Abstract
Measuring and managing data quality in healthcare has remained largely uncharted territory with few notable exceptions. A rules-based approach to data error identification was explored through compilation of over 6,000 data quality rules used with healthcare data. The rules were categorized based on topic and logic yielding twenty-two rule templates and associated knowledge tables used by the rule templates. This work provides a scalable framework with which data quality rules can be organized, shared among facilities and reused. The ten most frequent data quality problems based on the initial rules results are identified. While there is significant additional work to be done in this area, the exploration of the rule template and associated knowledge tables approach here shows rules-based data quality assessment and monitoring to be possible and scalable.
Keywords: Electronic health records, data quality, data quality assessment
1. Introduction
Data Quality Assessment (DQA) in healthcare and health-related research is not new. The earliest reports of data processing in clinical research included accounts of data checking [1–9]. In the therapeutic development industry, with the 1962 Kefauver Harris Amendment to the Food, Drug and Cosmetic act a New Drug Application (NDA) had to show that a new drug was both safe and effective and companies began to use rules to check data in support of NDAs for consistency. In fact, fear that notice of an errant data point would substantially delay a regulatory submission prompted a process in the therapeutic development industry of running often hundreds of rules for a clinical study and contacting the data provider in attempts to resolve each discrepancy against the source, i.e., the medical record [10]. The discrepancies often numbered in the tens of thousands for a small study of a few hundred patients. It is not uncommon for 10–30% of the cost of a clinical study to be spent on data cleaning [11]. This practice, albeit mediated by risk-based approaches2 continues today in therapeutic development and is the standard of practice3.
In healthcare, however, there is no source against which to resolve data discrepancies. With alert fatigue common for critical decision support algorithms, few would consider flagging data discrepancies as clinicians chart patient information. Further, aside from being used by physicians and other members of care teams in decision-making, widespread secondary use of routine clinical data is a fairly recent phenomenon. The current national emphasis on secondary use of healthcare data for research has been prompted by the large upswing in Electronic Health Record (EHR) adoption over the last decade4, and federal support for institutional clinical data repositories for research5 over the same period.
Today, the value of data quality assessment in healthcare has not been well studied or articulated. Though there have been reports of fixing data quality problems identified through attempts at data use, institutions have been hesitant to allocate even limited resources toward systematic DQA and improvement. For these reasons DQA in healthcare has received relatively little attention as an institutional priority or as a research agenda.
In early work, Carlson et al. (1995) successfully used rules to identify discrepancies in data used for clinical decision support in intensive care units (ICUs) however interventions based on the rules were not described [12]. In 2003, Brown et al. tested data quality rules to find data quality problems and improve data quality in EHRs. EHR information quality was then tracked and results were reported to clinicians, to encourage data quality improvement [13][14]. Around the same time, De Lusignan et al. reported a similar rules-based approach where data quality check results were collated and fed back to the participating general practitioners as an intervention to improve data quality [15]. Most recently, Hart and Kuo (2017) reported rule-based discrepancy identification and resolution in Canadian home health data with a few hundred data quality rules [16]. Records failing validation were reported back to the responsible staff for correction and re-submission. They reported a greater than 50% decrease in rejected records across three domains in six months [16]. Thus, in small studies of limited scope, rules-based approaches applied within a systems-theory feedback loop approach have been shown effective.
This research in healthcare DQA is motivated by (1) recent increases in national attention towards secondary use of healthcare data for research through broad programs such as the National Institutes of Health funded Healthcare Systems Research Collaboratory6 and the Patient-Centered Outcomes Research Institute7 funded through the Affordable Care Act, (2) national emphasis on use of healthcare data for organizational performance assessment and improvement, i.e., Accountable Care Organizations, (3) almost ubiquitous availability of rich healthcare data in most institutions, and (4) lack of methods for DQA, specifically assessment of data accuracy, demonstrated effective in healthcare. We seek to demonstrate and evaluate a rule-based data assessment and monitoring system in healthcare.
2. Methodology
A high-level implementation architecture diagram is provided as Figure 1. Though this is not the long-term architecture, the data quality assessment and monitoring system runs on our institutional data warehouse. The system has three main components, (1) rule templates, (2) knowledge tables for rules, and (3) rule results tables. Rule templates and knowledge tables are used to store and manage rules. Outputs of the system are stored in rule results tables and visualization monitoring reports are executed based on them.
Figure 1.

High level architecture.
2.1. Rule templates
To identify candidate rules, we first looked to existing rule sets. These included the publically available Observational Health Data Sciences and Informatics (OHDSI) formerly Observational Medical Outcomes Partnership (OMOP) rules, the Healthcare Systems Research Network (previously HMORN) rules [17], and the Sentinel data checking rules [18], and age and gender incompatible diagnosis and procedure lists from payers. We also utilized rules written for an internal project using multi-site EHR data [19]. Rules were also identified from data elements used in predictive analytics algorithm, data elements used in performance measures used by major payers. In an exploratory project, we also assessed Epic screens for anesthesiology to identify data elements for which rules could be written. All combined, these activities produced over 6,357 individual logic statements or rules.
Management of this many initial rules conflicted with our goal of scalable rule management and maintenance over time. Inspired by the rule abstraction in Brown’s work [14], we sorted the rules according to patterns in the rule logic. Rules sharing a topic and logic structure were abstracted into a single rule template. An example of such a rule template is Flag the record if GENDER is equal to some invalid gender and DIAGNOSIS is equal to a corresponding invalid diagnosis. The clinical information in the rules (in the example the list of gender – diagnosis incompatibilities) was extracted and compiled into a knowledge table against which the rule template runs. This categorization yielded twenty-two different rule templates. The twenty-two rule templates were further categorized into five higher-level types: incompatibility, value out of range, temporal sequence error, incompleteness and duplication (Figure 2). These correspond to the following Kahn 2016 criteria [20]: value conformance, relational conformance, completeness and plausibility. Incompatibility means one data value is logically incompatible with another data value, such as patient gender is incompatible with diagnoses. Value out of range is defined as the value of a record is out of the limits compatible with life or grossly incompatible with product labeling, such as drug dose, lab result, or date of birth is before 1880. Typos or wrong units could cause these errors. Temporal sequence templates focus on any two dates occurring in an invalid order. For example, date of encounter cannot be earlier than date of birth for an adult. Incompleteness is defined as occurrence of a data value that is expected but missing. Univariate checks for missing values were not included because they are easily quantified through data profiling approaches. The rules consist of multivariate and record-level incompleteness checks, i.e., when one record is present, but the other one is absent. For example, a procedure is present but there is no corresponding encounter record. Lastly, duplication, also a multivariate type data quality check, is defined as multiple occurrences of events that can physically happen only once, for example a patient with two hysterectomies.
Figure 2.
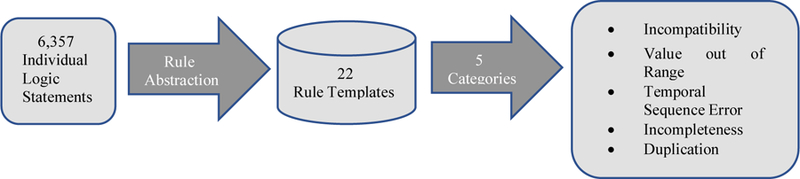
Rule templates schematic
2.2. Knowledge tables
As described above, we compiled or identified a knowledge table to support each rule template. The purpose of the knowledge table is to condense what may eventually be thousands of individual rules down to one template and a knowledge table that can be expanded or edited as medical coding systems change or new knowledge becomes available and shared among institutions. In this way, we purposely separated the rule logic from the knowledge. Twenty-two rule templates are easier to develop and maintain more than 6,000 rules.
3. Results
6,357 rules (eleven templates) have been programmed, tested and executed over our institutional data warehouse containing data from 1.46 million patients from nine facilities and four different EHR systems. A summary of results from the eleven rule templates implemented thus far is shown in Table 1.
Table 1.
Rule results summary
| Template Name | Number of Rules | Number of triggered rules | Discrepancies |
|---|---|---|---|
| Age and DIAGNOSIS (incompatibility) | 130 | 33 | 2,701 |
| Age and PROCEDURE (incompatibility) | 5,205 | 329 | 3,157 |
| Gender and DIAGNOSIS (incompatibility) | 79 | 18 | 3,710 |
| Gender and PROCEDURE (incompatibility) | 640 | 16 | 111 |
| Gender and clinical specialty (incompatibility) | 5 | 2 | 42 |
| DRUG and DIAGNOSIS (incompatibility) | 18 | 3 | 1,115 |
| DRUG and PROCEDURE (incompatibility) | 36 | 2 | 505 |
| DRUG and LAB (incompatibility) | 6 | 1 | 299 |
| LAB Result (value out of range) | 78 | 36 | 22,028 |
| Demographics data elements (value out of range) | 8 | 8 | 17,776 |
| Two Dates in invalid order (temporal sequence error) | 152 | 29 | 4,522 |
The number of rules is the number of records in a knowledge table supporting a rule template. The number of triggered rules is the number of knowledge table records that identified one or more discrepancies, the number of discrepancies is the count of the number of times the data were found to be in exception to the rule. Rule results grouped by rule template, however often does not directly inform action. For example, when considering invalid dates, the dates could come from anywhere in the health system. A way to partition rule results into groups meaningful to clinical leaders and information technology professionals was needed.
To explore this and frankly to use the rule results to investigate and inform interventions for data quality problems, we manually grouped rule results identifying similar data quality problems and calculated the frequency of distinct problems as the number of records fired across all rules for that group. For example, both ICD-9 code: V39.01(caesarean section) and ICD-10 code: Z38.5(twins) should be used for newborns. However, two rules from age and diagnoses incompatibility identify the two codes are used for some patients who are over 20 years old. So we grouped the results from the two rules as identifying the same problem: assignment of infant codes to mothers and vice versa. Based on our manual groupings, the ten most frequent data quality problems are presented in Table 2 ranked by frequency. Grouping rule results by category of problem provided lists that we could use to work with clinical leaders and IT professionals to investigate the problems.
Table 2.
Data Problems Ranked by Frequency
| Data Quality Problem | Number of Discrepancies |
|---|---|
| Lab results that are physically impossible or otherwise incompatible with life (36 different lab tests) | 22,028 |
| Dates of encounters, diagnoses, medications and procedures occurring more than one year before birth date | 3,325 |
| Age inconsistent procedures | 3,1328 |
| Assignment of infant codes to mothers and vice versa | |
| Infants with Adult codes | 2,666 |
| Adults with infant codes | 358 |
| Height greater than 3 meters. | 2,472 |
| Blood pressure values greater than 500 mmHg. | 2,292 |
| Weight greater than 300 kg. | 1,581 |
| Respiratory rate greater than 200 breaths per minute. | 587 |
4. Discussion
There are multiple possible causes of the 22,028 instances of invalid lab results occurring across 36 different lab tests. Possible causes include problems with the sample, problems with the instrumentation, recording mistakes, or incorrect units. Further grouping the rule results by lab test and data source would likely divulge the machine/s or process/s responsible for the discrepancies. Presentation of overall results and the groupings by lab test and data source as a trend-line over time may further inform troubleshooting and eventual remediation or intervention. We found this to be true for all rule results regarding measured physical quantities. Similarly, partitioning rule results for date-related discrepancies by data source and as a trend-line over time would facilitate troubleshooting and eventual remediation or intervention.
The 3,132 instances of age inconsistent procedures are likely problems in medical coding. In these cases, a patient’s age was inconsistent with ranges defined by CPT procedure codes. Many CPT codes represent same procedure but performed on different age ranges, for example, 99381, 99382, 99383, 99384, 99385, 99386 and 99387 (Initial comprehensive preventive medicine evaluation and management of an individual). Similarly, the 2,666 adults with infant diagnoses codes and the 358 babies with adult diagnoses codes, are likely coding problems. Presentation of code-related rule results by code would make these issues easy to investigate with Health Information Management professionals working in medical coding.
The rule templates for Diagnosis and corresponding Drug and Diagnosis and corresponding lab were programmed. For example the rule checking for Aspirin prescription in patients with ischemic heart disease identified 1,090 instances’ on exception. However, these patients may have had a contraindication to Aspirin therapy making these instances possibly valid. The rule checking for presence of an HbA1c lab test in patients with diabetes identified 178 instances of exception. However, HbA1c wasn’t commonly used till the turn of the century. Older data in the warehouse predate guidelines changes and may be valid. In these two examples, natural variation and changing practice respectively explain the exceptions as possible not data problems. Continued use of these rules for data quality monitoring would require customization. There are many examples where this is the case. For obvious reasons, to date, we have excluded rules such as these where exceptions could be conditionally valid. However, we note that the effort in including the additional conditions may be worth the increased relevance of data quality monitoring results to clinical practice and facility administration.
As previously reported [21], identification of knowledge sources was challenging. Knowledge sources did not exist and could not easily be identified for half of the twenty-two rule templates initially identified. While this remains a challenge today, collaborative approaches to building these knowledge sources are possible as evidenced by multiple publically identified knowledge sources. Through consolidation of existing rule sets and systematic approaches to identify clinical data elements and applicable multivariate data quality rules, we distilled a set of 6,357 rules across the eleven templates described here. Attempts to identify additional rules turned up few additions (64 for EHR predictive algorithms used in decision support and 73 from Anesthesiology EHR screens). Based on this, though the number of possible rules is combinatorially large, the actual combinations of interest and utility seen tractable.
5. Conclusions
Assessing the quality of EHR data is necessary to improve data quality yet doing so systematically represents uncharted territory in healthcare. This study illustrated a potentially scalable framework with which data quality rules can be organized, shared as rule templates and knowledge tables, and applied in healthcare facilities to identify data errors. Though the results reported here are preliminary, we have demonstrated that rule-based data quality assessment identifies real data problems. While there is significant additional work to be done in this area, the exploration of the rule template and associated knowledge tables approach here shows the approach to be possible, the number of rules likely tractable and their management scalable.
Footnotes
U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER) Center for Biologics Evaluation and Research (CBER), E6(R2) Good Clinical Practice: Integrated Addendum to ICH E6(R2) Guidance for Industry, March 2018. Available from https://www.fda.gov/downloads/Drugs/Guidances/UCM464506.pdf.
Society for Clinical Data Management (SCDM), Good Clinical Data Management Practices (GCDMP), 2013. Available from https://www.scdm.org.
Office of the National Coordinator for Health IT (ONC), Health IT Dashboard quick statistics, 2018. Available from https://dashboard.healthit.gov/quickstats/pages/.
Department of Health and Human Services, National Institutes of Health (NIH), National Center for Advancing Translational Sciences (NCATS), PAR-18–464 Clinical and Translational Science Award (U54). CFDA 93.350, posted December 5, 2017. Available from https://grants.nih.gov/grants/guide/pa-files/PAR-18-464.html accessed March 23, 2018.
Available from http://www.rethinkingclinicaltrials.org.
Available from http://www.pcori.org.
Assignment of infant codes to mothers and vice versa are not included.
References
- [1].FORREST WILLIAMH, and BELLVILLE JWELDON. “The use of computers in clinical trials.” British journal of anaesthesia 394 (1967): 311–319. [DOI] [PubMed] [Google Scholar]
- [2].Kronmal Richard A., et al. “Data management for a large collaborative clinical trial (CASS: Coronary Artery Surgery Study).” Computers and Biomedical Research 116 (1978): 553–566. [DOI] [PubMed] [Google Scholar]
- [3].Knatterud Genell L. “Methods of quality control and of continuous audit procedures for controlled clinical trials.” Controlled clinical trials 14 (1981): 327–332. [DOI] [PubMed] [Google Scholar]
- [4].Norton Susan L., et al. “Data entry errors in an on-line operation.” Computers and Biomedical Research 142 (1981): 179–198. [DOI] [PubMed] [Google Scholar]
- [5].Cato AE, Cloutier G, and Cook L. “Data entry design and data quality”
- [6].Bagniewska Anna, et al. “Data quality in a distributed data processing system: the SHEP pilot study.” Controlled clinical trials 71 (1986): 27–37. [DOI] [PubMed] [Google Scholar]
- [7].DuChene Alain G., et al. “Forms control and error detection procedures used at the Coordinating Center of the Multiple Risk Factor Intervention Trial (MRFIT).” Controlled clinical trials 73 (1986): 34–45. [DOI] [PubMed] [Google Scholar]
- [8].Crombie IK, and Irving JM. “An investigation of data entry methods with a personal computer.” Computers and Biomedical Research 196 (1986): 543–550. [DOI] [PubMed] [Google Scholar]
- [9].FORTMANN STEPHENP, et al. “Community surveillance of cardiovascular diseases in the Stanford Five-City Project: methods and initial experience.” American journal of epidemiology 1234 (1986): 656–669. [DOI] [PubMed] [Google Scholar]
- [10].Estabrook Ronald W., et al. , eds. Assuring data quality and validity in clinical trials for regulatory decision making: workshop report National Academies Press, 1999. [PubMed] [Google Scholar]
- [11].Eisenstein Eric L., et al. “Reducing the costs of phase III cardiovascular clinical trials.” American heart journal 1493 (2005): 482–488. [DOI] [PubMed] [Google Scholar]
- [12].Carlson Debra, et al. “Verification & validation algorithms for data used in critical care decision support systems.” Proceedings of the Annual Symposium on Computer Application in Medical Care. American Medical Informatics Association, 1995. [PMC free article] [PubMed] [Google Scholar]
- [13].Brown Philip, and Warmington Victoria. “Info-tsunami: surviving the storm with data quality probes.” Journal of Innovation in Health Informatics 114 (2003): 229–237. [DOI] [PubMed] [Google Scholar]
- [14].Brown Philip JB, and Warmington Victoria. “Data quality probes—exploiting and improving the quality of electronic patient record data and patient care.” International journal of medical informatics 681–3 (2002): 91–98. [DOI] [PubMed] [Google Scholar]
- [15].De Lusignan Simon, et al. “Does feedback improve the quality of computerized medical records in primary care?.” Journal of the American Medical Informatics Association 94 (2002): 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hart Robert, and Kuo Mu-Hsing. “Better Data Quality for Better Healthcare Research Results-A Case Study.” Studies in health technology and informatics 234 (2017): 161–166. [PubMed] [Google Scholar]
- [17].Bauck A, Bachman D, and Riedlinger K. “Developing a consistent structure for VDW QA checks. 2011” (2011).
- [18].Curtis LH, et al. “Mini-Sentinel year 1 common data model—data core activities. 2012” (2012).
- [19].Tenenbaum Jessica D., et al. “The MURDOCK Study: a long-term initiative for disease reclassification through advanced biomarker discovery and integration with electronic health records.” American journal of translational research 43 (2012): 291. [PMC free article] [PubMed] [Google Scholar]
- [20].Kahn Michael G., et al. “A harmonized data quality assessment terminology and framework for the secondary use of electronic health record data.” Egems 41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Zhan, et al. “Rule Templates and Linked Knowledge Sources for Rule-based Information Quality Assessment in Healthcare.” MIT ICIQ (2017) [Google Scholar]


