Abstract
Brown adipose tissue (BAT) has been considered beneficial for metabolic health by participating in the regulation of glucose homoeostasis. The browning factors that improve glucose uptake beyond normal levels are still unknown but glucose uptake is not affected in UCP1 knockout mice. Here, we demonstrate in human white adipocytes that basal/resting glucose uptake is improved by solely elevating UCP1 protein levels. Generating human white Simpson-Golabi-Behmel syndrome (SGBS) adipocytes with a stable knockout and overexpression of UCP1, we discovered that UCP1 overexpressing adipocytes significantly improve glucose uptake by 40%. Mechanistically, this is caused by higher glycolytic flux, seen as increased oxygen consumption, extracellular acidification and lactate secretion rates. The improvements in glucose handling are comparable to white-to-brown transitions, as judged by, for the first time, directly comparing in vitro differentiated mouse brown vs white adipocytes. Although no adipogenic, metabolic and mitochondrial gene expressions were significantly altered in SGBS cells, pharmacological inhibition of GLUT1 completely abrogated differences between UCP1+ and control cells, thereby uncovering GLUT1-mediated uptake as permissive gatekeeper. Collectively, our data demonstrate that elevating UCP1 levels is sufficient to improve human white adipocytes as a glucose sink without adverse cellular effects, thus not requiring the adrenergic controlled, complex network of browning which usually hampers translational efforts.
Keywords: Uncoupling protein 1, Human adipocytes, Glucose uptake
Abbreviations: BAT, brown adipose tissue; CT, computed tomography; FFA, free fatty acids; NST, non-shivering thermogenesis; PET, positron emission tomography; SGBS, Simpson-Golabi-Behmel syndrome; UCP1, uncoupling protein 1; WAT, white adipose tissue
Highlights
-
•
Basal glucose uptake in human adipocytes is improved by solely elevating UCP1 levels.
-
•
Adipogenic, metabolic and mitochondrial gene expressions were not affected by UCP1 overexpression.
-
•
UCP1-driven increase in glucose uptake is mediated by GLUT1.
1. Introduction
Brown adipose tissue (BAT) enables mammals to maintain their body temperature by non-shivering thermogenesis (NST). BAT is highly prevalent in newborns and essential for their survival in absence of other thermogenic means [1]. It is also detectable in adults, where its presence is associated with lower body weight [2]. Besides participating in the regulation of body temperature and energy homeostasis, BAT activity has beneficial effects on systemic metabolism [3,4]. As such, it increases glucose tolerance and insulin sensitivity, and decreases dyslipidemia and body weight [3,4]. Consequently, BAT is currently discussed as a target organ to treat obesity and obesity-associated diseases [5].
The key component mediating thermogenesis is uncoupling protein 1 (UCP1). UCP1 is located in the inner mitochondrial membrane, where it dissociates cellular respiration from the generation of ATP. Upon certain stimuli such as cold exposure, sympathetically-derived norepinephrine activates β3 adrenergic receptors on brown adipocytes, triggering the release of free fatty acids (FFA) and subsequent activation UCP1 [1]. The persistence of BAT from neonates into adulthood, however, is limited. The induction of “beige” or “brite” adipocytes in human white adipose tissue (WAT) represents another approach to combust excessive nutrient energy. In mice, cold-induced WAT browning improves systemic glucose and lipid homeostasis [6]. Mirabegron, a FDA-approved β3-adrenergic receptor agonist for overactive bladder, also induces in human WAT a complex regulatory network for thermogenesis, including UCP1, TMEM26, CIDEA and many other browning genes [7,8].
Data from in vitro studies suggest that glucose uptake and glycolysis are strictly required for adrenergically stimulated oxidative metabolism of murine brown adipocytes [9]. Ectopic UCP1 expression in murine WAT reduces plasma glucose levels [10]. Indeed, glucose uptake has been widely used to detect BAT activity in both mice and humans under basal and cold-exposed conditions with 18F-FDG-PET/CT being the gold standard of BAT detection and activity measurement [11]. This method is based on the observation that BAT is metabolically more active than other tissues [2,12,13]. It thus stands to reason that BAT takes up more glucose than WAT. Interestingly though, knockout of UCP1 in mice does not lead to diminished glucose uptake indicating that thermogenesis and glucose flux are dissociated in BAT and the molecular basis of elevated glucose uptake in BAT still has to be identified [14].
The aim of this study was to delineate whether UCP1 is a main driver of glucose uptake in adipocytes. To this end, we compared the cellular glucose uptake in murine white and brown adipocytes isolated and differentiated from inguinal white and interscapular brown adipose tissue. Furthermore, we knocked out and overexpressed UCP1 in Simpson-Golabi-Behmel (SGBS) cells, a unique human model system for white adipocytes to study adipogenesis [15,16].
2. Materials and methods
2.1. Materials
Standard chemicals were purchased from Merck (Darmstadt, Germany), media and buffers for cell culture were purchased from ThermoFisher Scientific (Waltham, USA).
2.2. Murine cell isolation and culture
6–8 week old C57BL/6J mice were sacrificed by cervical dislocation and inguinal white as well as interscapular brown adipose tissues were dissected. Stromal vascular cells (SVC) were isolated by collagenase digestion and subjected to adipogenic differentiation as described [17]. Briefly, tissues were digested using collagenase and stromal cells were harvested by centrifugation. Cells were seeded in DMEM:F12 containing 10% FCS until reaching subconfluency. For subsequent experiments, cells were trypsinized and reseeded into 12 well plates. Adipogenic differentiation was induced for 12 days DMEM:F12 medium supplemented with 10% FCS, 850 nM insulin, 1 nM T3, 125 μM indomethacin, and 1 μM rosiglitazone. For the first 2 days, 500 μM isobutylmethylxanthine, and 1 μM dexamethasone were added.
2.4. Generation of UCP1-overexpressing and UCP1-deficient cells
For UCP1 overexpression constructs, the UCP1 open reading frame was PCR amplified from SGBS adipocyte cDNA using specific primer pairs harboring attB sites (Supplemental table S2), inserted into pDONR221 using Gateway BP Clonase II (ThermoFisher Scientific), and was verified by Sanger sequencing. UCP1 overexpression cassettes were transferred from pDONR221-UCP1 to pLenti6.3/V5-DEST using Gateway LR Clonase II (ThermoFisher Scientific). Lentiviral particles were produced by calcium phosphate-mediated co-transfection of HEK293FT cells with pLenti6.3-UCP1 or pLenti6.3-empty, psPAX2, and pMD2. G (Addgene #12260 and #12259, kindly provided by Didier Trono). Lentivirus-containing supernatants were harvested 48 h post-transfection and titers were determined by standard colony assays. SGBS preadipocytes were transduced with UCP1 or empty control lentiviruses at an MOI of 1.5 and stable bulk cultures were generated by blasticidin selection. UCP1 overexpression was confirmed by qRT-PCR and Western Blot.
For UCP1 knockout constructs, sgRNA duplexes targeting the UCP1 gene and non-targeting controls were cloned into the pMuLE ENTR U6 stuffer sgRNA scaffold L1-L4 plasmid (Multiple Lentiviral Expression Kit, Addgene #1000000060, kindly provided by Ian Frew) [18]. Correct insertion of the sgRNA duplexes was verified by Sanger sequencing. A Gateway cloning-compatible SleepingBeauty transposon plasmid ‘pMuSE eGFP-P2A-PuroR DEST’ [19] was derived from pSBtet-GP (Addgene #60495, kindly provided by Eric Kowarz) and pLenti6.3/V5-DEST (ThermoFisher Scientific) using routine cloning methods. The specific pMuLE ENTR U6-sgRNA plasmids were recombined with pMuLE ENTR CMV-hCas9 R4-R3, pMuLE ENTR MCS L3-L2, and pMuSE eGFP-P2A-PuroR DEST plasmids using LR Clonase II Plus (ThermoFisher Scientific). SGBS preadipocytes were co-transfected with the resulting pMuSE U6-sgRNA + CMV-hCas9 + RPBSA-eGFP-P2A-PuroR plasmids and the SleepingBeauty-expressing pCMV(CAT)T7-SB100 plasmid (Addgene #34879, kindly provided by Zsuzsanna Izsvak) at a mass ratio of 19:1 using a Neon Transfection System (ThermoFisher Scientific) with 3 × 10 ms pulses of 1400 V. Stable bulk cultures were generated by puromycin selection. UCP1 knockout was confirmed by Western Blot. All essential sequences are listed in Supplemental Table S1.
SGBS cells were cultured and differentiated into adipocytes using a protocol described before [15]. Briefly, cells were seeded into cell culture vessels in DMEM:F12 containing 3.3 mM biotin, 1.7 mM panthotenate, and 10% FCS. Adipogenic differentiation was induced for 14 days in serum-free DMEM:F12 medium supplemented with 10 μg/ml apo-transferrin, 10 nM insulin, 200 pM T3, and 1 μM cortisol. For the first 4 days, 2 μM rosiglitazone, 250 μM isobutylmethylxanthine, and 25 nM dexamethasone were added.
2.5. Gene expression analysis
RNA was isolated using the Direct-Zol RNA Kit (Zymo Research, Irvine, USA). 0.5 μg of total RNA was reverse transcribed using SuperScript II Reverse Transcriptase (ThermoFisher Scientific). Relative expression of target genes was analyzed by quantitative real-time PCR using the ssoAdvanced Universal SYBR Green Supermix on a CFX Connect Real Time PCR Detection System (BioRad, Munich, Germany). Expression values were calculated using the dCt method with hypoxanthine-guanine phosphoribosyltransferase (HPRT) as a reference gene. Sequences of the primers are shown in Supplemental Table S2.
2.6. Western Blot
Cells were washed with ice-cold PBS and total protein was extracted by scraping them in a lysis buffer (10 mM Tris-HCl at pH 7.5, 150 mM sodium chloride, 2 mM EDTA, 1% TX-100, 10% glycerol, 1X cOmplete Protease Inhibitors (Roche, Mannheim, Germany)). After 30 min incubation on ice, cell debris were pelleted by centrifugation. Protein content in the supernatants was determined using the Bradford Protein Assay (BioRad). For immunodetection, 10–20 μg of protein were separated by SDS-PAGE on Bolt Bis-Tris Plus Gels (ThermoFisher Scientific) and transferred to nitrocellulose membranes by Western Blotting (ThermoFisher Scientific). The following antibodies were used: mouse anti-UCP1 (R&D Systems #536435, Minneapolis, UCA), mouse anti-OXPHOS protein cocktail (Abcam #ab110411, Cambridge, UK), rabbit anti-TIM23 (Abcam #ab116329), rabbit anti-perilipin A (Abcam #ab3526), rabbit anti-PPARG (Cell Signaling Technology #2443), rabbit anti-adiponectin (GeneTex #GTX112777), mouse anti-GAPDH (HyTest #5G4, clone 6C5, Turku, Finland), mouse anti-β-actin (Merck #A5441).
2.7. Glucose metabolism
In vitro differentiated adipocytes were washed with PBS and incubated in serum-free medium overnight. On the day of measurement, medium was replaced with glucose-free Krebs-Ringer buffer (130 mM NaCl, 10 mM MgSO4, 2.5 mM NaH2PO4, 4.6 mM KCl, 2.5 mM CaCl2, 2.5 mM sodium pyruvate, 5 mM HEPES, pH 7.4). To determine basal glucose uptake, [14C]-2-deoxy-d-glucose (0.2 μCi/well, PerkinElmer, Waltham, USA) was added and the cells were incubated for 15 min at 37 °C. Subsequently, cells were washed with ice-cold PBS and harvested with 100 mM NaOH. Incorporation of [14C]-2-deoxy-d-glucose was measured on a β-counter. In some experiments, the cells were pre-treated for 1 h with 10 μM Cytochalasin B or 100 nM of the selective GLUT1 inhibitor BAY-876 before addition of [14C]-2-deoxy-d-glucose. Data obtained from SGBS cells are given in counts per minute (cpm). As SVC isolated from iWAT and BAT were different in cell size and rate of adipogenic differentiation, we counted the number of adipocytes in the cell culture dishes. Raw cpm values were then normalized to adipocyte numbers per well (cpm/105 adipocytes).
For quantification of lactate secretion, the cells were incubated in serum-free medium overnight and then in DMEM medium supplemented with 10 mM glucose, 1 mM pyruvate, 2 mM GlutaMaxx, and 1% bovine serum albumin. Media supernatants were harvested after 16 h and assayed for lactate content using the Lactate Colorimetric Assay Kit II (Biovision, Milpitas, USA).
2.8. Mitochondrial quantification
Citric acid synthase activity was assayed as a measure for mitochondrial content and activity as described before [20]. In brief, 5 μg total protein were added to reaction buffer (100 mM Tris HCl pH 8.1, 100 μM 5,5’ dithiobis-2-nitrobenzoic acid (DTNB), 300 μM acetyl-CoA, 0.1% Triton X 100) and specific activity was determined by measuring conversion of DNTB to 2-nitro-5-benzoic acid (TNB) at a wavelength of 405 nm after adding 500 μM oxaloacetate as substrate.
DNA was isolated from cells using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). Content of both nuclear and mitochondrial DNA was quantified in by qRT-PCR using specific primers as described before [20]. Mitochondrial/nuclear ratio was calculated as a relative measure of mitochondrial DNA content.
2.9. Subcellular fractionation
Cells were scraped into fractionation buffer (20 mM HEPES pH 7.4, 250 mM sucrose, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM DTT, cOmplete Protease Inhibitors (Roche) and homogenized by passing them through a 25 G needle 10 times. Nuclei were separated by centrifugation at 750g for 5 min at 4 °C. The supernatant was centrifuged at 10,000 g for 20 min at 4 °C to obtain the mitochondrial fraction. The supernatant comprising the cytosolic fraction was precipitated by adding four volumes of ice-cold acetone, incubation for 60 min at −20 °C, and centrifugation at g for 30 min at 4 °C. The protein precipitates were dissolved in equal amounts of 1X Bolt LDS Sample Buffer (ThermoFisher Scientific) and subjected to Western Blot analysis.
2.10. Cellular respiration and extracellular acidification
Cells were plated in 96-well cell culture microplates (XFe96, Agilent Technologies, Santa Clara, USA) and differentiated into adipocytes for 14 days. One day before measurement, the culture medium was changed to insulin-free medium. On the day of measurement, the cells were incubated for 1 h in bicarbonate-free DMEM medium containing 5 mM HEPES, 10 mM glucose, 1 mM pyruvate, 2 mM GlutaMaxx, and 1% bovine serum albumin. Oxygen consumption and extracellular acidification rates (OCR and ECAR) were measured simultaneously using a Seahorse XFe96 Flux Analyzer (Agilent Technologies). To mimic thermogenic activation, the cells were treated with 0.5 mM dibutyryl cyclic adenosinmonophosphate (cAMP). Uncoupled (proton leak) respiration was profiled by injecting 2 μM oligomycin (inhibiting the ATP synthase), and full substrate oxidation capacity was determined by injecting 4 μM carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP, a chemical uncoupler). Non-mitochondrial respiration was determined by injecting 1.5 μM antimycin A and 1.5 μM rotenone (inhibiting electron flux through complex I and III). OCRs were determined by machine algorithms and plotted against time. Data were either normalized for lipid content by staining with NileRed or to cell content by staining with JanusGreen [21].
2.11. Mitochondrial membrane potential
To estimate mitochondrial membrane potential, the cells were stained with 30 nM MitoTracker Red CMXRos (ThermoFisher Scientific) for 30 min at 37 °C. Subsequently, the cells were washed with PBS, trypsinized, and centrifuged. The cell pellets were resuspended in PBS and the cells were analyzed with the FACSCalibur (BD Biosciences, Heidelberg, Germany).
2.12. Statistics
If not stated otherwise, data from three independent experiments are expressed as mean±standard error of the mean (SEM). For statistical comparison, analysis of variance (ANOVA) or t-tests were used as indicated. A p < 0.05 was considered statistically significant. GraphPad Prism version 7.01 (GraphPad Software, San Diego, USA) was used for all analyses.
3. Results
3.1. Elevated basal glucose uptake in mouse brown vs white adipocytes
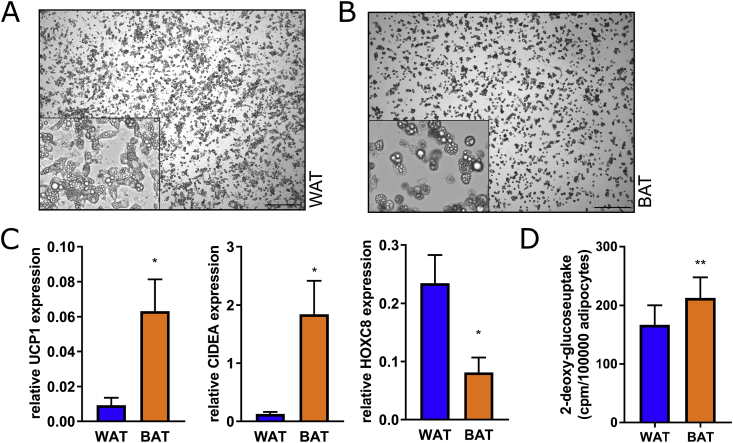
To determine a difference in the capacity to take up glucose on a cellular level, we isolated stromal-vascular cells from WAT and BAT and subjected them to adipogenic differentiation in vitro. After 10 days of differentiation, the murine cells showed histological signs of adipocytes, i.e. round shape and incorporated lipid droplets (Fig. 1A and B), significantly higher expression of UCP1 and CIDEA in BAT vs WAT cells, and higher expression of HOXC8 in WAT cells (Fig. 1C). To our knowledge, specific glucose uptake rates have never been experimentally compared between isolated BAT vs WAT cells. In line with previous assumptions on heterogeneous bulk tissue in vivo, brown adipocytes in vitro display the expected significantly higher basal glucose uptake rates per cell (cpm/105 adipocytes), compared to white adipocytes (Fig. 1D).
Fig. 1.
Comparison of glucose uptake in murine in vitro differentiated white and brown adipocytes. (A) Stromal-vascular cells isolated from inguinal WAT and interscapular BAT were differentiated for 10 days into mature adipocytes. (B) mRNA expression of the BAT-specific markers UCP1 and CIDEA and the WAT-specific marker HOXC8 was measured by qRT-PCR. (C) Basal uptake of 14C-deoxy-glucose was determined in adipocytes after serum starvation overnight, cpm values were normalized to adipocyte number. Data (n = 3) are shown as means + SEM, *p < 0.05, **p < 0.01 (Student's t-test). . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Generation of UCP1-deficient human adipocytes
To investigate whether UCP1 is involved in the regulation of cellular glucose uptake and utilization, we generated adipocytes with either loss or gain of UCP1-functionality. As model system we used human Simpson-Golabi-Behmel syndrome (SGBS) cells, which were derived from subcutaneous WAT of an infant affected by the eponymous syndrome [15]. These cells undergo adipogenic differentiation in the presence of a PPARγ agonist and express low levels of UCP1 at late adipogenesis [20].
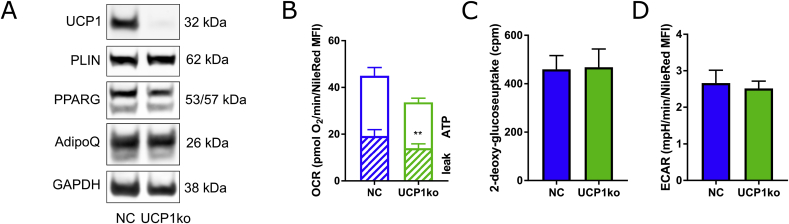
We generated UCP1-deficient cells using the CRIPSR-Cas9 system. UCP1 protein was completely absent in bulk cultures of knockout adipocytes as assessed by Western Blot analysis (Fig. 2A), with no difference in adipogenic differentiation, as judged by protein expression of adipogenic markers (Fig. 2A). UCP1-deficient adipocytes displayed a significantly reduced proton leak respiration by 17% compared to control cells (Fig. 2B). As expected from recent mouse data in vivo [14], the basal glucose uptake (Fig. 2C) as well as the extracellular acidification rates (ECARs) were not different between control and UCP1-deficient cells (Fig. 2D). Collectively, the knockout of UCP1 imposed only minor effects on the glucose metabolism of white adipocytes.
Fig. 2.
CRISPR/Cas9-mediated UCP1 knockout in human adipocytes. UCP1-deficient cells were generated by co-expressing Cas9 and specific sgRNAs targeting UCP1 in SGBS preadipocytes. (A) The cells were differentiated into mature adipocytes. The knockout of UCP1 and the protein levels of adipogenic markers adiponectin (AdipoQ), peroxisome proliferator-activated receptor gamma (PPARG), and perlipin A (PLIN) were confirmed by Western Blot. NC, non-targeting control. (B) Oxygen consumption rates were determined in a plate-based respirometer. ATP: oligomycin-sensitive OCR; leak: oligomycin resistant OCR (C) Basal glucose uptake was determined by 14C-deoxy-Glucose incorporation. (D) Extracellular acidification rates were determined as in (B). (B-D) Data obtained from both negative controls and UCP1 KO cell types were merged. Data of 3–4 independent experiments are shown as mean + SEM. **, p < 0.01 (Student's t-test).
3.3. Generation of UCP1-overexpressing human preadipocytes
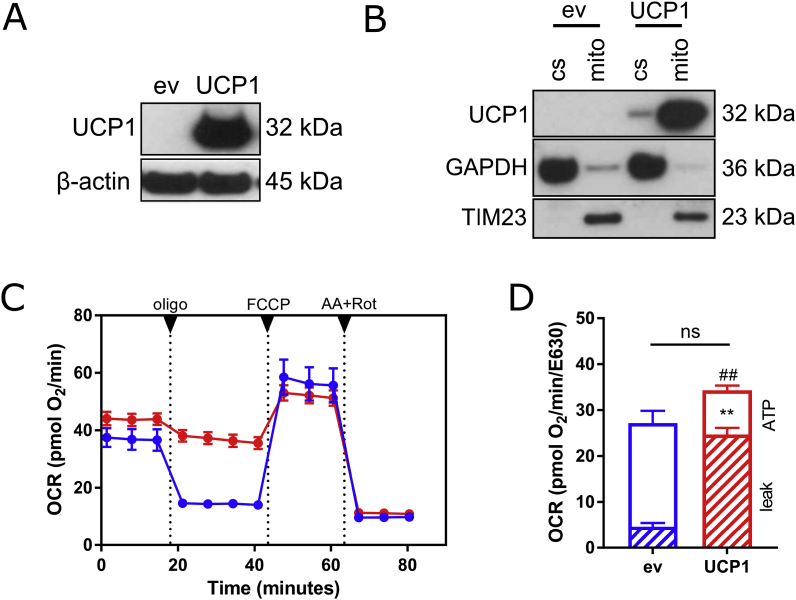
Next, we aimed to increase the expression of UCP1 in SGBS cells using a lentiviral system. Successful overexpression in the undifferentiated preadipocyte state was demonstrated by Western Blot (Fig. 3A), with no UCP1 detectable in empty vector-transduced control cells. Following subcellular fractionation, UCP1 was found to be localized in mitochondria as expected (Fig. 3B). UCP1 function was verified by assessing mitochondrial respiration by plate-based respirometry using a Seahorse extracellular flux analyzer. UCP1 overexpression had no statistically significant impact on basal respiration and FCCP-induced maximal substrate oxidation capacity (Fig. 3C). Importantly, UCP1-overexpressing cells have markedly increased proton leak respiration by 440% and reduced ATP-linked respiration rates by 57% (Fig. 3D). Taken together, this set of data demonstrates that UCP1 is fully functional, even in preadipocytes.
Fig. 3.
Lentivirus-mediated UCP1 overexpression in human preadipocytes. UCP1-overexpressing cells were generation by lentiviral transduction of SGBS preadipocytes. (A) UCP1 overexpression in preadipocytes was confirmed by Western Blot (n = 4 independent experiments). ev, empty vector control. (B) Mitochondrial localization of UCP1 in preadipocytes was confirmed by Western blot of cytosolic (cs) and mitochondrial (mito) subcellular fractions of (n = 3 independent experiments). TIM23 and GAPDH were used as markers for mitochondrial and cytosolic fractions. (C) Cellular respiration of UCP1-overexpressing and control preadipocytes was analyzed in a plate-based respirometer. During the measurement, the cells were sequentially treated with 2 μM oligomycin A, 4 μM FCCP and 1.5 μM antimycin A + 1.5 μM rotenone. (D) Basal, proton leak, and ATP-linked respiration rates were calculated from the respective OCR traces. Data are normalized to JanusGreen staining and are shown as mean + SEM. *, p < 0.05; **, p < 0.01 (Student's t-test).
3.4. UCP1 overexpression does not interfere with adipogenesis or mitochondrial biogenesis
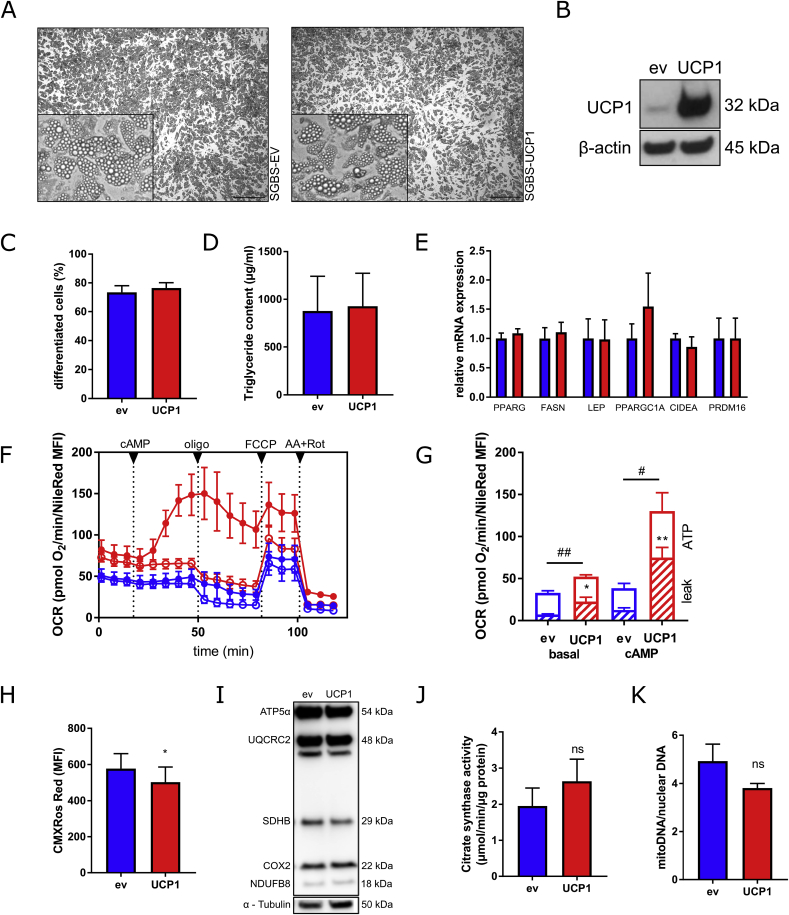
Control and UCP1-overexpressing preadipocytes were subjected to adipogenic differentiation. Both cell types displayed similar morphological features including cell shape, lipid droplet number, and lipid droplet size (Fig. 4A). In line, the rate of adipogenic differentiation (Fig. 4C) and triglyceride levels (Fig. 4D) were comparable. In day 14 adipocytes, UCP1 protein was still overexpressed compared to control cells (Fig. 4B), while the mRNA expression of the adipogenic marker genes PPARG, FASN, and LEP (Fig. 4E) were comparable. In addition, markers of adipocyte browning (e.g. PGC1A, PRDM16 and CIDEA) were not affected by UCP1 overexpression (Fig. 4E).
Fig. 4.
Adipogenic differentiation of human preadipocytes overexpressing UCP1. (A) The cells were subjected to adipogenic differentiation for 14 days. (B) UCP1 protein expression was determined by Western Blot. (C) The percentage of differentiated cells was determined by counting preadipocytes and adipocytes using a net micrometer. (D) Lipid accumulation was assessed by measuring the triglyceride content. (E) mRNA expression of key adipogenic marker genes as well as BAT associated genes was determined by qRT-PCR. (F) Oxygen consumption rates (OCR) of UCP1-overexpressing (red color) and control adipocytes (blue color) were determined using plate-based respirometer (media containing 1% BSA). To induce UCP1 activity, subsets of cells were stimulated with 0.5 mM dibutyryl-cAMP (cAMP, closed circles), while control cells were left untreated (open circles) (G) Sequential injections of oligomycin (2 μM), FCCP (4 μM), and antimycin A (1.5 μM) + rotenone (1.5 μM) were used to determine basal, proton leak (striped column part), and ATP-linked respiration rates (open column part). Data were normalized for cellular lipid content by NileRed staining. (H) Mitochondrial membrane potential was measured by CMXRos Mitotracker Red staining. (I) The protein levels of key electron transport chain components were determined by Western Blot. (J and K) To determine mitochondrial content, activity of citrate synthase (J) and relative mitochondrial DNA content (K) were assessed. Data of at least 4 independent experiments are shown as mean + SEM. */#, p < 0.05; **/##, p < 0.01; ns, not significant (Student's t-test). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Assessing mitochondrial activity revealed an increased basal respiration in UCP1-overexpressing adipocytes compared to control cells by 50% (Fig. 4F and G), mainly due to the increase in proton leak respiration, which was 200% higher compared to control cells. Treatment with dibutyryl-cAMP strongly enhanced respiration in UCP1-overexpressing adipocytes by 230%. In contrast, cAMP had only a minor effect (30% increase) on respiration in control cells (Fig. 4F and G). UCP1-overexpressing adipocytes revealed a reduced mitochondrial membrane potential (Fig. 4H), further corroborating UCP1 activity. Intriguingly, the mitochondrial content was not affected by UCP1 overexpression as determined by protein expression of components of the electron transport chain (Fig. 4I), the activity of citrate synthase (Fig. 4J), as well as the relative mitochondrial DNA content (Fig. 4K).
3.5. Elevated UCP1 levels enhance basal glucose uptake and glycolytic flux in adipocytes
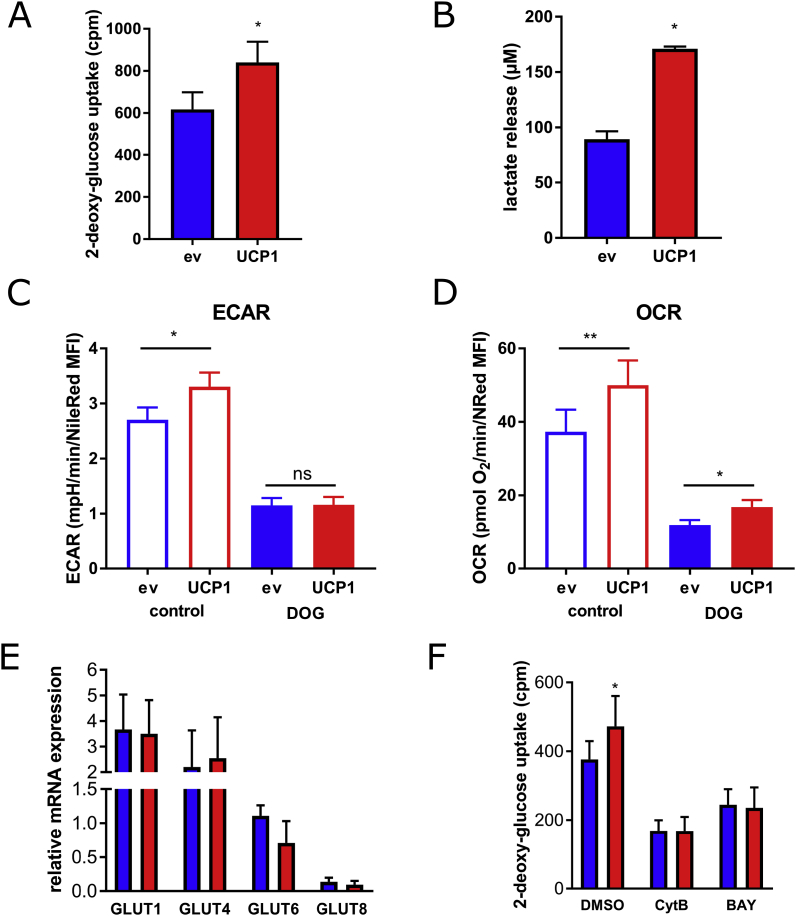
To investigate whether elevated levels of UCP1 are solely sufficient to drive cellular glucose uptake in adipocytes, cells were treated with 14C-deoxy-glucose in basal medium without further stimulation, followed by measurements of glucose uptake. Glucose uptake was significantly increased in UCP1-overexpressing compared to control adipocytes by 40% (Fig. 5A), accompanied by enhanced production of lactate (Fig. 5B). These data suggest that UCP1 overexpression is sufficient, without further exogenous stimulation, to induce glucose uptake and to enhance glycolytic flux in adipocytes.
Fig. 5.
Glucose uptake and glycolytic flux in adipocytes with elevated UCP1 levels. (A and B) Glycolytic activity of UCP1-overexpressing and control adipocytes were determined by measuring basal cellular 14C-deoxy-glucose incorporation (A) and lactate release (B). (C and D) Extracellular acidification rates (C) and oxygen consumption rates (D) upon inhibition of glycolysis by 50 mM 2-deoxy-glucose were determined in a plate-based respirometer. (E) Expression of glucose transporters 1, 4, 6, and 8 was determined by qRT-PCR. (F) 2-deoxy-glucose uptake was determined after preincubation with Cytochalasin B (CytB) or BAY-876. Data of at least 3 independent experiments are shown as mean + SEM.*, p < 0.05; **, p < 0.01 (Student's t-test).
We next tested whether inhibition of glucose metabolism inhibits UCP1 activity. Pretreatment with 2-deoxy-glucose blunted glycolytic rates, as judged by extracellular acidification rates (Fig. 5C). Interestingly, the corresponding OCR values were still significantly higher in UCP1-overexpressing adipocytes upon 2-deoxy-glucose treatment, suggesting that oxygen consumption partially relies on other carbon sources, most likely free fatty acids through beta oxidation (Fig. 5D). Despite the differences in glucose uptake, we did not detect differences in mRNA expression of GLUT1, the most important basal glucose transporter, or any other glucose transporters, including GLUT4, GLUT6, and GLUT8 (Fig. 4H). However, treatment of cells with cytochalasin B, a compound commonly used to inhibit glucose uptake, abolished the differences in glucose uptake between UCP1-overexpressing and control adipocytes (Fig. 5E). Strikingly, the same effect was observed by neutralizing the function of GLUT1 using the selective inhibitor BAY-876 [22] which completely inhibited the differences in glucose uptake between UCP1-overexpressing and control cells (Fig. 5F). Taken together, this set of data demonstrates that increasing UCP1 levels, independently of typical molecular browning networks, is sufficient to induce increased basal glucose uptake in human adipocytes; and that this effect is permitted by GLUT1.
4. Discussion
Since the early 2000s, 18F-FDG-PET/CT has been used for imaging BAT activity, clearly demonstrating that cold-activated BAT takes up glucose in amounts comparable to other high-glucose consuming tissues such as the brain [2,12,13,23]. It is commonly assumed that this high glucose uptake contributes to glycemic control, at least upon cold exposure. This is supported by studies demonstrating in subjects with higher BAT content lower glucose levels [24] and improved insulin sensitivity [4]. Weir et al. have recently shown that the increased glucose uptake by BAT does not necessarily require activation by cold, but that it also occurs at 25 °C. Using microdialysis, they convincingly demonstrated that BAT takes up significantly more glucose and releases more lactate than WAT [25], indicating a higher glycolytic activity even under basal conditions. These findings underline the widely accepted notion that BAT acts as a glucose sink. To the best of our knowledge, however, glucose uptake has never been assessed directly by comparing white and brown adipocytes in vitro. In confirmation of the prevalent notion, we exemplify for the first time that brown adipocytes take up significantly more glucose than white adipocytes.
As recently reported, inhibiting glucose uptake has pronounced effects on both basal and isoproterenol-stimulated respiration in brown adipocytes [9]. This suggests that glucose and its metabolic breakdown are essential for thermogenesis [9]. There are, however, also data from UCP1 knockout mice indicating that glucose uptake into BAT is independent of UCP1-dependent thermogenic activity [14,26]. In contrast, some indirect hints for UCP1-dependency of glucose metabolism were observed in mice with ectopic UCP1 expression in epididymal WAT [10]. Ectopic UCP1 expression significantly reduces blood glucose levels on both normal and high-fat diets. Interestingly, surgical and pharmacological denervation of WAT were ineffective at abrogating the effects of UCP1 overexpression, indicating that these effects persist independent of neuronal innervation [10].
Although mechanistic aspects of UCP1 function have been studied extensively in model systems such as isolated liposomes [27], yeast [28], mouse brown adipocytes [29,30], Chinese hamster ovary (CHO) cells [31], human embryonic kidney (HEK) cells [32], and HeLa cells [33], these studies were never focused on and thus, never revealed whether UCP1 activity per se is sufficient to enhance glycolytic flux, in particular in adipocytes. One study using in 3T3-L1 cells reports increased glucose uptake upon ectopic UCP1 expression, but did not experimentally prove UCP1 function [34].
Here, we introduce human SGBS preadipocytes and adipocytes that stably overexpress UCP1 as a unique, homologous model system to study physiological and pathophysiological relevant aspects of UCP1 function in detail. We demonstrate that UCP1 is functionally active in SGBS preadipocytes and adipocytes under basal as well as lipolytic conditions. This suggests that the activity of ectopically expressed UCP1 in intact white adipocytes is sufficient to increase proton leak, without further induced lipolysis by adrenergic induction. This is not intuitively expected considering that UCP1 per se is not active, at least not in isolated mitochondria in the absence of free fatty acids [30]. Moreover, while it was previously suggested that UCP1 overexpression affects mitochondrial integrity [30], no signs of mitochondrial dysfunction or cell viability were observed in our human adipocyte model system. Neither electron transport chain complexes, nor citrate synthase activity, nor relative mitochondrial DNA content were affected, emphasizing that forced UCP1 expression may be without adverse effects. Importantly, cellular proliferation (data not shown) and adipogenic differentiation were not affected in UCP1-overexpressing cells. Thus, we suggest that the constitutive activity of UCP1 in white adipocytes, probably due to free fatty acids from basal lipolysis, may offer the potential to improve glucose metabolism.
In line with previous publications [14], knocking out UCP1 did not alter glucose utilization, but overexpression impacts glucose uptake and lactate production in adipocytes, demonstrating enhanced glycolytic flux. Of note, the relative increase in glucose uptake rates is comparable to murine white vs brown adipocyte differences. Interestingly, increased lactate production supports a model of aerobic glycolysis that should operate independent of UCP1-induced glucose oxidation. Thus, the UCP1-dependent reduction of mitochondrial ATP production is possibly driving glycolytic ATP production to maintain cellular homeostasis. This integrates well with findings from human studies showing increased glycolytic activity in BAT [25]. We want to point out that undifferentiated preadipocytes overexpressing UCP1 may contribute to differences seen in glucose uptake. However, the majority of cells in the cell culture dish were differentiated adipocytes.
Inhibition of glycolysis by 2-deoxy-glucose partially inhibited respiration, but did not abolish the difference in oxygen consumption between UCP1-overexpressing and control adipocytes. This suggests that respiratory activity in our cell model is partially fueled by another substrate, such as lipids and amino acids. It has recently been proposed that glucose uptake in brown adipocytes is mediated either by GLUT1 [35] or both GLUT1 and GLUT4 [9]. In our model system of white adipocytes, selective inhibition of GLUT1 completely eradicated the differences in glucose uptake between UCP1-overexpressing and control cells identifying GLUT1 as an important gate keeper of glycolytic flux. The mRNA expression levels of glucose transporters were not altered by UCP1 overexpression suggesting an alternative level of regulation, e.g. an increased translocation of GLUT1 to the cell surface as described earlier [35].
Taken together, we demonstrate that brown adipocytes take up significantly more glucose under basal conditions than white adipocytes in vitro underlining that brown adipocytes are in general more active metabolically. We show that UCP1 and the insulin-independent glucose transporter GLUT1 are crucially involved in establishing this glucose-consuming phenotype of adipocytes. With this study we furthermore provide a unique cellular model system of UCP1-expressing human adipocytes. It allows detailed studies on human UCP1 function and its impact on cell metabolism, and is suitable to screen for UCP1 modulators, thus facilitating the discovery of new drugs to correct glucose metabolism.
Acknowledgements
D.T. is supported by the German Research Foundation (DFG, TE912/2-2). D.T. and P·F.-P. are supported by the Boehringer Ingelheim Ulm University Medical Center Ulm Biocenter (BIU). T.P. and J.-B.F. were supported by the International Graduate School in Molecular Medicine at the University of Ulm (GSC270). P.F.-P. is supported by the German Research Foundation (DFG, Fi1700/5-1; Heisenberg Professorship, Fi1700/7-1).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101286.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol. Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D., Schrauwen P., Teule G.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. 360/15/1500 [pii] [DOI] [PubMed] [Google Scholar]
- 3.Stanford K.I., Middelbeek R.J.W., Townsend K.L., An D., Nygaard E.B., Hitchcox K.M., Markan K.R., Nakano K., Hirshman M.F., Tseng Y.-H., Goodyear L.J. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J. Clin. Investig. 2013;123:215–223. doi: 10.1172/JCI62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chondronikola M., Volpi E., Børsheim E., Porter C., Annamalai P., Enerbäck S., Lidell M.E., Saraf M.K., Labbe S.M., Hurren N.M., Yfanti C., Chao T., Andersen C.R., Cesani F., Hawkins H., Sidossis L.S. Brown adipose tissue improves whole body glucose homeostasis and insulin sensitivity in humans. Diabetes. 2014 doi: 10.2337/db14-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chouchani E.T., Kazak L., Spiegelman B.M. New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metabol. 2019;29:27–37. doi: 10.1016/j.cmet.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 6.Cohen P., Levy J.D., Zhang Y., Frontini A., Kolodin D.P., Svensson K.J., Lo J.C., Zeng X., Ye L., Khandekar M.J., Wu J., Gunawardana S.C., Banks A.S., Camporez J.P.G., Jurczak M.J., Kajimura S., Piston D.W., Mathis D., Cinti S., Shulman G.I., Seale P., Spiegelman B.M. Ablation of {PRDM}16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cypess A.M., Weiner L.S., Roberts-Toler C., Elía E.F., Kessler S.H., Kahn P.A., English J., Chatman K., Trauger S.A., Doria A., Kolodny G.M. Activation of human Brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metabol. 2015;21:33–38. doi: 10.1016/j.cmet.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finlin B.S., Dupont-versteegden E.E., Philip A., Finlin B.S., Memetimin H., Confides A.L., Kasza I., Zhu B., Vekaria H.J., Harfmann B., Jones K.A., Johnson Z.R., Kern P.A. 2018. Human Adipose Beiging in Response to Cold and Mirabegron; p. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skjoldborg N., Isidor M.S., Cheung A., Winther S., Hansen J.B., Quistorff B., Basse A.L. Restricting glycolysis impairs brown adipocyte glucose and oxygen consumption. Am. J. Physiol. Metab. 2017;314:E214–E223. doi: 10.1152/ajpendo.00218.2017. [DOI] [PubMed] [Google Scholar]
- 10.Yamada T., Katagiri H., Ishigaki Y., Ogihara T., Imai J., Uno K., Hasegawa Y., Gao J., Ishihara H., Niijima A., Mano H., Aburatani H., Asano T., Oka Y. Signals from intra-abdominal fat modulate insulin and leptin sensitivity through different mechanisms: neuronal involvement in food-intake regulation. Cell Metabol. 2006;3:223–229. doi: 10.1016/j.cmet.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Chen K.Y., Cypess A.M., Laughlin M.R., Haft C.R., Hu H.H., Bredella M.A., Enerbäck S., Kinahan P.E., Lichtenbelt W. van M., Lin F.I., Sunderland J.J., Virtanen K.A., Wahl R.L. Brown adipose reporting criteria in imaging STudies (BARCIST 1.0): recommendations for standardized FDG-PET/CT experiments in humans. Cell Metabol. 2016;24:210–222. doi: 10.1016/j.cmet.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T., Taittonen M., Laine J., Savisto N.J., Enerback S., Nuutila P. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. 360/15/1518 [pii] [DOI] [PubMed] [Google Scholar]
- 13.Saito M., Okamatsu-ogura Y., Matsushita M., Watanabe K., Yoneshiro T., Nio-kobayashi J., Iwanaga T., Miyagawa M., Kameya T., Nakada K., Kawai Y., Tsujisaki M. High incidence of metabolically active Brown adipose effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hankir M.K., Kranz M., Keipert S., Weiner J., Andreasen S.G., Kern M., Patt M., Klöting N., Heiker J.T., Brust P., Hesse S., Jastroch M., Fenske W.K. Dissociation between Brown adipose tissue 18 F-FDG uptake and thermogenesis in uncoupling protein 1–deficient mice. J. Nucl. Med. 2017;58:1100–1103. doi: 10.2967/jnumed.116.186460. [DOI] [PubMed] [Google Scholar]
- 15.Wabitsch M., Brenner R.E., Melzner I., Braun M., Möller P., Heinze E., Debatin K.M., Hauner H. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. Relat. Metab. Disord. 2001;25:8–15. doi: 10.1038/sj.ijo.0801520. [DOI] [PubMed] [Google Scholar]
- 16.Fischer-Posovszky P., Newell F.S., Wabitsch M., Tornqvist H.E. Human {SGBS} cells - a unique tool for studies of human fat cell biology. Obes. Facts Eur. J. Obes. 2008;1:184–189. doi: 10.1159/000145784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y., Fromme T., Klingenspor M. Meaningful respirometric measurements of UCP1-mediated thermogenesis. Biochimie. 2016;134:56–61. doi: 10.1016/j.biochi.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Albers J., Danzer C., Rechsteiner M., Lehmann H., Brandt L.P., Hejhal T., Catalano A., Busenhart P., Gonçalves A.F., Brandt S., Bode P.K., Bode-Lesniewska B., Wild P.J., Frew I.J. A versatile modular vector system for rapid combinatorial mammalian genetics. J. Clin. Investig. 2015;125:1603–1619. doi: 10.1172/JCI79743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowarz E., Löscher D., Marschalek R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015;10:647–653. doi: 10.1002/biot.201400821. [DOI] [PubMed] [Google Scholar]
- 20.Tews D., Fischer-Posovszky P., Fromme T., Klingenspor M., Fischer J., Rüther U., Marienfeld R., Barth T.F., Möller P., Debatin K.M., Wabitsch M. FTO deficiency induces UCP-1 expression and mitochondrial uncoupling in adipocytes. Endocrinology. 2013;154:3141–3151. doi: 10.1210/en.2012-1873. [DOI] [PubMed] [Google Scholar]
- 21.Raspotnig G., Fauler G., Jantscher A., Windischhofer W., Schachl K., Leis H.J. Colorimetric determination of cell numbers by janus green staining. Anal. Biochem. 1999;275:74–83. doi: 10.1006/abio.1999.4309. [DOI] [PubMed] [Google Scholar]
- 22.Siebeneicher H., Cleve A., Rehwinkel H., Neuhaus R., Heisler I., Müller T., Bauser M., Buchmann B. Identification and optimization of the first highly selective GLUT1 inhibitor BAY-876. ChemMedChem. 2016;11:2261–2271. doi: 10.1002/cmdc.201600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B., Kuo F.C., Palmer E.L., Tseng Y.H., Doria A., Kolodny G.M., Kahn C.R. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. 360/15/1509 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsushita M., Yoneshiro T., Aita S., Kameya T., Sugie H., Saito M. Impact of brown adipose tissue on body fatness and glucose metabolism in healthy humans. Int. J. Obes. 2014;38:812–817. doi: 10.1038/ijo.2013.206. [DOI] [PubMed] [Google Scholar]
- 25.Weir G., Ramage L.E., Akyol M., Rhodes J.K., Kyle C.J., Fletcher A.M., Craven T.H., Wakelin S.J., Drake A.J., Gregoriades M.-L., Ashton C., Weir N., van Beek E.J.R., Karpe F., Walker B.R., Stimson R.H. Substantial metabolic activity of human Brown adipose tissue during warm conditions and cold-induced lipolysis of local triglycerides. Cell Metabol. 2018;27:1348–1355. doi: 10.1016/j.cmet.2018.04.020. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schweizer S., Oeckl J., Klingenspor M., Fromme T. Substrate fluxes in brown adipocytes upon adrenergic stimulation and uncoupling protein 1 ablation. Life Sci. Alliance. 2018;1 doi: 10.26508/lsa.201800136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Echtay K.S., Winkler E., Bienengraeber M., Klingenberg M. Site-directed mutagenesis identifies residues in uncoupling protein (UCP1) involved in three different functions. Biochemistry. 2000;39:3311–3317. doi: 10.1021/bi992448m. [DOI] [PubMed] [Google Scholar]
- 28.Stuart J.A., Harper J.A., Brindle K.M., Jekabsons M.B., Brand M.D. A mitochondrial uncoupling artifact can be caused by expression of uncoupling protein 1 in yeast. Biochem. J. 2001;356:779–789. doi: 10.1042/0264-6021:3560779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker N., Crichton P.G., Vidal-Puig A.J., Brand M.D. Uncoupling protein-1 (UCP1) contributes to the basal proton conductance of brown adipose tissue mitochondria. J. Bioenerg. Biomembr. 2009;41:335–342. doi: 10.1007/s10863-009-9232-8. [DOI] [PubMed] [Google Scholar]
- 30.Shabalina I.G., Ost M., Petrovic N., Vrbacky M., Nedergaard J., Cannon B. Uncoupling protein-1 is not leaky. Biochim. Biophys. Acta Bioenerg. 2010;1797:773–784. doi: 10.1016/j.bbabio.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Casteilla L., Blondel O., Klaus S., Raimbault S., Diolez P., Moreau F., Bouillaud F., Ricquier D. Stable expression of functional mitochondrial uncoupling protein in Chinese hamster ovary cells. Proc. Natl. Acad. Sci. 1990;87:5124–5128. doi: 10.1073/pnas.87.13.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jastroch M., Hirschberg V., Klingenspor M. Functional characterization of {UCP}1 in mammalian {HEK}293 cells excludes mitochondrial uncoupling artefacts and reveals no contribution to basal proton leak. Biochim. Biophys. Acta. 2012 doi: 10.1016/j.bbabio.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Li B., Holloszy J.O., Semenkovich C.F. Respiratory uncoupling induces δ-aminolevulinate synthase expression through a nuclear respiratory factor-1-dependent mechanism in HeLa cells. J. Biol. Chem. 1999;274:17534–17540. doi: 10.1074/jbc.274.25.17534. [DOI] [PubMed] [Google Scholar]
- 34.Si Y., Palani S., Jayaraman A., Lee K. Effects of forced uncoupling protein 1 expression in 3T3-L1 cells on mitochondrial function and lipid metabolism. J. Lipid Res. 2007;48:826–836. doi: 10.1194/jlr.M600343-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Olsen J.M., Sato M., Dallner O.S., Sandström A.L., Pisani D.F., Chambard J.-C., Amri E.-Z., Hutchinson D.S., Bengtsson T. Glucose uptake in brown fat cells is dependent on mTOR complex 2-promoted GLUT1 translocation. J. Cell Biol. 2014;207:365–374. doi: 10.1083/jcb.201403080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.