The cancer/testis antigen (CTA) genes were identified as human genes preferentially expressed in cancer cells and testis, but the contribution of CTAs to cancer and male germ cell development is unclear. In this study, we comprehensively examined mouse CTA functions and found that the majority of CTAs are involved in growth and/or survival of cancer cells. We focused on one mouse CTA gene, Tekt5, for its detailed functional analysis.
KEYWORDS: CTA, Tekt5, cancer cell, germ cell
ABSTRACT
The cancer/testis antigen (CTA) genes were identified as human genes preferentially expressed in cancer cells and testis, but the contribution of CTAs to cancer and male germ cell development is unclear. In this study, we comprehensively examined mouse CTA functions and found that the majority of CTAs are involved in growth and/or survival of cancer cells. We focused on one mouse CTA gene, Tekt5, for its detailed functional analysis. Tekt5 knockdown (KD) in ovarian cancer cells caused G1 arrest and apoptosis, and p27kip1 was concomitantly upregulated. Tekt5 KD also resulted in decreased levels of acetylated α-tubulin and subsequent fragmentation of β-III-tubulin, upregulation of HDAC6 that deacetylates α-tubulin, and nuclear accumulation of SMAD3 that induces p27kip1 expression. Because depolymerization of tubulin is known to cause translocation of SMAD3 to the nucleus, these results together suggested that TEKT5 negatively regulates Hdac6 expression and consequently maintains cell cycle via stabilization of tubulin. We also found that the number of spermatids was significantly decreased and acetylated α-tubulin levels were decreased in vivo by KD of Tekt5 in testis. Because acetylated α-tubulin is required for sperm morphogenesis, these results suggest that TEKT5 is necessary for spermiogenesis via maintenance of acetylated α-tubulin levels.
INTRODUCTION
Cancer/testis antigen (CTA) genes are a group of genes that are preferentially expressed in cancer and testis. De Smet et al. identified MAGE-1 as a gene encoding an antigen present on melanoma cells; the gene was then designated a CTA gene based on its specific expression in cancer cells and testis (1). Subsequently, examination of tumor specimens and patient sera have led to reports of approximately 270 human CTA genes to date (CTDatabase; http://www.cta.lncc.br/index.php). The roles of some CTA genes in cancer cells and germ cells have been defined. For instance, genes belonging to the MAGE-A and SSX family enhance the epithelial-mesenchymal transition (EMT) of cancer cells and the development of cancer stem cells and accelerate tumor development and metastasis (2). In addition, CTAs could be good biomarkers for cancer, and some CTAs, such as MAGE-A4, have been used for cancer immunotherapy (3). In germ cells, CTAs such as SYCP1 and SYCE2 are involved in formation of the synaptonemal complex in meiotic prophase (4, 5). Nevertheless, CTAs whose functions are critical in both cancer cells and germ cells are unknown. It is likely that some CTAs share related molecular mechanisms, playing distinct or similar functions in cancer cells and germ cells.
To find CTAs that function both in cancer cells and germ cells, we first selected mouse CTA genes that are highly expressed in cancer cells because use of the mouse model is expected to facilitate functional evaluation of the CTAs in testicular germ cells in vivo. We then carried out functional screening of mouse CTA genes by knockdown (KD) using RNA interference (RNAi) in mouse cancer cell lines in which the corresponding CTA genes are highly expressed. As a result, we identified the mouse Tektin 5 (Tekt5) gene, a homologue of a locus that originally was reported as a CTA gene highly expressed in human colon cancer and whose expression subsequently was detected in various cancer cells (6). TEKT5 is a member of the tektin protein family; some members of this family have been suggested to be components of cilia and flagella composed of microtubules (7). Among members of the tektin family, TEKT1, -2, -3, and -4 have been shown to localize within the entire sperm tail, while TEKT5 has been found to localize only in the mitochondrial sheath in the midpiece of the rat sperm tail (8). However, TEKT5’s function(s) in cancer cells, as well as in testicular germ cells, is unknown. In the present study, we showed that TEKT5 controls tubulin stability to enhance the growth and survival of cancer cells and to promote sperm morphogenesis.
RESULTS
Identification of mouse CTA genes.
CTA genes originally were identified as human genes, but systematic identification of mouse CTA genes has not been reported (to our knowledge). To facilitate examining the functions of CTAs in germ cells in vivo, we sought to identify mouse CTA genes and explored mouse homologues of human CTA genes by using a Web tool, Homologene Matcher (http://refdic.rcai.riken.jp/tools/matchom.cgi). Using this program, we identified 139 mouse homologue genes (see Table S1 in the supplemental material) from 277 human CTA genes. The smaller number of mouse genes is due, in part, to the existence of human-specific gene families, such as GAGE (9), XAGE (10), PAGE (11), and BAGE (12), among the human CTA genes.
Among the 139 mouse homologue genes, we excluded 13 genes (Ctage5, Cntn2, Spag9, Kif20b, 2610507B11Rik, Otoa, Lypd6b, Igsf11, Tmem108, RqCd1, 2410076I21Rik, Hemgn, and Stk38) that appear not to be expressed in testis, as judged by a Web tool, BioGPS (http://biogps.org/#goto). Suitable PCR primers were not designed for another 14 genes that also were excluded. Using quantitative reverse transcription-PCR (RT-qPCR), we examined the expression of the remaining 112 genes in mouse cancer cell lines and corresponding normal tissues, as well as in testis (Fig. 1 and 2; see Table S2 in the supplemental material). Most of the tested genes were highly expressed in testis compared with the tested cancer cells and normal tissues (Table S2). We selected 87 genes showing higher expression in at least one of the tested cancer cell lines compared with the corresponding normal tissues (Fig. 1; see Table S3 in the supplemental material).
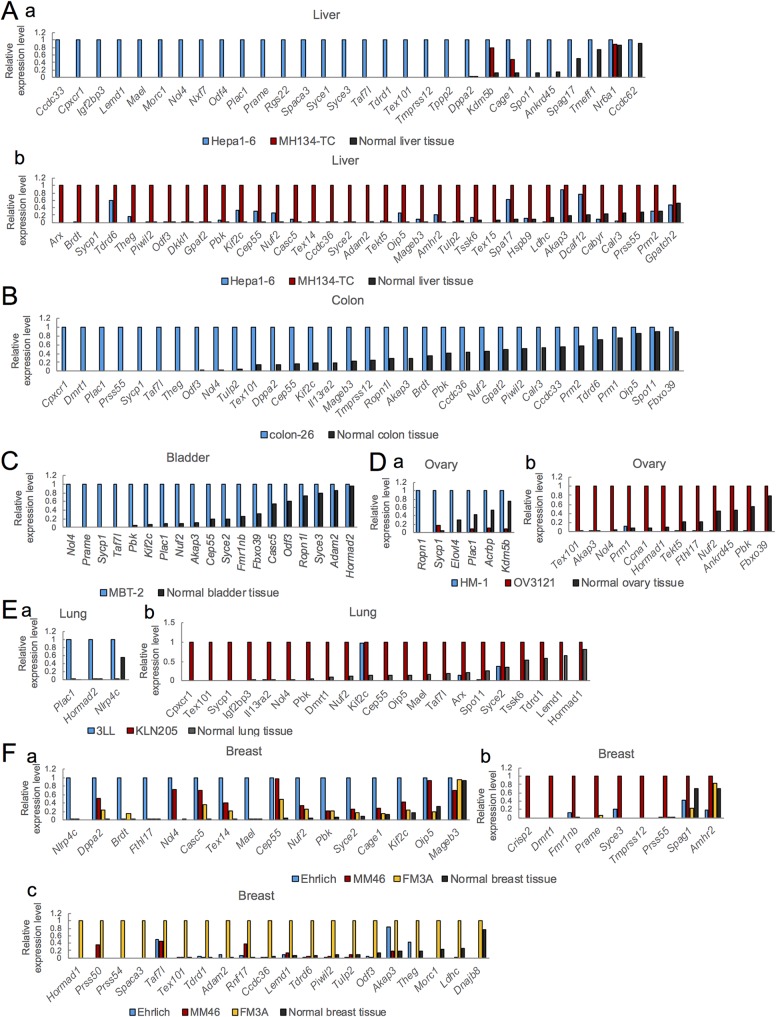
FIG 1.
Expression of mouse homologues of human CTA genes whose expression is >5% higher in cancer cell lines than in the corresponding normal tissues. The relative expression levels of CTA genes in liver (A), colon (B), bladder (C), ovary (D), lung (E), and breast (F) cancer cell lines and in the respective normal tissues are shown. The expression in Hepa1-6 (Aa), MH134-TC (Ab), colon-26 (B), MBT-2 (C), HM-1 (Da), OV3121 (Db), 3LL (Ea), KLN205 (Eb), Ehrlich (Fa), MM46 (Fb), and FM3A (Fc) was set as 1.0. Expression was determined by RT-qPCR using the dynamic arrays in the BioMark system.
FIG 2.
Expression of mouse homologues of human CTA genes whose expression is same or lower in cancer cell lines than in the corresponding normal tissues. The relative expression levels of CTA genes in liver (A), bladder (B), colon (C), ovary (D), lung (E), and breast (F) cancer cell lines and in the respective normal tissues are shown. The expression in normal tissues was set as 1.0. Expression was determined by RT-qPCR using the dynamic arrays on the BioMark system.
Screening of mouse CTAs involved in growth or survival of cancer cells.
To identify functionally important CTAs in cancer cells, we estimated the effects of the KD of mouse CTA genes on growth or survival of cancer cells. Among 87 genes highly expressed in mouse cancer cell lines, we excluded three genes (Sycp1, Spo11, and Spef2) for which siRNAs were not commercially available. We carried out KD of the remaining 84 genes by introducing two different siRNAs corresponding to each CTA gene into the cancer cell lines and estimating the changes in cell number. Those lines included melanoma (B16 [13] and B16C2W [14]), lung tumor (3LL [15] and KLN205 [16]), breast tumor (Ehrlich [17], MM46 [18], and FM3A [19]), liver tumor (Hepa1-6 [20] and MH134-TC [21]), bladder tumor (MBT-2 [22]), ovarian tumor (OV3121 [23] and OV2944-HM-1 [24]), and colon tumor (colon-26 [25]) cells. The combinations of genes and tested cell lines are shown in Table S4.
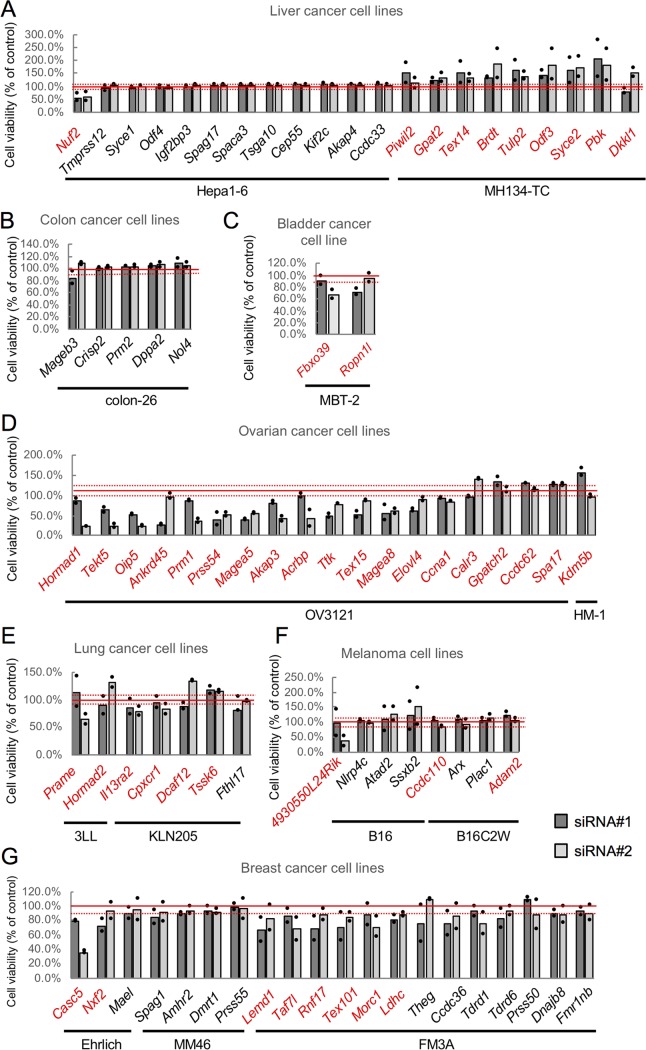
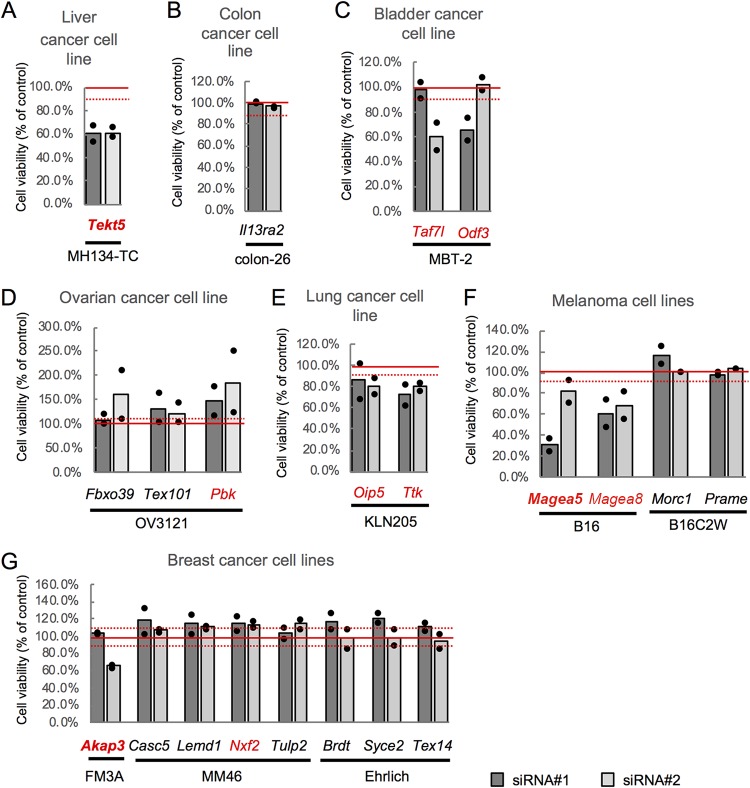
We first introduced the siRNAs into a first cell line, one in which the expression of the corresponding CTA gene was highest among the tested cell lines, and selected 47 genes whose KD reproducibly resulted in changes in cell number that exceeded 10% of control cell number (Fig. 3; Table S5). Among these 47 genes, we selected 21 genes whose expression was higher in more than one cancer cell line compared to that in the corresponding normal tissue (Fig. 1; Table S3). We then tested these 21 genes for KD in a second cell line, one that showed the next highest level of expression of the respective gene, and obtained 10 genes whose KD resulted in changes in cell number that exceeded 10% of control cell number (Fig. 4; Table S6). Among these 10 genes, KD of three genes (Tekt5, Akap3, and Magea5) caused changes in cell number of more than 30% in both the first and the second cell lines (Fig. 3 and 4). In subsequent work, we focused on Tekt5 for detailed analysis, because the function(s) of Tekt5 in both cancer cells and germ cells is unknown.
FIG 3.
siRNA screening of mouse CTA genes using the first cell lines. Changes in viability of liver (A), colon (B), bladder (C), ovary (D), lung (E), melanoma (F), and breast (G) cancer cell lines were tested using two different siRNAs (siRNA#1 and siRNA#2) corresponding to the CTA genes whose expression was highest in the respective tested cell lines. Cell viability was determined by MTS assay at 72 h after transfection with the siRNAs. The viability of KD cells relative to that of control cells is shown. Graphs represent data from two biological replicates. Dots indicate values from each of the two replicates. Solid red lines indicate 100% viability; broken red lines indicate 10% increased or decreased viability. Red text indicates genes for which KD (by at least one siRNA) caused a viability change of >10%.
FIG 4.
siRNA screening of mouse CTA genes using the second cell lines. Changes of viability of liver (A), colon (B), bladder (C), ovary (D), lung (E), melanoma (F), and breast (G) cancer cell lines were tested using two different siRNAs (siRNA#1 and siRNA#22) corresponding to the CTA genes selected by using the first cell lines. KD was carried out in the second cell lines, i.e., lines that showed the next highest level of expression of the tested genes. Cell viability was determined by MTS assay at 72 h after transfection with the siRNAs. Viability of KD cells relative to that of control cells is shown. Graphs represent data from two biological replicates. Dots indicate values from each of the two replicates. Solid red lines show 100% viability; broken red lines indicate 10% increased or decreased viability. Red text indicates genes for which KD (by at least one siRNA) caused a viability change of >10%.
Cell cycle enhancement of cancer cells by TEKT5 via regulation of the tubulin-SMAD axis.
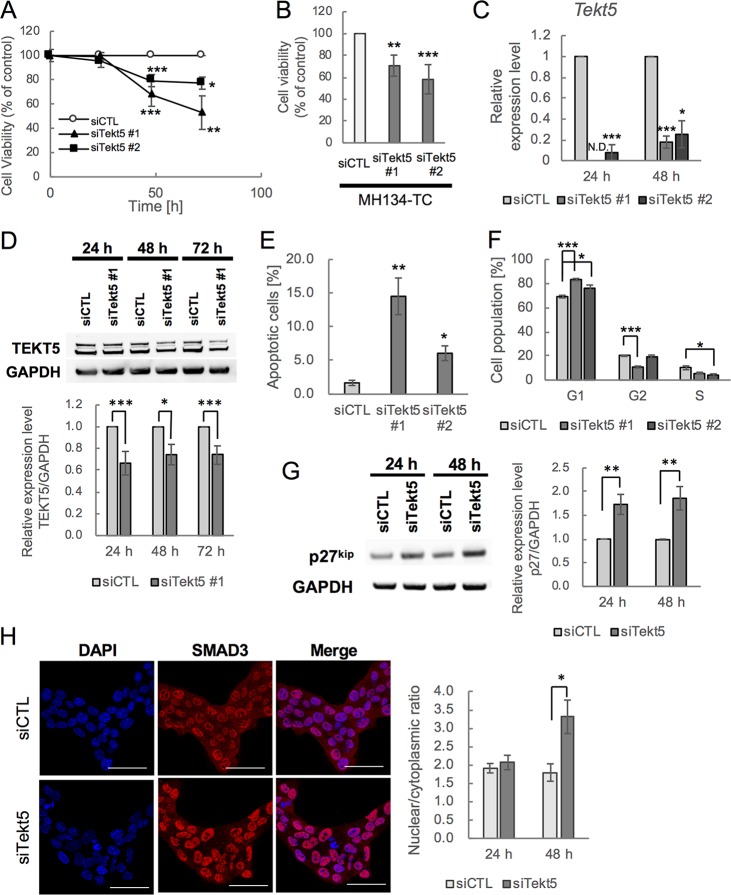
We first confirmed that cell number of OV3121 and MH134-TC was significantly decreased after 48 and 72 h of Tekt5-KD by either of two different siRNAs (Fig. 5A and B). This decrease in cell number was accompanied by decreases in the accumulation of both the Tekt5 mRNA and the TEKT5 protein (Fig. 5C and D). We then tested whether cell cycle progression and/or cell survival were affected by Tekt5-KD. We found increased numbers of apoptotic cells, as well as a prolonged G1 phase and a shortened G2 phase within the cell cycle (Fig. 5E and F). Because p27kip1, a cyclin-dependent kinase inhibitor, is known to repress the G1-S transition (26), we examined the expression of this protein and demonstrated that the p27kip1 protein accumulated to higher levels in Tekt5-KD cells (Fig. 5G).
FIG 5.
Repression of G1-S transition of cell cycle and cell survival and increased nuclear localization of SMAD3 by Tekt5-KD in OV3121 and MH134-TC cells. (A and B) Changes in viability of an ovarian cancer cell line, OV3121 (A), and of a liver cancer cell line, MH134-TC (B), by two different siRNA corresponding to Tekt5 (siTekt5#1 and siTekt5#2). siCTL, AllStars negative control siRNA. MH134-TC cells were assayed at 72 h after transfection with the siRNAs. Cell viability was determined by MTS assay. (C and D) Decreased accumulation of Tekt5 mRNA as determined by RT-qPCR (C) and of TEKT5 protein as determined by Western blotting (D, upper) after Tekt5-KD in OV3121 cells. For the Western blotting, the signal intensity of TEKT5 was quantified and then normalized against that of the housekeeping protein GAPDH (D, lower). (E and F) Ratios of active caspase-3-positive apoptotic cells (E) and of cells in each phase of the cell cycle (F) at 48 h after transfection of OV3121 cells with Tekt5 siRNA (siTekt5#1), as determined by flow cytometry. (G) Accumulation of p27kip 24 and 48 h after transfection of OV3121 cells with Tekt5 siRNA, as determined by Western blotting (left). The signal intensity of p27kip was quantified and then normalized against that of GAPDH (right). (H) Localization of SMAD3 24 and 48 h after transfection of OV3121 cells with Tekt5 siRNA, as determined by immunostaining (left). SMAD3 accumulation is shown in red at 48 h in the micrographs. Nuclear staining by DAPI (blue) also is shown. Scale bars, 50 μm. Signal intensity of SMAD3 was quantified by confocal laser scan microscopy, and nuclear-cytoplasmic ratios of the protein were determined. N.D., not detected. Graphs represent data from three biological replicates. Values are plotted as means ± the standard errors (SE). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed, nonpaired Student t test).
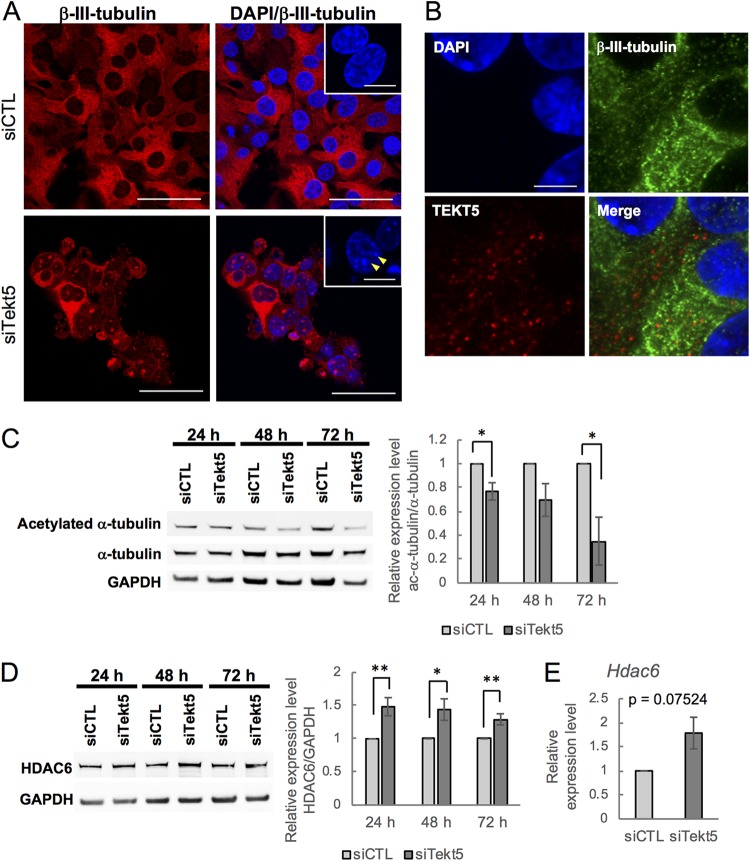
Previous studies have shown that the expression of p27kip1 is enhanced by SMADs, a family of TFG β-signaling molecule that includes SMAD3 (27, 28). In addition, some members of the tektin family of proteins have been suggested to be components of cilia and flagella consisting of microtubules (7, 29); dissociation of SMADs from microtubules after microtubule depolymerization permits the activation of SMADs, resulting in the transmission of a signal to the nucleus (30). We therefore tested whether localization of SMAD3 was affected by Tekt5-KD in OV3121; as expected, Tekt5-KD resulted in increased levels of nuclear SMAD3 (Fig. 5H). We also examined effects on tubulin and found that the fibrous appearance of β-III-tubulin was disrupted upon of Tekt5-KD (Fig. 6A). Meanwhile, we observed that TEKT5 did not colocalize with β-III-tubulin (Fig. 6B). Together, these results suggest that, unlike other members of tektin family, TEKT5 is not associated with tubulin and may affect tubulin stabilization only indirectly.
FIG 6.
Abnormality of microtubules in OV3121 cells subjected to Tekt5-KD. (A) Localization of β-III-tubulin (red) in Tekt5-KD (siTekt5) and control (siCTL) OV3121 cells, as detected by immunostaining. Fibrous β-III-tubulin is disrupted by Tekt5-KD. Insets show DAPI-staining in higher magnification. Arrows indicate DAPI-stained dots. Scale bars: 50 and 12.5 μm (insets). (B) Localization of TEKT5 in OV3121 cells. TEKT5 (red) is not colocalized with β-III-tubulin (green). DAPI staining (blue) is also shown. Scale bar, 10 μm. (C and D) The accumulation of α-tubulin (C) and HDAC6 (D) protein in Tekt5-KD and control OV3121 cells, as detected by Western blotting at 24, 48, and 72 h posttransfection. For both panels C and D, each panel consists of an image of the blot (left) and of results (ratios of acetylated [ac] α-tubulin to total α-tubulin [C, right] or of HDAC6 to GAPDH [D, right]) quantified by band intensity. (E) Accumulation of Hdac6 mRNA (D) in Tekt5-KD and control OV3121 cells, as detected by RT-qPCR at 48 h posttransfection. For panels C to E, the graphs represent data from three and five biological replicates for ac α-tubulin and HDAC6/Hdac6, respectively. Values are plotted as means ± the SE. *, P < 0.05; **, P < 0.01 (two-tailed, nonpaired Student t test).
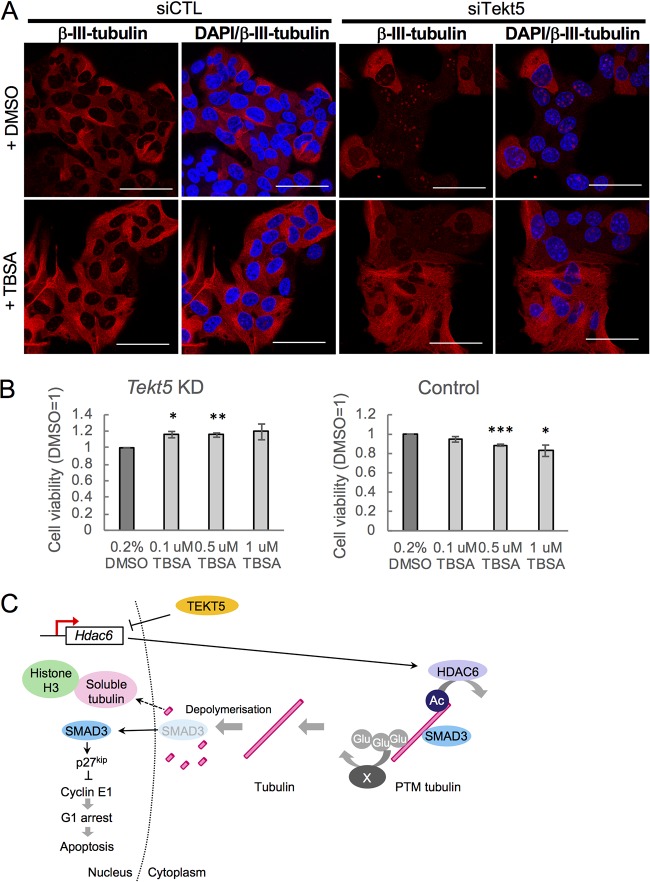
Because tubulin is stabilized by acetylation (31), we next examined levels of acetylated α-tubulin. The results showed that ratios of acetylated α-tubulin to total α-tubulin in OV3121 were decreased by Tekt5-KD (Fig. 6C). We also found that the expression of HDAC6, a protein known to deacetylate α-tubulin (32), was upregulated by Tekt5-KD (Fig. 6D). In addition, tubastatin A (TBSA), a specific inhibitor of HDAC6 (33), rescued Tekt5-KD-induced destabilization of β-III-tubulin in OV3121 (Fig. 7A). In addition, TBSA improved the cell viability of Tekt5-KD cells but not that of control cells (Fig. 7B). These results implied that TEKT5 is involved in the stabilization of tubulin via negative regulation of HDAC6 expression, which consequently results in cell cycle progression via attenuation of SMAD3 translocation and of p27kip1 induction (Fig. 7C).
FIG 7.
Effect of tubastatin A (TBSA), a specific inhibitor of HDAC6, in OV3121 cells. (A) Disruption of β-III-tubulin (red) by Tekt5-KD in control cells (+DMSO) was rescued by TBSA. DAPI staining (blue) is also shown. siCTL, AllStars negative control siRNA. Scale bars, 50 μm. (B) The cell viability of Tekt5-KD OV3121 cells, but not that of control OV3121 cells, at 72 h posttransfection was increased by TBSA exposure. Cell viability was determined by an MTS assay. (C) Possible mechanism for TEKT5 control of cancer cell viability. PTM, posttranslational modification; Glu, glutamyl side chain; X, unknown factor(s) for polyglutamylation of tubulin. Values are plotted as means ± the SE. **, P < 0.01; ***, P < 0.001 (two-tailed, nonpaired Student t test).
Roles of TEKT5 in spermiogenesis.
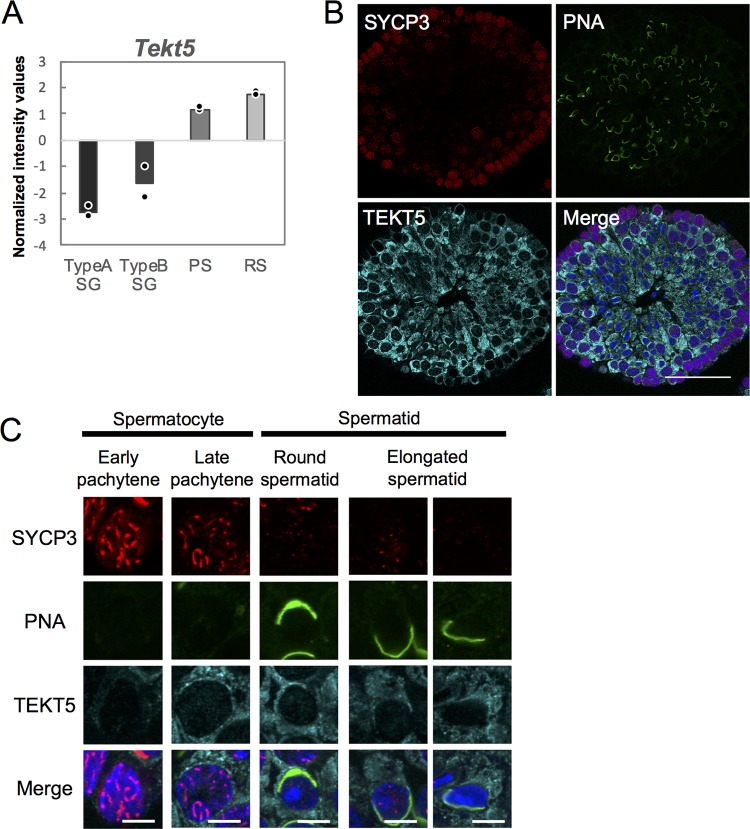
Reanalysis of transcriptome data from a public database (GenBank accession number GSE4193) (34) revealed that the expression of Tekt5 is gradually upregulated during spermatogenesis (Fig. 8A). Using immunostaining, we confirmed that TEKT5 protein expression accumulates in testis from the late pachytene stage on (Fig. 8B and C).
FIG 8.
Expression of Tekt5 in spermatogenic cells. (A) Graph representing signal intensity values normalized using global median scaling for Tekt5 gene expression. The graph is based on published microarray data (GSE4193). Dots show average values from each of two independent data sets. TypeA SG, type A spermatogonia; TypeB SG, type B spermatogonia; PS, pachytene spermatocyte; RS, round spermatid. (B and C) Accumulation of TEKT5 (cyan), SYCP3 (red), and PNA (green) in 3-month-old mouse testes, as detected by immunostaining. Merged images with DAPI (blue) are also shown. The images in panel C show higher magnifications of spermatogenic cells at different differentiation stages classified based on the staining patterns of SYCP3 and PNA. Scale bars, 5 μm.
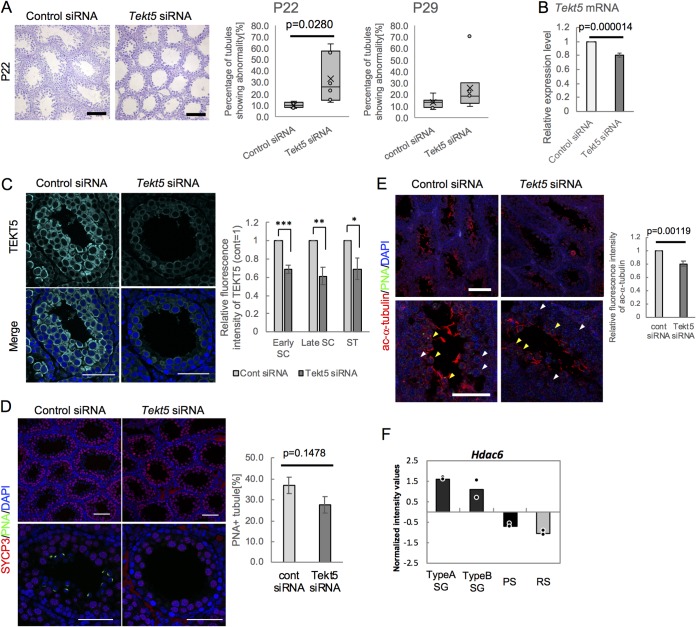
We then examined the in vivo functions of Tekt5 by performing KD in testis. We introduced siRNAs corresponding to Tekt5 and a control into the seminiferous tubules of the left and right testes, respectively, in 8-day-old mice (35) and evaluated the testes histologically at 14 and 21 days after injection of the siRNAs. Abnormal spermatogenesis was observed in some seminiferous tubules (Fig. 9A) in Tekt5 siRNA-injected testes. We confirmed decreased accumulation of both Tekt5 mRNA (Fig. 9B) and TEKT5 protein (Fig. 9C) in Tekt5 siRNA-injected testis (i.e., Tekt5-KD testis). The ratios of tubules with spermatids labeled by peanut agglutinin (PNA) were decreased in Tekt5-KD testis compared to those in control testis, while SYCP3-expressing spermatocytes were not affected (Fig. 9D). These results indicated that TEKT5 is involved in spermatid differentiation. We also observed decreased levels of acetylated α-tubulin in Tekt5-KD testis (Fig. 9E). Furthermore, the transcriptome analysis (in normal development) showed that Hdac6 transcript levels fell during spermatogenesis, concomitant with increased Tekt5 transcript accumulation (Fig. 8A and 9F). Together, these results implied that TEKT5 contributes to spermiogenesis by maintaining the levels of acetylated α-tubulin.
FIG 9.
Spermatogenic failure in Tekt5-KD testis at P22. (A) H&E staining of Tekt5-KD and control testes at 22 days of age (left). The graphs show ratios of abnormal seminiferous tubules in Tekt5-KD testis to those in control testis at P22 (left) and P29 (right). Error bars indicate the ranges of minimum and maximum values; Xs and horizontal lines in boxes indicate means and medians, respectively. (B and C) Decreased accumulation of Tekt5 mRNA (B) and TEKT5 protein (C) in Tekt5-KD testis compared to control testis. The expression of mRNA was determined by RT-qPCR (B). The signal intensity of TEKT5 immunostaining (C, left) in early spermatocytes (early SC), late spermatocytes (late SC), and spermatids (ST) (with stages determined based on the staining patterns for SYCP3 and PNA shown in Fig. 6C) was quantified; values were normalized to that in Leydig cells (C, right). Values in control germ cells were set as 1.0. (D) Decreased PNA-positive spermatids (green) in Tekt5-KD testis (left). SYCP3-expressing spermatocytes (red) are not affected by KD. The ratios of seminiferous tubules with PNA-positive cells in counted tubules were determined (right). (E) Decreased acetylated (ac) α-tubulin (red) in Tekt5-KD testis at P22. Yellow and white arrowheads indicate PNA (green)-positive spermatids and PNA-negative spermatocytes, respectively (left). The signal intensity of ac α-tubulin immunostaining was quantified, and values were normalized to those in Leydig cells (right). Values in control germ cells were set as 1.0. (F) Graph representing signal intensity values normalized using global median scaling for Hdac6 gene expression based on published microarray data (GSE4193). Dots indicate average values from each of two independent data sets. TypeA SG, type A spermatogonia; TypeB SG, type B spermatogonia; PS, pachytene spermatocyte; RS, round spermatid. Blue shows DAPI staining. Scale bars: 200 μm (A), 50 μm (C and D, lower), 75 μm (D, upper), 100 μm (E, upper), or 25 μm (E, lower). Graphs represent data from six and nine mice at P22 and P29, respectively, after injection of Tekt5 siRNA and control siRNA in left and right testes, respectively. Values in panels B, C, and D are plotted as means ± the SE. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed, nonpaired Student t test).
DISCUSSION
Among 139 mouse homologues of human CTA genes, we identified 87 genes as mouse CTA genes (Fig. 1; Table S3), loci that showed higher expression in at least one of the tested cancer cell lines compared to expression in corresponding normal tissues. Differences in the transcript levels (between the cancer cells and normal tissues) were not obvious for the rest of the genes (Fig. 2). It is likely that the correct counterparts of some human CTA genes were not selected by the Web tool or that the tested cancer cells express the tested genes only at low levels.
Our RNAi screening showed that KD of 47 of 84 genes caused >10% changes in cell number in cancer cell lines (Fig. 3 and 4; Table S5), suggesting that the majority of CTAs are involved in cancer cell development. It is worth noting that some of those CTA genes whose KD affected cancer cell number in culture are known to be mutated in human cancers. For instance, MORC1 typically is mutated in metastatic breast cancer, although the frequency of mutation of this gene is low in primary breast cancer (36). In a second example, a truncating mutation in the TEX15 open reading frame was identified as a possible risk factor for breast cancer (37). In a third example, frameshift mutations of TTK and TAF7L are commonly found in gastric and colorectal cancers (38, 39). Involvement of these mutations in cancer development is currently obscure, but our results suggest that various CTAs play a role in cancer cells, including instances other than those previously reported for members of the MAGE-A and SSX family.
KD of mouse CTA genes did not necessarily result in decrease in cell number; indeed, the KD of some CTA genes caused increases in cell number (Fig. 3 and 4), indicating that these genes may normally encode negative regulators of the growth and/or survival of cancer cells. This observation implies that, in some cases, these CTAs are involved in growth suppression of cancer cells, a process that might include metastasis-associated growth suppression (40). KD of some CTA genes did not yield consistent effects on cell number in different cancer cell lines (Fig. 3 and 4), suggesting that CTAs may serve functions or be required differentially in various cancer cell lines.
We found that Tekt5 KD caused accumulation of OV3121, an ovarian cancer cell line, in the G1 phase of cell cycle, with a concomitant increase in the fraction of apoptotic cells (Fig. 5E and F). These observations implied that TEKT5 positively regulates cell cycle progression. Consistent with this inference, p27kip1, a cyclin-dependent kinase inhibitor known to repress the G1-S transition, accumulated to higher levels in Tekt5 KD cells (Fig. 5G). Our results suggested that destabilization of tubulin by Tekt5 KD causes nuclear accumulation of SMAD3 (Fig. 5H and 6A), which is known to upregulate p27kip1 (27, 28). This inference also is consistent with a previous report showing that SMADs are activated by dissociating from microtubules after microtubule depolymerization (30). Meanwhile, previous studies indicated that destabilization of tubulin resulted in the translocation of some transcription factors, such as myc-interacting zinc finger protein (MIZ-1) (41) and NF-κB (42), from the cytoplasm to the nucleus, and we cannot exclude the possibility that additional unknown factors other than SMAD3 translocate into nucleus by Tekt5 KD via tubulin destabilization to repress the cell cycle.
In addition, a previous study also showed that the nuclear accumulation of α- and β-tubulins induced by their depolymerization by nocodazole treatment at 4°C or by inhibiting their nuclear export resulted in G0/G1 arrest and the apoptosis of cells and nuclear tubulins were associated with histone H3, which may affect chromatin organization (43). Because we found that a part of β-III-tubulin was ectopically localized in nucleus by Tekt5 KD (Fig. 6A), Tekt5 KD likely induces cell cycle arrest and apoptosis via nuclear tubulin in addition to nuclear SMAD3 (Fig. 7C). We also observed prominent DAPI (4′,6′-diamidino-2-phenylindole)-stained dots in nucleus by Tekt5 KD (Fig. 6A, inset images), which may reflect not only apoptosis but also changes in chromatin organization induced by Tekt5 KD.
In contrast to the known localization of other tektin family proteins in cilia and flagella (29), TEKT5 did not colocalize with microtubules (Fig. 6B), although TEKT5 is involved in Hdac6 gene expression (Fig. 6D and E). The mechanisms of possible transcriptional regulation of Hdac6 by TEKT5 are currently unclear, but cytoplasmic localization of TEKT5 implies its interaction with signaling molecules to transmit signals to the nucleus and subsequent downregulation of Hdac6 expression.
Although TBSA effectively rescued the disruption of tubulin by Tekt5 KD in OV3121 cells (Fig. 7A), TBSA’s effect in counteracting the attenuation of cell viability by Tekt5 KD was marginal (Fig. 7B). These observations suggested that additional modifications of tubulins are involved in their stabilization and subsequent enhancement of cell viability by TEKT5 (Fig. 7C). For instance, it was previously reported that hyperelongation of glutamyl side chains stabilized cytoplasmic microtubules in Tetrahymena (44). Consistently, a modest amount of β-III-tubulin was observed in the nucleus even in the presence of TBSA (Fig. 7A), which may explain why the effect of TBSA on the attenuation of cell viability by Tekt5 KD was marginal; further characterization of all these effects will be an important subject for future studies.
We observed spermatogenic failures, including abnormal spermiogenesis, upon in vivo KD of Tekt5 (Fig. 9). Notably, we observed significant downregulation of TEKT5 and the abnormal spermatogenesis at 14 days after siRNA injection into testicular tubules (Fig. 9A), but the influences of Tekt5 KD was not obvious at 21 days after injection (Fig. 9A). This distinction was presumed to reflect the transient nature of effects of the in vivo KD method, as reported previously (35). We injected siRNA into testicular tubules at postnatal day 8 (P8), in accord with the previous report, and observed the testes histologically at P22, when the first round of spermatogenesis is still in progress and mature sperm have not yet emerged. Although TEKT5 has been shown to localize to the midpiece of sperm, we did not observe sperm abnormalities at P29, 21 days after injection of siRNA, presumably due to diminished influence of the siRNA and recovery of spermiogenesis in unaffected spermatogenic cells. In addition, it is also likely that TEKT5 may not have functions in mature sperm. In this study, we showed a function of TEKT5 in spermiogenesis in the first-wave spermatogenesis in pubertal testis due to the limitation of the KD method, and further studies are needed to clarify its functions in adult testes.
When a round spermatid elongates to form a sperm in the final stage of spermiogenesis, tubulin is rearranged to form the manchette, a structure that likely is involved in nuclear elongation and the spiral arrangement of mitochondria in the midpiece of the sperm (45, 46). In Tekt5 KD testis, the level of acetylated α-tubulin was decreased not only in spermatids but also in PNA-negative late spermatocytes (Fig. 9E), implying that TEKT5 normally functions in manchette formation via tubulin stabilization. During spermatogenesis, the accumulation of Tekt5 transcript is increased (Fig. 8A), while that of Hdac6 transcript is decreased (Fig. 9F). Together, these observations suggest that TEKT5 negatively controls Hdac6 transcription in testis, as observed in cancer cells (Fig. 6D); however, immunostaining did not detect significant HDAC6 accumulation even in control testis (data not shown). It is likely that additional HDACs and/or histone acetylating enzymes are involved in controlling the acetylation of tubulin in spermatogenic cells.
Our results show that a single CTA gene, Tekt5, plays crucial roles in both cancer cells and testicular germ cells. Furthermore, our data imply that one or more other CTAs also function in both of these cell types. Analysis of the functions of these other CTAs may uncover distinct cellular regulation in cancer cells and testicular germ cells controlled by a related molecular mechanism.
MATERIALS AND METHODS
Cell culture.
Cancer cell lines obtained from Cell Resource Center for Biomedical Research (Tohoku University, Japan) included B16 (mouse melanoma), B16C2W (mouse melanoma), 3LL (mouse lung cancer), KLN205 (mouse lung cancer), Ehrlich (mouse breast cancer), MM46 (mouse breast cancer), FM3A (mouse breast cancer), MH134-TC (mouse liver cancer), and colon-26 (mouse colon cancer). Cancer cell lines obtained from Riken Cell Bank (Japan) included OV2944-HM-1 (HM-1; mouse ovarian cancer), Hepa1-6 (mouse liver cancer), and MBT-2 (mouse bladder cancer). OV3121, a mouse ovarian granulosa carcinoma cell line, was obtained from JCRB Cell Bank (Japan). B16, B16C2W, 3LL, Ehrlich, MM46, MH134-TC, MBT-2, OV3121, and colon-26 cells were cultured in RPMI 1640 (Sigma-Aldrich) medium containing 10% fetal bovine serum (FBS). KLN205 cells were cultured in minimal essential medium (MEM; Sigma-Aldrich) medium containing 10% FBS and nonessential amino acids. HM-1 and Hepa1-6 cells were cultured in α-MEM and Dulbecco modified Eagle medium containing 10% FBS, respectively. All cell lines were cultured at 37°C in a 5% CO2 environment.
Mouse strains.
C57BL/6J mice were purchased from Japan SLC. The mice were maintained and bred in the Animal Unit of the Institute of Development, Aging, and Cancer (Tohoku University), an environmentally controlled and specific-pathogen-free facility, according to the guidelines for experimental animals defined by the facility. Animal protocols were reviewed and approved by the Tohoku University Animal Studies Committee.
Antibodies.
The following primary antibodies were used for Western blotting: rabbit anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) at 1:2,000 (CST, catalog no. 2118), rabbit anti-TEKT5 at 1:1,000 (Thermo, PA5-21157), mouse anti-p27kip at 1:500 (Santa Cruz, sc-1641), mouse anti-acetylated-alpha-tubulin at 1:1,000 (Santa Cruz, sc-23950), mouse anti-alpha-tubulin at 1:1,000 (Wako, catalog no. 017-25031), and rabbit anti-HDAC6 at 1:1,000 (CST, catalog no. 7612). The following primary antibodies and reagent were used for immunofluorescence assays: rabbit anti-beta-III-tubulin at 1:200 (Sigma-Aldrich, T2200), rabbit anti-TEKT5 at 1:200 (Thermo, PA5-21157), rabbit anti-SMAD3 at 1:400 (Abcam, ab40854), mouse anti-SYCP3 at 1:400 (Abcam, ab97672), mouse anti-alpha-tubulin at 1:400 (Wako, catalog no. 017-25031), and Alexa Fluor 488-conjugated lectin PNA at 1:500 (Molecular Probes, L-21409).
In vitro RNAi.
The identities of the siRNAs used in screening are listed in Table S3 in the supplemental material. siRNAs for Tekt5-KD in cancer cells were prepared as follows: Tekt5 siRNA#1 (Qiagen, SI00831887), Tekt5 siRNA#2 (Qiagen, SI00831894), and AllStars negative control siRNA (Qiagen, SI03650318). siRNAs were transfected into cells using Lipofectamine RNAiMAX (Invitrogen) and the reverse method according to the manufacturer’s instructions. Briefly, transfection was performed in a 24-well format as follows. Lipofectamine RNAiMAX (2 μl) and siRNA (20 pmol) were diluted with 100 μl of Opti-MEM, followed by incubation for 20 min. The mixtures of cells and Lipofectamine were seeded onto a 24-well plate. The cells were incubated for 24 h, and the medium was then replaced.
Real-time PCR.
Total RNA was extracted from paraffin-embedded tissues or cells using the RNeasy Plus minikit (Qiagen) or RNeasy FFPE kit (Qiagen) according to the manufacturer’s instructions. RNAs were reverse transcribed using SuperScript III and random primers. Real-time PCR was performed using the Power SYBR green PCR Master Mix (Applied Biosystems). Thermal conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 45 cycles of 15 s at 95°C and 60 s at 60°C. The sequences of the primers used for the PCR are shown in Table S7. The housekeeping gene Arbp (which encodes a mouse ribosomal protein) was used as an internal control. The relative expression was analyzed using the comparative cycle threshold (CT) method. If a CT value was not obtained in 45 cycles of amplification, the expression level was considered “0.”
High-throughput RT-qPCR.
cDNA was mixed with PreAmp Master Mix (Fluidigm) and the STA Multiplex Primer Pool. Subsequently, the cDNA was amplified by adding primers and subjecting the mixture to 14 cycles of 95°C for 15 s (denaturing) and 60°C for 4 min (annealing and amplifying). Nested PCR was carried out on the primary PCR products to ensure specific detection of the transcripts. A list of the primers used for gene expression analyses is provided in Table S1 in the supplemental material. The preamplified products were diluted 5-fold before further analysis and then mixed with 2× SsoFast EvaGreen Super mix with Low ROX (Bio-Rad) and 20× DNA binding dye sample loading reagent (Fluidigm). The mixtures containing the samples, primer pair mix, and 2× assay loading reagent (Fluidigm) were used as probes against 48.48 dynamic arrays on a BioMark system (Fluidigm). CT values were calculated by using the system software (BioMark real-time PCR analysis; Fluidigm). The expression value of each gene was calculated by the comparative CT method using the housekeeping gene Hrpt (which encodes hypoxanthine guanine phosphoribosyl transferase 1) as an internal control.
Western blotting.
For extraction, cells were resuspended in lysis buffer (50 mM Tris-HCl, 1% sodium dodecyl sulfate [SDS], 1× cOmplete [Roche], and 1× PhosStop [Roche]) and sonicated using a Bioruptor (Sonic Bio) for five cycles of 30-s on and off at the “high” setting. The resulting protein extracts were subjected to electrophoresis on NuPAGE 10% Bis-Tris gels (Invitrogen) and then transferred to polyvinylidene difluoride membranes (Millipore Immobilon-P). The membranes were washed in Tris-buffered saline containing 0.05% Tween 20 (TBST) and blocked for 1 h with TBST containing 5% skim milk or 5% bovine serum albumin (BSA). The membranes then were incubated overnight at 4°C with the indicated primary antibodies diluted in TBST containing 1% skim milk or 5% BSA. After a washing step, the membranes were incubated for 1 h at room temperature with horseradish peroxidase-labeled secondary antibodies. Immunoblotting was visualized using a Clarity Western ECL substrate kit (Bio-Rad) and the LAS-3000 system (Fujifilm). After the images were captured, the membranes were washed for 20 min at 50°C with stripping buffer (100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl [pH 6.8]), washed with TBST, blocked as described above, and incubated for 1 h at room temperature with the next antibody (as indicated). The intensity of the target bands was assessed using Multi-Gauge software (Fujifilm).
MTS assay.
Cell proliferation rates were assessed by using an MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay. Briefly, after culture in 96 well-plates, the cells were incubated for 3 h at 37°C with 20 μl/well of MTS (Promega, G3580). The level of cellular proliferation was determined by MTS conversion to formazan using a SpectraMax Me2 (Molecular Devices) at 490 nm.
Active caspase analysis.
Analysis for active caspase was performed using a caspase-3, active-form, monoclonal antibody apoptosis kit and fluorescein isothiocyanate (BD Pharmingen) according to the manufacturer’s instructions. The cells were dissociated to yield a single-cell suspension, washed, and fixed with BD Cytofix/Cytoperm. The cells then were washed and incubated for 30 min at room temperature with the kit-supplied antibody. Fluorescence-activated cell-sorting (FACS) analysis of these stained cells was performed on a Cytomics FC500 cell analyzer (Beckman Coulter).
Cell cycle analysis.
Cell cycle analysis was performed using a APC BrdU flow kit (BD Pharmingen) according to the manufacturer’s instructions. The cells were dissociated to yield a single-cell suspension, washed, and fixed with BD Cytofix/Cytoperm. The cells then were incubated with DNase and RNase A in BD Perm/Wash solution for 1 h at 37°C, washed, and incubated with 7-amino-actinomycin D (7AAD) for 1 h at 37°C. FACS analysis of these cells was performed on a Cytomics FC500 cell analyzer.
Immunofluorescence.
Cells were grown on poly-l-lysine-coated, glass-bottom dishes. After a washing step, the cells were fixed in 4% paraformaldehyde buffer solution or iced methanol and then permeabilized for 15 min at room temperature with phosphate-buffered saline (PBS) containing 0.1% Triton X-100. The cells then were washed with PBS, blocked for 1 h at room temperature with 10% FBS, washed again, and incubated for 1 h at room temperature with a secondary antibody diluted in 10% FBS. The resulting samples were mounted with Vectashield mounting medium (Vector Laboratories), and all images were captured with Leica TSC SP8 (Leica Microsystems). The intensity of the fluorescence was assessed using Leica Imaging software (Leica Microsystems).
In vivo knockdown.
The Mouse Accell SMART pools of siRNAs used for the in vivo KD experiments were prepared using either Tekt5 siRNAs (Dharmacon, E-051627-00-0020) or nontargeting siRNAs (Dharmacon, D-001910-10-05). siRNA solutions were formulated in Dulbecco PBS containing 100 μM Accell siRNAs, 1× siRNA buffer (Dharmacon), and 0.01% trypan blue solution (Sigma-Aldrich) (as a tracer). Solutions of Tekt5 siRNA or control siRNA were microinjected into the seminiferous tubules of the left and right testes, respectively, in live mouse pups, using the method described by Dai et al. (35). Briefly, each mouse (8 days old) was anesthetized with ice. Testes were pulled from the abdominal cavity, and (working under a binocular microscope) approximately 3 μl of siRNA solution was injected into the rete testis using a glass capillary. The testes were then returned to the abdominal cavity, and the abdominal wall and skin were closed with sutures. The mouse was placed on a hot plate at 37°C until awake.
H&E staining and IHC staining.
Testes were fixed in 4% paraformaldehyde buffer solution and embedded in paraffin. Sections (5-μm thicknesses) were cut and subjected to hematoxylin and eosin (H&E) staining. For immunohistochemical (IHC) staining, sections were deparaffinized, and antigen retrieval was performed by incubation at 95°C for 15 min in an antigen retrieval solution. Next, the sections were washed with PBS, blocked for 1 h at room temperature in 10% FBS, and stained as described above.
Reanalysis of published microarray data.
The data obtained from published microarray data (GenBank accession number GSE4193 [34]) were normalized by using global median scaling and reanalyzed using GeneSpring (Agilent).
Statistical analysis.
Statistical analysis was performed using a two-tailed, nonpaired Student t test. A P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Cell Resource Center for Biomedical Research for helpful discussions. We thank the Center of Research Instruments at the Institute of Development, Aging, and Cancer (IDAC) of Tohoku University and the Biomedical Research Core of the Tohoku University Graduate School of Medicine for the use of instruments and for technical support.
N.A. was supported by JSPS KAKENHI (grant JP17J02028) and the Division for Interdisciplinary Advanced Research and Education at Tohoku University. Y.M. was supported by KAKENHI, Innovative Area Mechanisms Regulating Gamete Formation in Animals (grant 16H06530) from MEXT, and by AMED-CREST (grant JP17gm0510017h) from the Japan Agency for Medical Research and Development.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00154-19.
REFERENCES
- 1.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. 1996. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A 93:7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang P, Huo Z, Zhou HL, Zhou Q. 2015. Cancer/testis antigens trigger epithelial-mesenchymal transition and genesis of cancer stem-like cells. CPD 21:1292–1300. doi: 10.2174/1381612821666141211154707. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi N, Ohkuri T, Homma S, Ohtake J, Wakita D, Togashi Y, Kitamura H, Todo S, Nishimura T. 2012. First clinical trial of cancer vaccine therapy with artificially synthesized helper/killer-hybrid epitope long peptide of MAGE-A4 cancer antigen. Cancer Sci 103:150–153. doi: 10.1111/j.1349-7006.2011.02106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Vries FAT, De Boer E, Van Den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, Van Zeeland AA, Heyting C, Pastink A. 2005. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolcun-Filas E, Costa Y, Speed R, Taggart M, Benavente R, De Rooij DG, Cooke HJ. 2007. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol 176:741–747. doi: 10.1083/jcb.200610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanafusa T, Mohamed AEA, Domae S, Nakayama E, Ono T. 2012. Serological identification of Tektin5 as a cancer/testis antigen and its immunogenicity. BMC Cancer 12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amos LA. 2008. The tektin family of microtubule-stabilizing proteins. Genome Biol 9:229. doi: 10.1186/gb-2008-9-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murayama E, Yamamoto E, Kaneko T, Shibata Y, Inai T, Iida H. 2008. Tektin5, a new TEKTIN family member, is a component of the middle piece of flagella in rat spermatozoa. Mol Reprod Dev 75:650–658. doi: 10.1002/mrd.20804. [DOI] [PubMed] [Google Scholar]
- 9.Gjerstorff MF, Ditzel HJ. 2008. An overview of the GAGE cancer/testis antigen family with the inclusion of newly identified members. Tissue Antigens 71:187–192. doi: 10.1111/j.1399-0039.2007.00997.x. [DOI] [PubMed] [Google Scholar]
- 10.Zendman AJW, Van Kraats AA, Weidle UH, Ruiter DJ, Van Muijen G. 2002. The XAGE family of cancer/testis-associated genes: alignment and expression profile in normal tissues, melanoma lesions, and Ewing’s sarcoma. Int J Cancer 99:361–369. doi: 10.1002/ijc.10371. [DOI] [PubMed] [Google Scholar]
- 11.Brinkmann U, Vasmatzis G, Lee B, Yerushalmi N, Essand M, Pastan I. 1998. PAGE-1, an X chromosome-linked GAGE-like gene that is expressed in normal and neoplastic prostate, testis, and uterus. Proc Natl Acad Sci U S A 95:10757–10762. doi: 10.1073/pnas.95.18.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boel P, Wildmann C, Sensi ML, Brasseur R, Renauld JC, Coulie P, Boon T, van der Bruggen P. 1995. BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity 2:167–175. doi: 10.1016/S1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 13.Hu F, Lesney PF. 1964. The isolation and cytology of two pigment cell strains from B16 mouse melanomas. Cancer Res 24:1634–1643. [PubMed] [Google Scholar]
- 14.Oikawa A, Nakayasu M, Claunch C, Tchen TT. 1972. Two types of melanogenesis in monolayer cultures of melanoma cells. Cell Differ 1:149–155. doi: 10.1016/0045-6039(72)90024-3. [DOI] [PubMed] [Google Scholar]
- 15.Bertram JS, Janik P. 1980. Establishment of a cloned line of Lewis lung carcinoma cells adapted to cell culture. Cancer Lett 11:63–73. doi: 10.1016/0304-3835(80)90130-5. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko T, LePage GA. 1978. Growth characteristics and drug responses of a murine lung carcinoma in vitro and in vivo. Cancer Res 38:2084–2090. [PubMed] [Google Scholar]
- 17.Klein G. 1950. Use of the Ehrlich ascites tumor of mice for quantitative studies on the growth and biochemistry of neoplastic cells. Cancer 3:1052–1061. doi:. [DOI] [PubMed] [Google Scholar]
- 18.Nishioka K, Irie RF, Inoue M, Chang S, Takeuchi S. 1969. Immunological studies on mouse mammary tumors. I. Induction of resistance to tumor isograft in C3H/He mice. Int J Cancer 4:121–129. doi: 10.1002/ijc.2910040202. [DOI] [PubMed] [Google Scholar]
- 19.Yamane I, Nakano N. 1966. Long-term culture of the tumor ascites form of a mouse medium mammary in serum-free medium. Tohoku J Exp Med 88:203–214. doi: 10.1620/tjem.88.203. [DOI] [PubMed] [Google Scholar]
- 20.Darlington G. 1987. Liver cell lines. Methods Enzymol 151:19–38. doi: 10.1016/S0076-6879(87)51006-0. [DOI] [PubMed] [Google Scholar]
- 21.Kataoka S, Konishi Y, Nishio Y, Fujikawa-Adachi K, Tominaga A. 2004. Antitumor activity of eosinophils activated by IL-5 and eotaxin against hepatocellular carcinoma. DNA Cell Biol 23:549–560. doi: 10.1089/dna.2004.23.549. [DOI] [PubMed] [Google Scholar]
- 22.Soloway MS. 1977. Intravesical and systemic chemotherapy in the management of superficial bladder cancer. Cancer Res 37:2918–2929. [PubMed] [Google Scholar]
- 23.Yanagihara K, Nii M, Tsumuraya M, Numoto M, Seito T, Seyama T. 1995. A radiation-induced murine ovarian granulosa cell tumor line: introduction of v-ras gene potentiates a high metastatic ability. Jpn J Cancer Res 86:347–356. doi: 10.1111/j.1349-7006.1995.tb03063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hashimoto M, Niwa O, Nitta Y, Takeichi M, Yokoro K. 1989. Unstable expression of E-cadherin adhesion molecules in metastatic ovarian tumor cells. Jpn J Cancer Res 80:459–463. doi: 10.1111/j.1349-7006.1989.tb02336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbett TH, Roberts BJ, Peckham JC, Schabel FM. 1975. Tumor induction relationships in development of transplantable cancers of the colon in mice for chemotherapy assays, with a note on carcinogen structure. Cancer Res 35:2434–2439. [PubMed] [Google Scholar]
- 26.Toyoshima H, Hunter T. 1994. p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78:67–74. doi: 10.1016/0092-8674(94)90573-8. [DOI] [PubMed] [Google Scholar]
- 27.Kanska J, Zakhour M, Taylor-Harding B, Karlan BY, Wiedemeyer WR. 2016. Cyclin E as a potential therapeutic target in high-grade serous ovarian cancer. Gynecol Oncol 143:152–158. doi: 10.1016/j.ygyno.2016.07.111. [DOI] [PubMed] [Google Scholar]
- 28.Cooley A, Zelivianski S, Jeruss JS. 2010. Impact of cyclin E overexpression on Smad3 activity in breast cancer cell lines. Cell Cycle 9:4900–4907. doi: 10.4161/cc.9.24.14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linck RW, Albertini DF, Kenney DM, Langevin GL. 1982. Tektin filaments: chemically unique filaments of sperm flagellar microtubules. Cell Motil 1:127–132. doi: 10.1002/cm.970020724. [DOI] [PubMed] [Google Scholar]
- 30.Dong C, Li Z, Alvarez R, Feng XH, Goldschmidt-Clermont PJ. 2000. Microtubule binding to Smads may regulate TGFβ activity. Mol Cell 5:27–34. doi: 10.1016/S1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- 31.Takemura R, Okabe S, Umeyama T, Kanai Y, Cowan NJ, Hirokawa N. 1992. Increased microtubule stability and alpha tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2, or tau. J Cell Sci 103:953–964. [DOI] [PubMed] [Google Scholar]
- 32.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang X-F, Yao TP. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 33.Butler KV, Kalin J, Brochier C, Vistoli G, Langley B, Kozikowski AP. 2010. Rational design and simple chemistry yield a superior, neuroprotective HDAC6 inhibitor, tubastatin A. J Am Chem Soc 132:10842–10846. doi: 10.1021/ja102758v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. 2006. Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 35.Dai J, Voloshin O, Potapova S, Camerini-Otero RD. 2017. Meiotic knockdown and complementation reveals essential role of RAD51 in mouse spermatogenesis. Cell Rep 18:1383–1394. doi: 10.1016/j.celrep.2017.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah SP, Sun M, Turashvili G, Leung G, Tse K, Senz J, Marra MA, Severson T, Guliany R, Watson P, Morin RD, Steidl C, Teschendorff AE, Moore R, Jones S, Hirst M, Caldas C, Warren RL, Varhol R, Burleigh A, Huntsman D, Zhao Y, Pugh T, Taylor GA, Aparicio S, Gelmon K, Prentice L, Holt RA, Khattra J, Delaney A. 2009. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 37.Mantere T, Tervasmäki A, Nurmi A, Rapakko K, Kauppila S, Tang J, Schleutker J, Kallioniemi A, Hartikainen JM, Mannermaa A, Nieminen P, Hanhisalo R, Lehto S, Suvanto M, Grip M, Jukkola-Vuorinen A, Tengström M, Auvinen P, Kvist A, Borg Å, Blomqvist C, Aittomäki K, Greenberg RA, Winqvist R, Nevanlinna H, Pylkäs K. 2017. Case-control analysis of truncating mutations in DNA damage response genes connects TEX15 and FANCD2 with hereditary breast cancer susceptibility. Sci Rep 7:681. doi: 10.1038/s41598-017-00766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn CH, Kim YR, Kim SS, Yoo NJ, Lee SH. 2009. Mutational analysis of TTK gene in gastric and colorectal cancers with microsatellite instability. Cancer Res Treat 41:224–228. doi: 10.4143/crt.2009.41.4.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh HR, An CH, Yoo NJ, Lee SH. 2015. Frameshift mutations of TAF7L gene, a core component for transcription by RNA polymerase II, in colorectal cancers. Pathol Oncol Res 21:849–850. doi: 10.1007/s12253-014-9832-4. [DOI] [PubMed] [Google Scholar]
- 40.Matus DQ, Lohmer LL, Kelley LC, Schindler AJ, Kohrman AQ, Barkoulas M, Zhang W, Chi Q, Sherwood DR. 2015. Invasive cell fate requires G1 cell cycle arrest and histone deacetylase-mediated changes in gene expression. Dev Cell 35:162–174. doi: 10.1016/j.devcel.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ziegelbauer J, Shan B, Yager D, Larabell C, Hoffmann B, Tjian R. 2001. Transcription factor MIZ-1 is regulated via microtubule association. Mol Cell 8:339–349. doi: 10.1016/S1097-2765(01)00313-6. [DOI] [PubMed] [Google Scholar]
- 42.Rosette C, Karin M. 1995. Cytoskeletal control of gene expression: depolymerization of microtubules activates NF-xB. J Cell Biol 128:1111–1119. doi: 10.1083/jcb.128.6.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akoumianaki T, Kardassis D, Polioudaki H, Georgatos SD, Theodoropoulos PA. 2009. Nucleocytoplasmic shuttling of soluble tubulin in mammalian cells. J Cell Sci 122:1111–1118. doi: 10.1242/jcs.043034. [DOI] [PubMed] [Google Scholar]
- 44.Wloga D, Dave D, Meagley J, Rogowski K, Jerka-Dziadosz M, Gaertig J. 2010. Hyperglutamylation of tubulin can either stabilize or destabilize microtubules in the same cell. Eukaryot Cell 9:184–193. doi: 10.1128/EC.00176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mochida K, Tres LL, Kierszenbaum AL. 1999. Structural and biochemical features of fractionated spermatid manchettes and sperm axonemes of the Azh/Azh mutant mouse. Mol Reprod Dev 52:434–444. doi:. [DOI] [PubMed] [Google Scholar]
- 46.Kazarian E, Son HY, Sapao P, Li W, Zhang Z, Strauss JF, Teves ME. 2018. SPAG17 is required for male germ cell differentiation and fertility. Int J Mol Sci 19:1252. doi: 10.3390/ijms19041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.