ABSTRACT
HOXA11, which is a member of the homeobox (HOX) gene family, and its natural antisense transcript (NAT) HOXA11-AS have been reported to be closely related to the development of lung cancer. We aimed to investigate their specific roles in cisplatin (DDP) resistance in lung adenocarcinoma (LUAD). First, we found that HOXA11 is hypermethylated and significantly downregulated in a DDP-resistant A549 cell line (A549/DDP) and LUAD tissues, while the HOXA11-AS expression level is elevated. Although HOXA11 and HOXA11-AS mRNA overlap in the 5ʹ-untranslated region (5ʹ UTR) and share two CpG islands, DNA methylation only regulates the expression of HOXA11. Then, we found that HOXA11 and HOXA11-AS have an inverse interaction by transfecting their siRNAs and overexpression vectors into A549 and A549/DDP cells. A dual-luciferase reporter assay further confirmed that the overlapping 5ʹUTR is essential for the bidirectional regulation between HOXA11 and HOXA11-AS. Functional analysis showed that knockdown of HOXA11 expression in A549 cells induced DDP resistance and activated Akt/β-catenin signaling, while overexpression of HOXA11 in A549/DDP cells increased DDP sensitivity and inhibited Akt/β-catenin signaling. Moreover, HOXA11-AS knockdown in A549 cells increased DDP sensitivity and inhibited Akt/β-catenin signaling, while the overexpression of HOXA11-AS in A549/DDP cells induced DDP resistance and activated Akt/β-catenin signaling. In conclusion, our study demonstrates that the inverse interaction between HOXA11 and HOXA11-AS promotes DDP resistance in LUAD.
KEYWORDS: Lung adenocarcinoma, resistance, methylation, HOXA11, HOXA11-AS
Introduction
Lung cancer leads to the most cancer-related deaths worldwide, and non-small cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancer cases [1]. Lung adenocarcinoma (LUAD) has been the fastest growing subtype of NSCLC in recent years. Platinum-based doublets are widely used as a first-line NSCLC treatment and improve survival rates compared to placebo treatments [2]. Although many innovative drugs, including epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitors (TKIs) and checkpoint inhibitors, have made substantial progress in the treatment of LUAD, chemotherapy remains the standard treatment, and the combination of chemotherapy and molecular targeted therapy or immunotherapy has become a new therapeutic strategy [3,4]. Cisplatin (DDP), which disrupts the structure and function of DNA, is the most commonly used platinum agent for lung cancer [5]. However, resistance to DDP eventually develops and then induces recurrence, invasion and therapeutic failure [6]. Several cellular events involved in chemoresistance have been revealed, such as DNA methylation, noncoding RNA, stem cells and autophagy [7–9], but these events are not yet clearly defined. Therefore, an improved understanding of the molecular mechanism of DDP resistance is required for the advancement of LUAD treatment.
Our previous studies have demonstrated a panel of candidate genes that are downregulated by DNA methylation-induced DDP resistance in NSCLC using high-throughput microarrays [10,11]. We also found that HOXA11, a member of the homeobox (HOX) gene family, is hypermethylated and significantly downregulated in the resistant A549/DDP cell line, while in the parental A549 cells, HOXA11 is hypomethylated and overexpressed. Many other studies have confirmed that HOXA11 expression is downregulated in human gastric cancer [12], renal cell carcinoma [13], glioblastoma [14] and LUAD [15]. Furthermore, hypermethylation may cause HOXA11 inactivation and contribute to the progression of NSCLC by promoting cell proliferation or migration, suggesting the tumor-suppressive function of HOXA11 [16].
Interestingly, HOXA11-AS, which is the natural antisense transcript (NAT) of HOXA11, has been reported to be upregulated in various types of carcinomas and is generally associated with increased lymph node metastasis, advanced tumor stage, poor tumor differentiation, and poor prognosis [17,18]. In vitro and in vivo assays have revealed that HOXA11-AS acts as an oncogenic long noncoding RNA (lncRNA) that promotes cell growth and metastasis by recruiting multiple chromosome-modifying enzymes to target genes or by sponging specific microRNAs (miRNA) [19,20]. A latest study also found that HOXA11-AS promotes cisplatin resistance in human LUAD cells by modulating miR-454-3p/Stat3 [21].
Because HOXA11 and HOXA11-AS have a complementary overlap in the 5ʹ-untranslated region(5ʹ UTR) in a head-to-head (5ʹ-5ʹ) manner and share two CpG islands that are sites of hypermethylation, we aimed to determine the interactions between HOXA11 and HOXA11-AS. Thus, in the present study, we explored the roles of HOXA11 and HOXA11-AS in DDP resistance in LUAD.
Results
The expression status of HOXA11 and HOXA11-AS in DDP-resistant LUAD cell lines and tissues
Information from the National Center for Biotechnology Information gene database (http://www.ncbi.nlm.nih.gov/gene) shows that HOXA11 and HOXA11-AS mRNA overlap at the 5ʹUTR in a head-to-head manner (Figure 1) and that HOXA11 and HOXA11-AS mRNA share two CpG islands (CpG 1, chr7:27225050–27225629; CpG 2, chr7:27224267–27224596).
Figure 1.
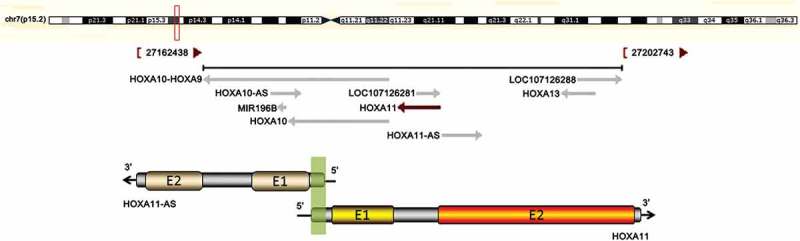
The correlation between HOXA11 and HOXA11-AS mRNA. The upper chart in this panel shows the genomic locus of HOXA11 indicated on the UCSC site. The lower chart in this panel is a schematic of HOXA11-AS and HOXA11 mRNA. ‘E’ indicates exons. The green shadow indicates the overlapping region of HOXA11-AS and HOXA11 mRNA. The black arrows show the direction of transcription.
Real-time PCR was used to analyze the expression of HOXA11 and HOXA11-AS, and qMSP and BSP were used to evaluate their methylation status. The results showed that HOXA11-AS expression was upregulated and that HOXA11 expression was downregulated in the DDP-resistant A549/DDP cell line, with hypermethylation of CpG 1 and CpG 2 compared to the parental A549 cell line. After 5-aza-2ʹ-deoxycytidine (5-aza-CdR) treatment (1 μM), HOXA11 expression was restored in A549/DDP cells, and the methylation statuses of CpG 1 and CpG 2 were reversed, while the expression of HOXA11-AS was not affected (Figure 2(a–c)). Thus, HOXA11 expression is mainly regulated by DNA methylation, whereas its antisense RNA HOXA11-AS is regulated by other mechanisms.
Figure 2.

The expression status of HOXA11-AS and HOXA11 in cisplatin (DDP)-resistant lung adenocarcinoma cell lines and tissues. Real-time PCR (a) was used to analyze the expression of HOXA11-AS and HOXA11. qMSP (b) and BSP (c) were used to evaluate their methylation statuses. HOXA11-AS expression was upregulated and HOXA11 expression was downregulated in the A549/DDP cell line, with hypermethylation of CpG 1 and CpG 2. Treatment with 5-aza-CdR (1 μM) restored HOXA11 expression and reversed the hypermethylation of the CpG islands. The HOXA11-AS and HOXA11 expression status (d) and methylation (e) were also analyzed in primary tumor cells; 20 LUAD samples were considered DDP-sensitive samples (IC50 < 5 mg/L), and 20 samples were considered DDP-resistant samples (IC50 > 10 mg/L). PMR, percentage of methylation reference.
Using primary tumor cell culture and drug susceptibility testing, 20 LUAD samples were considered DDP-sensitive samples (IC50 < 5 μg/mL), and 20 samples were considered DDP-resistant samples (IC50 > 10 μg/mL). The results showed that the expression of HOXA11-AS was upregulated and that the expression of HOXA11 was downregulated in the DDP-resistant tissues (Figure 2(d)), with a higher percentage of methylation reference (PMR) of CpG 1 and CpG 2 (Figure 2(e)) compared to sensitive tissues.
The inverse interaction between HOXA11 and HOXA11-AS through the overlapping 5ʹUTR
The common function of antisense transcripts is the regulation of the expression of sense transcripts. To investigate the relationship between HOXA11 and HOXA11-AS, specific siRNAs that target their nonoverlapping regions were transfected into the A549 cell line, and HOXA11 and HOXA11-AS overexpression vectors were transfected into A549/DDP cells. The results showed that the knockdown of HOXA11 and HOXA11-AS mRNA expression by siRNA increased the expression levels of their respective counterparts in A549 cells, while the overexpression of HOXA11 an HOXA11-AS mRNA led to a significant decrease in the expression levels of their respective counterparts in A549/DDP cells (Figure 3(a,b)). siRNA targeting HOXA11-AS also increased the protein expression level of HOXA11 in A549 cells, and the overexpression of HOXA11-AS reduced the protein expression level of HOXA11 in A549/DDP cells (Figure 3(c)). Furthermore, the mRNA expression levels of HOXA11 and HOXA11-AS were negatively correlated in primary cultured tumor cells, regardless of DDP sensitivity or resistance (Figure 3(d)). Finally, we performed a dual-luciferase reporter assay to investigate the role of the overlapping 5ʹUTR, and the result showed that HOXA11-AS overexpression decreased the 5ʹUTR luciferase activity of HOXA11 in A549 cells, while the mutant overlapping sequence abrogated this effect (Figure 3(e)). Thus, the overlapping 5ʹUTR is essential for the bidirectional regulation between HOXA11 and HOXA11-AS.
Figure 3.
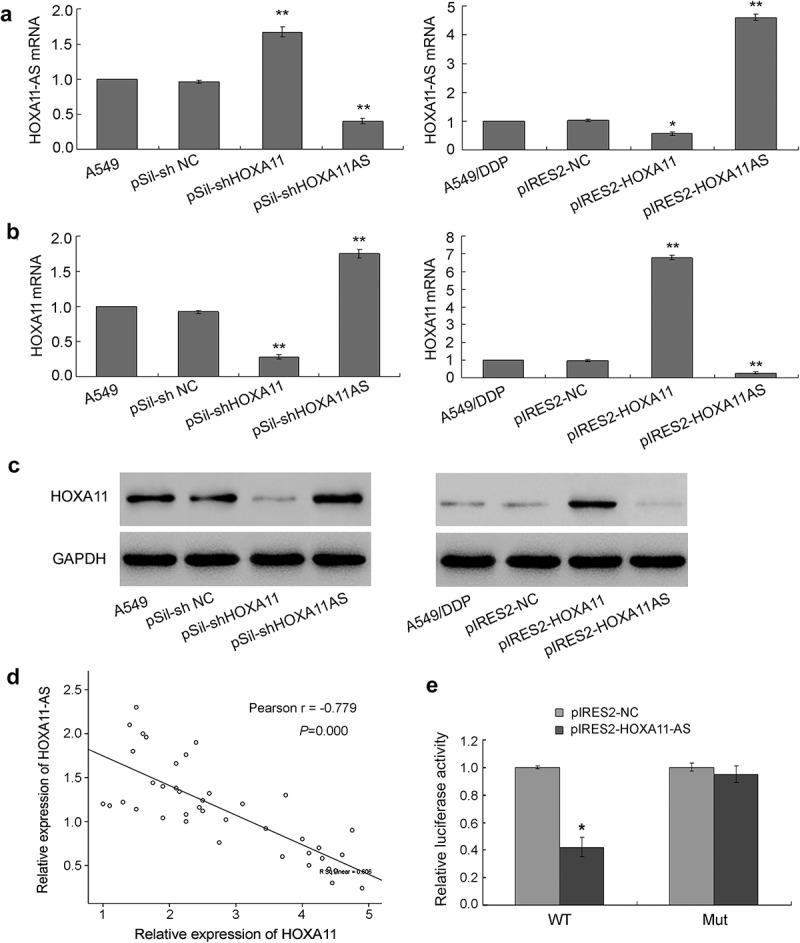
The inverse interaction between HOXA11 and HOXA11-AS. (a) The effects of HOXA11 and HOXA11-AS siRNA on the mRNA expression of HOXA11-AS in A549 cells. The effects of the HOXA11 and HOXA11-AS overexpression vectors on the mRNA expression of HOXA11-AS in A549/DDP cells. (b) The effects of HOXA11 and HOXA11-AS siRNA on the mRNA expression of HOXA11 in A549 cells. The effects of the HOXA11 and HOXA11-AS overexpression vectors on the mRNA expression of HOXA11 in A549/DDP cells. (c) The effects of HOXA11 or HOXA11-AS siRNA on the protein expression of HOXA11 in A549 cells. The effects of the HOXA11 and HOXA11-AS overexpression vectors on the protein expression of HOXA11 in A549/DDP cells. (d) The mRNA expression levels of HOXA11 and HOXA11-AS were negatively correlated in primary cultured tumor cells regardless of cisplatin sensitivity or resistance. (e) The luciferase activity in A549 cells cotransfected with luciferase reporters containing wild-type (WT) or mutant (mut) 5ʹUTR overlapping sequence and pIRES2-HOXA11-AS or negative control. The data are presented as the relative ratio of firefly luciferase activity to Renilla luciferase activity. * P < 0.05 vs. NC; **P < 0.01 vs. NC.
HOXA11 and HOXA11-AS function in DDP resistance in vitro
To investigate the function of HOXA11 and HOXA11-AS in DDP resistance, their specific siRNAs were transfected or cotransfected into A549 cells, and their overexpression vectors were transfected or cotransfected into A549/DDP cells. Then, a CCK-8 assay, flow cytometry and TUNEL assay were performed. The knockdown of HOXA11 expression caused cell proliferation to increase significantly (Figure 4(a)), and the IC50 values of DDP increased in A549 cells (Figure 4(b)). Additionally, the G1 phase of the cell cycle was decreased and the S phase was increased, but cell apoptosis did not decrease significantly (Figure 4(c)). Conversely, the knockdown of HOXA11-AS expression caused cell proliferation to be significantly inhibited (Figure 4(a)), and the IC50 values of DDP decreased (Figure 4(b)) in A549 cells; besides, the G1 phase of the cell cycle and cell apoptosis were significantly increased (Figure 4(c)). When the specific siRNAs of HOXA11 and HOXA11-AS were cotransfected into A549 cells, their respective effects on cell proliferation, cell cycle and apoptosis were weakened (Figure 4(a–c)), indicating that HOXA11 and HOXA11-AS biologically antagonize each other.
Figure 4.

The functions of HOXA11 and HOXA11-AS in cisplatin (DDP) resistance in vitro. HOXA11- and HOXA11-AS-specific siRNAs were transfected or cotransfected into A549 cells, and HOXA11 and HOXA11-AS overexpression vectors were transfected or cotransfected into A549/DDP cells. Using a CCK-8 assay, cell proliferation was analyzed (a), and the IC50 values of DDP were calculated (b). Flow cytometry and TUNEL assays revealed the cell cycle and apoptosis in A549 (c) and A549/DDP cells (d).
Similarly, the overexpression of HOXA11 in A549/DDP cells resulted in a significant inhibition of cell proliferation (Figure 4(a)), decrease of the IC50 values of DDP (Figure 4(b)), and increase of cell cycle G1 phase and cell apoptosis (Figure 4(d)). However, the overexpression of HOXA11-AS caused cell proliferation and the IC50 values of DDP to increase significantly in A549/DDP cells (Figure 4(a,b)). Additionally, the S phase of the cell cycle was increased, but cell apoptosis was not significantly affected (Figure 4(d)). When the overexpression vectors of HOXA11 and HOXA11-AS were cotransfected into A549/DDP cells, their respective effects on cell proliferation, cell cycle and apoptosis were weakened (Figure 4(a,b,d)).
Furthermore, the knockdown of HOXA11 expression in A549 cells caused the downregulation of p21 (negative regulatory protein of cell cycle), bax, cleaved caspase 3 (apoptotic protein), and VEGF (vascular endothelial growth factor) expression and the upregulation of cyclin D1 (positive regulatory protein of cell cycle), bcl-2 (antiapoptotic protein), β-catenin and p-AKT (activated signaling pathway) expression, while the knockdown of HOXA11-AS expression caused the opposite effects in A549 cells. The overexpression of HOXA11 in A549/DDP cells caused the upregulation of p21, bax, cleaved caspase 3, and VEGF expression and the downregulation of cyclin D1, bcl-2, β-catenin and p-AKT expression, while the overexpression of HOXA11-AS caused the opposite effects in A549/DDP cells (Figure 5).
Figure 5.
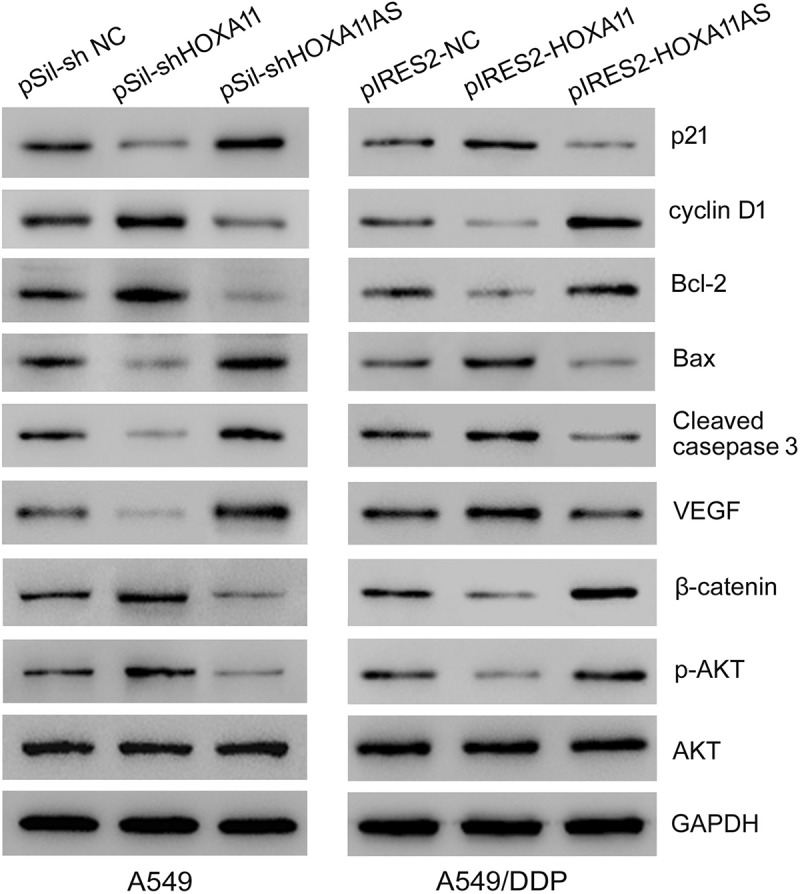
The effects of HOXA11 and HOXA11-AS on cellular function-related proteins. Western blot analysis of the protein expression levels in A549 cells with HOXA11 or HOXA11-AS knockdown and the protein expression levels in A549/DDP cells with HOXA11 or HOXA11-AS overexpression.
HOXA11 and HOXA11-AS function in DDP resistance in vivo
Mice were separated into eight groups (five mice per group) according to the inoculated cells. In mice inoculated with A549 cells, the knockdown of HOXA11 expression caused cell proliferation to increase significantly, while the knockdown of HOXA11-AS expression inhibited cell proliferation, and A549 cells cotransfected with siRNAs of HOXA11 and HOXA11-AS attenuated their respective effects (Figure 6(a,b)). In mice inoculated with A549/DDP cells, the overexpression of HOXA11 significantly inhibited cell proliferation, while the overexpression of HOXA11-AS increased cell proliferation, and A549/DDP cells cotransfected with vectors of HOXA11 and HOXA11-AS attenuated their respective effects (Figure 6(a,c)). These results further indicated the tumor-suppressive role of the HOXA11 gene and the oncogenic role of HOXA11-AS in DDP resistance of LUAD.
Figure 6.
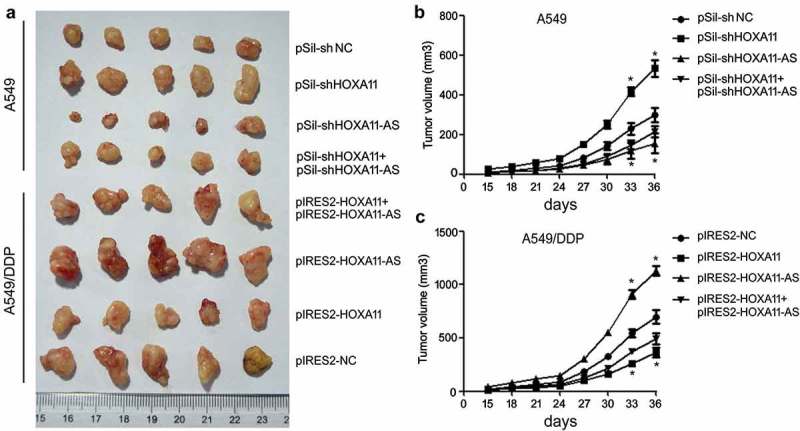
The functions of HOXA11 and HOXA11-AS in cisplatin (DDP) resistance in vivo. Cells (2 × 106 cells/100 μL PBS) were subcutaneously inoculated into the right flank of BALB/c nu/nu mice, and the animals were randomly separated into eight groups (five per group) according to the inoculated cells. The mice were sacrificed, and the tumors were isolated after 36 days (a). The tumor size was monitored every 3 days after A549 (b) or A549/DDP (c) cell implantation.
Discussion
In the present study, we first found that HOXA11 is hypermethylated and significantly downregulated in the DDP-resistant A549 cell line and LUAD tissues and that the 5-aza-CdR treatment restores HOXA11 expression and reverses the hypermethylation of CpG islands. HOX genes encode transcription factors that play crucial roles in a wide range of processes, including apoptosis, differentiation, motility, and angiogenesis. In humans, HOX genes are arranged into four clusters (A, B, C, and D) on different chromosomes, and 39 HOX genes have been identified [22]. HOX genes are also known to play an essential role in lung development and are expressed in the normal human adult lungs [23]. The HOXA cluster, which is located on chromosome 7p15–7p14.2, consists of 12 genes, including HOXA11 [24]. Highly dense CpG islands are prevalent in most HOXA promoters, and HOXA11 hypermethylation has recently been reported in breast [25], gastric [12,26], ovarian [27] and cervical cancers [28] and in renal cell carcinoma [13]. The methylation of HOXA11 has also been proposed as an early detection marker and as a poor prognostic marker in certain types of cancers [25,27,28], and functional studies have shown that HOXA11 acts as a tumor suppressor gene [12,13]. In lung cancer, HOXA11 has been verified as one of the most frequently hypermethylated genes using high-throughput DNA methylation arrays [29,30]. Furthermore, Hwang et al [6]. revealed that HOXA11 hypermethylation occurred in 218 (69%) of 317 primary NSCLC cases, 5-Aza-dC treatment restores the expression of HOXA11 in lung cancer cell lines, and transient transfection of HOXA11 into H23 cells results in the inhibition of cell proliferation and migration. Li et al [5]. also found aberrant hypermethylation, and the methylation-induced downregulation of HOXA11 might be a diagnostic and prognostic marker in patients with LUAD.
However, a meta-analysis of data gathered from The Cancer Genome Atlas (TCGA) and Oncomine microarrays of 934 LUAD and 319 normal control tissues (the normal controls included tissues from healthy individuals combined with adjacent tumor samples and only 32 pairs of LUAD tissues and adjacent nontumorous tissues) revealed that HOXA11 is significantly overexpressed in LUAD tissues and that the expression level of HOXA11 is significantly higher in patients with lymphoid metastases and an advanced clinical stage, indicating the potential oncogenic role of HOXA11 in the genesis of LUAD [31]. Similar results have also been reported in lung squamous cell carcinoma (LUSC) by bioinformatics analysis [32]. Thus, the molecular mechanisms of HOXA11 in lung cancer pathogenesis mostly remain elusive and require further investigation.
Investigations of HOXA11 in DDP-resistant lung cancer are even more deficient. Se et al [1]. have indicated that a reduction in the expression level HOXA11 induces decreased anticancer effects of radiotherapy and/or temozolomide and leads to a poor prognosis in glioblastoma. In a phase II clinical trial, the demethylation reagent decitabine was shown to alter the DNA methylation of HOXA11, restoring sensitivity to carboplatin in patients with platinum-resistant ovarian cancer [33]. Consistent with these results, our research confirmed that the knockdown of HOXA11 expression in A549 cells increases cell proliferation, decreases the G1 phase of the cell cycles, increases the S phase of the cell cycle, induces DDP resistance in vitro and in vivo. Western blot analysis showed activation of the Akt and β-catenin signaling proteins, accompanied by corresponding changes in the cell cycle and apoptosis-related proteins. Conversely, the overexpression of HOXA11 in A549/DDP cells was shown to inhibit cell proliferation, induce G1 phase cell cycle arrest and apoptosis, increase DDP sensitivity in vitro and in vivo, and inhibit the Akt and β-catenin signaling proteins, indicating that HOXA11 acts as a tumor suppressor gene against DDP-resistant human LUAD cells likely by inhibiting the Akt/β-catenin signaling pathway.
Furthermore, we found that HOXA11-AS, the NAT of HOXA11, was upregulated in the DDP-resistant A549 cell line and LUAD tissues. Functional analysis showed that the knockdown of HOXA11-AS expression in A549 cells inhibits cell proliferation, induces G1 phase cell cycle arrest and apoptosis, increases DDP sensitivity in vitro and in vivo, and inhibits the Akt and β-catenin signaling proteins and that the overexpression of HOXA11-AS in A549/DDP cells caused the opposite effects, indicating that HOXA11-AS acts as an oncogene to promote DDP resistance in human LUAD cells likely by activating the Akt/β-catenin signaling pathway. Growing evidences have demonstrated that HOXA11-AS lncRNA plays key roles in the development and progression of cancers [34–36]. Recently, a total of eight eligible studies consisting of 1320 patients with cancer were subjected to a meta-analysis, and the results revealed that an increased expression level of HOXA11-AS is significantly associated with increased lymph node metastasis, advanced tumor stage, poor tumor differentiation, and shorter overall survival (OS) and progression-free survival (PFS) [18]. Bioinformatics analyses have further indicated that HOXA11-AS is significantly overexpressed in both LUAD and LUSC tissues based on the TCGA database and have predicted that HOXA11-AS functions by regulating various pathways and genes [37]. Moreover, the knockdown of HOXA11-AS expression inhibits the proliferation, migration, invasion, and tumorigenic and angiogenic abilities of NSCLC cell lines and induces apoptosis and cell cycle arrest at the G0/G1 or G2/M phase [20]. Chen et al [8,38]. also found that HOXA11-AS expression is significantly upregulated in NSCLC tissues compared with adjacent normal tissues and that higher HOXA11-AS expression levels are associated with a poor prognosis in patients with NSCLC. The mechanistic findings showed that HOXA11-AS recruits the histone methyltransferase enhancer of zeste homologue 2 (EZH2) and DNA methyltransferase 1 (DNMT1) to the miR-200b promoter regions to repress miR-200b expression in NSCLC cells, promoting the epithelial-mesenchymal transition (EMT) in NSCLC cells. A recent study demonstrated that HOXA11-AS acts as a competing endogenous RNA (ceRNA) to promote DDP resistance in human LUAD cells via the miR-454-3p/Stat3 axis through the knockdown of HOXA11-AS expression in DDP-resistant A549 and H157 cell lines [21]. These results are all consistent with the results from our present study.
NATs, which are also known as natural antisense RNA, are endogenous transcripts containing sequences that are complementary to their sense RNAs. They encode either mRNA or noncoding RNA and are mainly divided into the following two types: cis-NATs and trans-NATs. Cis-NATs are further classified according to their relative orientation and degree of overlap as follow: head-to-head (5′ to 5′), tail-to-tail (3′ to 3′) and fully overlapping [39]. Recently, NATs have been found to play important roles in the development of cancers by directly interacting with sense transcripts. For example, KRT7-AS promotes gastric cancer cell proliferation and migration by forming an RNA-RNA duplex to stabilize KRT7 mRNA [40]. ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1 [41], while FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression level of FOXC2 [42].
HOXA11-AS and its sense RNA HOXA11 overlap in the 5ʹ UTR region in a head‐to‐head manner and share two CpG islands. However, in our study, the expression statuses of HOXA11 and HOXA11-AS in LUAD cells were completely opposite, and their regulation mechanisms were different. HOXA11 expression is mainly regulated by DNA methylation, whereas HOXA11-AS is not affected by methylation. Additionally, their biological functions in LUAD cells were antagonistic, and cotransfection of HOXA11- and HOXA11-AS-specific siRNAs into A549 cells or cotransfection of HOXA11 and HOXA11-AS overexpression vectors into A549/DDP cells weaken their individual roles. The dual-luciferase reporter assay confirmed that HOXA11 and HOXA11-AS were inversely and bidirectionally regulated via the overlapping 5ʹUTR. However, the specific interaction mechanisms between HOXA11 and HOXA11-AS require further investigation.
Overall, our study demonstrates that the inverse interaction between HOXA11 and HOXA11-AS promotes DDP resistance, providing new insight into the mechanisms underlying HOXA11-AS lncRNA through overlapping regions with sense transcripts and contributing to developments in LUAD treatment because increasing the sensitivity to DDP has been considered as a potentially effective strategy to overcome cancer.
Materials and methods
Cell culture
The A549 human LUAD cell line was purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Cell Resource Center. The construction and culture of the DDP-resistant A549/DDP cell line were based on our previous study [8]. For treatment with 5-aza-CdR (Sigma Aldrich, St. Louis, MO), which is a specific DNA methyltransferase inhibitor, the cell culture medium was changed every 24 h. Primary LUAD cell isolation from fresh tumors, culture and identification of DDP sensitivity have been described previously [8].
Real-time quantitative PCR
Total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA). First-strand cDNA was synthesized using 2 μg of total RNA with a reverse transcription kit (TaKaRa, Dalian, China). For amplification, cDNA was initially denatured at 95°C for 20 s, followed by 40 cycles at 95°C for 5 s, and annealing at 60°C for 30 s in an ABI 7300 thermocycler (Applied Biosystems, Foster City, CA) using Power SYBR Green (TaKaRa). The specific primer sequences for each gene are shown in Table 1. The relative expression levels of the genes were calculated by the 2-ΔΔCt method.
Table 1.
List of primers.
| Gene | Forward(5ʹ-3ʹ) | Reverse (5ʹ-3ʹ) | Product (bp) |
|---|---|---|---|
| HOXA11 | ggaaatgctcagaggcagtc | attttccttgtgcccagttg | 209 |
| HOXA11-AS | tctcctggagtctcgcattt | tcggaagtgaccatgaatga | 197 |
| CpG1 (MSP) | gggaagcgtttttgttttc | tcaaatcaccgtacaaatcg | 106 |
| CpG1 (USP) | tttgggaagtgtttttgttttt | tcaaatcaccatacaaatcaaac | 106 |
| CpG1 (BSP) | gtgtaatttatgttggttgggg | aacaaaccaccaaacaaacac | 387 |
| CpG2 (MSP) | gggtgtcgtaggttgtttc | cgacgtacctaacgacgtac | 152 |
| CpG2 (USP) | ggggtgttgtaggttgtttt | ccaacatacctaacaacatac | 152 |
| CpG2 (BSP) | gggtagtttttaatatttgggg | cctccttctacccaactacata | 364 |
Analysis of the methylation status
After genomic DNA extraction and spectrophotometric quantification, 1 μg of genomic DNA was treated with bisulfite using an EZ-DNA Methylation Gold Kit (Zymo Research, Orange, CA). Quantitative methylation-specific PCR (qMSP) was then performed in an ABI 7300 thermocycler using a SYBR Premix Ex Taq kit (TaKaRa). The specific primer sequences for each gene were designed by Methyl Primer Express® Software v1.0 and are shown in Table 1. β-actin (ACTB) was used to normalize the expression levels of the input DNA. The amount of methylated DNA was determined by the threshold cycle number (Ct) of each sample and presented as the percentage of methylation reference (PMR). For bisulfite sequencing (BSP), the bisulfite-treated DNA was amplified by PCR with BSP primers, and the PCR products were cloned into a pUC57 vector (GenScript, Nanjing, China). Five clones were selected and sequenced for each sample.
Transient transfection
The gene-expressing plasmids (pIRES2-HOXA11 and pIRES2-HOXA11AS) and gene-interfering plasmids (pSIL-HOXA11 and pSIL-HOXA11AS) were all constructed by GenePharma Corp (Suzhou, China). pIRES2-EGFP and pSIL-eGFP control vectors were purchased from Addgene (Cambridge, MA, USA). These recombinant vectors were transfected into cells using Lipofectamine 3000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. After 48 h, the cells were collected for subsequent experiments.
Western blotting
Cell protein lysates were separated by 10% sodium salt (SDS)-Polyacrylamide gel electrophoresis (PAGE) and blotted onto a polyvinylidene fluoride (PVDF) membrane (Roche Diagnostics, Mannheim, Germany). After soaking the membrane in 10 mL of 5% nonfat milk in Tris-buffered saline with tween 20 (TBST) solution for 1 h, the membrane was incubated with primary antibodies specific to HOXA11, p21, cyclin D1, bcl-2, bax, cleaved caspase 3, VEGF, β-catenin, p-AKT, AKT and GAPDH (Univ-bio Inc., Shanghai, China). Horseradish peroxidase-conjugated goat anti-rabbit IgG was used as a secondary antibody. The results were observed following a treatment with an enhanced chemiluminescent (ECL) substrate (Merck Millipore, Hong Kong, China).
Dual-luciferase reporter assay
Luciferase reporter gene vectors were constructed with HOXA11 mRNA 5ʹUTR sequences and mutant 5ʹUTR sequences in pGL3 plasmids (GenePharma). The HOXA11-AS1 sequence was subcloned into a pcDNA3.1 vector (GenePharma), and an empty pcDNA3.1 vector was used as a control. A549 cells were cotransfected with the wild-type (or mutant-type) pGL3 plasmid and pcDNA3.1 plasmid containing the HOXA11-AS1 sequence (or the control pcDNA3.1 plasmid) by using Lipofectamine 3000 (Invitrogen). Luciferase activity was measured with a Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI) after 48 h and presented as the ratio between firefly and Renilla luciferase activity (Fluc/Rluc).
Cell viability and proliferation analysis
Cell viability was assessed using a cell counting kit-8 (CCK-8) assay. Briefly, cells were seeded into 96-well plates at an initial density of 2 × 103 cells/well for 1–3 days. Then, 90 µl of fresh serum-free medium and 10 µl of CCK-8 reagent (Beyotime, Shanghai, China) were added to each well after decanting the old medium, and the culture was continued at 37°C for 1 h. The optical density was determined by scanning with a microplate reader (Promega) at a wavelength of 450 nm. IC50 values were calculated by a DDP concentration-response curve (concentration gradient: 0, 2, 5, 10 and 20 μg/mL for a 48-h treatment period) using GraphPad Prism 5.0 (GraphPad Software, La Jolla, CA, USA).
Flow cytometric analysis
Cells were harvested directly or 48 h after transfection and washed with ice-cold phosphate-buffered saline (PBS). The PI/RNase staining kits (Multisciences Biotech, Hangzhou, China) and annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kits (Keygene Biotech, Nanjing, China) were used to detect cell cycle and apoptosis in a FACScan instrument (Becton Dickinson, Mountain View, CA), respectively.
In situ dUTP nick end labeling (TUNEL) assay
Apoptosis was analyzed in situ using a TUNEL assay mediated by oligonucleotide-end deoxyribonucleotidyl transferase (TdT) (Beyotime) based on the manufacturer’s instructions. Slides were evaluated under a light microscope. Nuclei stained with a blue dye were labeled as ‘−’, indicating normal cells, and nuclei that were stained green were labeled as ‘+’, indicating apoptotic cells. Cells in five fields of view were randomly counted to calculate the apoptotic rate.
In vivo xenograft model
Six-week-old male BALB/c nude mice were purchased from the Laboratory Animal Center of Nanjing Medical University and maintained under pathogen-free conditions. Tumor xenografts were established by a subcutaneous injection of 0.1 mL of mock-transfected or transfected cell suspension (2 × 106 cells/mL) into nude mice on the right side of the posterior flank (n = 5 mice per group). Tumor growth was examined every other day. After 5-7 days, the tumor volume grew to ≈100 mm3, and the mice were intraperitoneally injected with a suspension of PBS containing DDP (2.5 mg/kg) or PBS alone twice per week. The xenograft tumors were harvested after 4 weeks. Then, the tumor volumes were measured according to the following formula: tumor volume = length×width2 × 0.52. The entire experimental protocol was conducted in accordance with the guidelines of the local institutional animal care and use committee.
Statistical analysis
SPSS version 16.0 (SPSS, Chicago, IL) was used for statistical analysis. The data are presented as the mean ± standard error. Differences between groups were analyzed using Student’s t-test for comparisons between 2 groups or one-way analysis of variance for comparisons between more than 2 groups. All tests were two-sided. P < 0.05 was considered statistically significant.
Funding Statement
This study was supported by the National Natural Science Foundation of China [81472615].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Siegel RL, Miller KD, Jemal A.. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Fennell DA, Summers Y, Cadranel J, et al. Cisplatin in the modern era: the backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat Rev. 2016;44:42–50. [DOI] [PubMed] [Google Scholar]
- [3].Wen M, Xia J, Sun Y, et al. Combination of EGFR-TKIs with chemotherapy versus chemotherapy or EGFR-TKIs alone in advanced NSCLC patients with EGFR mutation. Biologics. 2018;12:183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Shen K, Cui J, Wei Y, et al. Effectiveness and safety of PD-1/PD-L1 or CTLA4 inhibitors combined with chemotherapy as a first-line treatment for lung cancer: a meta-analysis. J Thorac Dis. 2018;10:6636–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ghosh S. Cisplatin: the first metal based anticancer drug. Bioorg Chem. 2019;88:102925. [DOI] [PubMed] [Google Scholar]
- [6].Galluzzi L, Vitale I, Michels J, et al. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis. 2014;5:e1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fang S, Shen Y, Chen B, et al. H3K27me3 induces multidrug resistance in small cell lung cancer by affecting HOXA1 DNA methylation via regulation of the lncRNA HOTAIR. Ann Transl Med. 2018;6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang X, Meng Q, Qiao W, et al. miR-181b/Notch2 overcome chemoresistance by regulating cancer stem cell-like properties in NSCLC. Stem Cell Res Ther. 2018;9:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Huang FX, Chen HJ, Zheng FX, et al. LncRNA BLACAT1 is involved in chemoresistance of non-small cell lung cancer cells by regulating autophagy. Int J Oncol. 2019;54:339–347. [DOI] [PubMed] [Google Scholar]
- [10].Zhang Y, Wang X, Han L, et al. Green tea polyphenol EGCG reverse cisplatin resistance of A549/DDP cell line through candidate genes demethylation. Biomed Pharmacother. 2015;69:285–290. [DOI] [PubMed] [Google Scholar]
- [11].Zhang YW, Zheng Y, Wang JZ, et al. Integrated analysis of DNA methylation and mRNA expression profiling reveals candidate genes associated with cisplatin resistance in non-small cell lung cancer. Epigenetics. 2014;9:896–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cui Y, Gao D, Linghu E, et al. Epigenetic changes and functional study of HOXA11 in human gastric cancer. Epigenomics. 2015;7:201–213. [DOI] [PubMed] [Google Scholar]
- [13].Wang L, Cui Y, Sheng J, et al. Epigenetic inactivation of HOXA11, a novel functional tumor suppressor for renal cell carcinoma, is associated with RCC TNM classification. Oncotarget. 2017;8:21861–21870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Se YB, Kim SH, Kim JY, et al. Underexpression of HOXA11 is associated with treatment resistance and poor prognosis in glioblastoma. Cancer Res Treat. 2017;49:387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li Q, Chen C, Ren X, et al. DNA methylation profiling identifies the HOXA11 gene as an early diagnostic and prognostic molecular marker in human lung adenocarcinoma. Oncotarget. 2017;8:33100–33109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hwang JA, Lee BB, Kim Y, et al. HOXA11 hypermethylation is associated with progression of non-small cell lung cancer. Oncotarget. 2013;4:2317–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lu CW, Zhou DD, Xie T, et al. HOXA11 antisense long noncoding RNA (HOXA11-AS): A promising lncRNA in human cancers. Cancer Med. 2018;7:3792–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mu S, Ai L, Fan F, et al. Prognostic and clinicopathological significance of long noncoding RNA HOXA11-AS expression in human solid tumors: a meta-analysis. Cancer Cell Int. 2018;18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xue JY, Huang C, Wang W, et al. HOXA11-AS: a novel regulator in human cancer proliferation and metastasis. Onco Targets Ther. 2018;11:4387–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang Y, Chen WJ, Gan TQ, et al. Clinical significance and effect of lncRNA HOXA11-AS in NSCLC: a study based on bioinformatics, in vitro and in vivo verification. Sci Rep. 2017;7:5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhao X, Li X, Zhou L, et al. LncRNA HOXA11-AS drives cisplatin resistance of human LUAD cells via modulating miR-454-3p/Stat3. Cancer Sci. 2018;109:3068–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bhatlekar S, Fields JZ, Boman BM. HOX genes and their role in the development of human cancers. J Mol Med (Berl). 2014;92:811–823. [DOI] [PubMed] [Google Scholar]
- [23].Golpon HA, Geraci MW, Moore MD, et al. HOX genes in human lung: altered expression in primary pulmonary hypertension and emphysema. Am J Pathol. 2001;158:955–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rauch T, Wang Z, Zhang X, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc Natl Acad Sci U S A. 2007;104:5527–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Xia B, Shan M, Wang J, et al. Homeobox A11 hypermethylation indicates unfavorable prognosis in breast cancer. Oncotarget. 2017;8:9794–9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bai Y, Fang N, Gu T, et al. HOXA11 gene is hypermethylation and aberrant expression in gastric cancer. Cancer Cell Int. 2014;14:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fiegl H, Windbichler G, Mueller-Holzner E, et al. HOXA11 DNA methylation–a novel prognostic biomarker in ovarian cancer. Int J Cancer. 2008;123:725–729. [DOI] [PubMed] [Google Scholar]
- [28].Apostolidou S, Hadwin R, Burnell M, et al. DNA methylation analysis in liquid-based cytology for cervical cancer screening. Int J Cancer. 2009;125:2995–3002. [DOI] [PubMed] [Google Scholar]
- [29].Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profling using universal bead arrays. Genome Res. 2006;16:383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Nelson HH, Marsit CJ, Christensen BC, et al. Key epigenetic changes associated with lung cancer development: results from dense methylation array profling. Epigenetics. 2012;7:559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang X, Deng Y, He RQ, et al. Upregulation of HOXA11 during the progression of lung adenocarcinoma detected via multiple approaches. Int J Mol Med. 2018;42:2650–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang R, Zhang TT, Zhai GQ, et al. Evaluation of the HOXA11 level in patients with lung squamous cancer and insights into potential molecular pathways via bioinformatics analysis. World J Surg Oncol. 2018;16:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Matei D, Fang F, Shen C, et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Xu C, He T, Li Z, et al. Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth, migration and invasion of glioma cells. Biomed Pharmacother. 2017;95:1504–1513. [DOI] [PubMed] [Google Scholar]
- [35].Qu L, Jin M, Yang L, et al. Expression of long non-coding RNA HOXA11-AS is correlated with progression of laryngeal squamous cell carcinoma. Am J Transl Res. 2018;10:573–580. [PMC free article] [PubMed] [Google Scholar]
- [36].Liu Z, Chen Z, Fan R, et al. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer. 2017;16:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang Y, He RQ, Dang YW, et al. Comprehensive analysis of the long noncoding RNA HOXA11-AS gene interaction regulatory network in NSCLC cells. Cancer Cell Int. 2016;16:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chen JH, Zhou LY, Xu S, et al. Overexpression of lncRNA HOXA11-AS promotes cell epithelial-mesenchymal transition by repressing miR-200b in non-small cell lung cancer. Cancer Cell Int. 2017;17:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Latgé G, Poulet C, Bours V, et al. Natural antisense transcripts: molecular mechanisms and implications in breast cancers. Int J Mol Sci. 2018;19:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Huang B, Song JH, Cheng Y, et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Liu C, Lin J. Long noncoding RNA ZEB1-AS1 acts as an oncogene in osteosarcoma by epigenetically activating ZEB1. Am J Transl Res. 2016;8:4095–4105. [PMC free article] [PubMed] [Google Scholar]
- [42].Zhang CL, Zhu KP, Ma XL. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017;396:66–75. [DOI] [PubMed] [Google Scholar]


