Summary
Lung cancer is closely associated with chronic inflammation, but the causes of inflammation and the specific immune mediators have not been fully elucidated. The lung is a mucosal tissue colonized by a diverse bacterial community, and pulmonary infections commonly present in lung cancer patients are linked to clinical outcomes. Here we provide evidence that local microbiota provoke inflammation associated with lung adenocarcinoma by activating lung-resident γδ T cells. Germ-free or antibiotic-treated mice were significantly protected from lung cancer development induced by Kras mutation and p53 loss. Mechanistically, commensal bacteria stimulated Myd88-dependent IL-1β and IL-23 production from myeloid cells, inducing proliferation and activation of Vγ6+Vδ1+ γδ T cells that produced IL-17 and other effector molecules to promote inflammation and tumor cell proliferation. Our findings clearly link local microbiota-immune crosstalk to lung tumor development, and thereby define key cellular and molecular mediators that may serve as effective targets in lung cancer intervention.
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide, with lung adenocarcinoma (LUAD) being the most common form of the disease. Despite the recent development of targeted therapies for some genetic subtypes of human LUAD, overall survival rates for this cancer remain very low (Herbst et al., 2018). Although the genomic landscape of LUAD has been extensively characterized (Campbell et al., 2016; Cancer Genome Atlas Research, 2014), the tumor-cell extrinsic factors that control lung cancer development are less well understood. Importantly, as a mucosal tissue harboring the largest surface area in the body, the lung is exposed to a variety of air-borne microbes and environmental insults through inhalation. Therefore, the relationship between the host response to microbes and lung tumorigenesis is of particular interest.
A network of lung-resident immune cells maintain pulmonary tissue homeostasis in the steady state and provide immune surveillance in response to invading pathogens (Lloyd and Marsland, 2017). The development of lung cancer is closely associated with chronic inflammation characterized by infiltration of inflammatory cells and accumulation of pro-inflammatory factors including cytokines, chemokines and prostaglandins that stimulate cell proliferation, angiogenesis, tissue remodeling or metastasis (Palucka and Coussens, 2016). However, the underlying mechanisms responsible for the tumor-associated inflammation in lung cancer have not been clearly defined. In particular, it remains to be elucidated how the developing tumor lesions exploit the lung-resident immune system to create the tumor-promoting microenvironment, and how this process is influenced by non-host derived factors.
Mucosal surfaces exposed to the external environment are colonized by a vast number of microbes, collectively referred to as the commensal microbiota, and their primary habitat is the gastrointestinal tract. Interestingly, while the intestinal microbiota can promote local inflammation and development of gastrointestinal cancers (Elinav et al., 2013; Garrett, 2015), it can exert an opposing effect on transplanted tumors at distal sites by priming the host immune system and enhancing the systemic anti-tumor immune response (Dzutsev et al., 2017). Recent data have shown that commensal microbiota can affect the outcomes of immunotherapeutic interventions in human cancers (Gopalakrishnan et al., 2018; Routy et al., 2018). Taken together, the microbiota may function to orchestrate the balance between tumor-promoting inflammation and anti-tumor immunity, depending on the specific context of the tumor microenvironment. Despite accumulating evidence linking microbiota to cancer, the role of distal (gut) or local (lung) microbiota in lung cancer has not been elucidated.
Healthy lungs were traditionally believed to be sterile due to the inability to culture bacteria from lower airway samples using routine microbiological approaches. However, recent culture-independent sequencing studies have identified the presence of microbial communities containing a complex diversity of bacteria in the lower respiratory tract (Dickson et al., 2016). Changes in this local microbial community have been associated with the exacerbation of several pulmonary disorders, including chronic obstructive pulmonary disease, asthma and cystic fibrosis (Dickson et al., 2016; Marsland and Gollwitzer, 2014). Multiple lines of clinical evidence have linked lung microbiota to cancer. The course of lung cancer is frequently complicated by pulmonary infections (Akinosoglou et al., 2013), and post-obstructive pneumonia can negatively affect the efficacy of lung cancer therapy and overall survival of cancer patients (Qiao et al., 2015). However, these associations are not understood mechanistically.
In the current study, we investigated the host-microbiota interaction in lung cancer development using a genetically-engineered mouse (GEM) model driven by an activating point mutation of Kras and loss of p53, which are frequently associated with human lung adenocarcinoma. This autochthonous model of LUAD not only faithfully mimics the genetic and histopathological features of the human disease but also enables us to study the complex crosstalk between the commensal microbiota and host immune system in the physiological context of developing lung cancer. Using this model, we found that local microbiota associated with tumor growth could promote inflammation and cancer progression via lung-resident γδ T cells. Depleting microbiota or inhibiting γδ T cells or their downstream effector molecules all effectively suppressed lung cancer development. These findings provide conceptually novel insights regarding the pathogenesis of lung cancer by revealing the role of commensal microbiota in shaping the tumor-associated immune response, and shed light on cellular and molecular targets for therapeutic intervention in lung cancer.
Results
Commensal microbiota promote tumor growth in an autochthonous GEM model of lung adenocarcinoma
We have previously established a GEM model of human LUAD using conditional alleles of KrasLSL-G12D; p53flox/flox in mice (KP mice) (Jackson et al., 2001). In this model, intratracheal infection with adenoviral vectors expressing Cre recombinase under the control of the Sftpc promoter activates oncogenic KrasG12D and deletes the tumor suppressor p53 in lung epithelial cells to induce lung adenoma and adenocarcinoma (DuPage et al., 2009; Sutherland et al., 2014). To evaluate the functional importance of commensal bacteria in the initiation and progression of tumors in this model, we first compared tumor development between GF KP mice and their counterparts maintained under specific-pathogen-free (SPF) conditions (Figure 1A).
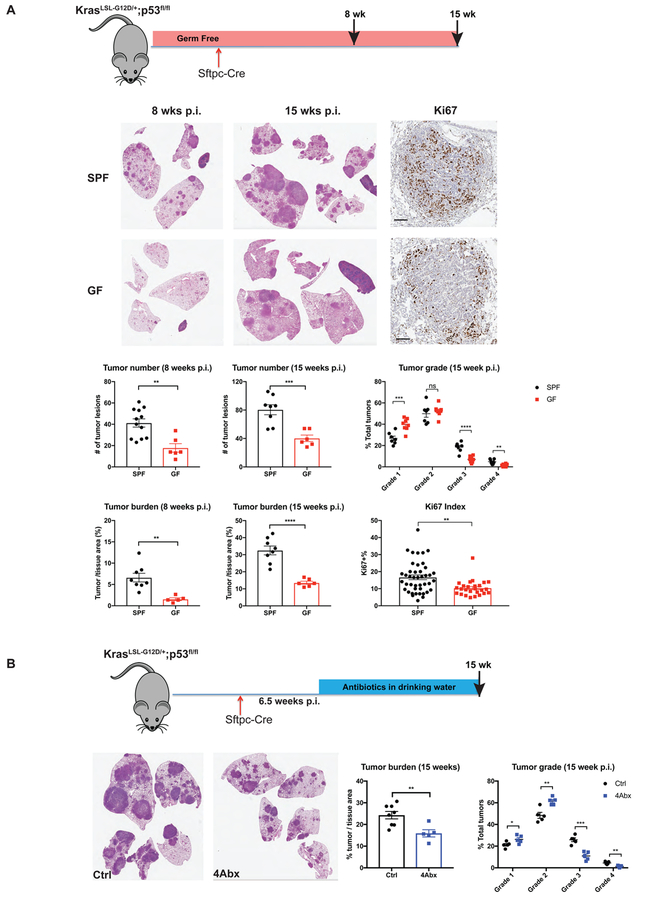
Figure 1. Commensal microbiota promote development of lung adenocarcinoma.
(A) KrasLSL-G12D; p53fl/fl mice in germ-free (GF) and specific-pathogen-free (SPF) conditions were infected with adenovirus expressing Cre under the control of the alveolar type II cell specific promoter SPC (Sftpc-Cre). Eight or fifteen weeks post infection (p.i.), lung tissues were collected for histopathological analysis to determine the tumor number, tumor burden and tumor grade. Immunohistochemistry staining of Ki67 was performed to examine tumor cell proliferation. Representative H&E pictures and corresponding quantification are shown. For Ki67 index, ~50 tumors from 4–6 mice/group were analyzed. Scale bar = 100 μm.
(B) SPF KP mice were treated with combined antibiotics (4Abx) in drinking water starting at 6.5 weeks post tumor initiation. Tumor burden and grade were analyzed at 15 weeks p.i..
Results are expressed as the mean ± SEM. n.s.= not significant, ** p<0.01, *** p<0.001, **** p<0.0001 by Student’s t test. For each experiment, n= 5–12 mice/group; data represent 3 independent experiments. See also Figure S1.
As shown in Figure 1A, GF mice were significantly protected against lung cancer development induced by Kras mutation and p53 deletion. Compared with age-matched SPF controls, GF KP mice exhibited substantially delayed tumor growth, with diminished tumor burden and decreased tumor numbers at both 8 weeks and 15 weeks post tumor initiation, and lower percentages of high-grade lesions (Figure 1A). At the cellular level, GF mice displayed reduced tumor cell proliferation as demonstrated by immunohistochemistry (IHC) analysis of Ki67 staining (Figure 1A). To further confirm that the absence of microbiota in GF mice is responsible for their decreased tumor burden, we examined tumor development in GF KP mice exposed to the microbiome via cohousing with SPF mice (referred to as ex-GF mice). Ex-GF mice displayed significantly increased tumor burden compared to their counterparts maintained in the GF conditions (Figure S1A).
To determine the effects of microbiota on different stages of lung tumor progression, we treated SPF KP mice with an antibiotic cocktail (4Abx) of ampicillin, neomycin, metronidazole and vancomycin at different time points after tumor initiation (2 weeks post infection, Figure S1B; 6.5 weeks post infection, Figure 1B). We found that this treatment robustly suppressed tumor growth in both early and advanced stages, resulting in significant reduction of high-grade tumors (Figure 1B). Notably, antibiotic treatment did not affect the proliferation of tumor cells in vitro (Figure S1C), arguing against the possibility of direct cytotoxic effects of these antibiotics. Together with the findings in GF KP mice, these results demonstrate that the commensal bacteria play a profound role in promoting tumor development in the autochthonous GEM model of lung adenocarcinoma.
Lung tumor development is associated with altered local microbiota and increased pro-inflammatory cytokine expression
To examine the relationship between the local microbiota and lung tumor development, we performed 16S rDNA-based qPCR and sequencing to analyze the bacterial abundance and composition in bronchoalveolar lavage fluid (BALF). Our data showed that the lungs from tumor-bearing KP mice harbored unique commensal bacterial communities as compared to the gut (Figure S2A). The most common bacterial genera present in lung microbiome included Staphylococcus (~15%), Streptococcus (~15%), and Lactobacillus (~15%), along with the family Pasteurellaceae (~10%) in healthy and tumor-bearing mice. These bacteria have also been reported in other mouse as well as human lung microbiome studies (Kostric et al., 2018; Yun et al., 2014). Compared to healthy (non-tumor bearing control) mice, the development of lung cancer was associated with a significant increase in the total bacterial burden as well as reduced bacterial diversity in the airway, as measured by the total amount of 16S DNA and the Shannon index of the bacterial community in the BALF (Figure 2A, B). Moreover, we identified several bacterial taxa, including Herbaspirillum and Sphingomonadaceae, that were significantly over-represented in tumor-bearing lungs, as well as a number of other taxa such as Aggregatibacter and Lactobacillus that are enriched in healthy lungs through LEfSe (linear discriminate analysis coupled with effect size measurements) (Figure S2B). Of particular importance, the correlation between the microbiome and tumor development was local; in mice treated with antibiotics that partially eliminated the commensal bacteria, the local bacterial burden in the lung, but not the fecal bacterial load, was significantly correlated with the tumor burden (Figure 2C). Moreover, treating mice with metronidazole alone effectively suppressed tumor growth in the lung without substantially reducing the overall bacterial abundance in the gut (Figure S2C). As an additional test of the importance of the local lung microbiota in tumor development, we isolated and cultured a number of bacterial species from late-stage lung tumors in SPF mice (see EXPERIMENTAL MODEL AND SUBJECT DETAILS). These bacteria were then intratracheally inoculated into to a separate cohort of KP mice shortly after tumor initiation. The introduction of this consortium of bacterial isolates dramatically accelerated disease development in these mice in comparison to untreated controls (Figure 2D). Furthermore, PathSeq analysis of human lung cancer datasets in The Cancer Genome Atlas (TCGA) identified a number of bacterial taxa that were significantly enriched or depleted in non-small cell lung cancer samples (including squamous cell carcinoma and adenocarcinoma) as compared to normal lungs. Consistent with our data from the mouse model, these results demonstrate the association between local dysbiosis and lung cancer (Figure S2D).
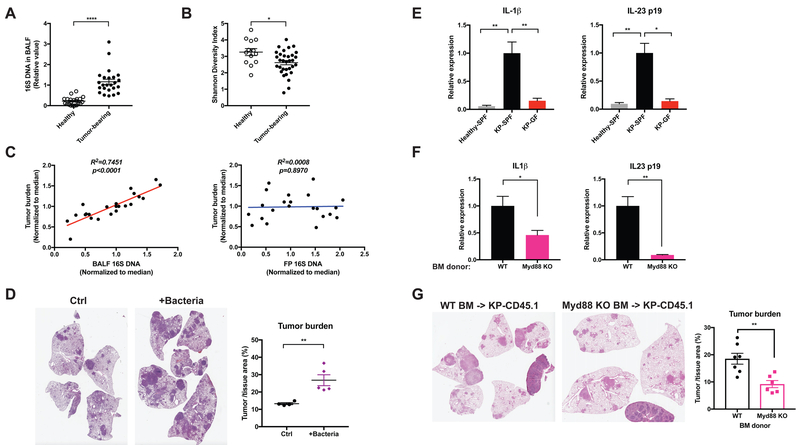
Figure 2. Lung tumor development is associated with altered local microbiota and increased pro-inflammatory cytokine expression.
(A, B) BALF samples were collected from tumor-bearing lungs from KP mice or healthy controls. Data were pooled from 5 independent cohorts.
(A) Total bacterial burden in BALF determined by 16S rDNA based qPCR analysis. Data were normalized to the corresponding median value of the tumor-bearing samples in each cohort.
(B) Shannon diversity index of the bacterial communities present in BALF.
(C) Linear regression curves illustrating the correlation between tumor burden and bacterial load in BALF or fecal pellet (FP) samples. Bacterial load in fecal pellets and BALF from untreated and antibiotic-treated SPF KP mice was measured by 16S rDNA-based qPCR; tumor burden was quantified at 15 weeks post tumor initiation. Data were normalized to the corresponding median value of each cohort, and the plots represent pooled data from 3 experimental cohorts.
(D) KP mice were infected with a lower dose of Sftpc-Cre adenovirus than the other experiments. 3.5 weeks post tumor initiation, they were left untreated as controls (Ctrl) or intratracheally inoculated with a consortium of bacteria that were isolated and cultured from late-stage lung tumors in a separate cohort of SPF mice (+Bacteria, see Experimental Model and Subject Details). Tumor burden was analyzed 8 weeks following inoculation; representative H&E pictures and quantifications are shown. n= 5 mice/group.
(E) RT-qPCR analysis of IL-1β and IL-23 p19 mRNA expression in the lung tissues from healthy SPF mice, tumor-bearing SPF mice and tumor-bearing GF mice.
(F) KP mice on the CD45.1 background were lethally irradiated and reconstituted with bone marrow from either wild-type (WT) or Myd88-deficient donors. Expression of IL-1β and IL-23 p19 mRNA were analyzed in FACS-purified alveolar macrophages and neutrophils respectively. Tumor burden was quantified 15 weeks post tumor initiation and representative H&E pictures are shown.
Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, **** p<0.0001 by Student’s t test. For each experiment, n= 6–7 mice/group; data represent 3 independent experiments. See also Figure S2.
Microbial products derived from intestinal microbiota are known to induce pro-inflammatory cytokines and mediate tumor-associated inflammation in colon cancer (Grivennikov et al., 2012; Kostic et al., 2013). Compared to healthy lungs, we noted a significant increase in IL-1β and IL-23 p19 gene expression associated with the increased local bacterial burden in the tumor-bearing lung tissues, and this increased cytokine expression was abrogated in GF mice (Figure 2E). To examine the upstream signaling pathway mediating this response, we reconstituted KP-CD45.1 mice with bone marrow derived from Myd88 knockout donors. Hematopoietic deficiency of Myd88, an essential adaptor molecule downstream of multiple toll-like receptors (TLRs), inhibited the production of IL-1β and IL-23 from alveolar macrophages and neutrophils (Figure 2F), and was associated with significantly reduced tumor growth (Figure 2G).
Microbiota drive the expansion of IL-17 producing γδ T cells associated with lung adenocarcinoma.
To further probe the cellular mechanism(s) of microbiota-mediated inflammation and tumor promotion, we analyzed the immune cells in tumor-bearing lungs from GF and SPF mice by flow cytometry. Our data showed a dramatic increase in γδ T cell abundance associated with lung adenocarcinoma in SPF mice (Figure 3A). While γδ T cells only accounted for a minor fraction (~3–5%) of total CD3+ lymphocytes in the circulation (Figure 3B) or peripheral lymphoid tissues (the spleen and the draining mediastinal lymph node, Figure S3A), they were highly enriched in the tumor-bearing lung tissue of SPF mice (approximately 20% of total CD3+ lymphocytes) (Figure 3B). Importantly, tumor-associated γδ T cell expansion was completely abrogated in the GF mice (Figure 3A, B), suggesting that the commensal microbiota are critical for driving this response. Consistent with the findings from the KP mouse model, we also observed significantly increased infiltration of γδ T cells in human LUAD samples as compared to normal lung tissues by IHC (Figure S3B).
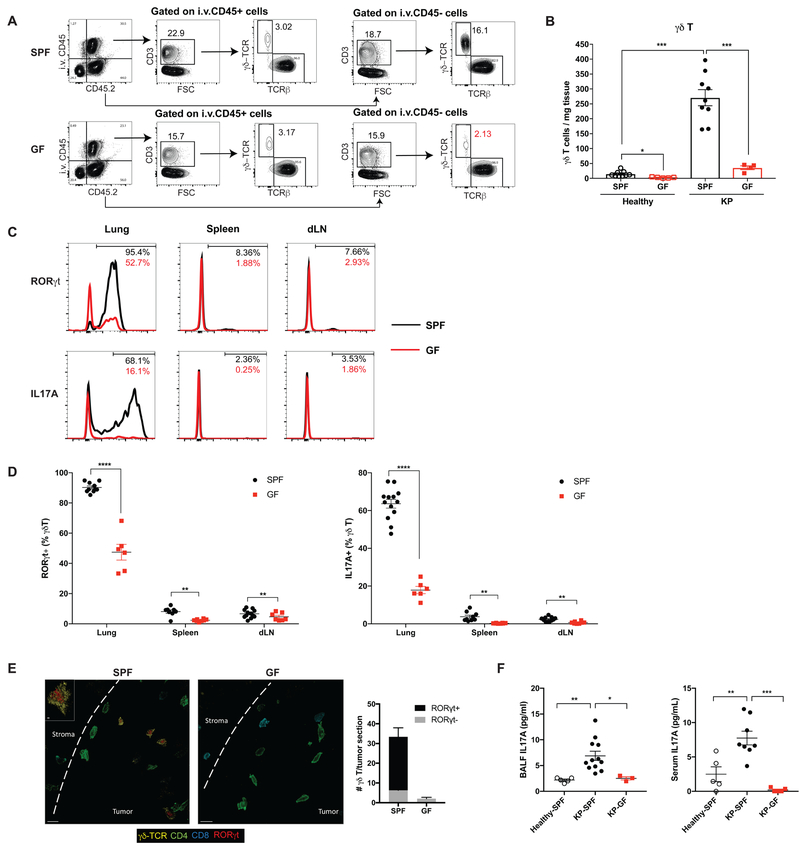
Figure 3. Microbiota are required for the expansion of γδ T17 cells associated with tumor development.
(A, B) Flow cytometry analysis of γδ T cells in the tumor-bearing lungs from SPF and GF mice. Samples were first gated on total live cells, then gated on the immune cell compartments in the circulation (positive for intravascular CD45 labeling, i.v.CD45+) and in the lung tissue (i.v. CD45−) separately with subsequent gating on CD3+ lymphocytes. The numbers of γδ T cells in the lung tissue (i.v. CD45−) are calculated (A) and representative plots are shown (B).
(C, D) RORγt and IL-17A expression in γδ T cells from the tumor-bearing lung, spleen and draining lymph node examined by flow cytometry and compared between SPF mice (black) and GF mice (red). Representative histograms are shown (C) and the frequencies of RORγt+ or IL-17A+ γδ T cells are quantified (D).
(E) Confocal immunofluorescent images of tumor-bearing lungs from SPF and GF mice with quantification. Yellow: γδ-TCR; Red: RORγt; Green: CD4; Aqua: CD8; dashed line: tumor border. Scale bar =10 μm. Inset: optical slice of γδ T cells with RORγt expression (z-depth different than larger image), scale bar = 2 μm.
(F) IL-17A levels in BALF and serum samples collected from healthy SPF mice, tumor-bearing SPF mice and tumor-bearing GF mice as measured by ELISA.
Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by Student’s t test. For each experiment, n= 6–12 mice/group; data represent at least 4 independent experiments. See also Figure S3.
Among the γδ T cells that accumulated in the tumor-bearing lungs from SPF mice, ~90% expressed the transcriptional factor RORγt and up to 75% produced the pro-inflammatory cytokine IL-17A (defining them as γδ T17 cells); by contrast, only around 5% of the total γδ T cells in the spleen or draining lymph node were IL-17 producers (Figure 3C, D). In addition to the reduction in γδ T cell abundance, GF mice also exhibited dramatically decreased RORγt expression and IL-17 production in γδ T cells (Figure 3C, D). These results imply that commensal microbiota not only impact the abundance of γδ T cells, but also play a critical role in determining their functionality. To examine the topographic distribution of these γδ T cells with respect to the developing tumors, we employed multi-parameter confocal immunofluorescence microscopy, which revealed infiltration of tumor lesions by γδ T17 cells in the SPF lungs that was absent in GF lungs (Figure 3E).
It is notable that γδ T cells are a major source of IL-17 in the tumor-bearing lungs from SPF mice in this KP model. They accounted for more than 60% of RORγt+ or IL-17+ lymphocytes whereas the other source of IL-17 was conventional CD4 T cells (i.e. Th17 cells) (Figure S3C). While no substantial difference in the Th17 compartment was observed between SPF and GF mice (Figure S3D), IL-17A protein levels in the BALF and serum were significantly diminished in GF mice (Figure 3F). Collectively, these data demonstrate that γδ T cells are the predominant IL-17 producing cells that accumulate in the lung in response to tumor development in a microbiota-dependent manner.
Commensal microbiota induce proliferation and activation of tissue-resident Vγ6+Vδ1+ T cells
One of the key features distinguishing γδ T cells from αβ T cells is their limited TCR diversity, with preferential usage of specific TCR Vγ chains depending on their effector function and tissue localization (Vantourout and Hayday, 2013). Vγ1+, Vγ4+ and Vγ6+ γδ T cells have all been reported to exist in the lung, and their relative abundance seems to be context dependent during different types of infection or inflammatory conditions (Papotto et al., 2017). The expanded γδ T17 population in KP lungs consisted primarily of Vγ6+Vδ1+ T cells (Figure 4A). These cells accounted for an average of 80% of total γδ T cells, and more than 90% of the γδ T17 population. By contrast, only a small fraction of γδ T17 cells were Vγ4+, whereas Vγ1+ γδ T cells made up fewer than 5% of total γδ T cells and most of them did not express RORγt (Figure 4A). Vγ6+ γδ T cells are long-lived, tissue-resident cells that colonize the lung during embryonic development and undergo self-renewal under homeostatic conditions (Haas et al., 2012). Consistent with this, the IL17-producing Vγ6+Vδ1+ T cells that expanded in tumor-bearing lungs were highly resistant to radiation and reconstitution. Twenty weeks after transplantation of lethally irradiated KP-CD45.1 mice with bone marrow from CD45.2 donors, γδ T cells were readily detected in tumor-bearing lungs. However, despite the robust reconstitution of Tbet+ γδ T cells or the Vγ4+ γδ T cell compartment by the donor bone marrow, RORγt+ γδ T cells were largely of the recipient origin in these chimeras and donor-derived Vγ6+Vδ1+ T cells were virtually absent (Figure S4A).
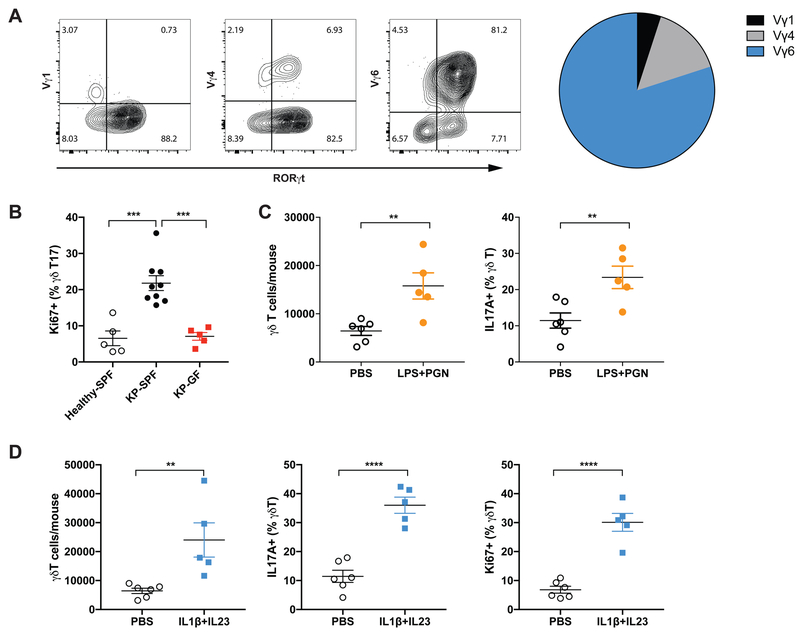
Figure 4. Commensal microbiota induce proliferation and activation of tissue-resident Vγ6+Vδ1+ T cells.
(A) γδ T cells in the tumor-bearing lungs were stained with monoclonal antibodies specific for Vγ1, Vγ4 and Vγ6/Vδ1 TCRs as well as RORγt. Representative FACS plots are shown and the relative percentage of these subsets in total γδ T cells is depicted in the pie chart. Data represent 20 mice.
(B) Proliferation of γδ T17 cells in the lungs from healthy SPF mice, tumor-bearing SPF mice and tumor-bearing GF mice was assessed by flow cytometric analysis of Ki67 expression.
(C) The number and IL-17 expression of γδ T cells in the lungs analyzed 36 hours post local delivery of LPS and PGN.
(D) Recombinant mouse IL-1β and IL-23 were administered to healthy mice through intratracheal instillation. The number of total γδT cells and the frequency IL-17+ or Ki67+ γδT cells were analyzed 36 hours post dosing.
(B–D) Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by Student’s t test. For each experiment, n= 5–9 mice/group. Data represent at least 2 independent experiments.
See also Figure S4.
The lung-resident origin of these γδ T cells strongly indicated that tumor-associated γδ T expansion was largely due to local proliferation rather than recruitment from the circulation. Indeed, we observed increased proliferation of γδ T cells, particularly those expressing IL-17, in tumor-bearing lungs compared to healthy lungs, which was largely abrogated in the GF mice (Figure 4B). By contrast, no apparent changes in the proliferation of RORγt- γδ T cells or Th17 cells were observed in GF mice (Figure S4B). Moreover, associated with the increased bacterial load in the airway, IL-17 production from γδ T cells in the tumor-bearing lungs was substantially enhanced as compared to that from cells in healthy lungs (Figure S4C). Reducing the commensal microbiota with the 4Abx cocktail dramatically decreased the abundance of γδ T17 cells in the tumor-bearing lung, resulting in lower IL-17A levels in the BALF or serum (Figure S4D). In the GF reconstitution experiment described above, we found that microbiome exposure robustly restored γδ T17 cell compartment in ex-GF mice (Figure S4E). Further supporting a model in which tumor-associated increase in the local bacterial burden induces proliferation and activation of tissue-resident γδ T17 cells, local administration of TLR ligands (such as lipopolysaccharide, LPS and peptidoglycan, PGN) triggered the expansion and IL-17 production of γδ T cells in the lung (Figure 4C). In addition, direct stimulation of the lung γδ T17 cells via intratracheal delivery of IL-1β and IL-23, the pro-inflammatory mediators produced by myeloid cells upon microbial exposure, potently induced their proliferation and activation (Figure 4D).
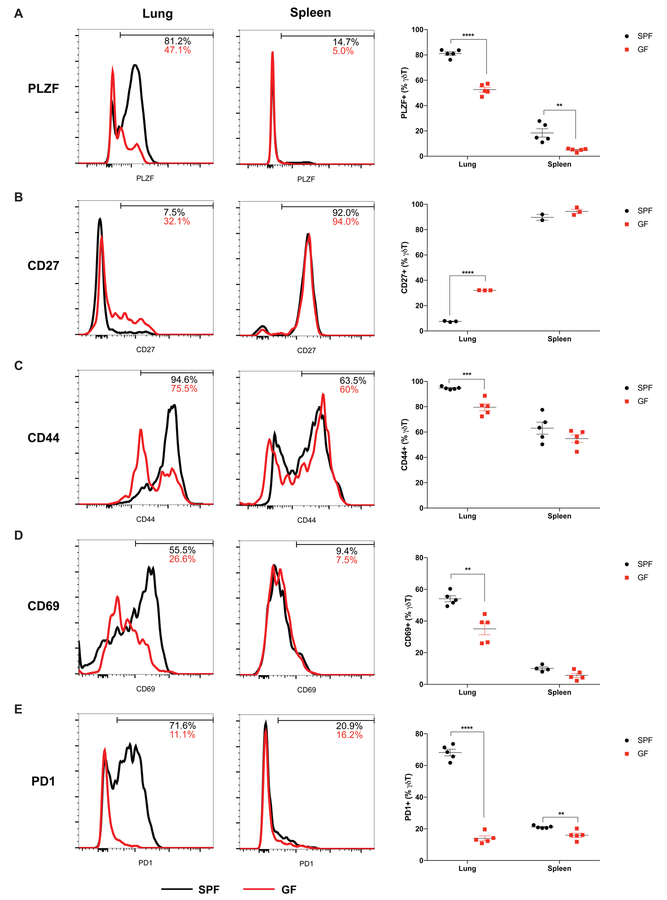
γδ T cells in the tumor-bearing lungs from SPF mice showed minimal capacity for IFNγ or TNFα production coupled with low expression of the transcription factor Tbet (Figure S5). In contrast to splenic γδ T cells, γδ T cells in the tumor-bearing lungs were largely positive for PLZF (promyelocytic leukemia zinc finger) (Figure 5A), which is a transcription factor essential for the development of tissue-resident IL17-producing Vγ6+ γδ T cells (Lu et al., 2015). In addition, they displayed low surface expression of CD27 (Figure 5B), which is critical for the differentiation of IFNγ-producing γδ T cells (Ribot et al., 2009). γδ T cells in the tumor-bearing SPF lungs exhibited elevated surface expression of activation/maturity markers including CD44, CD69 and PD1 compared to those in the spleen (Figure 5C, D, E). However, γδ T cells from the tumor-bearing lungs of the GF mice displayed a different phenotype: they had higher expression of IFNγ, Tbet and CD27, and significantly lower expression of PLZF as well as CD44, CD69 and PD1(Figure 5A–E; Figure S5). These results suggest commensal microbiota are critical for the differentiation and activation/maturation of tumor-associated γδ T cells in the lung.
Figure 5. Commensal microbiota are important for the differentiation and activation of tumor-associated γδ T cells in the lung.
The expression of PLZF (A), CD27 (B), CD44 (C), CD69 (D), PD1 (E) in γδ T cells in the tumor-bearing lungs and spleens were analyzed and compared between SPF and GF mice.
Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by Student’s t test. n= 2–5 mice/group; data represent 2–3 independent experiments. See also Figure S5.
Microbiota-induced γδ T cells promote neutrophil infiltration and tumor development.
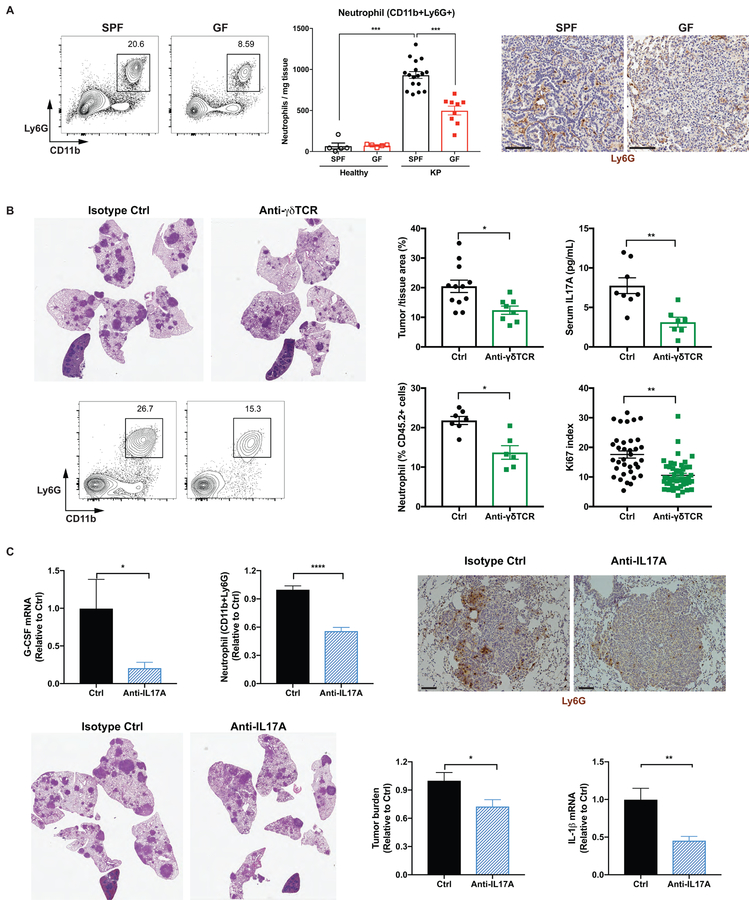
Accompanying the expansion of γδ T cells, neutrophil infiltration was also dramatically increased in tumor-bearing lungs compared to healthy lungs in SPF mice, and this response was suppressed in GF mice (Figure 6A). By comparison, we did not observe major changes in the abundance of other myeloid cell populations between SPF and GF mice (Figure S6A). While neutrophils are known to promote tumorigenesis and cancer metastasis in various tumor models including lung cancer (Coffelt et al., 2016), the upstream signal inducing the expansion of neutrophils in the KP tumors has not been clearly identified. To assess the functional importance of γδ T cells in mediating the microbiota-dependent neutrophil expansion and tumor promotion, we treated KP mice with monoclonal antibodies against γδ T cells (UC7–13D5 or GL3) 4 weeks after tumor initiation and evaluated their effects on tumor development. Both of these antibodies effectively inhibited γδ T cells in the lung (UC7–13D5: Figure S6B, C; GL3: Figure S6G, H), and significantly suppressed the progression of lung tumors (UC7–13D5: Figure 6B; GL3: Figure S6F). Mechanistically, inhibition of γδ T cells substantially reduced IL-17A levels, neutrophil infiltration and tumor cell proliferation (Figure 6B). On the other hand, no significant difference was observed in the anti-tumor activities of CD4 or CD8 T cell compartments between SPF and GF mice (Figure S6K), nor did γδ T cell inhibition affect the proliferation or type 1 effector cytokine production of CD4 or CD8 T cells (Figure S6D, E, I, J).
Figure 6. Microbiota-induced γδ T cells promote neutrophil infiltration and tumor development.
(A) The frequency, absolute number and tissue localization of neutrophils in the lungs from SPF and GF mice were examined by flow cytometry and IHC analysis. Scale bar = 100 μm.
(B) KP mice were treated with the monoclonal antibody UC7–13D5 to block γδ T cells starting at 4 weeks post tumor initiation. Tumor burden was quantified at 15 weeks p.i. and representative H&E sections are shown. Serum IL-17A levels was determined by ELISA, neutrophil abundance by flow cytometry, and tumor cell proliferation by Ki67 index (50–60 tumors from 5 mice/group),
(C) KP mice were treated with a monoclonal antibody to neutralize IL-17A starting at 4 weeks post tumor initiation. Tumor burden was quantified at 13–15 weeks p.i. and representative H&E pictures are shown; neutrophil infiltration was assessed by flow cytometry and IHC analysis (scale bar = 100 μm); G-CSF and IL-1β mRNA expression in the lung tissue were measured by RT-qPCR.
Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by Student’s t test. n= 6–12 mice/group.
See also Figure S6.
IL-17A has been implicated both in tumor-promoting inflammation and anti-tumor immunity, with contradictory roles being reported for IL-17A in different models of lung cancer (Chang et al., 2014; Cheng et al., 2014). Therefore, we used an IL-17A neutralizing antibody to evaluate the relevance of IL-17A in the context of the KP model. Our results showed that IL-17A blockade led to significantly decreased G-CSF expression and neutrophil infiltration in the lung, as well as a significant reduction in tumor growth and IL-1β expression (Figure 6C).
γδ T cells associated with lung tumors exhibit a distinct transcriptional profile
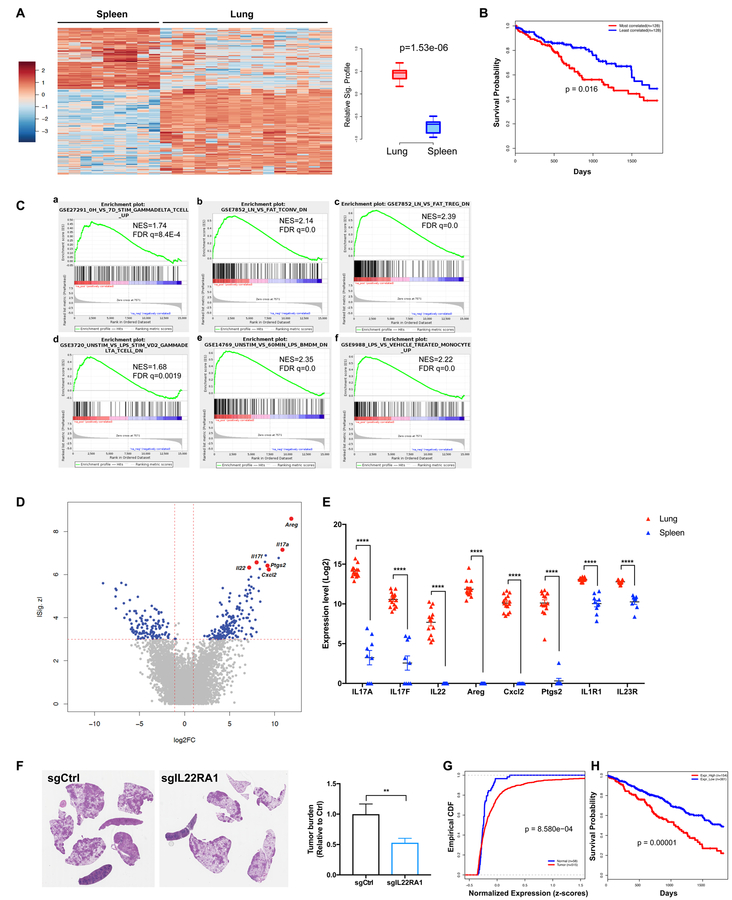
Beyond their production of IL-17A, microbiota-induced γδ T cells might promote lung tumor development through other effector mechanisms. To further understand the biology of tumor-associated γδ T cells in the lung, we characterized their transcriptional profiles in comparison to splenic γδ T cells using RNA-seq. Our data demonstrated that γδ T cells in the tumor-bearing KP lungs exhibited a distinct gene expression signature as compared to their splenic counterparts (Figure 7A). We further explored the prognostic value of this signature in a human lung adenocarcinoma patient cohort from TCGA. Using RNA-seq data from TCGA, individual patient samples were scored for their correlation with the KP-lung γδ T cell signature. High-scoring patients (top 25%, n=128), whose gene expression profiles most correlated with the KP-lung γδ T cell signature, exhibited significantly poorer survival compared to patients with lower scores (bottom 25%, n=128) (Figure 7B). Of note, this result is not merely a reflection of the extent of immune infiltration, as indicated by the lack of correlation between the γδ T cell signature score and ESTIMATE (Yoshihara et al., 2013) immune signature score for individual patients (See METHOD DETAILS, Figure S7A).
Figure 7. γδ T cells associated with lung tumors exhibit a distinct transcriptional profile.
(A–E) γδ T cells were FACS purified from tumor-bearing lungs or spleens of KP mice and their gene expression was analyzed by RNA-seq. A total of 9 spleen and 15 lung samples from three independent experiments were sequenced.
(A) Heatmap showing differential expression of driver genes of the KP-lung versus spleen γδ T signature (|Z| > 3, fold change > 2). Boxplot illustrating the significant difference between standardized signature scores of sample groups.
(B) Kaplan-Meier (KM) survival curves comparing subjects in the TCGA LUAD cohort stratified by correlation with the mouse-derived KP-lung γδ T signature. The top 25% most correlated patients (n = 128, red) exhibited significantly decreased survival as compared to the 25% least-correlated patients (n = 128, blue) from the TCGA LUAD cohort (p=0.016, log-rank test).
(C) Relevant gene set enrichment plots from GSEA of the KP lung γδ T signature (NES: Normalized Enrichment Score; FDR: False discovery rate).
(D) Volcano plot illustrating the magnitude of fold-change (x-axis, lung/spleen log2 fold change) for all genes ranked by their absolute z-score in the KP lung γδ T signature (y-axis). Some of the driver genes highly-upregulated in lung samples are highlighted.
(E) Pairwise comparison of the expression levels of 8 relevant genes between KP-lung and spleen samples (**** denotes FDR<1.63E-12).
(F) KP-R26LSL-cas9 mice were infected with lentiviral vectors co-expressing Cre and sgRNA against IL-22RA1 or an irrelevant locus (control). Tumor burden was quantified at 10 weeks post tumor initiation and representative H&E pictures are shown.
(G) Empirical cumulative distribution function (CDF) plots showing expression of IL22RA1 in human LUAD samples (n=515) in comparison to normal lung tissues (n=58) from the TCGA cohort (p = 8.58E-04, Kolmogorov-Smirnov test).
(H) KM survival curves comparing subjects in the TCGA LUAD cohort stratified by IL22RA1 expression. The patients with high IL22RA1 expression (top 30%) exhibit significantly worse survival when compared to the rest of the patients in the cohort (p=0.00001, log-rank test).
See also Figure S7.
Gene set enrichment analysis (GSEA) further revealed that the γδ T cells in the tumor-bearing lungs exhibited a gene expression pattern similar to activated human γδ T cells (Figure 7C, a). Additionally, as tissue-resident lymphocytes, γδ T cells in KP lungs also shared common gene signatures enriched in tissue-resident αβ T cells (Figure 7C, b, c). The robust upregulation of LPS-responsive genes in these KP-lung associated γδ T cells provided further evidence at the transcriptional level that they underwent microbial stimulation from airway microbiota (Figure 7C, d, e, f).
This transcriptional signature showed a number of genes to be highly upregulated in KP-lung associated γδ T cells compared to splenic γδ T cells (Figure 7D, E), and these data were further validated at the mRNA and/or protein levels (Figure S7B, C, D). In addition to IL-17A and IL-17F, γδ T cells in the KP lungs strongly expressed other effector molecules that might contribute to tumor-promoting inflammation. For example, CXCL2 is a neutrophil chemoattractant, whereas PTGS2 (prostaglandin-endoperoxide synthase 2) is essential for the biosynthesis of prostaglandin, a potent inflammatory mediator. IL-1R1 and IL-23R expression were also elevated in γδ T cells from tumor-bearing lungs (Figure 7E), indicative of their enhanced capability of responding to IL-1β and IL-23 induced by microbial products as compared to splenic γδ T cells.
Among the top differentially expressed genes, IL-22 is an epithelial growth factor significantly up-regulated in lung-tumor associated γδ T cells. Although known to exert a tumor-promoting effect in colorectal cancer (Kirchberger et al., 2013; Wang et al., 2017), the role of IL-22 in lung cancer has not been well studied. We tested the effects of IL-22 receptor deletion in developing lung cancer cells by introducing a lentivirally-encoded sgRNA targeting IL-22RA1 in KP mice harboring a LSL-Cas9 allele. As shown in Figure 7, targeting IL-22RA1 led to a significant reduction in tumor burden (Figure 7F). Moreover, recombinant IL-22 stimulated tumor cell proliferation in vitro (Figure S7E). As IL-22RA1 determines cellular sensitivity towards IL-22, we further examined the IL22RA1 expression in human lung cancer patients and found IL-22RA1 expression was substantially elevated in LUAD samples as compared to normal lung tissues (Figure 7G; Figure S7F). When patients were stratified by their levels of IL22RA1 expression, we found that those with high expression (top 30%) exhibited significantly worse survival as compared to the rest of the cohort (Figure 7H). These data are consistent with a role for IL-22 signaling in human lung cancer progression. Another top-scoring gene in the KP-lung γδ T cell signature was amphiregulin (Areg), a ligand for the epidermal growth factor receptor (EGFR). Increased Areg levels have been correlated with a poor prognostic outcome and poor response to therapy in lung cancer patients (Busser et al., 2011). We observed a robust expression of Areg in lung tumor-associated γδ T cells at both the mRNA and the protein levels (Figure S7B, D). KP tumor-derived cell lines showed dose-dependent increases in proliferation in response to recombinant Areg (Figure S7G). These findings, together with the data on the IL-22 pathway, provide strong evidence that microbiota-activated γδ T cells may promote lung cancer progression by directly stimulating tumor cell proliferation, in addition to stimulating inflammation in the lung microenvironment.
Discussion
The lung is a mucosal tissue exposed to a vast number of air-borne microbes and colonized by a diverse bacterial community in normal physiological conditions. Thus, the development of lung cancer may be influenced not only by the distal intestinal microbiota indirectly through its regulation of the systemic immune response, but also by direct interaction of the local airway microbiota with developing tumor cells and local immune system. Our findings demonstrate that lung tumor growth in the KP model is associated with increased total bacterial load and reduced bacterial diversity in the airway, suggesting that tumor-associated barrier defects and airway obstruction might result in impaired bacterial clearance and altered growth conditions for microbes. Our data support a model in which this dysregulation of local microbiota stimulates tissue-resident γδ T cells, which in turn produce IL-17 and other pro-inflammatory mediators to promote neutrophil expansion and tumor cell proliferation. This may further potentiate inflammation and dysbiosis, establishing a vicious cycle that exacerbates tumor growth.
Although increasing evidence has linked commensal microbiota to tumorigenesis and cancer response to therapy, the current literature has been mainly focused on the intestinal microbiota. How the commensal microbiota interact with developing tumor cells during malignant transformation at other mucosal sites and how the distal gut microbiota and local microbiota act together to regulate the balance between tumor-promoting inflammation and anti-tumor immunity remains unclear. Our data showed that lung tumor burden was highly correlated with local bacterial abundance in the airway, but not the intestinal tract, indicating a relatively more significant contribution of the local microbiota in lung cancer development. In addition, we demonstrated that intratracheal inoculation of a consortium of bacteria isolated from late-stage lung tumors significantly accelerated tumor growth, and that local administration of bacterial products (LPS and PGN) robustly stimulated γδ T cells in the lung. These results strongly suggest the importance of lung microbiota in driving local inflammation and tumor promotion. In mouse studies, it has proven to be technically challenging to selectively manipulate the airway microbiota (for example, recolonizing the lungs of GF mice or depleting the lung microbiota in SPF mice without affecting their gut microbiota). Although the low biomass of microbial cells in the lung also makes the characterization of lung microbiota difficult (Dickson et al., 2016; Lloyd and Marsland, 2017), future studies will focus on defining the role of specific composition of the lung microbiota in tumor development.
γδ T cells constitute a major tissue-resident T cell component of mucosal barrier tissues such as the gut, skin and lung. They display ‘innate-like’ characteristics and respond rapidly to infections and tissue damage (Papotto et al., 2017; Vantourout and Hayday, 2013). In particular, IL-17-producing γδ T cells are involved in the pathogenesis of autoimmune and inflammatory diseases, but their most prominent function is thought to provide protective immunity to infections at mucosal sites, including the lung (Papotto et al., 2017). Our study reveals an unappreciated role of lung-resident γδ T cells in promoting tumorigenesis upon activation by local commensal bacteria in the airway. We observe a baseline difference in the γδ T cell abundance between healthy SPF and GF animals. Although this difference is quite modest compared to that seen in tumor-bearing SPF and GF mice, it indicates that the presence of commensal bacteria maintains tissue-resident γδ T cells in homeostatic conditions. Our data support a model in which early tumor development upon Kras activation and p53 loss acts via the local microbiota to amplify the γδ T cell response by stimulating their proliferation, which in turn promotes tumor progression. In this case, both the microbiome and genes responsible for tumor initiation are key to the observed changes in γδ T cell numbers. In addition to their increased abundance, γδ T cells also exhibited up-regulated production of IL-17A and elevated surface expression of PD-1 and CD103 in tumor-bearing lungs as compared to normal lungs, whereas they expressed similar levels of PLZF and CD27 reflective of the Vγ6+ γδ T17 lineage (data not shown). Taken together, our data show that lung cancer development in the KP model is associated with the expansion and phenotypic alteration of tissue-resident γδ T cells in the tumor microenvironment. While it has been known from previous studies that intestinal microbiota can influence cancer outcomes by modulating systemic immune response and regulating activation/recruitment of inflammatory cells from the circulation, our findings describe a distinct mechanism whereby developing tumors in the lung hijack tissue-resident lymphocytes to establish a pro-tumorigenic microenvironment in a fashion dependent on the local microbiota.
γδ T cells can have either protective roles or tumor-promoting functions in cancer; tumor-promoting activities of γδ T cells have been ascribed to distinct mechanisms in different types of cancers (Daley et al., 2016; Silva-Santos et al., 2015). We found elimination of commensal microbiota or inhibition of γδ T cells in this lung cancer model did not lead to any measurable change in the anti-tumor immune response of αβ T cells, but microbiota-activated γδ T cells did play a critical role in promoting neutrophil expansion and local inflammation through the expression of IL-17 and other pro-inflammatory factors such as CXCL2 and PTGS2. Moreover, our data suggested that IL-22 and Areg may be important mediators via which γδ T cells directly promote lung cancer cell proliferation.
These new findings have clinical relevance for human lung cancer. Bacterial infections occur in up to 70% lung cancer patients and markedly influence the clinical outcome and patent survival (Hsu-Kim et al., 2013). These post-obstructive pneumonias are often polymicrobial, suggesting commensal bacteria, which are otherwise non-pathogenic and colonize pulmonary tissue at a much lower density in healthy individuals, could provoke chronic inflammation and disease exacerbation in the context of lung cancer. Consistent with our data from the mouse model, we also demonstrated the association between local dysbiosis and human non-small cell lung cancer via PathSeq analysis, and we showed increased infiltration of γδ T cells in LUAD as compared to normal lung tissues through IHC analysis of patient samples. Increased abundance of γδ T17 cells was observed to be positively associated with tumor invasion and metastasis in LUAD patients, although the underlying mechanism was not defined (Bao et al., 2016). Our study provides strong experimental evidence to establish a link between local microbial dysbiosis and tumor-promoting inflammation mediated by γδ T cells. Mechanistically, TLR ligands derived from the lung microbiota may stimulate the production of IL-1β and IL-23 from alveolar macrophages and neutrophils in a Myd88-dependent manner, and thereby induce proliferation and activation of lung-resident γδ T cells to further augment the inflammatory response. Interestingly, neutralization of IL-17, one of the key effector molecules produced by γδ T cells, led to decreased neutrophil infiltration and tumor burden, as well as a significant reduction in IL-1β level in the lung, implicating an amplification loop involving myeloid cells/IL-1β/γδ T cells/IL-17 in tumor promotion. Of note, a recent human clinical study reported IL-1β inhibition with the neutralizing antibody canakinumab significantly reduced lung cancer incidence and mortality in a cohort of high-risk individuals (Ridker et al., 2017). Given the importance of IL-1β in γδ T biology as shown by our data and studies in other systems (Sutton et al., 2009), it is conceivable that the effect might be at least partly mediated through the IL-1β/γδ T cells axis. Altogether, these results indicated that strategies for disrupting pathways that mediate the microbiota-induced γδ T cell activation may provide therapeutic benefits for lung cancer patients.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tyler Jacks (tjacks@mit.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and In Vivo Procedures
KrasLSL-G12D/+; p53flox/flox (KP) mice have been described (DuPage et al., 2009). R26LSL-cas9 and CD45.1 mice were purchased from Jackson Laboratory (Bar Harbor, ME) and crossed to KP mice to generate KP-R26LSL-cas9 or KP-CD45.1 mice. Both male and female mice were used in all experimental groups in all mouse experiments. In most experiments, KP mice were infected with 2.5×108 plaque-forming units (pfu) of Sftpc-Cre expressing adenovirus (Viral Vector Core, University of Iowa) or 30,000 transforming units of lentiviral vectors co-expressing Cre and specific sgRNAs through intratracheal instillation between 8–12 weeks of age to initiate tumors, as previously described (DuPage et al., 2009). In the bacterial inoculation experiment, KP mice were infected with 2.0×108 plaque-forming units (pfu) of Sftpc-Cre expressing adenovirus. Mice were randomized before treatments. For antibiotic treatment, mice were given a cocktail of ampicillin (1g/L, American Bioanalytical), neomycin trisulfate (1g/L, Sigma), metronidazole (1g/L, Sigma) and vancomycin (500 mg/L, Goldbio) (4Abx) or metronidazole alone in drinking water at indicated time points. For antibody-mediated depletion experiments, animals were treated every 2–3 days with i.p. injection of neutralizing monoclonal antibodies (200 μg/mouse) directed against γδ-TCR (UC7–13D5, BioXCell; or GL3, purified from the hybridoma generously provided by Dr. O’Brien, National Jewish Health, Denver) or IL-17A (17F3, BioXCell), and their isotype controls (BioXCell). For in vivo stimulation of γδ T cells in the lung, recombinant mouse IL-1β (R&D Systems, 100 ng/mouse) plus IL-23 (R&D Systems, 100 ng/mouse), or LPS (Lipopolysaccharide from E. coli O111:B4, Invivogen, 2 μg/mouse) plus PGN (Peptidoglycan from S. aureus, Invivogen, 5 μg/mouse) were administered intratracheally to wild-type C57BL/6J mice. For bacterial inoculation experiment, 14 bacterial strains were isolated and cultured from late-stage lung tumors from SPF mice. They were mixed and intratracheally administered at a total dose of 107 CFU to a separate cohort of KP mice 3.5 weeks post tumor initiation. For bone marrow chimera experiments, KP mice on the CD45.1 background were irradiated with a Gammacell-40 Irradiator (5.5 Gy × 2, 3.5 hours apart), and then transplanted with bone marrow collected from wild type or Myd88 knockout donors (Jackson Laboratory) on the CD45.2 background. Seven weeks post bone marrow reconstitution, mice were infected with 2.5×108 pfu of Sftpc-Cre expressing adenovirus for tumor initiation. All studies were performed under a Massachusetts Institute of Technology Committee on Animal Care-approved animal protocol, or an ICUC approved animal study protocol at NIAID, NIH (LSB-1E and LSB-4E). Mice were assessed for morbidity according to MIT Division of Comparative Medicine guidelines and were humanely euthanized when ill.
The gnotobiotic facility at MIT has been in continuous operation for the past 12 years. The germ-free KP mice used in this study were derived by embryo transfer using sterile techniques. Briefly, blastocysts were harvested from hormone-primed specific pathogen-free donor KP females and surgically implanted in hormone-primed, pseudo-pregnant germ-free Swiss Webster recipient females. The re-derived KP mice were maintained in germ-free conditions by following stringent standard operating procedures to prevent contamination. The germ-free status was confirmed by microbiological monitoring by culture and fecal PCR, alternatively every two weeks. Composite samples for culture consisted of swabbing the interior of the isolator, samples from the food trap, bedding and water bottles. Composite samples were plated on blood, MacConkey and chocolate agar plates and inoculated in thioglycollate broth to identify aerobic and anaerobic bacterial contaminants. The composite samples were also plated on Sabouraud dextrose agar to identify fungal contaminants. For fecal PCR, fresh fecal pellets were collected from each cage in the isolator and DNA was extracted from the pooled fecal pellets. PCR was performed using universal 16S rRNA primers 341F: 5’-CCTACGGGAGGCAGCAG-3’ and 806R: 5’-GGACTACHVGGGTWTCTAAT-3’. In addition to the routine testing and screening carried out at the gnotobiotic facility described above, we also inspected the mice for the typical appearance of GF animals such as the enlarged cecum at the time of tissue collection. Moreover, we collected stool samples from the small and large intestine, performed DNA extraction and 16S real-time qPCR analysis. Fecal DNA of GF mice didn’t give any signal in 40 cycles, whereas those from SPF mice had Ct values of 6–8 cycles.
In vitro cell proliferation
Tumor cell lines were derived from primary lung tumors of KP mice, and maintained in DMEM medium (Gibco) supplemented with 10% FBS (Sigma). Cells were serum-starved for 24 hours (1% FBS) before treated with recombinant human amphiregulin (Sigma-Aldrich) or recombinant mouse IL-22 (R&D Systems) at the indicated dose, or left untreated as controls. The number of live cells was determined 72 hours post treatment by counting the Trypan Blue (Gibco) negative cells or through the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions.
Bacterial culture
Late-stage lung tumors were collected from SPF KP mice, cut and plated using aseptic techniques for bacterial culture under both aerobic and anaerobic conditions (Bullman et al., 2017). Fastidious Anaerobic Agar (FAA, Acumedia) and Brain-Heart Infusion Agar (BHA, Acumedia) were used for anaerobes; Tryptone Soya Agar (TSA, Sigma-Aldrich) and BHA were used for aerobes. All media were supplemented with 8% defibrinated horse blood. Aerobes grew well within 24–48 hours, and anaerobes grew well within 72 hours. All organisms were identified using universal 16s rRNA gene primers and sanger sequencing, including Aneurinibacillus aneurinilyticus, Lactobacillus murinus, Lactobacillus reuteri, Streptococcus acidominimus, Corynebacterium, Propionibacterium acnes, Brevibacterium massiliense, Microbacterium lacticum, Lactococcus, Bifidobacterium (potentially pseudolongum), Acinetobacter radioresistens and 3 strains of Staphylococcus.
METHOD DETAILS
Flow cytometry, FACS sorting
Because the lung tissue is highly vascularized, mice were injected retro-orbitally with a PE-CF594 conjugated anti-CD45 antibody (30-F11, BD Bioscience) 2 minutes prior to euthanasia. This procedure labeled all hematopoietic cells in the circulation but not those localized inside the tissue (Anderson et al., 2014). Lungs were collected, cut into small pieces, and digested with 125U/mL collagenase IV (Worthington Biochemical) and 40U/mL DNase I (Roche) for 30 minutes at 37°C. Dissociated lung tissue was then filtered with a 100 μm cell strainer. Spleen and lymph node samples were grinded up and passed through a 100 μm cell strainer. After blocking FcgRIII/II with an anti-CD16/CD32 mAb (eBioscience), single cell suspensions were stained with the following antibodies: CD45.2 (104), CD45.1 (A20), Ly6G (1A8), CD11b (M1/70), CD11c (N418), CD103 (2E7), Ly6C (HK1.4), TCRβ (H57–597), TCRγδ (GL3 or UC7–13D5), CD3 (17A2), CD4 (RM4–5), CD8 (53–6.7), Vγ1 (Vg1.1, 2.11), Vγ4 (Vγ2, UC3–10A6), IFN-γ (XMG1.2), IL-17A (TC11–18H10.1), TNF-α (MP6-XT22), IL-17F (9D3.1C8), CD44 (IM7), CD27 (LG.3A10), CD69 (H1.2F3), PD-1 (29F. 1A12), T-bet (eBio4B10), RORγt (B2D), PLZF (Mags.21F7) and Ki67 (B56) purchased from eBioscience, BD or Biolegend. Biotinylated anti-amphiregulin antibody was purchased from R&D Systems. For staining of the Vγ6Vδ1+ TCR, cells were labeled first with the anti-TCRγδ mAb (GL3), and then incubated with the supernatant from the 17D1 hybridoma (kindly provided by Dr. Robert Tigelaar and Julia Lewis, Yale University) followed by staining with the goat-anti-rat IgM (Jackson Immunoresearch Laboratories) secondary antibody as previously described (Roark et al., 2004). For intracellular cytokine staining, cells were pre-incubated with the Cell Stimulation Cocktail and the Protein Transport Inhibitor Cocktail (both from eBioscience) for 4 hours at 37°C before surface staining. Transcription factor staining was performed using the Foxp3 Fixation/Permeabilization Solution Kit (eBioscience). Flow cytometry was performed on the LSR II or LSRFortessa (BD), and data were analyzed by the FlowJo software (Treestar). FACS sorting was performed on a FACSAria III (BD).
ELISA, RT-qPCR
IL-17 levels in serum or bronchoalveolar lavage fluid were measured using a mouse IL-17A ELISA kit (eBioscience) according to the manufacturer’s instructions. Total RNA was isolated from lung tissues or FACS-purified cells using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s instructions. cDNA was synthesized from RNA with the Superscript III Reverse Transcription Kit (Thermo Fisher). For real-time qPCR analysis, Il1b, Il23, Gcsf expression were analyzed in triplicates and normalized to b-actin or Hprt with Taqman probes (Thermo Fisher) on a LightCycler 480 (Roche).
Histology, Immunohistochemistry (IHC)
For the mouse samples, tumor-bearing lung lobes and a portion of the spleen were fixed in 4% paraformaldehyde overnight and embedded in paraffin. Hematoxylin and eosin stain (H&E stain) was performed with a standard method (KI Histology Facility). IHC staining of Ki67 (Cell Signaling, 12202) and Ly6G (BD Pharmingen, 1A8) were performed on 5 μm unstained sections after antigen retrieval with citrate buffer or proteinase-K (Sigma) respectively (Rickelt and Hynes, 2018). Digitally scanned images of H&E or Ki67-stained slides were created with the Aperio ScanScope AT2 at 20X magnification and analyzed with the Aperio’s WebScope software. For tumor burden quantification, tumor regions were outlined and the percentage of tumor area relative to total lung area was calculated for each mouse. For Ki67 index, tumor cells were classified based on the intensity of the nuclear Ki67 staining from 0 to 3+ by using a built-in Nuclear algorithm in the Webscope, and the percentage of nuclei with intense nuclear staining (3+) in total nuclei was calculated. All tumor burden/grading and IHC analyses were done in a blinded fashion, and at least five mice per groups were included in the analyses.
For the human samples, formalin-fixed and paraffin embedded (FFPE) tissues of primary NSCLC and normal lung were for used IHC analysis of TCRδ (H-41, Santa Cruz Biotechnology). Some of the LUAD samples and normal lung controls were from human tissue arrays (Genvelop Life Science, LLC); others were from patients who underwent surgical resection at Boston Medical Center (Boston, MA) between 2011– 2014 and consented to have tissue become part of the Biospecimen Archive Research Core (BARC). The BARC study was approved by the Boston Medical Center and Moston University Medical School institution review board to collect tissue for future research purposes. All samples were de-identified and limited clinical information provided.
Immunofluorescence and Confocal imaging
Lung tissues were perfused and then incubated in a fixation and permeabilization solution (BD Bioscience, 554722) overnight followed by dehydration in 30% sucrose before embedding in OCT compound (Sakura Finetek). 18-μm sections were cut on a CM3050S cryostat (Leica) and adhered to Superfrost Plus slides (VWR). Frozen sections were permeabilized and blocked in PBS containing 0.3% Triton X-100 (Sigma-Aldrich) and Fc-blocker (CD16/CD32 Monoclonal Antibody, eBioscience) followed by staining with antibodies diluted in the blocking buffer. The following antibodies were used for staining: anti-CD3 (17A2; Biolegend), anti-CD4 (GK 1.5, Biolegend), anti-CD8 (53–6.7, BD), anti-RORγt (Q31–378, BD) and anti-TCRγδ (GL3, Biolegend). After staining, slides were mounted with Fluormount G (Southern Biotech), and examined on a Leica TCS SP8 confocal microscope. Images were analyzed with Imaris software (Bitplane).
Lung and fecal microbiome analysis
Bronchoalveolar lavage fluid (BALF) was collected with sterile PBS and centrifuged at 16,000g for 10 minutes to pellet down bacterial cells. Fecal pellets were collected from the colon at the time of tissue harvest. BALF and fecal samples were processed for DNA isolation using a previously established protocol (Palm et al., 2014). For quantification of total bacterial burden, qPCR was performed with universal 16S primers targeting the V1–2 region (27F: AGAGTTTGATCMTGGCTCAG, 338R: TGCTGCCTCCCGTAGGAGT) and the Kapa SYBR Fast 2X master mix (Kapa Biosystems). E. coli genomic DNA was used as a standard, with samples at 1, 0.1, 0.01, and 0.001 ng per reaction used to create a standard curve. For 16S based sequencing, we utilized two rounds of PCR to prepare sequencing libraries (Caporaso et al., 2011). The first PCR amplified the V1–2 region of the 16S rRNA gene with 27F/338R, and added adapters at each end, followed by the second PCR adding full sequencing adapters and barcodes. Agencourt AMPure XP beads (Beckman Coulter) were used to purify the PCR products following the manufacturer’s protocol and Kapa HiFi HotStart 2x ReadyMix (Kapa Biosystems) was used for PCR reaction. The final PCR products were quantified using the Qubit DNA assay (Thermo Fisher Scientific), and all samples were normalized and pooled at equal mass for the final sequencing pool. Samples were sequenced on the Illumina MiSeq with 2 × 250 bp paired-end reads.
RNA-seq sample preparation
γδ T cells were isolated from tumor bearing lungs or spleens of KP mice by FACS. Approximately 1000 cells/condition were sorted directly into TCL buffer (Qiagen), and the sequencing libraries were prepared using a modified Smartseq V2 protocol as previously described (Picelli et al., 2014; Singer et al., 2017). A total of 9 spleen and 15 lung samples from three independent experiments were sequenced on the Illumina HiSeq 2000 platform.
The graphical abstract image is created with BioRender.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
GraphPad Prism 7 was used for statistical analyses with the histology/IHC/16S quantification and flow cytometry data. P-values from unpaired two-tailed student’s t-tests were used for comparisons between two groups and one-way ANOVA with Bonferroni’s post hoc test was used for multiple comparisons. For tumor burden and bacterial load correlation analysis, linear regression was performed with GraphPad Prism 7. Figure legends specify the statistical analysis used.
RNA-seq analysis
Illumina HiSeq 2000 40-nt single-ended reads were mapped to the UCSC mm9 mouse genome build (genome.ucsc.edu) using RSEM (Li and Dewey, 2011). Raw estimated expression counts were upper-quartile normalized to a count of 1000 (Bullard et al., 2010). Genes with an upper-quartile expression distribution value less than 20 in both conditions were considered lowly expressed and dropped from downstream analyses. The expression dataset was log (base 2) transformed to stabilize variance. KP-lung (n = 15) and KP spleen (n = 9) samples were jointly analyzed to derive a murine signature of lung γδ T gene expression. A high-resolution signature discovery approach (Independent Component Analysis, ICA) was employed to characterize changes in gene expression profiles as described previously (Romero et al., 2017). Briefly, this unsupervised blind source separation technique was used on this discrete count-based expression dataset to elucidate statistically independent and biologically relevant signatures. ICA is a signal processing and multivariate data analysis technique in the category of unsupervised matrix factorization methods (Hyvarinen and Oja, 2000). The R implementation of the core JADE algorithm (Joint Approximate Diagonalization of Eigenmatrices) (Biton et al., 2014) was used along with custom R utilities. Statistical significance of biologically relevant signatures was assessed using the Mann-Whitney-Wilcoxon test (alpha = 0.05). Signature genes with |z-score| > 3 and |fold-change| > 2 were used to generate a heatmap illustrating changes in gene expression, with the HeatPlus package in R. Gene set enrichment analyses (GSEA) were carried out using the signature scores per gene (z-scores) in pre-ranked mode with default settings (Subramanian et al., 2005). A volcano plot was used to illustrate the magnitude of fold-change for top-scoring (z-scores) genes in the signature. The expression levels of 8 biologically relevant top-scoring genes in the signature were illustrated using a pairwise dot-plot between lung and spleen samples. False discovery rate (FDR) values for differential expression status of these genes were calculated using a pairwise comparison in EBSeq (Leng et al., 2013). All RNA-seq analyses were conducted in the R Statistical Programming language (http://www.r-project.org/).
Clinical data analysis
IL-22 receptor (IL22RA1) expression levels were compared between normal and tumor tissue within the TCGA ( cancergenome.nih.gov) Lung Adenocarcinoma (LUAD) cohort. Matched normal and tumor tissue IL22RA1 standardized expression levels were illustrated using an Empirical Cumulative Distribution Function plot (ECDF) and significance was assessed using a Kolmogorov-Smirnov test. Similarly, the set of normal samples was compared with the entire set of tumor samples (either with or lacking matched normal samples). For survival analysis, RNA-seq gene expression profiles of primary tumors and relevant clinical data of 515 lung adenocarcinoma (LUAD) patients were obtained from TCGA and survival analyses were conducted as described previously (Romero et al., 2017; Tammela et al., 2017). The murine KP-lung derived γδ T gene expression signature (|z| > 3 genes) was translated to human symbols (homology information from MGI, informatics.jax.org) and was used to score individual TCGA tumor expression profiles using ssGSEA (Barbie et al., 2009). Patients were stratified based on standardized ssGSEA scores and Kaplan-Meier survival analyses were conducted to compare high-scoring patients (top quartile) with low-scoring patients (bottom quartile) and significance was assessed using the log-rank test. Additionally, the TCGA LUAD patients were also scored by the ESTIMATE immune signature to evaluate their global immune cell infiltration (Yoshihara et al., 2013). Pearson correlation analysis was performed between patient scores from the two signatures. For Kaplan-Meier survival analysis based on IL22RA1 expression, TCGA LUAD cohort patients were similarly stratified based on standardized expression values of IL22RA1 and the top 30% of patients were compared with the rest of the cohort. All survival analyses were conducted using the survival package in R.
16S sequencing analysis
16S sequencing reads were processed using the QIIME analysis pipeline. OTUs were clustered at 97% identity using QIIME pick_open_reference_otus.py with the Greengenes 13.8 reference database (Caporaso et al., 2010). OTUs that did not appear in at least 3 samples and have at least 50 sequence counts were removed. On average, BAL samples had 50,000 sequencing reads and FP samples had 10,000 sequencing reads. A minimum threshold of 500 reads was used to retain samples after filtering. For alpha diversity, we calculated the Shannon index of the 97% identity OTUs using alph_diversity.py, with samples rarefied to 1000 reads. Beta diversity measurements were calculated using beta_diversity.py to determine the Bray-Curtis and weighted UniFrac metrics. PCoA plots were made using make_2d_plots.py. To test for differences in the community composition between sample groups, we performed Permutational Multivariate Analysis of Variance (PERMANOVA) based on the Bray-Curtis and weighted UniFrac distance matrices using QIIME compare_categories.py. The LEfSe method (Segata et al., 2011) first utilizes the non-parametric Kruskal-Wallis rank-sum test to compare relative abundances of all bacterial taxa between tumor-bearing and healthy mice (at α = 0.05). Subsequently, linear discriminant analysis (LDA) is used to estimate the effect size of each differentially abundant taxa. Taxa are included if present at ⩾ 0.01% total relative abundance and in at least two samples. Taxa with LDA score > 2.0 were included in the plot.
Microbial Detection in RNA Sequencing Datasets
The PathSeq (Kostic et al., 2011) algorithm was used to perform computational subtraction of human reads, followed by alignments of residual reads to human reference genomes / transcriptomes and microbial reference genomes (which include bacterial, viral, archaeal, and fungal sequences - downloaded from NCBI in October, 2015). These alignments resulted in taxonomical classification of reads into bacterial, viral, archaeal, and fungal sequences in RNA sequencing (RNA-Seq) data. The human reference genome/transcriptome sequences were downloaded from the Ensembl database (ftp://ftp.ensembl.org/pub/current_fasta/homo_sapiens/cdna/Homo_sapiens.GRCh37.74.cdna.all.fa.gz AND ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/RNA/rna.fa.gz), female human reference genome (ftp://ftp.ncbi.nih.gov/1000genomes/ftp/technical/reference/human_g1k_v37.fasta.gz), human genome plus transcriptome database (ftp://ftp.ncbi.nih.gov/blast/db/human_genomic_transcript.tar.gz) and the human reference genome from NCBI (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/Assembled_chromosomes/seq/). Bacterial, fungal and archaeal reference sequences were downloaded from NCBI reference sequence database (ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/). Viral and phage sequences were downloaded from NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore) using query strings “Viruses[Organism] AND srcdb_refseq[PROP] NOT wgs[prop] NOT cellular organisms[ORGN] NOT AC_000001:AC_999999[pacc]” and “Viruses[Organism] NOT srcdb_refseq[PROP] NOT cellular organisms[ORGN] AND nuccore genome samespecies[Filter] NOT nuccore genome[filter] NOT gbdiv syn[prop]”. The curated databases in FASTA format are available at http://software.broadinstitute.org/pathseq/Downloads.html webpage.
Briefly, PathSeq (Kostic et al., 2011) is used to remove low quality reads followed by subtraction of human reads by mapping reads to a database of human genomes (downloaded from NCBI in November 2011) using BWA (Li and Durbin, 2009) (Release 0.6.1, default settings), MegaBLAST (Release 2.2.25, cut-off E-value 10−7, word size 16) and BLASTN (Altschul et al., 1997) (Release 2.2.25, cut-off E-value 10−7, word size 7, nucleotide match reward 1, nucleotide mismatch score −3, gap open cost 5, gap extension cost 2). Sequences with perfect or near perfect matches to the human genome are removed in the subtraction process. In addition, low complexity and highly repetitive reads are removed using Repeat Masker (version open-3.3.0, libraries dated 2011–04–19).
Taxonomic classification is performed by residual reads alignment using MegaBLAST (Release 2.2.25, cut-off E-value 10−7, word size 16) to a database of bacterial sequences and followed by BLASTN (Release 2.2.25, cut-off E-value 10−7, word size 7) to the human reference genome and microbial reference genomes.
Following the taxonomic classification of non-human DNA sequencing reads, the relative abundance value for each bacterial organism is calculated by using reads that maps with >=90% sequence identity and >=90% query coverage. Classifications are performed at the domain, then phylum, then genus, then species level, requiring unique alignments (i.e., reads with equivalent E-values to multiple taxa were removed from the analysis).
Species level relative abundance (RA) for each organism was calculated as follows: relative abundance of a given organism in a sample= (number of unique alignment positions across the genome × 1,000,000)/ (number of total aligned bacterial reads × bacterial genome size). The RA values were then “per-sample” normalized such that the total relative abundance for each sample sums to one (or 100% for percentage relative abundance).
Comparative Microbial Analysis
Following PathSeq analysis of lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC) samples from TCGA, comparative analysis was carried out using LEfSe (Segata et al., 2011) (online version available on July 2018), with default parameters, expect for the linear discriminant score (LDA score) which was increased >=3 to identify bacterial organisms significantly enriched or depleted in LUAD tumors (n=248) vs. a subset of adjacent normal tissues (n=56), and LUSC tumors (n=221) vs. a subset of adjacent normal tissues (n=17). The GENE-E analysis tool (https://software.broadinstitute.org/GENE-E/), marker selection analysis (default settings) was used to further identify enriched/depleted microbes at the genus level in the LUAD and LUSC samples and to confirm LEfSe results.
DATA AND SOFTWARE AVAILABILITY
The RNA-seq data for γδ T cells from tumor-bearing lungs or spleens of KP mice have been deposited in the GEO repository under the ID code GSE114340.
The 16S rDNA sequencing data have been deposited to the SRA under the ID code PRJNA492954.
Supplementary Material
Figure S1. Related to Figure 1.
(A) GF KP mice were exposed to the microbiome via cohousing with SPF mice (ex-GF) or maintained in GF conditions as controls. Tumor burden was analyzed 12 weeks post tumor initiation and representative pictures were shown. n=5 mice/group.
(B) SPF KP mice were left untreated or treated with combined antibiotics (4Abx) in drinking water starting at 2 weeks post tumor initiation. Tumor burden and number were analyzed at 15 weeks p.i. Results are expressed as the mean ± SEM. * p<0.05, *** p<0.001 by Studenťs t test. For each experiment, n= 11–13 mice/group; data represent 2 independent experiments.
(C) Tumor cell lines derived from different primary KP tumors were treated with combined antibiotics or left untreated as controls. Cell number was determined at every passage by counting the Trypan Blue negative cells at indicated time points. Data are expressed as the mean values of triplicates.
Figure S2. Related to Figure 2.
(A) Principal components analysis (PCoA) using Bray-Curtis distance metric (left) and weighted UniFrac metric (right). The lung microbiota community is significantly different from the gut microbiota community (p<0.01 for both Bray-Curtis and weighted UniFrac distance metrics, PERMANOVA).
(B) LEfSe plots showing differentially abundant taxa in the lung microbiome of healthy mice (red) and tumor-bearing mice (blue). Linear discriminant analysis (LDA) scores were calculated using LEfSe, with higher scores indicating greater effect size (significance determined by LDA score > 2.0 and p < 0.05 for Kruskal-Wallis test). Taxonomic categories include p = phyla, c = class, o = order, f = family, and g = genus. Taxa present at ≥ 0.01% total relative abundance and in at least two samples were included.
(C) SPF KP mice were left untreated or treated with metronidazole (1g/L) in drinking water starting 5 weeks post tumor initiation. Tumor burden was quantified 15 weeks post tumor initiation and representative H&E pictures were shown; fecal bacteria burden was determined by 16S based qPCR analysis. n=7–9 mice/group.
(D) LEfSe plots showing the differentially abundant taxa in the lung microbiome of normal lungs (red) and lung adenocarcinoma (LUAD) or lung squamous cell carcinoma (LUSC) samples (green) based on PathSeq analysis of the TCGA cohort.
Figure S3. Related to Figure 3.
(A) The frequency of γδ T cells in total CD3+ lymphocytes in the lung, blood, spleen or draining lymph node from SPF and GF KP mice as determined by flow cytometry.
(B) Representative pictures and quantification of immunohistochemistry staining of human TCRδ on formalin-fixed paraffin-embedded normal lung (NL) and lung adenocarcinoma (LUAD) tissue samples. Positively stained cells are in purple.
(C, D) RORγt and IL-17A expression in total CD3+ lymphocytes from the tumor-bearing lungs from SPF mice and GF mice. Representative flow cytometric plots are shown (C) and the frequency of IL-17A+ CD4 T cells (Th17) is quantified (D).
Results are expressed as the mean ± SEM. ** p<0.01, *** p<0.001 by Studenťs t test. For each experiment, n= 8–15 mice/group; data represent 3 independent experiments.
Figure S4. Related to Figure 4.
(A) KP mice on the CD45.1 background were lethally irradiated and transplanted with bone marrow from CD45.2 donors. Seven weeks after reconstitution, mice were infected with adenovirus expressing Sftpc-Cre, and 15 weeks after tumor initiation, γδ T cells in the tumor-bearing lungs were analyzed by flow cytometry. The percentage of donor vs. recipient derived cells was quantified in the Vγ4+ and Vγ6+ subsets, as well as the RORγt+ and Tbet+ compartments. Representative plots are shown and data represent 15 mice.
(B) The proliferation of RORγt- γδ T cells and Th17 cells in the tumor-bearing lungs from tumor-bearing SPF mice and GF mice was assessed by flow cytometric analysis of Ki67 expression.
(C) IL-17A expression in lung-infiltrating γδ T cells from healthy SPF mice, tumor-bearing SPF mice and tumor-bearing GF mice was analyzed by flow cytometry.
(D) SPF KP mice were treated with combined antibiotics (4Abx) starting 6.5 weeks after tumor initiation. The frequency of IL-17A-producing γδ T cells and IL-17A concentration in BALF were analyzed by flow cytometry and ELISA respectively.
(E) The abundance of γδ T cells, and the expression of RORγt and IL-17A in γδ T cells were analyzed by flow cytometry in GF mice and ex-GF mice that were exposed to the microbiome via cohousing with SPF mice.
(B-E) Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by Studenťs t test. For each experiment, n= 5–13 mice/group.
Figure S5. Related to Figure 5.
The expression of Tbet, IFNγ and TNFα in the tumor-bearing lungs from SPF and GF mice were analyzed by flow cytometry.
Results are expressed as the mean ± SEM. *p<0.05, *** p<0.001 by Studenťs t test. n= 3–5 mice/group. Data represent 3 independent experiments.
Figure S6. Related to Figure 6.
(A) Flow cytometry analysis of different types of myeloid cells in the tumor-bearing lungs from SPF and GF mice. The frequencies of each cell type in total tissue-resident immune cells are quantified. Alveolar macrophages: CD11c+, CD11b-intermediate, autofluorescence-high; monocytes: Ly6C+, CD11b-high or intermediate; DC: CD11c+, MHCII+, CD11b+ or CD103+.
(B-E) KP mice were treated with the monoclonal antibody UC7–13D5 staring at 4 weeks post tumor initiation. (B, C) Blockade of γδT cells was verified by the loss of cells that were stained positively with a different monoclonal antibody against γδ-TCR (GL3), as well as the reduction of TCRγ- CD3+ lymphocytes. (D, E) Flow cytometry analysis of Ki67 and IFNγ expression in CD4 and CD8 T cells in the tumor-bearing lungs from mice treated with UC7–13D5 or isotype control.
(F-J) KP mice were treated with the monoclonal antibody GL3 starting at 4 weeks post tumor initiation. Tumor burden was quantified at 15 weeks p.i. and representative H&E pictures are shown.
(G, H) Blockade of γδ T cells was verified by the loss of cells that were stained positively with a different monoclonal antibody against γδ-TCR (UC7–13D5), as well as the reduction of TCRβ- CD3+ lymphocytes. (I, J) Flow cytometry analysis of Ki67 and IFNγ expression in CD4 and CD8 T cells in the tumor-bearing lungs from mice treated with GL3 or isotype control.
(K) IFNγ and/or TNFα expression in CD4 and CD8 T cells from the tumor-bearing lungs were analyzed by flow cytometry and compared between SPF and GF mice.
Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by Studenťs t test. n= 6–12 mice/group.
Figure S7. Related to Figure 7.
(A) LUAD patients in the TCGA cohort were scored for their gene expression correlation with the KP-lung γδ T cell signature as well as the ESTIMATE immune infiltration signature. Pearson correlation analysis of the scores from the two signatures for individual patients does not suggest a significant correlation (r= 0.15, n=515 primary tumors).
(B) RT-qPCR analysis of the candidate genes in FACS-purified γδ T cells from tumor-bearing lungs and spleens of KP mice.
(C) Flow cytometric analysis of IL-17A and IL-17F in γδ T cells. Plots representative of 8 mice.
(D) Flow cytometric analysis of amphiregulin in γδ T cells. Plots representative of 8 mice.
(E) Tumor cell lines derived from different primary KP tumors were treated with 1ng/ml recombinant mouse IL-22 for 72 hours or left untreated as controls; cell number was determined by counting the Trypan Blue negative cells.
(F) Empirical cumulative distribution function (CDF) plots showing expression of IL22RA1 in human LUAD samples, the matched normal tissues from the same patients, as well as other adenocarcinoma samples in the TCGA cohort (p values shown by Kolmogorov-Smirnov test).
(G) Tumor cell lines derived from different primary KP tumors were treated with the indicated dose of recombinant amphiregulin for 72 hours; cell number was determined by the CellTiter-Glo assay.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| In vivo mAb UC7–13D5 | BioXCell | UC7–13D5 Cat# BE0070 |
| In vivo mAb IL-17A | BioXCell | 17F3 Cat# BE0173 |
| In vivo mAb GL3 | Dr. O’Brien (National Jewish Health, Denver) | |
| FC: CD45 | BD Bioscience | 30-F11 Cat#562420 |
| FC: CD16/CD32 | BD Bioscience | 2.4G2 Cat#553142 |
| FC: CD45.2 | Biolegend | 104 Cat#109841 |
| FC: CD45.1 | Biolegend | A20 Cat#110743 |
| FC: Ly6G | Biolegend | 1A8 Cat#127622 |
| FC: CD11b | eBioscience | M1/70 Cat# 101243 |
| FC: TCRβ | Biolegend | H57 −597 Cat# 109220 |
| FC: TCRγδ | Biolegend | GL3 Cat# 118118 |
| FC: TCRγδ | Biolegend | UC7–13D5 Cat# 107504 |
| FC: CD3 | Biolegend | 17A2 Cat# 100228 |
| FC: CD4 | Biolegend | RM4–5 Cat# 100552 |
| FC: CD8 | BD Bioscience | 53–6.7 Cat# 563786 |
| FC: CD11c | eBioscience | N418 Cat# 25–0114–82 |
| FC: CD103 | Biolegend | 2E7 Cat# 121414 |
| FC: Ly6C | Biolegend | HK1.4 Cat# 128014 |
| FC: Vγ1 (Vg1.1) | Biolegend | 2.11 Cat# 141104 |
| FC: Vγ4 (Vγ2) | Biolegend | UC3–10A6 Cat# 137706 |
| FC: IFN-γ | BD Bioscience | XMG1.2 Cat# 554412 |
| FC: IL-17A | Biolegend | TC11–18H10.1 Cat# 506916 |
| FC: TNFa | eBioscience | MP6-XT22 Cat# 11–7321–82 |
| FC: IL-17F | Biolegend | 9D3.1C8 Cat# 517008 |
| FC: CD44 | Biolegend | IM7 Cat# 103039 |
| FC: CD27 | eBioscience | LG.3A10 Cat# 11–0271–85 |
| FC: CD69 | Biolegend | H1.2F3 Cat# 104527 |
| FC: PD-1 | Biolegend | RMP1–30 Cat# 109110 |
| FC: T-bet | eBioscience | eBio4B10 Cat# 12–5825–82 |
| FC: RORγt | eBioscience | B2D Cat# 17–6981–82 |
| FC: PLZF | eBioscience | Mags.21F7 Cat# 12–9320–82 |
| FC: Ki67 | BD Bioscience | B56 Cat# 561277 |
| FC: 17D1 | Dr. Robert Tigelaar and Julia Lewis (Yale University, CT) | 17D1 |
| IHC: Purified Rat Anti-Mouse Ly-6G | BD Pharmingen | 1A8 Cat# 551459 |
| IHC: Mouse mAb against human TCRδ | Santa Cruz | H-41 Cat# sc-100289 |
| IHC: Ki-67 (D3B5) Rabbit mAb (Mouse Preferred) | Cell Signaling | #12202 |
| Bacterial and Virus Strains | ||
| Adenovirus (Sftpc-Cre) | Viral Vector Core, University of Iowa | Ad5mSPC-Cre |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant mouse IL-1β | R&D Systems | 401-ML-005 |
| Recombinant mouse IL-23 | R&D Systems | 1887-ML-010/CF |
| Recombinant mouse IL-22 | R&D Systems | 582-ML-010 |
| Recombinant human amphiregulin | Sigma | A7080 |
| Lipopolysaccharide from E. coli O111:B4 (LPS) | Invivogen | tlrl-eblps |
| Peptidoglycan from S. aureus (PGN) | Invivogen | tlrl-pgns2 |
| Critical Commercial Assays | ||
| Mouse IL-17A (homodimer) ELISA | eBioscience | 88–7371–86 |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | G7571 |
| Deposited Data | ||
| RNA-seq | GEO | GSE114340 |
| 16S sequencing | SRA | PRJNA492954 |
| Experimental Models: Organisms/Strains | ||
| B6 CD45.2: C57BL/6J | Jackson Laboratory | 000664 |
| Myd88 Null: B6.129P2(SJL)-Myd88tm1.1Defr/J | Jackson Laboratory | 009088 |
| B6J.129(B6N)-Gt(ROSA)26Sortm1(CAG-cas9*,-EGFP)Fezh/J | Jackson Laboratory, crossed to KP mice | 026175 |
| B6 CD45.1: B6.SJL-Ptprca Pepcb/BoyJ | Jackson Laboratory, crossed to KP mice | 002014 |
Acknowledgements
We thank Dr. Rebecca O’Brien (National Jewish Health, CO) and Dr. Robert Tigelaar and Julia Lewis (Yale University, CT) for kindly providing the GL3 and 17D1 hybridomas respectively. We thank the germ-free facility staff for assistance with the GF experiments, Mingqi Dong for technical assistance, Steffen Rickelt for help with IHC and Sjoerd Klarenbeek (Netherlands Cancer Institute) for the Ly6G IHC protocol. We thank the Histology, Flow Cytometry, Genomics and Animal Imaging Core Facilities at the Swanson Biotechnology Center. C.J. is supported by the Helen Hay Whitney Foundation-Merck postdoctoral fellowship, the Margaret A. Cunningham Immune Mechanisms in Cancer Research Fellowship Award, and a K99 Award (CA226400) from the National Cancer Institute (NCI). G.L. was supported by a Broad’s Next10 Award and the Hugh Hampton Young Fellowship. S.B. is funded by a K99 (CA229984) from NCI and B.H is a CRI fellow. This work was supported by a Lung Cancer Research Program Concept Award (W81XWH-15–1–0623) from Department of Defense (C.J.), a R01 grant (CA185020) and a Cancer Center Support (core) grant (P30-CA14051) from NCI (T.J.), and support from the Howard Hughes Medical Institute (T.J.). P.C.B was supported by Broad Institute and a Career Award at the Scientific Interface from the Burroughs Wellcome Fund. This work was also supported in part by P30-ES002109 (J.F.), and the Intramural Research Programs of NIAID and NHLBI, NIH (C.Z. and R. G.).
Declaration of Interests
T.J. and S.M. receive research support from the J&J Lung Cancer Initiative. P.C.B. is a consultant to and equity holder in GALT and 10X Genomics. M.M. is an inventor on patent applications on Fusobacterium and the inventor on a patent for pathogen discovery. A full list of all of authors’ interests can be found in Supplemental Materials.
Footnotes
Comprehensive Declaration of Interests
The authors wish to list to following outside interests.
T.J. is a member of the Board of Directors of Amgen and Thermo Fisher Scientific and an equity holder in both companies. He is co-Founder and Scientific Advisory Board member of Dragonfly Therapeutics, a co-founder of T2 Biosystems, and a Scientific Advisory Board member of SQZ Biotech; he is an equity holder in all three companies. His laboratory currently receives funding from the Johnson & Johnson Lung Cancer Initiative and Calico. P.C.B. is a consultant to and equity holder in GALT and 10X Genomics, and a consultant to Insitro. S. M. receives funding from the Johnson & Johnson Lung Cancer Initiative. M.M. is a Scientific Advisory Board member for OrigiMed and receives consulting fees for this role. He was a consultant for Foundation Medicine and held equity, some of which was sold to Roche. He receives research support from Bayer and is an inventor of several joint patents and patent applications, none of which have been licensed at the time of this disclosure (12–17–18). In addition, he is an inventor on several patents on EGFR mutations in lung cancer diagnosis, licensed to LabCorp, for which he receives royalties. He is also an inventor on several patent applications on Fusobacterium, none issued or licensed, and the inventor on a patent for pathogen discovery, not licensed, for none of which he receives royalties.
References
- Akinosoglou KS, Karkoulias K, and Marangos M (2013). Infectious complications in patients with lung cancer. Eur Rev Med Pharmacol Sci 17, 8–18. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, et al. (2014). Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc 9, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Lu G, Cui D, Yao Y, Yang G, and Zhou J (2016). IL-17A-producing T cells are associated with the progression of lung adenocarcinoma. Oncol Rep 36, 641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. (2009). Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462, 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton A, Bernard-Pierrot I, Lou Y, Krucker C, Chapeaublanc E, Rubio-Perez C, Lopez-Bigas N, Kamoun A, Neuzillet Y, Gestraud P, et al. (2014). Independent component analysis uncovers the landscape of the bladder tumor transcriptome and reveals insights into luminal and basal subtypes. Cell Rep 9, 1235–1245. [DOI] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, and Dudoit S (2010). Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, et al. (2017). Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busser B, Sancey L, Brambilla E, Coll JL, and Hurbin A (2011). The multiple roles of amphiregulin in human cancer. Biochim Biophys Acta 1816, 119–131. [DOI] [PubMed] [Google Scholar]
- Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. (2016). Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 48, 607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N. (2014). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, and Knight R (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108 Suppl 1, 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SH, Mirabolfathinejad SG, Katta H, Cumpian AM, Gong L, Caetano MS, Moghaddam SJ, and Dong C (2014). T helper 17 cells play a critical pathogenic role in lung cancer. Proc Natl Acad Sci U S A 111, 5664–5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M, Qian L, Shen G, Bian G, Xu T, Xu W, Shen G, and Hu S (2014). Microbiota modulate tumoral immune surveillance in lung through a gammadeltaT17 immune cell-dependent mechanism. Cancer Res 74, 4030–4041. [DOI] [PubMed] [Google Scholar]
- Coffelt SB, Wellenstein MD, and de Visser KE (2016). Neutrophils in cancer: neutral no more. Nat Rev Cancer 16, 431–446. [DOI] [PubMed] [Google Scholar]
- Daley D, Zambirinis CP, Seifert L, Akkad N, Mohan N, Werba G, Barilla R, Torres-Hernandez A, Hundeyin M, Mani VR, et al. (2016). gammadelta T Cells Support Pancreatic Oncogenesis by Restraining alphabeta T Cell Activation. Cell 166, 1485–1499 e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RP, Erb-Downward JR, Martinez FJ, and Huffnagle GB (2016). The Microbiome and the Respiratory Tract. Annu Rev Physiol 78, 481–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, and Jacks T (2009). Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4, 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzutsev A, Badger JH, Perez-Chanona E, Roy S, Salcedo R, Smith CK, and Trinchieri G (2017). Microbes and Cancer. Annu Rev Immunol 35, 199–228. [DOI] [PubMed] [Google Scholar]
- Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, and Flavell RA (2013). Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 13, 759–771. [DOI] [PubMed] [Google Scholar]
- Garrett WS (2015). Cancer and the microbiota. Science 348, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu GY, Osterreicher CH, Hung KE, et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas JD, Ravens S, Duber S, Sandrock I, Oberdorfer L, Kashani E, Chennupati V, Fohse L, Naumann R, Weiss S, et al. (2012). Development of interleukin-17-producing gammadelta T cells is restricted to a functional embryonic wave. Immunity 37, 48–59. [DOI] [PubMed] [Google Scholar]
- Herbst RS, Morgensztern D, and Boshoff C (2018). The biology and management of non-small cell lung cancer. Nature 553, 446–454. [DOI] [PubMed] [Google Scholar]
- Hsu-Kim C, Hoag JB, Cheng GS, and Lund ME (2013). The microbiology of postobstructive pneumonia in lung cancer patients. J Bronchology Interv Pulmonol 20, 266–270. [DOI] [PubMed] [Google Scholar]
- Hyvarinen A, and Oja E (2000). Independent component analysis: algorithms and applications. Neural Netw 13, 411–430. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, and Tuveson DA (2001). Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 15, 3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, and Powrie F (2013). Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med 210, 917–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic AD, Ojesina AI, Pedamallu CS, Jung J, Verhaak RG, Getz G, and Meyerson M (2011). PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nature biotechnology 29, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostric M, Milger K, Krauss-Etschmann S, Engel M, Vestergaard G, Schloter M, and Scholer A (2018). Development of a Stable Lung Microbiome in Healthy Neonatal Mice. Microb Ecol 75, 529–542. [DOI] [PubMed] [Google Scholar]
- Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, and Kendziorski C (2013). EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CM, and Marsland BJ (2017). Lung Homeostasis: Influence of Age, Microbes, and the Immune System. Immunity 46, 549–561. [DOI] [PubMed] [Google Scholar]
- Lu Y, Cao X, Zhang X, and Kovalovsky D (2015). PLZF Controls the Development of Fetal-Derived IL-17+Vgamma6+ gammadelta T Cells. J Immunol 195, 4273–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland BJ, and Gollwitzer ES (2014). Host-microorganism interactions in lung diseases. Nat Rev Immunol 14, 827–835. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucka AK, and Coussens LM (2016). The Basis of Oncoimmunology. Cell 164, 1233–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papotto PH, Ribot JC, and Silva-Santos B (2017). IL-17(+) gammadelta T cells as kick-starters of inflammation. Nat Immunol 18, 604–611. [DOI] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181. [DOI] [PubMed] [Google Scholar]
- Qiao D, Wang Z, Lu Y, Wen X, Li H, and Zhao H (2015). A retrospective study of risk and prognostic factors in relation to lower respiratory tract infection in elderly lung cancer patients. Am J Cancer Res 5, 423–432. [PMC free article] [PubMed] [Google Scholar]
- Ribot JC, deBarros A, Pang DJ, Neves JF, Peperzak V, Roberts SJ, Girardi M, Borst J, Hayday AC, Pennington DJ, et al. (2009). CD27 is a thymic determinant of the balance between interferon-gamma- and interleukin 17-producing gammadelta T cell subsets. Nat Immunol 10, 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickelt S, and Hynes RO (2018). Antibodies and methods for immunohistochemistry of extracellular matrix proteins. Matrix Biol. [DOI] [PubMed] [Google Scholar]
- Ridker PM, MacFadyen JG, Thuren T, Everett BM, Libby P, Glynn RJ, and Group CT (2017). Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet 390, 1833–1842. [DOI] [PubMed] [Google Scholar]
- Roark CL, Aydintug MK, Lewis J, Yin X, Lahn M, Hahn YS, Born WK, Tigelaar RE, and O’Brien RL (2004). Subset-specific, uniform activation among V gamma 6/V delta 1+ gamma delta T cells elicited by inflammation. J Leukoc Biol 75, 68–75. [DOI] [PubMed] [Google Scholar]
- Romero R, Sayin VI, Davidson SM, Bauer MR, Singh SX, LeBoeuf SE, Karakousi TR, Ellis DC, Bhutkar A, Sanchez-Rivera FJ, et al. (2017). Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat Med 23, 1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, and Huttenhower C (2011). Metagenomic biomarker discovery and explanation. Genome biology 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Santos B, Serre K, and Norell H (2015). gammadelta T cells in cancer. Nat Rev Immunol 15, 683–691. [DOI] [PubMed] [Google Scholar]
- Singer M, Wang C, Cong L, Marjanovic ND, Kowalczyk MS, Zhang H, Nyman J, Sakuishi K, Kurtulus S, Gennert D, et al. (2017). A Distinct Gene Module for Dysfunction Uncoupled from Activation in Tumor-Infiltrating T Cells. Cell 171, 1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. P Natl Acad Sci USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, and Berns A (2014). Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A 111, 4952–4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton CE, Lalor SJ, Sweeney CM, Brereton CF, Lavelle EC, and Mills KH (2009). Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity 31, 331–341. [DOI] [PubMed] [Google Scholar]
- Tammela T, Sanchez-Rivera FJ, Cetinbas NM, Wu K, Joshi NS, Helenius K, Park Y, Azimi R, Kerper NR, Wesselhoeft RA, et al. (2017). A Wnt-producing niche drives proliferative potential and progression in lung adenocarcinoma. Nature 545, 355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantourout P, and Hayday A (2013). Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol 13, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Gong G, Sheh A, Muthupalani S, Bryant EM, Puglisi DA, Holcombe H, Conaway EA, Parry NAP, Bakthavatchalu V, et al. (2017). Interleukin-22 drives nitric oxide-dependent DNA damage and dysplasia in a murine model of colitis-associated cancer. Mucosal Immunol 10, 1504–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA, et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4, 2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Y, Srinivas G, Kuenzel S, Linnenbrink M, Alnahas S, Bruce KD, Steinhoff U, Baines JF, and Schaible UE (2014). Environmentally determined differences in the murine lung microbiota and their relation to alveolar architecture. PLoS One 9, e113466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Related to Figure 1.
(A) GF KP mice were exposed to the microbiome via cohousing with SPF mice (ex-GF) or maintained in GF conditions as controls. Tumor burden was analyzed 12 weeks post tumor initiation and representative pictures were shown. n=5 mice/group.
(B) SPF KP mice were left untreated or treated with combined antibiotics (4Abx) in drinking water starting at 2 weeks post tumor initiation. Tumor burden and number were analyzed at 15 weeks p.i. Results are expressed as the mean ± SEM. * p<0.05, *** p<0.001 by Studenťs t test. For each experiment, n= 11–13 mice/group; data represent 2 independent experiments.
(C) Tumor cell lines derived from different primary KP tumors were treated with combined antibiotics or left untreated as controls. Cell number was determined at every passage by counting the Trypan Blue negative cells at indicated time points. Data are expressed as the mean values of triplicates.
Figure S2. Related to Figure 2.
(A) Principal components analysis (PCoA) using Bray-Curtis distance metric (left) and weighted UniFrac metric (right). The lung microbiota community is significantly different from the gut microbiota community (p<0.01 for both Bray-Curtis and weighted UniFrac distance metrics, PERMANOVA).
(B) LEfSe plots showing differentially abundant taxa in the lung microbiome of healthy mice (red) and tumor-bearing mice (blue). Linear discriminant analysis (LDA) scores were calculated using LEfSe, with higher scores indicating greater effect size (significance determined by LDA score > 2.0 and p < 0.05 for Kruskal-Wallis test). Taxonomic categories include p = phyla, c = class, o = order, f = family, and g = genus. Taxa present at ≥ 0.01% total relative abundance and in at least two samples were included.
(C) SPF KP mice were left untreated or treated with metronidazole (1g/L) in drinking water starting 5 weeks post tumor initiation. Tumor burden was quantified 15 weeks post tumor initiation and representative H&E pictures were shown; fecal bacteria burden was determined by 16S based qPCR analysis. n=7–9 mice/group.
(D) LEfSe plots showing the differentially abundant taxa in the lung microbiome of normal lungs (red) and lung adenocarcinoma (LUAD) or lung squamous cell carcinoma (LUSC) samples (green) based on PathSeq analysis of the TCGA cohort.
Figure S3. Related to Figure 3.
(A) The frequency of γδ T cells in total CD3+ lymphocytes in the lung, blood, spleen or draining lymph node from SPF and GF KP mice as determined by flow cytometry.
(B) Representative pictures and quantification of immunohistochemistry staining of human TCRδ on formalin-fixed paraffin-embedded normal lung (NL) and lung adenocarcinoma (LUAD) tissue samples. Positively stained cells are in purple.
(C, D) RORγt and IL-17A expression in total CD3+ lymphocytes from the tumor-bearing lungs from SPF mice and GF mice. Representative flow cytometric plots are shown (C) and the frequency of IL-17A+ CD4 T cells (Th17) is quantified (D).
Results are expressed as the mean ± SEM. ** p<0.01, *** p<0.001 by Studenťs t test. For each experiment, n= 8–15 mice/group; data represent 3 independent experiments.
Figure S4. Related to Figure 4.
(A) KP mice on the CD45.1 background were lethally irradiated and transplanted with bone marrow from CD45.2 donors. Seven weeks after reconstitution, mice were infected with adenovirus expressing Sftpc-Cre, and 15 weeks after tumor initiation, γδ T cells in the tumor-bearing lungs were analyzed by flow cytometry. The percentage of donor vs. recipient derived cells was quantified in the Vγ4+ and Vγ6+ subsets, as well as the RORγt+ and Tbet+ compartments. Representative plots are shown and data represent 15 mice.
(B) The proliferation of RORγt- γδ T cells and Th17 cells in the tumor-bearing lungs from tumor-bearing SPF mice and GF mice was assessed by flow cytometric analysis of Ki67 expression.
(C) IL-17A expression in lung-infiltrating γδ T cells from healthy SPF mice, tumor-bearing SPF mice and tumor-bearing GF mice was analyzed by flow cytometry.
(D) SPF KP mice were treated with combined antibiotics (4Abx) starting 6.5 weeks after tumor initiation. The frequency of IL-17A-producing γδ T cells and IL-17A concentration in BALF were analyzed by flow cytometry and ELISA respectively.
(E) The abundance of γδ T cells, and the expression of RORγt and IL-17A in γδ T cells were analyzed by flow cytometry in GF mice and ex-GF mice that were exposed to the microbiome via cohousing with SPF mice.
(B-E) Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by Studenťs t test. For each experiment, n= 5–13 mice/group.
Figure S5. Related to Figure 5.
The expression of Tbet, IFNγ and TNFα in the tumor-bearing lungs from SPF and GF mice were analyzed by flow cytometry.
Results are expressed as the mean ± SEM. *p<0.05, *** p<0.001 by Studenťs t test. n= 3–5 mice/group. Data represent 3 independent experiments.
Figure S6. Related to Figure 6.
(A) Flow cytometry analysis of different types of myeloid cells in the tumor-bearing lungs from SPF and GF mice. The frequencies of each cell type in total tissue-resident immune cells are quantified. Alveolar macrophages: CD11c+, CD11b-intermediate, autofluorescence-high; monocytes: Ly6C+, CD11b-high or intermediate; DC: CD11c+, MHCII+, CD11b+ or CD103+.
(B-E) KP mice were treated with the monoclonal antibody UC7–13D5 staring at 4 weeks post tumor initiation. (B, C) Blockade of γδT cells was verified by the loss of cells that were stained positively with a different monoclonal antibody against γδ-TCR (GL3), as well as the reduction of TCRγ- CD3+ lymphocytes. (D, E) Flow cytometry analysis of Ki67 and IFNγ expression in CD4 and CD8 T cells in the tumor-bearing lungs from mice treated with UC7–13D5 or isotype control.
(F-J) KP mice were treated with the monoclonal antibody GL3 starting at 4 weeks post tumor initiation. Tumor burden was quantified at 15 weeks p.i. and representative H&E pictures are shown.
(G, H) Blockade of γδ T cells was verified by the loss of cells that were stained positively with a different monoclonal antibody against γδ-TCR (UC7–13D5), as well as the reduction of TCRβ- CD3+ lymphocytes. (I, J) Flow cytometry analysis of Ki67 and IFNγ expression in CD4 and CD8 T cells in the tumor-bearing lungs from mice treated with GL3 or isotype control.
(K) IFNγ and/or TNFα expression in CD4 and CD8 T cells from the tumor-bearing lungs were analyzed by flow cytometry and compared between SPF and GF mice.
Results are expressed as the mean ± SEM. *p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001 by Studenťs t test. n= 6–12 mice/group.
Figure S7. Related to Figure 7.
(A) LUAD patients in the TCGA cohort were scored for their gene expression correlation with the KP-lung γδ T cell signature as well as the ESTIMATE immune infiltration signature. Pearson correlation analysis of the scores from the two signatures for individual patients does not suggest a significant correlation (r= 0.15, n=515 primary tumors).
(B) RT-qPCR analysis of the candidate genes in FACS-purified γδ T cells from tumor-bearing lungs and spleens of KP mice.
(C) Flow cytometric analysis of IL-17A and IL-17F in γδ T cells. Plots representative of 8 mice.
(D) Flow cytometric analysis of amphiregulin in γδ T cells. Plots representative of 8 mice.
(E) Tumor cell lines derived from different primary KP tumors were treated with 1ng/ml recombinant mouse IL-22 for 72 hours or left untreated as controls; cell number was determined by counting the Trypan Blue negative cells.
(F) Empirical cumulative distribution function (CDF) plots showing expression of IL22RA1 in human LUAD samples, the matched normal tissues from the same patients, as well as other adenocarcinoma samples in the TCGA cohort (p values shown by Kolmogorov-Smirnov test).
(G) Tumor cell lines derived from different primary KP tumors were treated with the indicated dose of recombinant amphiregulin for 72 hours; cell number was determined by the CellTiter-Glo assay.
Data Availability Statement
The RNA-seq data for γδ T cells from tumor-bearing lungs or spleens of KP mice have been deposited in the GEO repository under the ID code GSE114340.
The 16S rDNA sequencing data have been deposited to the SRA under the ID code PRJNA492954.