ABSTRACT
DNA molecules are highly compacted in the eukaryotic nucleus where distal regulatory elements reach their targets through three-dimensional chromosomal interactions. G-quadruplexes, stable four-stranded non-canonical DNA structures, can change local chromatin organization through the exclusion of nucleosomes. However, the relationship between G-quadruplexes and higher-order genome organization remains unknown. Here, we found that G-quadruplexes are significantly enriched at boundaries of topological associated domains (TADs). Architectural protein occupancy, which plays critical roles in the formation of TADs, was highly correlated with the content of G-quadruplexes at TAD boundaries. Moreover, adjacent boundaries containing G-quadruplexes frequently interacted with each other because of the high enrichment of architectural protein binding sites. Similar to CCCTC-binding factor (CTCF) binding sites, G-quadruplexes also showed strong insulation ability in the separation of adjacent regions. Additionally, the insulation ability of CTCF binding sites and TAD boundaries was significantly reinforced by G-quadruplexes. Furthermore, G-quadruplex motifs on different strands were associated with the orientation of CTCF binding sites. These findings suggest a potential role for G-quadruplexes in loop extrusion. The enrichment of transcription factor binding sites (TFBSs) around regulatory elements containing G-quadruplexes led to frequent interactions between regulatory elements containing G-quadruplexes. Intriguingly, more than 99% of G-quadruplexes overlapped with TFBSs. The binding sites of CTCF and cohesin proteins were preferentially located surrounding G-quadruplexes. Accordingly, we proposed a new mechanism of long-distance gene regulation in which G-quadruplexes are involved in distal interactions between enhancers and promoters.
KEYWORDS: Chromatin structure, enhancer-promoter interaction, G-quadruplex, Hi-C, loop extrusion, transcription factor
Introduction
G-quadruplexes, the non-canonical secondary structures formed in guanine-rich nucleic acid sequences, have been shown to carry out critical roles in a diverse range of biological processes [1]. To form G-quadruplexes, at least two square planes where four guanines are located in the same plane, should be stabilized by a monovalent cation. Most DNA sequences capable of forming G-quadruplexes can be recognized by a canonical motif pattern, G≥3N1–7G≥3N1–7G≥3N1–7G≥3, where N represents the G-quadruplex loops and can be any nucleotide [2]. All DNA sequences that conform to this pattern are known as potential G-quadruplex structures (PG4). PG4s tend to distribute around transcription start sites (TSSs), 5′-untranslated regions, the 5′ ends of first exons, and regulatory elements, and are depleted in coding regions [3,4]. Therefore, PG4 motifs have a powerful bias toward particular genomic regions rather than being randomly distributed in the genome. Previous studies have investigated the existence and function of G-quadruplexes in biologically relevant contexts [5]. Taking advantage of polymerase stalling at G-quadruplexes, Chambers et al. established a high-throughput method to determine the accurate positions of G-quadruplexes in vitro [6]. By adopting the single chain variable fragment of an antibody, Schaffitzel et al. first reported the visualization of G-quadruplexes in vivo [7]. Hoffmann et al. found that the mouse monoclonal antibody 1H6 could bind synthetic G-quadruplexes enabling immune electron microscopy to observe G-quadruplexes in different organisms [8]. Moreover, Hansel-Hertsch et al. used G-quadruplex chromatin immunoprecipitation sequencing (G4 ChIP-seq) with the BG4 antibody to demonstrate the existence of G-quadruplexes in vivo [9]. More recently, this same group described the detailed G4 ChIP-seq method that robustly determined genome-wide G-quadruplexes formation in chromatin [10].
Having been observed in some eukaryotic genomes [7–9], G-quadruplexes were initially only found at telomere where their disruption allowed telomere extension [11,12]. Hansel-Hertsch et al. reported that G-quadruplexes were primarily found in regulatory and nucleosome-depleted regions and co-localized with active genes [9]. Hegyi et al. proposed that G-quadruplexes forming between enhancers and promoters could facilitate enhancer–promoter interactions [13]. Today, it is generally believed that G-quadruplexes have both negative and positive effects on transcription [4,14]. Mao et al. found that DNA G-quadruplexes can sequester DNA methyltransferase 1, thereby preventing CpG islands undergoing methylation [15]. The unfolding of G-quadruplexes, which requires interaction with helicases, has been implicated in transcriptional and post-transcriptional regulation [16], and a deficiency of human G-quadruplex helicases was shown to significantly upregulate the transcription levels of genes containing G-quadruplexes [9].
Most eukaryotic genomes are tightly folded inside the nucleus. The recently developed powerful Hi-C technology [17–19] was used to show that chromosomes are partitioned into topologically associated domains (TADs) that are highly correlated with gene regulation [20–23]. The formation of TAD boundaries plays critical roles in aging [24], cell differentiation [25,26], and cell fate [24,27]. Chromatin interactions within TADs were shown to be quite frequent, while interactions across different TADs were limited because of the insulation ability of TAD boundaries. Although TADs are normally conserved across different cell types, TAD boundaries vary because of the existence of architectural proteins such as CCCTC-binding factor (CTCF) [28–30]. Gong et al. demonstrated that higher TAD boundary insulation was associated with elevated CTCF levels, and that the insulation ability of TAD boundaries varied across different cell types [31]. Moreover, the disruption of TAD boundaries led to unexpected interactions between regulatory elements and genes [30,32]. For example, by imposing temperature stress on Drosophila chromosomes, Li et al. observed a decline in the TAD boundary strength, leading to an increase in long-distance inter-TAD interactions [33]. Several studies reported the close correlations between TAD boundaries and different genomic elements [17,34,35]. Gorkin et al. concluded that in most cases the formation of a TAD boundary requires more than one sequence element, such as architectural proteins, TSSs of active genes, and transcription factor binding sites (TFBSs) [36].
There is considerable evidence that G-quadruplexes are capable of excluding nucleosomes [37,38], but the relationship between G-quadruplexes and higher-order chromatin structures such as TADs remains unclear. Our study aimed to answer the following questions: 1) whether G-quadruplexes are associated with TADs; 2) whether G-quadruplexes can potentially impact on long-range interactions between regulatory elements; and 3) what is the relationship between G-quadruplexes and TFBSs. In this study, the accurate positions of G-quadruplexes in vivo were achieved from G4 ChIP-seq generated by Mao et al. [15]. G-quadruplex sequences (G4 sequences), which can form G-quadruplexes in vitro, were derived from Chambers et al. [6]. Our analysis revealed that G-quadruplexes are correlated with three-dimensional chromatin structures and transcription factors (TFs) including some key architectural proteins.
Results
TAD boundaries are rich in G-quadruplexes
TADs, which are identified by computational algorithms measuring the directionality of interactions in the genome, are a fundamental unit of genome organization [20]. Since G-quadruplexes could change local chromatin structure through exclusion of nucleosomes [38], we wondered that G-quadruplexes would be related to the higher-order genome organization such as the domain-wide level structures.
Hi-C sequencing data of K562 cell lines generated by Rao et al. were used to call TADs [17], and G-quadruplexes were identified by G4 ChIP-seq in K562 cell lines [15]. We observed a high accumulation of G-quadruplexes at TAD boundaries, with the G-quadruplex profile found to gradually decline towards the central regions of TADs (Figure 1a). Global analysis showed that G-quadruplexes are more enriched at TAD boundaries (Student’s t-test, p = 8.21 × 10−56). Likewise, G4 sequences (Figure 1b) and PG4 motifs (Figure 1c) preferentially located at TAD boundaries rather than inside TADs (Student’s t-test, p = 6.95 × 10−68 and 4.03 × 10−80, respectively).
Figure 1.
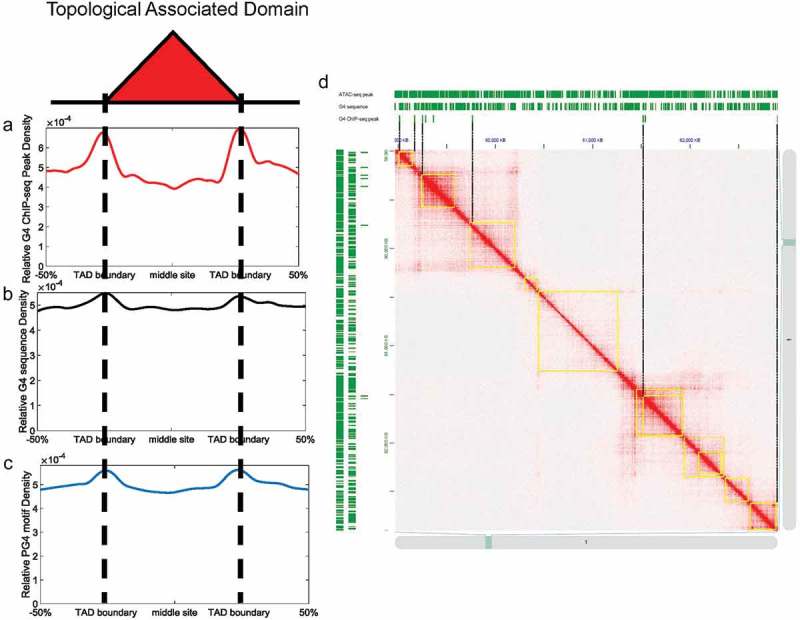
G-quadruplexes are highly enriched at TAD boundaries. (a-c) Relative density of G4 ChIP-seq peaks/G4 sequence/PG4 motifs across TADs. The x-axes indicate genomic distance from middle sites of TADs. The y-axes indicate relative peak density across TADs. (d) Hi-C contact matrices at 10 kb resolution from chromosome 1 of K562 cell lines. The TADs are highlighted in yellow rectangles. ATAC-seq peaks/G4 sequences/G4 ChIP-seq peaks are shown in upper/left panels of contact matrices.
Previous research demonstrated that most G4 ChIP-seq peaks overlapped with ATAC-seq peaks [9]. Because TAD boundaries also contain large amounts of ATAC-seq peaks (Figure. S1A, Student’s t-test, p = 9.36 × 10−79), we considered that the enrichment of G-quadruplexes at TAD boundaries may be simply caused by more accessible chromatin. The proportion of ATAC-seq peaks containing G-quadruplexes is 2.71% (8,953/330,208). However, 3.92% (3,612/92,180) of ATAC-seq peaks at TAD boundaries contained G-quadruplexes, suggesting that chromatin accessibility alone is not sufficient for the enrichment of G-quadruplexes at TAD boundaries. Intriguingly, the relative density of G-quadruplexes at TAD boundaries is 1.7 times higher than that at central regions of TADs, compared with 1.3 and 1.2 times higher for G4 sequences and PG4 motifs, respectively. This represents a significantly higher relative peak density for G4 ChIP-seq than G4 sequences and PG4 motifs at TAD boundaries (Student’s t-test, p = 1.39 × 10−308 for G4 ChIP-seq peaks versus G4 sequences; p = 1.05 × 10−269 for G4 ChIP-seq peaks versus PG4 motifs). Therefore, we concluded that the enrichment of G-quadruplexes at TAD boundaries not only depends on the sequence property but also depends on the in-vivo chromatin context (Figure 1d). We also found that G-quadruplexes are depleted in non-TAD regions, and that the relative density of G-quadruplexes inside TADs is significantly higher than in non-TAD regions (Figure S1B and Figure S1C, Student’s t-test, p = 8.14 × 10−66). The heatmap of contact matrices is generated by JuiceBox [39,40].
The recruitment of architectural proteins, such as CTCF, RAD21, and SMC3, to a particular region is essential for the formation of TADs [17,34]. In this study, we divided all TAD boundaries into two groups: G4-containing boundaries and non-G4-containing boundaries. The TAD boundaries containing G-quadruplexes were defined as G4-containing boundaries; and those boundaries without G-quadruplexes were defined as non-G4-containing boundaries. A total of 7,250 boundaries were identified, including 2,489 G4-containing boundaries and 4,761 non-G4-containing boundaries. CTCF and cohesin protein ChIP-seq peaks around G4-containing and non-G4-containing boundaries were shown in Figure 2, which revealed them to be highly enriched around TAD boundaries (Figure 2a–c, Figure S2A–C). This is in agreement with findings that TAD boundaries are rich in architectural protein binding sites [33,41].
Figure 2.
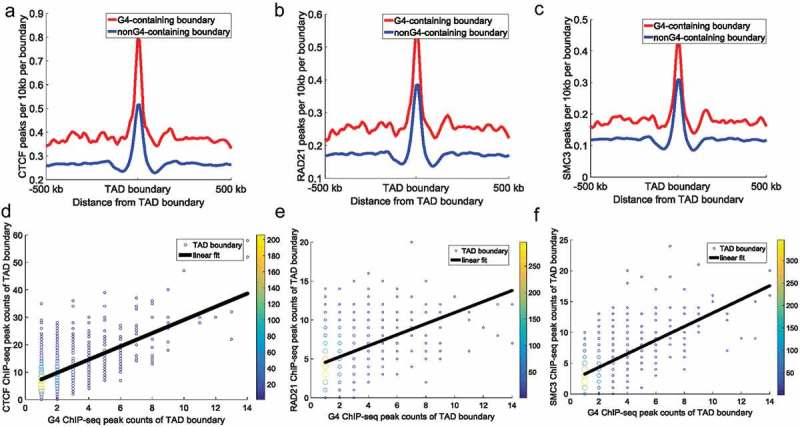
CTCF, RAD21, and SMC3 ChIP-seq peak counts at TAD boundaries. All TAD boundaries were divided into: G4-containing boundaries and non-G4-containing boundaries. (a–c) The top panels represent CTCF (a), RAD21 (b), and SMC3 (c) peak counts around TAD boundaries. Red lines and blue lines indicate G4-containing boundaries and non-G4-containing boundaries, respectively. The y-axes indicate ChIP-seq peak counts per 10 kb per boundary. The Student’s t-test p-value for CTCF, RAD21, and SMC3 ChIP-seq peak counts between the two types of boundaries was 9.20 × 10−71, 1.94 × 10−22, and 1.20 × 10−41, respectively. (d–f) The bottom panels represent the relationship between architectural protein (CTCF [d], RAD21 [e], and SMC3 [f]) and G-quadruplexes at G4-containing boundaries. The data points represent different TAD boundaries. The x-coordinates of the plots represent the number of G4 ChIP-seq peaks overlapping with the TAD boundaries, and the y-coordinates of the plots represent the number of architectural protein ChIP-seq peaks overlapping with the TAD boundaries. We calculated G4 ChIP-seq peak counts and architectural protein ChIP-seq peak counts in a 50 kb (±25 kb around TAD boundary) window around each TAD boundary. Different colours show the counts of overlapping boundaries. The black continuous line indicates the linear fit between architectural protein ChIP-seq peak counts and G-quadruplex counts at TAD boundaries (the Pearson correlation coefficient between G4 ChIP-seq peak counts and CTCF, RAD21, and SMC3 ChIP-seq peak counts at TAD boundaries was 0.62, 0.40, and 0.63, respectively [p = 3.10 × 10−82, 5.77 × 10−65, and 6.98 × 10−77, respectively]).
CTCF and cohesin protein ChIP-seq peak counts around G4-containing boundaries were significantly higher than those around non-G4-containing boundaries (Figure 2a–c, Student’s t-test, p = 9.20 × 10−71, 1.94 × 10−22, and 1.20 × 10−41 for CTCF, RAD21, and SMC3 ChIP-seq peak counts between two types of boundaries, respectively). These results indicate that G-quadruplexes are associated with the binding of architectural proteins at TAD boundaries. Furthermore, TAD boundaries containing more G-quadruplexes overlapped with more architectural protein ChIP-seq peaks (Figure 2d–f). As reported in previous studies [17,34], TAD boundaries can separate adjacent TADs by the binding of CTCF and cohesin proteins. The disruption of CTCF and cohesin protein in TAD boundaries can lead to unexpected interactions across adjacent TADs, thus causing pathogenicity [32]. Our results indicate that G-quadruplexes relate to the TAD boundaries because of the correlation between G-quadruplexes and architectural protein binding.
Because of the close relationship between architectural proteins and TAD boundaries, we hypothesized that TADs consisting of G4-containing boundaries would have different properties to those consisting of non-G4-containing boundaries. To test this, we checked corner scores which indicate the possibility that a pixel is at the corner of a contact domain [17], and boundary–boundary interactions in K562 cell lines. The corner scores of TADs in K562 cell lines were calculated by Rao et al. [17]. Figure 3a shows that TADs with low corner scores normally consist of non-G4-containing boundaries, while TADs with high corner scores normally consist of G4-containing boundaries. From Figure 3a, it can be seen that there is almost no difference in the corner score between TADs with one or more G-quadruplexes; however, there are significant differences between TADs with no and one G-quadruplex. The same phenomenon is evident in Figure 3d. We found that adjacent boundaries containing G-quadruplexes could contact each other significantly more frequently than those without G-quadruplexes (Figure 3b–c, Student’s t-test, p = 4.11 × 10−32). Only interactions between two adjacent boundaries were calculated because the boundary insulation ability would affect interactions across boundaries. Combined with earlier findings (Figure 2), these results suggest that abundant CTCF and cohesin protein binding sites around G4-containing boundaries lead to more stable TADs and a high frequency of boundary-boundary interactions. The relationship between G-quadruplexes and TAD corner scores and boundary–boundary interactions is illustrated in Figure 3d for a representative genome region in K562 cell lines. We speculated that G-quadruplexes are associated with the high enrichment of CTCF and cohesin at G4-containing boundaries, which further influence boundary-boundary interactions (Figure 3e).
Figure 3.
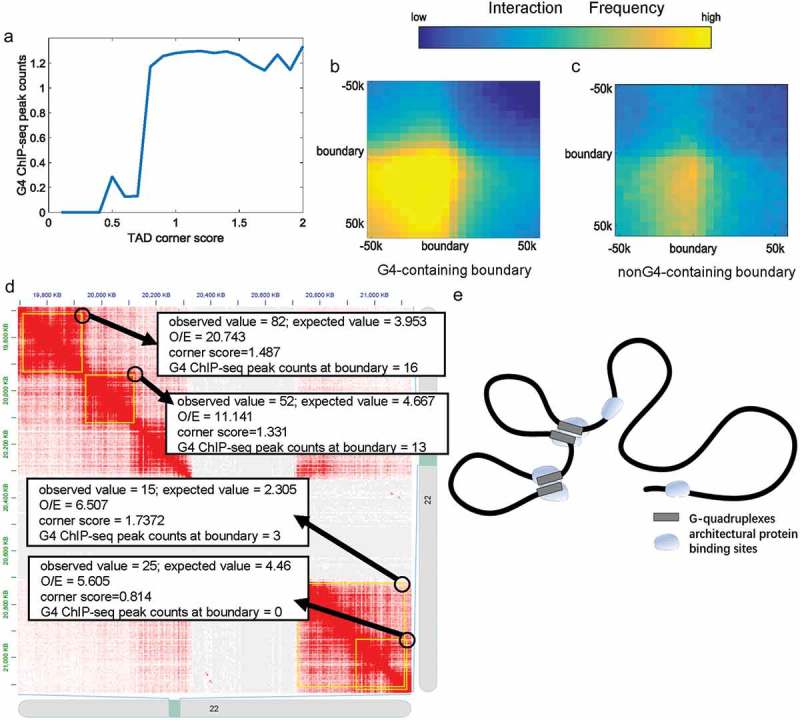
The relationship between G-quadruplexes and TADs. (a) TAD corner score plot used to identify the link between G-quadruplexes and TAD corner score. All TADs were categorized into groups by TAD corner scores in equal intervals. The y-axes indicate the average G4 ChIP-seq peak counts of the group. (b) Interactions between adjacent G4-containing boundaries. (c) Interactions between adjacent non-G4-containing boundaries. (d) The Hi-C contact matrix for representative genomic regions of chromosome 22 in K562 cell lines. TADs are marked by yellow rectangles. The black circles indicate the corner/boundary–boundary interactions of TADs. When TADs consist of non-G4-containing boundaries, the corner score and boundary–boundary interaction frequency were quite low. (e) Adjacent G4-containing boundaries with many architectural protein binding sites strongly interact with each other; as a contrast, adjacent non-G4 containing boundaries with few architectural protein binding sites weakly interact with each other.
G-quadruplexes are correlated with the role of CTCF in chromatin structure
We applied an insulation score metric, which estimates the ability of a given locus to separate adjacent regions [21,42], to show the insulation ability of G-quadruplexes. The insulation score of one locus equals the interaction strength between adjacent genome regions, with lower insulation scores indicating fewer interactions between two neighbouring regions of the locus.
Insulation scores around G-quadruplexes and CTCF binding sites were calculated. Because G-quadruplexes are highly correlated with CTCF binding sites, we only selected G4 ChIP-seq peaks located distal (>10 kb) to CTCF binding sites. G-quadruplexes in vivo displayed a strong insulation ability in the separation of adjacent regions (Figure 4a–b), suggesting that G-quadruplexes are capable of separating adjacent regions similar to CTCF binding sites. And there was no significant difference in insulation scores between CTCF binding sites and CTCF-distal G-quadruplexes (Figure 4a, Student’s t-test, p > 0.001). Furthermore, almost all G-quadruplexes displayed high insulation abilities (Figure 4b). To test whether the insulation ability of G-quadruplexes was affected by the CTCF binding sites, we showed the insulation scores of G-quadruplexes with respect to their distance to the nearest CTCF binding sites (Figure 4c). Regardless of the distance to the nearest CTCF binding sites, the insulation scores of G-quadruplexes were significantly lower than those of random selected regions (Student’s t-test, p = 2.79 × 10−150), As shown in Figure 4e, to exclude the influence of different proximal CTCF densities we only selected isolated CTCF binding sites locate distally (>10 kb) from other CTCF binding sites, then classified CTCF binding sites into two groups: those overlapping G-quadruplexes (1,253) and those without G-quadruplexes (26,549). CTCF binding sites overlapping G-quadruplexes showed significantly stronger insulation abilities than those without G-quadruplexes (Figure 4e, Student’s t-test, p = 6.29 × 10−36), suggesting that G-quadruplexes can strengthen the insulation ability of CTCF binding sites.
Figure 4.
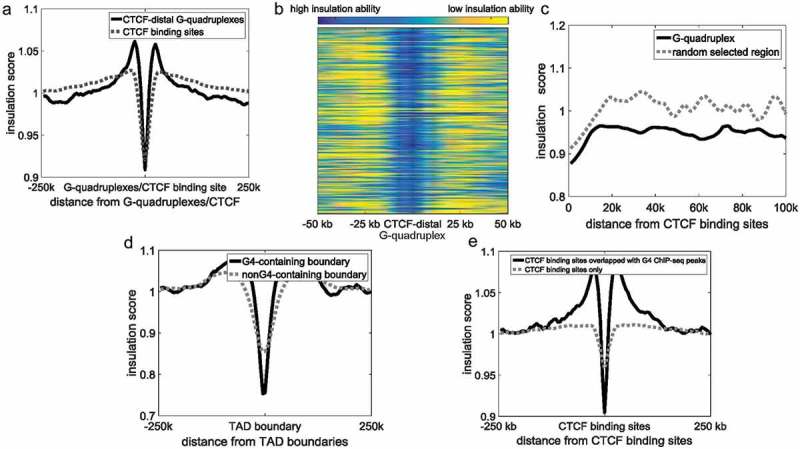
The relationship between G-quadruplexes and CTCF binding sites. (a) Insulation scores around CTCF binding sites and CTCF-distal G-quadruplexes. The black continuous line and grey dotted line indicate CTCF-distal G-quadruplexes and CTCF binding sites, respectively. (b) Heatmap of CTCF distal-G-quadruplexes insulation score. Each row represents a G4 ChIP-seq peak located distal (>10 kb) to CTCF binding sites. Blue represents a high insulation ability and low insulation score, and yellow indicates a low insulation ability and high insulation score. (c) Insulation scores of G-quadruplexes. The x-axes indicate the distance between G-quadruplexes and their closest CTCF ChIP-seq peaks. (d) Insulation scores around CTCF binding sites. The black continuous line and grey dotted line indicates CTCF binding sites overlapped with G-quadruplexes and CTCF binding sites only, respectively. (e) Insulation scores of G4-containing boundaries (black) and non-G4-containing boundaries (grey).
A strong relationship between TAD boundaries and CTCF was previously reported, with CTCF binding sites shown to be highly enriched at TAD boundaries to separate two adjacent TADs [41]. We suspected that G-quadruplexes enriched in TAD boundaries are also involved in separating adjacent TADs. We calculated the insulation scores of G4-containing boundaries and non-G4-containing boundaries. The insulation abilities of G4-containing boundaries were significantly higher than non-G4-containing boundaries (Figure 4e, Student’s t-test, p = 1.98 × 10−174), suggesting that G-quadruplexes potentially relate to the insulation ability of TAD boundaries similar to CTCF binding sites.
It is widely accepted that CTCF forms loops must be capable of discriminating between CTCF sites in forward and reverse orientations [43]. The homer, a software for motif finding, was used to find the genomic positions of CTCF motifs in the whole human genome [44]. As suggested by Rao et al. [17], we designated the consensus DNA sequence, which is written as 5'–CCACNAGGTGGCAG–3', for CTCF binding sites as the forward orientation. We found that 5' loop anchor sites are rich in forward CTCF motifs, while 3' loop anchor sites are rich in reverse CTCF motifs (Figure 5a–b), consistent with previous findings demonstrating that the CTCF motifs that anchor a loop should be in the convergent orientation [17,45]. The PG4 motifs on the plus strand are enriched at the forward CTCF motifs (Figure 5c). In contrast, the reverse CTCF motifs are rich in the PG4 motifs on the minus strand (Figure 5d). Likewise, the G4 sequences on the plus strand were also enriched surrounding the forward CTCF motifs (Figure 5e), and the G4 sequences on the minus strand preferred to locate around the reverse CTCF motifs (Figure 5f). The results suggested PG4 motifs on different strands potentially characterize CTCF binding sites with different orientations.
Figure 5.
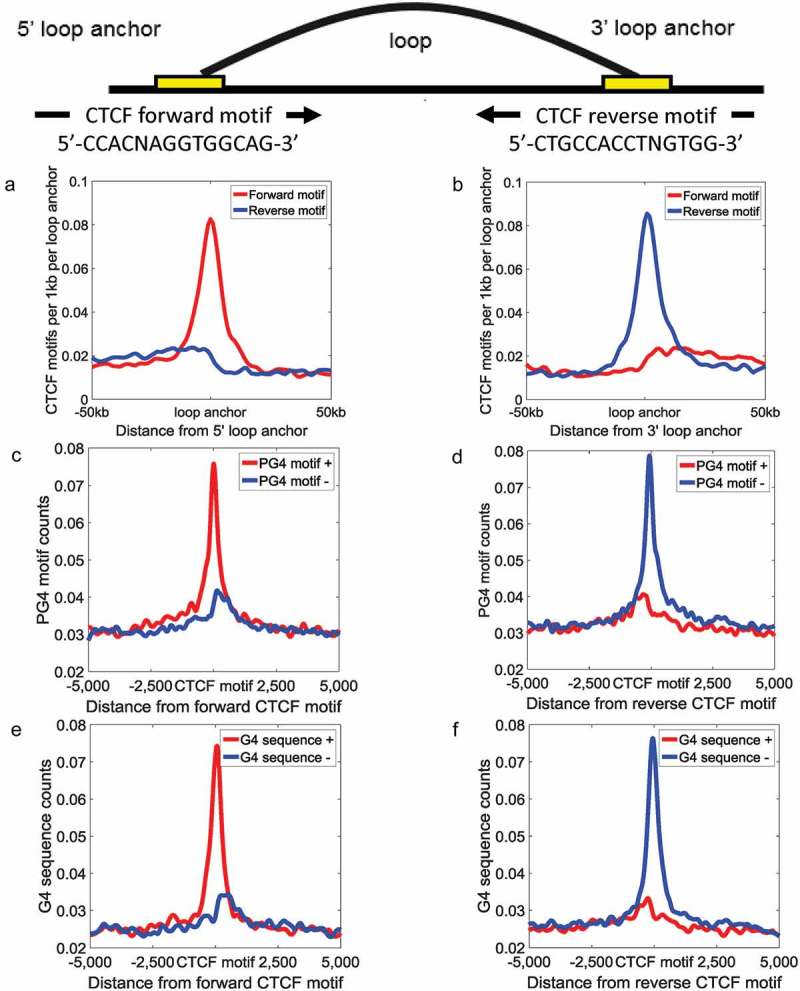
PG4 motifs are correlated with the orientation of CTCF motifs. The distribution of CTCF motifs surrounding 5' (a) and 3' (b) loop anchors. The red lines and the blue lines indicate forward CTCF motifs and reverse CTCF motifs, respectively. The distribution of PG4 motifs based on matching strand around forward (c) and reverse (d) CTCF motifs. The distribution of G4 sequence based on matching strand around forward (e) and reverse (f) CTCF motifs. The red lines and the blue lines indicate PG4 motfs/G4 sequences on the plus strand and the minus strand, respectively.
Although previous studies proved that the CTCF motif orientations play critical roles during loop extrusion, the mechanism by which cohesin proteins recognized the orientations of CTCF motifs remains unclear. Herein, we proposed a possibility that G-quadruplexes on different strands, in term of their relative position to the different orientations of CTCF motifs, participate in loop extrusion through stall the slide of cohesin proteins. We calculated the distance between G-quadruplexes/CTCF/RAD21/SMC3 ChIP-seq peaks and forward/reverse CTCF motifs (Figure S3A–B). In line with previous study, we found a characteristic shift pattern between CTCF motifs and cohesin protein ChIP-seq peaks [46]. Surprisingly, similar to cohesin protein ChIP-seq peaks, G-quadruplexes tend to distribute downstream of forward CTCF motifs (average distance = 3.15 bp, 2.18 bp, and 5.21 bp for G-quadruplexes, SMC3, and RAD21, respectively) and upstream of reverse CTCF motifs (average distance = −4.49 bp, −2.31 bp, and −5.79 bp for G-quadruplexes, SMC3, and RAD21, respectively). In contrast, CTCF ChIP-seq peaks prefer to locate upstream of forward CTCF motifs (average distance = −1.56 bp) and downstream of reverse CTCF motifs (average distance = 0.75 bp). We further calculated the distance between cohesin protein binding sites and G-quadruplexes/CTCF binding sites (Figure S3C, average distance = 9.19 bp, 10.80 bp, 9.52 bp, and 10.77 bp for G-quadruplexes to RAD21, CTCF to RAD21, G-quadruplexes to SMC3, and CTCF to SMC3, respectively). There was almost no difference between G-quadruplexes and CTCF binding sites. We speculated that at least in some cases G-quadruplexes can help CTCF binding sites to prevent cohesin flow-through.
G-quadruplexes are correlated with the strong interactions between enhancers and promoters
G-quadruplexes in promoters are capable of regulating gene expression in both positive way and negative way [14,47]. However, it was unclear whether G-quadruplexes potentially relate to long-distance regulation.
In K562 cell lines, 3.31% (1,034/31,237) of enhancers and 29.43% (5,245/17,819) of promoters overlapped with G-quadruplexes. We defined these enhancers and promoters as G4-containing enhancers and G4-containing promoters, respectively. Using Hi-C sequencing data, we identified an enhancer–promoter pair as an interaction pair if the false discovery rate (FDR) of the interaction was lower than 0.01. The FDR of regulatory element interactions was calculated by Fit-Hi-C [48]. Only enhancers and promoters located on the same chromosomes were considered enhancer–promoter pairs. If either end of an interaction pair contained G-quadruplexes, the pair was defined as a G4-containing interaction. If neither end of an interaction contained G-quadruplexes, the pair was defined as a non-G4-containing interaction. A total of 15,657 G4-containing enhancer–promoter interactions and 24,161 non-G4-containing enhancer–promoter interactions were identified. We found that the interaction frequency of G4-containing enhancer–promoter pairs (interaction frequency = 15.01) was significantly higher than that of non-G4-containing pairs (interaction frequency = 11.92) (Figure 6a, Student’s t-test, p = 1.71 × 10−44). Likewise, G4-containing enhancer–enhancer (interaction frequency = 17.21)/promoter–promoter interactions (interaction frequency = 12.86) were also significantly stronger than non-G4-containing enhancer–enhancer (interaction frequency = 11.77)/promoter–promoter (interaction frequency = 10.61) interactions (Figure 6a, Student’s t-test, p = 5.08 × 10−33 and 9.16 × 10−21, respectively). Moreover, genes with G4-containing promoters (average FPKM = 16.89) were expressed at significantly higher levels than other genes (average FPKM = 9.32, Student’s t-test, p = 2.92 × 10−22).
Figure 6.
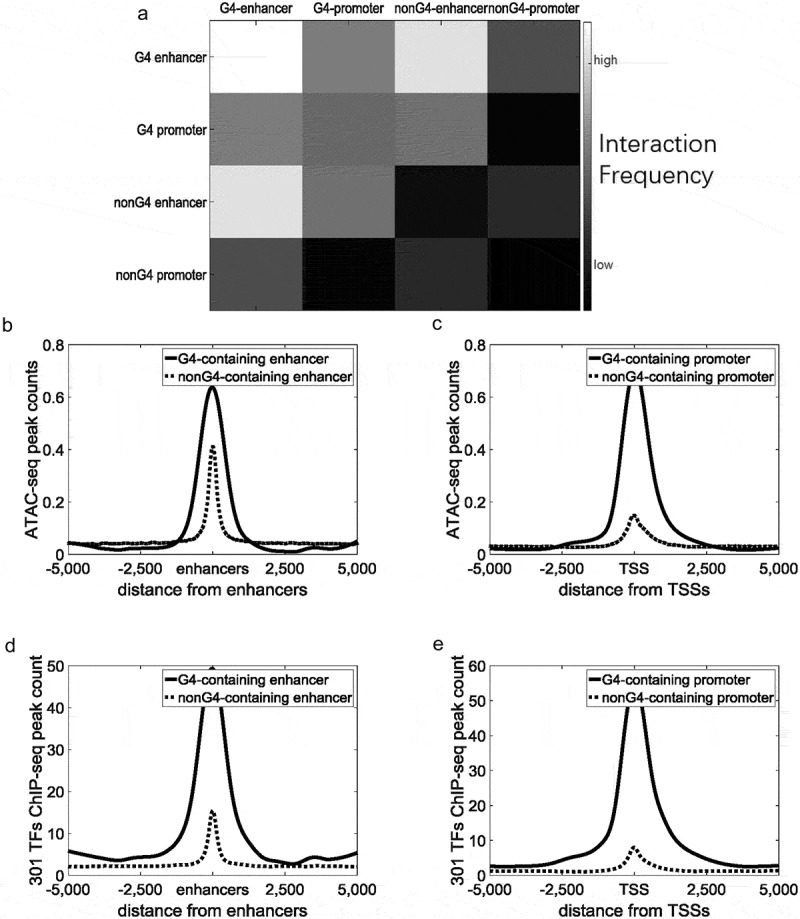
G-quadruplexes are correlated with the strong interaction between regulatory elements. Black continuous lines indicate G4-containing enhancers/promoters, and grey dotted lines indicate non-G4-containing enhancers/promoters. (a) Heatmaps of interaction frequencies between different types of regulatory elements. (b) Distribution of ATAC-seq peak counts around enhancers. (c) Distribution of ATAC-seq peak counts around TSSs. (d) Distribution of 301 TFs ChIP-seq peak counts around enhancers. (e) Distribution of 301 TFs ChIP-seq peak counts around TSSs.
We speculated that G-quadruplexes are associated with protein binding to enable stable interactions of regulatory elements. To investigate this, we calculated the distribution of ATAC-seq peaks and some known protein binding sites around enhancers and promoters. ATAC-seq peaks on G4-containing enhancers and promoters were significantly higher than non-G4-containing enhancers and promoters (Figure 6b–c, Student’s t-test, p = 2.66 × 10−96 and 1.54 × 10−108). A total of 301 ChIP-seq datasets of K562 cell lines were combined by the online analysis tool, ReMap [49]. ChIP-seq peaks of TFs on G4-containing enhancers were also significantly higher than those on non-G4-containing enhancers (Figure 6d, Student’s t-test, p < 7.71 × 10−151), and the same was observed for promoters (Figure 6e, Student’s t-test, p < 5.58 × 10−176). Because of the high enrichment of ATAC-seq peaks on G4-containing enhancers/promoters, we wondered whether these G-quadruplexes are functional or merely a reflection of chromatin accessibility on regulatory elements. We selected 8,955 most accessible ATAC-seq peaks, as many as G4 ChIP-seq peaks, according to ChIP-seq peak values. We compared the interaction strength of G4-containing regulatory pairs with that of the regulatory pairs overlapped with the most accessible ATAC-seq peaks. The average interaction strength of G4-containing regulatory pairs (15.00) is significantly higher than that of the pairs overlapped with the most accessible ATAC-seq peaks (12.95, Student’s t-test, p = 8.92 × 10−21). The results indicated that G-quadruplexes are more than merely a reflection of chromatin accessibility, but also play a role in the regulation of long-range interactions.
G-quadruplexes are highly correlated with TFBSs
We next used ReMap to check the overlapping regions between G-quadruplexes and some known TFBSs [49], and observed a significant overlap between G-quadruplexes and known TFBSs. Using the bedtools [50], we generated a control dataset containing 8,955 randomly selected human genomic regions with the same length as G-quadruplexes. A total of 99.55% (8,915/8,955) of G-quadruplexes overlapped with TFBSs compared with no random control sequences (0/8,955). PHF8 was the most enriched TF in the intersection (Figure 7a). PHF8 functions as a histone lysine demethylase, and removes mono-methyl marks at H4K20, leading to chromatin opening for transcription [51]. E2F4 is a member of the E2F family that plays an important role in the suppression of proliferation-associated genes. However, similar to G-quadruplexes, E2F4 can function as an activator as well as a repressor [52]. The observed high intersection between E2F4 binding sites and G-quadruplexes suggest a potential role for G-quadruplexes in tumours (Figure 7a). G-quadruplexes were rich in the binding sites of NEUROD1, which regulates expression of the insulin gene and acts as a transcriptional activator (Figure 7a). NEUROD1 was also previously shown to alter chromatin structures at enhancers and promoters [53]. Likewise, the binding sites of CTCF, RAD21, and SMC3 were also enriched in G-quadruplexes according to ReMap calculation [49]. ChIP-seq peak counts of architectural proteins around G-quadruplexes were shown in Figure S4A–C, suggesting that G-quadruplexes are associated with architectural proteins binding sites. These proteins form the base of loop structures and TAD boundaries.
Figure 7.
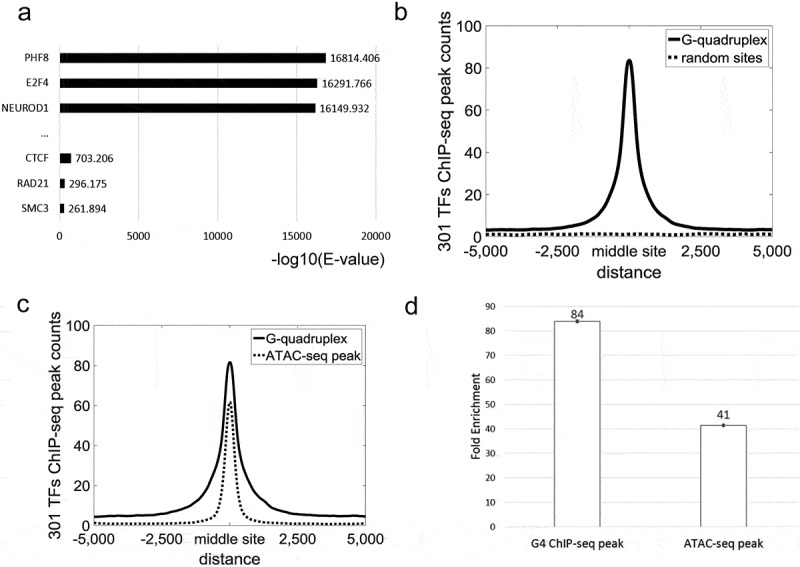
The relationship between TFBSs and G-quadruplexes. (a) Most enriched TFs on G-quadruplexes. (b) Peaks of 301 ChIP-seq datasets around G-quadruplexes. (c) TF ChIP-seq peak counts around G-quadruplexes/ATAC-seq peaks. The ATAC-seq peaks were the most accessible peaks according to peak values. The count of ATAC-seq peaks was equal to that of G4 ChIP-seq peaks. (d) Enrichment analysis of G-quadruplexes and ATAC-seq peaks. Error bars, s.d. (n = 1).
To characterize the link between G-quadruplexes and TF binding, ChIP-seq peaks of all known TFs in K562 cell lines around G-quadruplexes are shown in Figure 7b. TF ChIP-seq peaks were abundant around G-quadruplexes, suggesting that G-quadruplexes tend to co-localize with TFs to change chromatin structures. However, because most G-quadruplexes overlapped with ATAC-seq peaks, it remains unclear whether the enrichment of TFBSs is caused by ATAC-seq or G-quadruplexes. To exclude factors caused by accessible regions, we obtained the 8,955 most accessible ATAC-seq peaks, as many as G4 ChIP-seq peaks, according to ChIP-seq peak values calculated by MACS [54]. Overlapped peaks (3,784/8,955) of G4 ChIP-seq and ATAC-seq were discarded. TF ChIP-seq peak counts on G-quadruplexes were significantly higher than those on the most accessible ATAC-seq peaks, suggesting that G-quadruplexes highly relate to the binding of TFs (Figure 7c–d, Student’s t-test, p = 5.12 × 10−94).
Discussion
In this work, we characterized the critical roles of G-quadruplexes in chromatin structures. G-quadruplexes were thought to regulate local chromatin structures through exclusion of nucleosome [37,38], gene transcription [9,55], and co-binding with proteins [56]. However, the relationship between G-quadruplexes and three-dimensional chromosome organization was unknown.
We found that G-quadruplexes relate not only to local structures but also to the domain-wide level of organization. At TAD boundaries, frequent transcription of genes is known to generate large amounts of single-stranded DNA [28,57]. We speculated that abundant PG4 motifs (Figure 1c) and continual transcriptional events would facilitate the formation of G-quadruplexes at TAD boundaries. Consistent with our expectations, we observed a significant accumulation of G-quadruplexes TAD boundaries (Figure 1a–d).
TAD boundaries are rich in architectural proteins required for the formation of TADs [28,34]. To identify links between G-quadruplexes and TAD boundaries, we divided all boundaries into: G4-containing and non-G4-containing boundaries. We investigated the distribution of architectural proteins, including CTCF, RAD21, and SMC3, which play key roles in the formation of TADs (Figure 2) [43,58,59]. G4-containing boundaries were found to harbour more architectural protein binding sites (Figure 2), suggesting that G-quadruplexes might relate to the formation of TADs by co-binding with architectural proteins. Our calculation suggested that G-quadruplexes highly relate to the boundary–boundary interactions (Figure 3). Adjacent G4-containing boundaries were shown to strongly interact with each other, while weak interactions were seen for adjacent non-G4-containing boundaries.
We found that G-quadruplexes have insulation ability to block interactions between flanking regions (Figure 4a–c). Because of this, G-quadruplexes could aid the separation of adjacent TADs by CTCF binding sites (Figure 4d). Furthermore, because of the high enrichment of architectural protein binding sites and strong insulation abilities of G-quadruplexes, G4-containing boundaries have stronger insulation abilities than non-G4-containing boundaries (Figure 4e).
From these findings, we proposed a model to characterize the relationship between G-quadruplexes and TADs (Figure 8a). In our model, G4-containing boundaries harboured abundant architectural protein binding sites which lead to frequent boundary–boundary interactions. Additionally, G4-containing boundaries have significantly stronger insulation ability.
Figure 8.
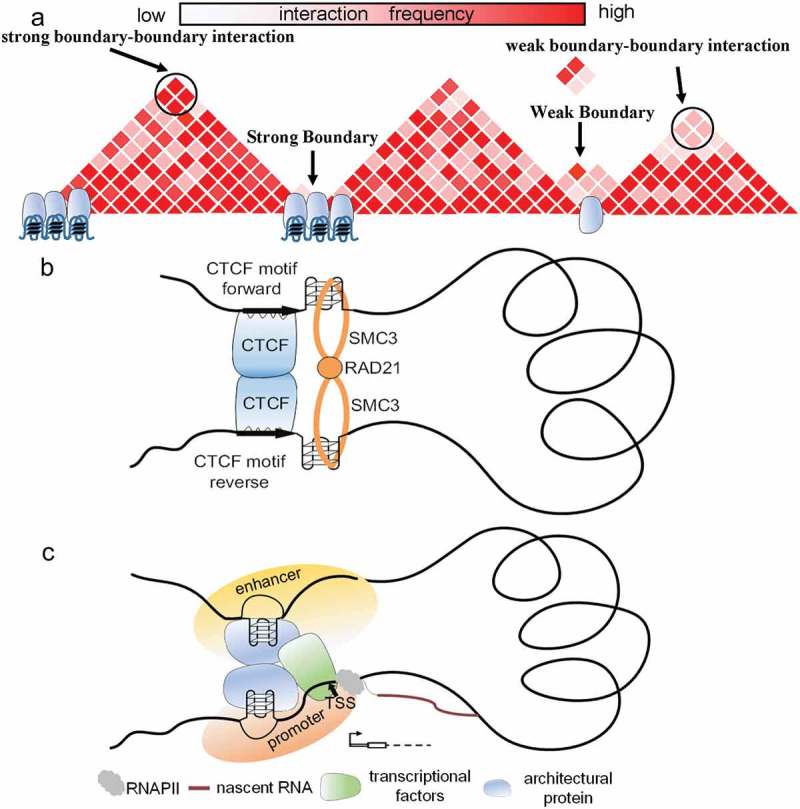
The relationship between G-quadruplexes and chromatin structures. (a) In model A, architectural protein binding sites are abundant at TAD boundaries containing G-quadruplexes. Such boundaries can frequently interact with each other. And G4-containing boundaries have strong insulation ability. (b) G-quadruplexes tend to form downstream of forward CTCF motifs and upstream of reverse CTCF motifs. G-quadruplexes surrounding CTCF motifs can characterize the orientation of CTCF motifs and prevent the sliding of cohesin protein. (c) In model C, G-quadruplexes highly relate to the binding of TF, especially for CTCF, RAD21, and SMC3. Accordingly, G4-containing enhancers interact with G4-containing promoters more stably through protein–protein interaction.
The SMC complex can extrude DNA at a high speed to form higher-order genome organization by which cohesin proteins promote the generation of TADs [58,59]. In the chromatin extrusion model, CTCF can stop the sliding of the SMC complex to form loop structures and TADs [60]. Moreover, the vast majority of CTCF motif pairs in loop anchors are convergent [17,60]. Nevertheless, it remains unknown that how cohesin protein recognized the context and is capable of discriminating between CTCF sites in convergent and divergent orientations. Because high accumulation of cohesin protein ChIP-seq peaks is observed around G-quadruplexes, we hypothesized that G-quadruplexes are capable of preventing the sliding of the SMC complex to form loop structures and TADs. Furthermore,we found that PG4 motifs on different strands displayed quite different patterns surrounding forward/reverse CTCF motifs, suggesting a potential role for PG4 motifs in characterization the direction of looping.
Surprisingly, cohesin protein binding sites and G-quadruplexes are normally locate downstream of forward CTCF motifs and upstream of reverse CTCF motifs (Figure S3A–B). In contrast, CTCF binding sites are normally locate upstream of forward CTCF motifs and downstream of reverse CTCF motifs (Figure S3A–B). Additionally, we found that the cohesin protein ChIP-seq peaks are normally close to G-quadruplexes (Figure S3C). We speculated that at least in some cases G-quadruplexes are capable of preventing cohesin flow-through. Accordingly, we proposed a potential role of G-quadruplexes in loop extrusion (Figure 8b). In this model, CTCF may promote G-quadruplexes formation and G-quadruplexes are further involved in loop extrusion. G-quadruplexes are formed in a specific orientation related to the orientation of CTCF binding. Then G-quadruplexes can prevent the sliding of cohesin protein, which further facilitate the loop extrusion. In line with previous study, we used the cohesin ring model to represent extrusion complex [60].
Prior to our work, some studies indicated that G-quadruplexes impair the initiation of transcription by RNA polymerase, or inhibit transcription when G-quadruplexes were present in the antisense strand [61]. However, other studies found that G-quadruplexes can positively regulated gene expressions [9]. Regardless of the viewpoint, these previous investigations mainly focused on local G-quadruplexes in promoters. We found that the interaction frequency of G4-containing enhancer–promoter pairs was significantly higher than that of non-G4-containing pairs (Figure 6a). In addition, we observed significant overlap between G-quadruplexes and known TF binding sites. More than 99% of G-quadruplexes overlapped with TFBSs, suggesting G-quadruplexes are highly associated with TF binding sits. Moreover, architectural proteins including CTCF, RAD21, and SMC3 were significantly enriched in intersections. We herein proposed a new mechanism in which G-quadruplexes regulate genes from afar through three-dimensional interactions. In model C, G-quadruplexes are associated with the frequent interaction of enhancers and promoters (Figure 8c). Abundant TFBSs around G4-containing regulatory elements can strengthen enhancer–promoter interactions by protein–protein interactions.
Materials and methods
Identification of genomic elements
The sequences of all human genes were downloaded from RefSeq Genes of GRCh37/hg19 datasets [62]. Only protein coding genes were selected as our candidate sets. The regions around TSSs (–2,000 bp to 500 bp) were designated as promoters as suggested by He et al. [63]. In the case of genes with more than one TSS, we selected the TSS closest to the 5′ end. Protein coding genes with TSSs located within the border of another gene were excluded. A total of 17,819 promoters of protein coding genes were retained.
Enhancers of K562 cell lines were derived from research by Yip [64], which identified enhancers from the intersection of gene-distal regulatory modules (DRMs) and predicted enhancers of ChromHMM and Segway. Enhancers overlapping with promoter regions were excluded. A total of 31,237 enhancers were retained in K562 cell lines.
G4 sequence data and G4 ChIP-seq data
G4 sequence data were originally derived by Chambers et al. [6], and were downloaded from the Gene Expression Omnibus (GEO) repository under accession number GSE63874. Across the genome, 716,311 observed G4 sequences were found in G4-stabilizing ligand pyridostatin (PDS) liquid and 525,908 observed G4 sequence were found in K+ liquid. Of these, 409,365 G4 sequences (K+: 78%, PDS: 57%) occurring in both experimental conditions were selected as our G4 sequences data.
G4 ChIP-seq data in K562 cell lines generated by Mao et al. were adopted to identify exact positions of G-quadruplexes in vivo [15]. G4 ChIP-seq data were achieved from the GEO repository under accession number GSE107690. To identify exact G-quadruplex regions, we performed peak calling by MACS v2.0 with default parameters [54].
Hi-C and TAD boundaries
Hi-C data produced by Rao et al. [17] were obtained from the GEO repository under accession number GSE63525. Five kb-resolution contact matrixes of K562 cell lines were used to call TADs. To normalize the Hi-C matrices, the matrix balancing algorithm proposed by Knight and Ruiz was adopted [65]. This algorithm based on inner–outer iteration schemes efficiently balance a matrix when the original matrix is not too sparse. To avoid an increased number of interactions at short distances, we used the methods of Rao et al. [17] in which each entry (Mi,j) of the matrix is divided by the expected value corresponding to the distance i-j.
From the 5,985 previous defined TADs of K562 cell lines [17], all TADs shorter than 200 kb were excluded as suggested by Hong et al. [34]. This left a total of 4,457 TADs in K562 cell lines. The start/end sites of retained TADs were chosen as candidate boundaries. For overlapping TADs, the midpoint of the overlap region was designated as the boundary. When a gap occurred the two ends, we merged them into a boundary if the spacing was less than 100 kb, as suggested by Hong et al. [34].
Relative density of G4 ChIP-seq peak/G4 sequences/PG4 motifs along TADs
We calculated relative peak density in the range of 50% ×L upstream and downstream of the TADs. L indicates the length of TADs. Each TAD was divided into 2000 equal-sized bins. Next the number of G4 ChIP-seq peaks/G4 sequences/PG4 motifs per bin was counted. Conducting this analysis across all TADs yielded a matrix. The sum of each column was taken. To account for differences in total peak count for different data (G4 ChIP-seq peaks/G4 sequences/PG4 motifs), normalization was conducted by taking sum of peak/motif counts in all TADs and dividing the bin sums by this normalizing factor.
Insulation score
The insulation score metric was proposed by Crane et al. to estimate the ability of a given locus in the separation of adjacent regions [21,42]. The insulation score indicates the interaction strength between adjacent regions of the locus. Lower insulation scores indicate higher insulation, representing fewer interactions between adjacent regions of the locus. The score is calculated by sliding a square window along the diagonal of the contact matrix and recording the interactions within the window. If C is the binned contact matrix and w is the window size in bins, then the insulation score I for bin number i can be calculated as follows:
To normalize the insulation score, we divide I(i,w) by the central moving average (300 bins window).
ChIP-seq data
We downloaded the ATAC-seq data and ChIP-seq datasets of CTCF, RAD21, and SMC3 of K562 cell lines from the UCSC ftp server (http://genome.ucsc.edu) [66–69]. Then we performed peak calling by MACS v2.0 with default parameters [54].
Determination of distances between G-quadruplexes and CTCF motifs
We used the middle points of ChIP-seq peaks to represent the binding sites. And the distances between G-quadruplexes and their closest CTCF motifs were calculated by BEDtools [50]. Only the pairs with distance < 100 bp were retained. We used the same strategy on calculation of the distances between CTCF binding sites and cohesin protein binding sites.
Interaction of regulatory elements
To assess the interaction strength between a pair of regulatory elements (enhancer-promoter, enhancer-enhancer, and promoter-promoter), we used Hi-C interaction reads counts to represent the interaction strength. For example, for an enhancer (X) and a promoter (Y), their interaction strength can be calculated as follows:
where Hx,y is the interaction strength of X and Y; and hi,j is the Hi-C reads of which one end should be located in gene promoter regions (Y) and the other end should be located in enhancer regions (X). Then we used Fit-HiC to evaluate the FDR of the interaction pairs (interaction pairs with FDR<0.01 were retained) [48].
RNA-seq data
The paired-end RNA-seq data for K562 cell lines, generated by the ENCODE/Caltech group, were downloaded from the UCSC ftp server [68,69]. All the RNA-seq reads were mapped to the human reference genome (GRCh37/hg19) using Tophat [70]. We used cufflinks to generate transcriptome assembly [71].
Enrichment analysis
The method of enrichment analysis is proposed by Hansel-Hertsch et al. [9]. The most accessible genomic regions according to ATAC-seq peaks were selected. The count of ATAC-seq peaks is equal to that of G4 ChIP-seq peaks. We exclude the overlapped data of ATAC-seq peaks and G4 ChIP-seq peaks. All TF ChIP-seq peak file were randomly shuffled (N = 6) across the human genome. We counted the overlap peaks between the TF ChIP-seq peaks and G4 ChIP-seq peaks/ATAC-seq peaks. Enrichment between TF ChIP-seq and G4 ChIP-seq/ATAC-seq data sets were calculated from the ratio of the direct overlaps with the randomly shuffled overlaps.
Funding Statement
This work was supported by the National Natural Science Foundation of China under grant number 61472078; Key Research and Development Program of Jiangsu province under grant number BE2016002-3.
Acknowledgments
We thank Sarah Williams, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn), for editing the English text of a draft of this manuscript
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental materials data for this article can be accessed here.
References
- [1].Kwok CK, Merrick CJ.. G-Quadruplexes: prediction, characterization, and biological application. Trends Biotechnol. 2017. October;35(10):997–1013. PubMed PMID: 28755976. [DOI] [PubMed] [Google Scholar]
- [2].van Wietmarschen N, Merzouk S, Halsema N, et al. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat Commun. 2018. January 18;9(1):271 10.1038/s41467-017-02760-1. PubMed PMID: 29348659; PubMed Central PMCID: PMC5773480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Maizels N, Gray LT.. The G4 genome. PLoS Genet. 2013. April;9(4):e1003468 PubMed PMID: 23637633; PubMed Central PMCID: PMC3630100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Du Z, Zhao Y, Li N.. Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res. 2009. November;37(20):6784–6798. PubMed PMID: 19759215; PubMed Central PMCID: PMC2777415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hansel-Hertsch R, Di Antonio M, Balasubramanian S. DNA G-quadruplexes in the human genome: detection, functions and therapeutic potential. Nat Rev Mol Cell Biol. 2017. May;18(5):279–284. PubMed PMID: 28225080. [DOI] [PubMed] [Google Scholar]
- [6].Chambers VS, Marsico G, Boutell JM, et al. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol. 2015. August;33(8):877–881. PubMed PMID: 26192317. [DOI] [PubMed] [Google Scholar]
- [7].Schaffitzel C, Berger I, Postberg J, et al. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A. 2001. July 17;98(15):8572–8577. PubMed PMID: 11438689; PubMed Central PMCID: PMC37477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hoffmann RF, Moshkin YM, Mouton S, et al. Guanine quadruplex structures localize to heterochromatin. Nucleic Acids Res. 2016. January 08;44(1):152–163. PubMed PMID: 26384414; PubMed Central PMCID: PMC4705689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hansel-Hertsch R, Beraldi D, Lensing SV, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016. October;48(10):1267–1272. PubMed PMID: 27618450. [DOI] [PubMed] [Google Scholar]
- [10].Hansel-Hertsch R, Spiegel J, Marsico G, et al. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018. March;13(3):551–564. PubMed PMID: 29470465. [DOI] [PubMed] [Google Scholar]
- [11].Henderson E, Hardin CC, Walk SK, et al. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987. December 24;51(6):899–908. PubMed PMID: 3690664. [DOI] [PubMed] [Google Scholar]
- [12].Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci U S A. 2005. August 2;102(31):10864–10869. PubMed PMID: 16043710; PubMed Central PMCID: PMC1180509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hegyi H. Enhancer-promoter interaction facilitated by transiently forming G-quadruplexes. Sci Rep. 2015;5:9165 PubMed PMID: 25772493; PubMed Central PMCID: PMC4360481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Renciuk D, Rynes J, Kejnovska I, et al. G-quadruplex formation in the Oct4 promoter positively regulates Oct4 expression. Biochim Biophys Acta. 2017. February;1860(2):175–183. PubMed PMID: 27863263. [DOI] [PubMed] [Google Scholar]
- [15].Mao SQ, Ghanbarian AT, Spiegel J, et al. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol. 2018. October;25(10):951–957. 10.1038/s41594-018-0131-8. PubMed PMID: 30275516; PubMed Central PMCID: PMC6173298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen MC, Tippana R, Demeshkina NA, et al. Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature. 2018. June;558(7710):465–469. 10.1038/s41586-018-0209-9. PubMed PMID: 29899445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rao SS, Huntley MH, Durand NC, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014. December 18;159(7):1665–1680. PubMed PMID: 25497547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lin D, Hong P, Zhang S, et al. Digestion-ligation-only Hi-C is an efficient and cost-effective method for chromosome conformation capture. Nat Genet. 2018. May;50(5):754–763. 10.1038/s41588-018-0111-2. PubMed PMID: 29700467. [DOI] [PubMed] [Google Scholar]
- [19].Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009. October 9;326(5950):289–293. PubMed PMID: 19815776; PubMed Central PMCID: PMC2858594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ke Y, Xu Y, Chen X, et al. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell. 2017. July 13;170(2):367–381 e20. PubMed PMID: 28709003. [DOI] [PubMed] [Google Scholar]
- [21].Hug CB, Grimaldi AG, Kruse K, et al. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell. 2017. April 6;169(2):216–228 e19. PubMed PMID: 28388407. [DOI] [PubMed] [Google Scholar]
- [22].Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012. May 17;485(7398):376–380. PubMed PMID: 22495300; PubMed Central PMCID: PMC3356448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sexton T, Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015. March 12;160(6):1049–1059. PubMed PMID: 25768903. [DOI] [PubMed] [Google Scholar]
- [24].Sun L, Yu R, Dang W. Chromatin architectural changes during cellular senescence and aging. Genes (Basel). 2018. April 16;9(4). PubMed PMID: 29659513; PubMed Central PMCID: PMC5924553 DOI: 10.3390/genes9040211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Went M, Sud A, Forsti A, et al. Identification of multiple risk loci and regulatory mechanisms influencing susceptibility to multiple myeloma. Nat Commun. 2018. September 13;9(1):3707 PubMed PMID: 30213928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choy MK, Javierre BM, Williams SG, et al. Promoter interactome of human embryonic stem cell-derived cardiomyocytes connects GWAS regions to cardiac gene networks. Nat Commun. 2018. June 28;9(1):2526 10.1038/s41467-018-04931-0. PubMed PMID: 29955040; PubMed Central PMCID: PMC6023870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Rivera-Mulia JC, Dimond A, Vera D, et al. Allele-specific control of replication timing and genome organization during development. Genome Res. 2018. June;28(6):800–811. PubMed PMID: 29735606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ramirez F, Bhardwaj V, Arrigoni L, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018. January 15;9(1):189 10.1038/s41467-017-02525-w. PubMed PMID: 29335486; PubMed Central PMCID: PMC5768762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Canela A, Maman Y, Jung S, et al. Genome organization drives chromosome fragility. Cell. 2017. July 27;170(3):507–521 e18. PubMed PMID: 28735753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Rao SSP, Huang SC, Glenn St Hilaire B, et al. Cohesin loss eliminates all loop domains. Cell. 2017. October 05;171(2):305–320 e24. PubMed PMID: 28985562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gong Y, Lazaris C, Sakellaropoulos T, et al. Stratification of TAD boundaries reveals preferential insulation of super-enhancers by strong boundaries. Nat Commun. 2018. February 7;9(1):542 10.1038/s41467-018-03017-1. PubMed PMID: 29416042; PubMed Central PMCID: PMC5803259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lupianez DG, Kraft K, Heinrich V, et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015. May 21;161(5):1012–1025. PubMed PMID: 25959774; PubMed Central PMCID: PMC4791538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Li L, Lyu X, Hou C, et al. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol Cell. 2015. April 16;58(2):216–231. PubMed PMID: 25818644; PubMed Central PMCID: PMC4402144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hong S, Kim D. Computational characterization of chromatin domain boundary-associated genomic elements. Nucleic Acids Res. 2017. October 13;45(18):10403–10414. PubMed PMID: 28977568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li A, Yin X, Xu B, et al. Decoding topologically associating domains with ultra-low resolution Hi-C data by graph structural entropy. Nat Commun. 2018. August 15;9(1):3265 10.1038/s41467-018-05691-7. PubMed PMID: 30111883; PubMed Central PMCID: PMC6093941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gorkin DU, Leung D, Ren B. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014. June 5;14(6):762–775. PubMed PMID: 24905166; PubMed Central PMCID: PMC4107214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Halder K, Halder R, Chowdhury S. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol Biosyst. 2009. December;5(12):1703–1712. PubMed PMID: 19587895. [DOI] [PubMed] [Google Scholar]
- [38].Foulk MS, Urban JM, Casella C, et al. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015. May;25(5):725–735. PubMed PMID: 25695952; PubMed Central PMCID: PMC4417120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jt R, Turner D, Durand NC, et al. Juicebox.js provides a cloud-based visualization system for Hi-C data. Cell Syst. 2018. February 28;6(2):256–258 e1. PubMed PMID: 29428417; PubMed Central PMCID: PMC6047755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Durand NC, Robinson JT, Shamim MS, et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016. July;3(1):99–101. PubMed PMID: 27467250; PubMed Central PMCID: PMC5596920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Van Bortle K, Nichols MH, Li L, et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 2014;15(6):R82 PubMed PMID: 24981874; PubMed Central PMCID: PMC4226948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Crane E, Bian Q, McCord RP, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015. July 9;523(7559):240–244. PubMed PMID: 26030525; PubMed Central PMCID: PMC4498965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nichols MH, Corces VG. A CTCF Code for 3D genome architecture. Cell. 2015. August 13;162(4):703–705. PubMed PMID: 26276625; PubMed Central PMCID: PMC4745123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010. May 28;38(4):576–589. PubMed PMID: 20513432; PubMed Central PMCID: PMC2898526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Benner C, Isoda T, Murre C. New roles for DNA cytosine modification, eRNA, anchors, and superanchors in developing B cell progenitors. Proc Natl Acad Sci U S A. 2015. October 13;112(41):12776–12781. PubMed PMID: 26417104; PubMed Central PMCID: PMC4611620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nagy G, Czipa E, Steiner L, et al. Motif oriented high-resolution analysis of ChIP-seq data reveals the topological order of CTCF and cohesin proteins on DNA. BMC Genomics. 2016. August 15;17(1):637 10.1186/s12864-016-2940-7. PubMed PMID: 27526722; PubMed Central PMCID: PMC4986361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012. November;13(11):770–780. PubMed PMID: 23032257; PubMed Central PMCID: PMC3725559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Ay F, Bailey TL, Noble WS. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 2014. June;24(6):999–1011. PubMed PMID: 24501021; PubMed Central PMCID: PMC4032863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cheneby J, Gheorghe M, Artufel M, et al. ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Res. 2018. January 4;46(D1):D267–D275. PubMed PMID: 29126285; PubMed Central PMCID: PMC5753247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010. March 15;26(6):841–842. PubMed PMID: 20110278; PubMed Central PMCID: PMC2832824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Qi HH, Sarkissian M, Hu GQ, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010. July 22;466(7305):503–507. PubMed PMID: 20622853; PubMed Central PMCID: PMC3072215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lee BK, Bhinge AA, Iyer VR. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res. 2011. May;39(9):3558–3573. PubMed PMID: 21247883; PubMed Central PMCID: PMC3089461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Pataskar A, Jung J, Smialowski P, et al. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. Embo J. 2016. January 4;35(1):24–45. PubMed PMID: 26516211; PubMed Central PMCID: PMC4718003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9(9):R137 PubMed PMID: 18798982; PubMed Central PMCID: PMC2592715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Prioleau MN. G-Quadruplexes and DNA replication origins. Adv Exp Med Biol. 2017;1042:273–286. PubMed PMID: 29357063. [DOI] [PubMed] [Google Scholar]
- [56].Moriyama K, Yoshizawa-Sugata N, Masai H. Oligomer formation and G-quadruplex binding by purified murine Rif1 protein, a key organizer of higher-order chromatin architecture. J Biol Chem. 2018. March 9;293(10):3607–3624. PubMed PMID: 29348174; PubMed Central PMCID: PMC5846147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Saito TL, Hashimoto S, Gu SG, et al. The transcription start site landscape of C. elegans. Genome Res. 2013. August;23(8):1348–1361. PubMed PMID: 23636945; PubMed Central PMCID: PMC3730108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ganji M, Shaltiel IA, Bisht S, et al. Real-time imaging of DNA loop extrusion by condensin. Science. 2018. April 6;360(6384):102–105. PubMed PMID: 29472443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Wang X, Brandao HB, Le TB, et al. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017. February 3;355(6324):524–527. PubMed PMID: 28154080; PubMed Central PMCID: PMC5484144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Sanborn AL, Rao SS, Huang SC, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015. November 24;112(47):E6456–65. PubMed PMID: 26499245; PubMed Central PMCID: PMC4664323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rhodes D, Lipps HJ. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015. October 15;43(18):8627–8637. PubMed PMID: 26350216; PubMed Central PMCID: PMC4605312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Pruitt KD, Tatusova T, Brown GR, et al. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. (Database issue):D130–5. PubMed PMID: 22121212; PubMed Central PMCID: PMC3245008 2012. January;40:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].He B, Chen C, Teng L, et al. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci U S A. 2014. May 27;111(21):E2191–9. PubMed PMID: 24821768; PubMed Central PMCID: PMC4040567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yip KY, Cheng C, Bhardwaj N, et al. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol. 2012. September 26;13(9):R48 PubMed PMID: 22950945; PubMed Central PMCID: PMC3491392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Knight PA, Ruiz D. A fast algorithm for matrix balancing. Ima J Numer Anal. 2013. July;33(3):1029–1047. PubMed PMID: WOS:000321450400011; English [Google Scholar]
- [66].Ep C. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012. September 6;489(7414):57–74. PubMed PMID: 22955616; PubMed Central PMCID: PMC3439153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008. December;26(12):1351–1359. PubMed PMID: 19029915; PubMed Central PMCID: PMC2597701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Casper J, Zweig AS, Villarreal C, et al. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res. 2018. January 4;46(D1):D762–D769. PubMed PMID: 29106570; PubMed Central PMCID: PMC5753355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Speir ML, Zweig AS, Rosenbloom KR, et al. The UCSC genome browser database: 2016 update. Nucleic Acids Res. 2016. January 4;44(D1):D717–25. PubMed PMID: 26590259; PubMed Central PMCID: PMC4702902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009. May 1;25(9):1105–1111. PubMed PMID: 19289445; PubMed Central PMCID: PMC2672628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012. March 1;7(3):562–578. PubMed PMID: 22383036; PubMed Central PMCID: PMC3334321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


