfection (Figure 3B). MALAT1 expression was downregulated by miR-503-5p overexpression, whereas upregulated by miR-503-5p inhibition (Figure 3C). Noncoding RNAs have been shown to target mRNAs via direct or indirect RNA–RNA interaction, and so we suspected that there may be a direct interaction between MALAT1 and miR-503-5p. To clarify the interaction between miR-503-5p and MALAT1, at first, starBase and RegRNA 2.0 analyses were used in our study to predict the interaction between MALAT1 and miR-503-5p. To clarify the relationship between miR-503-5p and MALAT1, a wild-type MALAT1 luciferase reporter vector (named WT-MALAT1) and a mutant-type MALAT1 luciferase reporter vector (named MT-MALAT1) containing a 10-bp mutation in the predicted miR-503-5p binding site was constructed (Figure 3D). After that, we co-transfected indicated vector with miR-503-5p mimics or miR-503-5p inhibitor in OVCAR3 cells, respectively. After co-transfection, the luciferase activity was examined using Dual Luciferase assays. A decreased reporter activity was shown in WT-MALAT1 and miR-503-5p mimics co-transfection, and increased reporter activity was exhibited when WT-MALAT1 and miR-503-5p inhibitors were transfected together into OVCAR3 cells (Figure 3E). However, after the mutation in the predicted binding site of miR-503-5p, the changes of the luciferase activity were totally abolished (Figure 3E). The binding of lncRNA-MALAT1 and miR-503-5p was further verified by RNA pull-down assay. The results showed that substantial miR-503-5p can be pulled down by MALAT1 probe (Figure 3F). MALAT1 enrichment by miR-503-5p probe was also found by Northern blot (Figure 3G). Taken together, miR-503-5p can directly bind with MALAT1.
Figure 3.
MALAT1 directly targets miR-503-5p.
Notes: (A) The expressions of miR-503-5p in SKOV3 and OVCAR3 cells were determined in response to overexpression or knockdown of MALAT1 using reverse transcription-quantitative polymerase chain reaction analysis. (B) MALAT1 expression in response to miR-503-5p overexpression or miR-503-5p inhibition was determined by using real-time PCR. (C) Mimics NC/miR-503-5p mimics or inhibitor NC/miR-503-5p inhibitor was transfected into SKOV3 and OVCAR3 cells to achieve miR-503-5p overexpression or inhibition, verified by using real-time PCR. (D) A WT-MALAT1 luciferase reporter vector (WT-MALAT1) and a MT-MALAT1 luciferase reporter vector (MT-MALAT1) with mutations on miR-503-5p binding sites of the MALAT1 were constructed. (E) The WT-MALAT1/MT-MALAT1 vectors and miR-503-5p NC/miR-503-5p mimics/miR-503-5p inhibitor were co-transfected into SKOV3 and OVCAR3 cells. The luciferase activity of the MALAT1 luciferase reporter vector was determined. (F) Verification of the binding of MALAT1 and miR-503-5p determined by the pull-down assay using MALAT1 probe. (G) Verification of the binding of MALAT1 and miR-503-5p determined by the pull-down assay using miR-503-5p probe. Scram probe was used as negative control. Data are presented as the mean ± standard error of the mean (n=3). *P<0.05, **P<0.01, ***P<0.001 vs NC or blank control.
Abbreviations: MALAT1, metastasis-associated lung adenocarcinoma transcript 1; si-MALAT1, MALAT1 small interfering RNA; si-NC, noncoding small interfering RNA; NB, Northern blot.
MALAT1 regulates OC cell proliferation and apoptosis through interaction with miRNA-503-5p
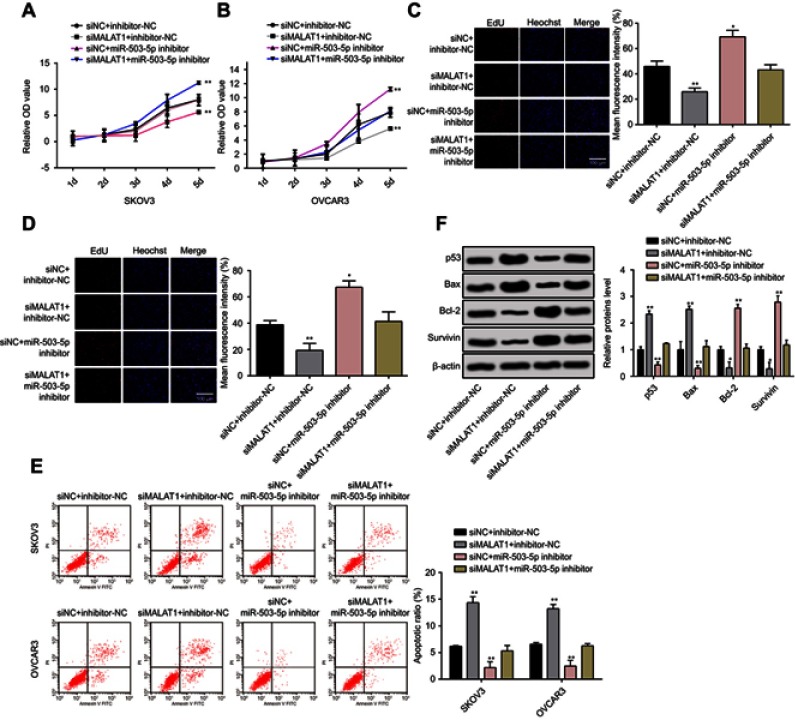
Cell proliferation and apoptosis were monitored in response to co-effect of MALAT1 and miR-503-5p. Cell proliferation by MTT assay showed that proliferation rate was inhibited after MALAT1 was knockdown and this inhibition was reversed by transfection of miR-503-5p inhibitor (Figure 4A and B). EdU staining on SKOV3 and OVCAR3 cells demonstrated that cells transfection with si-MALAT1 had significantly deceased positive cells. Transfection of miR-503-5p inhibitor could increase the number of positive cells and even partially reverse the inhibition of si-MALAT1 on positive cells. Moreover, flow cytometry assay demonstrated that apoptotic rate was increased in SKOV3 and OVCAR3 cells after transfection of MALAT1 knockdown, but transfection of miR-503-5p inhibition can suppress the cell apoptotic rate in tumor cells (Figure 4E). Furthermore, the co-effect of MALAT1 and miR-503-5p on apoptotic-related proteins was evaluated. The differential p53, Bax, Bcl-2, and Survivin protein levels were examined using Western blot assays. Co-transfected si-MALAT1 and inhibitor NC markedly increased the levels of p53 and Bax proteins, and on the contrary, the levels of Bcl-2 and Survivin proteins were significantly decreased. Expressions of proapoptotic proteins (p53 and Bax) were elevated after cells were transfected with MALAT1 knockdown plasmid, while cells transfected with miR-503-5p inhibition had decreased apoptotic rate. miR-503-5p inhibition can partially reverse the inhibitory effect of si-MALAT1 on expressions of anti-apoptotic proteins (Bcl-2 and Survivin) (Figure 4F). Taken together, MALAT1 regulates OC cell growth through interaction with miR-503-5p.
Figure 4.
MALAT1 regulated OC cell proliferation and apoptosis through interaction with miR-503-5p.
Notes: (A and B) The cell growth was monitored in response to co-processing MALAT1 knockdown and miR-503-5p inhibition using MTT assay in both SKVO3 and OVCAR3 cells. (C and D), Edu staining was applied to observe the co-effect of MALAT1 knockdown and miR-503-5p inhibition on cell proliferation on SKVO3 and OVCAR3 cells (200×). (E) Apoptosis of SKOV3 and OVCAR3 cells co-transfected si-MALAT1/si-NC and miR-503-5p inhibitor/inhibitor NC was analyzed by flow cytometry. The percentage displayed on the histograms indicates apoptotic rate. Comparison of the apoptotic rates of cells co-transfected with si-MALAT1/si-NC and miR-503-5p inhibitor/inhibitor NC. (F) Apoptotic or antiapoptotic proteins were detected by Western blot. β-Actin was used as internal control to ensure equal loadings. Graphs are represented as the mean density of p53, Bax, Bcl-2, and Survivin bands normalized against the mean density of β-actin band from three independent experiments (presented as relative density of individual protein). Data are presented as the mean ± standard error of the mean (n=3). *P<0.05, **P<0.01 vs si-NC or blank control.
Abbreviations: MALAT1, metastasis-associated lung adenocarcinoma transcript 1; si-MALAT1, MALAT1 small interfering RNA; si-NC, noncoding small interfering RNA.
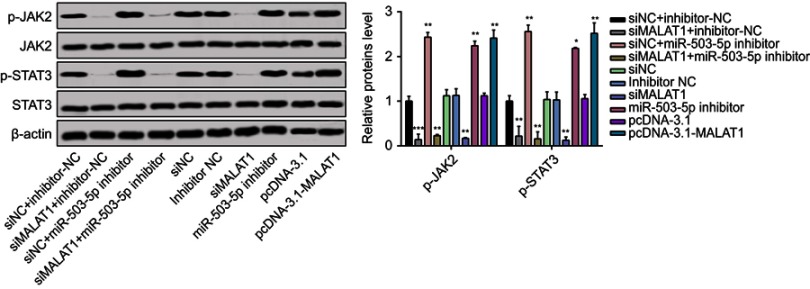
MALAT1 activates JAK2/STAT3 pathways
miRNAs were recently recognized to adjust JAK-STAT signaling in cancers.25 Phosphorylation of the JAK2/STAT3 pathway is linked to antiapoptosis.26 Western blot suggested that the expressions of JAK2 and STAT3 did not change in OVCAR3 cells after miR-503-5p inhibitor treatment or si-MALAT1 transfection (Figure 5). However, compared to control groups, p-JAK and p-STAT3 were both upregulated by miR-503-5p inhibitor and downregulated by si-MALAT1. miR-503-5p inhibitor could partially offset the inhibitory effect of si-MALAT1 on p-JAK and p-STAT3. On the basis of these results, MALAT1 mediates the activation of JAK2/STAT3 signal pathway through regulating miR-503-5p expression.
Figure 5.
MALAT1 activated JAK2/STAT3 pathway.
Notes: (A) Total cell lysates were collected and immunoblotted with the indicated antibodies. β-Actin was used as a loading control. Graphs are presented as the mean density of p-JAK2 and p-STAT3 normalized against the mean density of JAK2 or STAT3 from three independent experiments (presented as relative density of phosphoprotein vs total protein). Data are presented as the mean ± standard error of the mean (n=3). *P<0.05, **P<0.01, ***P<0.001 compared to untreated control cells at 0 min.
Abbreviations: MALAT1, metastasis-associated lung adenocarcinoma transcript 1; si-MALAT1, MALAT1 small interfering RNA; si-NC, noncoding small interfering RNA.
Discussion
The regulatory roles of lncRNAs in carcinogenesis were reported in many studies, while some studies implied some lncRNAs could be served as notable therapeutic targets in OC.27,28 The evidence showed MALAT1 is regarded as a new oncogene in OC by modulating cell proliferation, migration, and apoptosis.29 The effect of targeted sponge/decoy lncRNAs on miRNAs was recently discovered.23,30 Despite these results, the possible mechanisms of MALAT1 in OC are still fully unknown. On the basis of current research, we learned that MALATI through negatively targeting miR-503-5p in OC can promote cell proliferation and inhibit cell apoptosis.
Cells with MALAT1 overexpression together with si-MALAT1 transfection were used for cell proliferation and apoptosis assays. After MALAT1 was knockdown by transfected siRNA, cell proliferation rates of OC cells were suppressed while cell apoptosis rates were increased. In addition, overexpression of MALAT1 remarkably promoted the cell proliferation rate and decreased apoptosis compared to control group. Evidence reported that the interaction of lncRNA and miRNA was implicated in disease pathogenesis. MALAT1 was reported to promote the development of aggressive renal cell carcinoma by the chromatin methyltransferase enhancer of zeste homolog 2. Meanwhile, the interaction between MALAT1 and microRNA-205 leads to oncogenic activity.31 The research on tongue cancer showed that through direct targeting, MALAT1 can influence cancer growth by interacting with miR-124.32 This inspired us to assume that MALAT1 might interact with miRNAs to exert its functions in OC. In our study, miR-503-5p was predicted as a target gene of MALAT1 in OC. We observed that miR-503-5p was significantly decreased in OC cells and indicated its role as a tumor suppressive gene for OC. These results suggest that MALAT1 exerts its biologic functions via directly binding with miR-503-5p. We reported a novel mechanism for MALAT1 in OC, and our results revealed that MALAT1/miR-503-5p was involved in OC and could act as a therapeutic indicator.
It is reported that MALAT1 participates in cancer cell modulation through several possible approaches, including EMT regulation33,34 and cancer-related downstream pathways such as PI3K/AKT,35 miR-200s/ZEB2,36 ATM-CHK2,37 and EZH2.38 In this study, the activation of JAK2/STAT3 factors was inhibited once MALAT1 was knockdown, while overexpression of MALAT1 could induce the phosphorylation of JAK2/STAT3. The results demonstrated that MALAT1 can regulate the activation of JAK2/STAT3 pathway in OC. We hypothesized that the regulatory effect of MALAT1 may be an important mechanism in cancer modulation. More and more evidence proved miRNAs was important in cancer pathogenesis, while it was clarified that miRNAs regulate the JAK/STAT3 signaling pathway in malignant tumors.39–41 For example, through suppression on IL-6-JAK-STAT3 signaling pathway, Let-7 was shown to inhibit inflammation-related transformation event.42 Moreover, miR-9 secreted from tumor cells can activate JAK-STAT3 signaling pathway by suppressing the expression of SOCS5, consequently leading to deterioration of tumor progression.43 In this study, we highlighted that MALAT1 could interact with miR-503-5p, resulting in the activation of JAK2/STAT3 signaling pathway. Although we clarified that JAK2/STAT3 signaling pathway plays a key role in OC cells, the detailed mechanism of MALAT1 and miR-503-5p to activate JAK2/STAT3 signaling pathway has not been completely understood. This may serve as one major limitation of our study. However, this shortcoming also provides a future direction for our further studies.
From our research, we found that as an oncogene, MALAT1 can regulate OC cell proliferation and apoptosis. Meanwhile, this study also offers experimental data on the original mechanism of MALAT1 via interacting with miR-503-5p, thereby inducing cell proliferation and inhibiting cell apoptosis in OC cells. In conclusion, MALAT1 can induce cell proliferation and impede cell apoptosis in OC cells by negatively regulating miR-503-5p and activating JAK2/STAT3 signaling pathway.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 2.Suh DH, Kim JW, Kim K, Kim HJ, Lee KH. Major clinical research advances in gynecologic cancer in 2012. J Gynecol Oncol. 2013;24:66–82. doi: 10.3802/jgo.2013.24.1.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jelovac D, Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. 2011;61:183–203. doi: 10.3322/caac.20113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shapira I, Oswald M, Lovecchio J, et al. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br J Cancer. 2014;110:976–983. doi: 10.1038/bjc.2013.795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010;177:1053–1064. doi: 10.2353/ajpath.2010.100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao X, Zhao R, Chen Q, et al. MALAT1 might be a predictive marker of poor prognosis in patients who underwent radical resection of middle thoracic esophageal squamous cell carcinoma. Cancer Biomark. 2015;15:717–723. doi: 10.3233/CBM-150513 [DOI] [PubMed] [Google Scholar]
- 8.Furi I, Kalmar A, Wichmann B, et al. Cell free DNA of tumor origin induces a ‘metastatic’ expression profile in HT-29 cancer cell line. PLoS One. 2015;10:e0131699. doi: 10.1371/journal.pone.0131699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846 [DOI] [PubMed] [Google Scholar]
- 10.Luo F, Sun B, Li H, et al. A MALAT1/HIF-2alpha feedback loop contributes to arsenite carcinogenesis. Oncotarget. 2016;7:5769–5787. doi: 10.18632/oncotarget.6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liz J, Esteller M. lncRNAs and microRNAs with a role in cancer development. Biochim Biophys Acta. 2016;1859:169–176. doi: 10.1016/j.bbagrm.2015.06.015 [DOI] [PubMed] [Google Scholar]
- 12.Lei L, Jin S, Gonzalez G, Behringer RR, Woodruff TK. The regulatory role of dicer in folliculogenesis in mice. Mol Cell Endocrinol. 2010;315:63–73. doi: 10.1016/j.mce.2009.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D, Lu P, Mi X, Miao J. Downregulation of miR-503 contributes to the development of drug resistance in ovarian cancer by targeting PI3K p85. Arch Gynecol Obstet. 2018;297:699–707. doi: 10.1007/s00404-018-4649-0 [DOI] [PubMed] [Google Scholar]
- 14.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35:600–604. doi: 10.3109/10799893.2015.1030412 [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Liang K, Hu Q, et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J Clin Invest. 2017;127:4498–4515. doi: 10.1172/JCI91553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu YR, Qi HJ, Deng DF, Luo YY, Yang SL. MicroRNA-21 promotes cell proliferation, migration, and resistance to apoptosis through PTEN/PI3K/AKT signaling pathway in esophageal cancer. Tumour Biol. 2016;37:12061–12070. doi: 10.1007/s13277-016-5074-2 [DOI] [PubMed] [Google Scholar]
- 18.Abubaker K, Luwor RB, Zhu H, et al. Inhibition of the JAK2/STAT3 pathway in ovarian cancer results in the loss of cancer stem cell-like characteristics and a reduced tumor burden. BMC Cancer. 2014;14:317. doi: 10.1186/1471-2407-14-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosen DG, Mercado-Uribe I, Yang G, et al. The role of constitutively active signal transducer and activator of transcription 3 in ovarian tumorigenesis and prognosis. Cancer. 2006;107:2730–2740. doi: 10.1002/cncr.22293 [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Li Y, Su Z, et al. Identification of miR125a5p as a tumor suppressor of renal cell carcinoma, regulating cellular proliferation, migration and apoptosis. Mol Med Rep. 2015;11:1278–1283. doi: 10.3892/mmr.2014.2848 [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Wang K, Jiang YZ, Chang XW, Dai CF, Zheng J. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) inhibits human ovarian cancer cell proliferation. Cell Oncol (Dordr). 2014;37:429–437. doi: 10.1007/s13402-014-0206-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21 [DOI] [PubMed] [Google Scholar]
- 24.Shi D, Zhang Y, Lu R, Zhang Y. The long non-coding RNA MALAT1 interacted with miR-218 modulates choriocarcinoma growth by targeting Fbxw8. Biomed Pharmacother. 2018;97:543–550. doi: 10.1016/j.biopha.2017.10.083 [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- 26.Kim HC, Kim E, Bae JI, et al. Sevoflurane postconditioning reduces apoptosis by activating the JAK-STAT pathway after transient global cerebral ischemia in rats. J Neurosurg Anesthesiol. 2017;29:37–45. doi: 10.1097/ANA.0000000000000331 [DOI] [PubMed] [Google Scholar]
- 27.Qiu JJ, Lin YY, Ye LC, et al. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134:121–128. doi: 10.1016/j.ygyno.2014.03.556 [DOI] [PubMed] [Google Scholar]
- 28.Silva JM, Boczek NJ, Berres MW, Ma X, Smith DI. LSINCT5 is over expressed in breast and ovarian cancer and affects cellular proliferation. RNA Biol. 2011;8:496–505. doi: 10.4161/rna.8.3.14800 [DOI] [PubMed] [Google Scholar]
- 29.Wu L, Wang X, Guo Y. Long non-coding RNA MALAT1 is upregulated and involved in cell proliferation, migration and apoptosis in ovarian cancer. Exp Ther Med. 2017;13:3055–3060. doi: 10.3892/etm.2017.4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8:e53823. doi: 10.1371/journal.pone.0053823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75:1322–1331. doi: 10.1158/0008-5472.CAN-14-2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang TH, Liang LZ, Liu XL, et al. Long non-coding RNA MALAT1 interacts with miR-124 and modulates tongue cancer growth by targeting JAG1. Oncol Rep. 2017;37:2087–2094. doi: 10.3892/or.2017.5445 [DOI] [PubMed] [Google Scholar]
- 33.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0 [DOI] [PubMed] [Google Scholar]
- 34.Xu S, Sui S, Zhang J, et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int J Clin Exp Pathol. 2015;8:4881–4891. [PMC free article] [PubMed] [Google Scholar]
- 35.Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol. 2015;36:1477–1486. doi: 10.1007/s13277-014-2631-4 [DOI] [PubMed] [Google Scholar]
- 36.Xiao H, Tang K, Liu P, et al. LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget. 2015;6:38005–38015. doi: 10.18632/oncotarget.5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu L, Wu Y, Tan D, et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Ding L, Wang L, et al. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6:41045–41055. doi: 10.18632/oncotarget.5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du L, Subauste MC, DeSevo C, et al. miR-337-3p and its targets STAT3 and RAP1A modulate taxane sensitivity in non-small cell lung cancers. PLoS One. 2012;7:e39167. doi: 10.1371/journal.pone.0039167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo L, Chen C, Shi M, et al. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene. 2013;32:5272–5282. doi: 10.1038/onc.2012.573 [DOI] [PubMed] [Google Scholar]
- 41.Sugimura K, Miyata H, Tanaka K, et al. Let-7 expression is a significant determinant of response to chemotherapy through the regulation of IL-6/STAT3 pathway in esophageal squamous cell carcinoma. Clin Cancer Res. 2012;18:5144–5153. doi: 10.1158/1078-0432.CCR-12-0701 [DOI] [PubMed] [Google Scholar]
- 42.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuang G, Wu X, Jiang Z, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. Embo J. 2012;31:3513–3523. doi: 10.1038/emboj.2012.183 [DOI] [PMC free article] [PubMed] [Google Scholar]