CREB mediates effects of cyclic AMP on cellular gene expression. Ubiquitous CREB target genes are induced following recruitment of CREB and its coactivators to promoter proximal binding sites. We found that CREB stimulates the expression of pancreatic beta cell-specific genes by targeting CBP/p300 to promoter-distal enhancer regions.
KEYWORDS: BRD4, CBP, CREB, CRTC2, NeuroD1, PKA, beta cell, cAMP, insulin
ABSTRACT
CREB mediates effects of cyclic AMP on cellular gene expression. Ubiquitous CREB target genes are induced following recruitment of CREB and its coactivators to promoter proximal binding sites. We found that CREB stimulates the expression of pancreatic beta cell-specific genes by targeting CBP/p300 to promoter-distal enhancer regions. Subsequent increases in histone acetylation facilitate recruitment of the coactivators CRTC2 and BRD4, leading to release of RNA polymerase II over the target gene body. Indeed, CREB-induced hyperacetylation of chromatin over superenhancers promoted beta cell-restricted gene expression, which is sensitive to inhibitors of CBP/p300 and BRD4 activity. Neurod1 appears critical in establishing nucleosome-free regions for recruitment of CREB to beta cell-specific enhancers. Deletion of a CREB-Neurod1-bound enhancer within the Lrrc10b-Syt7 superenhancer disrupted the expression of both genes and decreased beta cell function. Our results demonstrate how cross talk between signal-dependent and lineage-determining factors promotes the expression of cell-type-specific gene programs in response to extracellular cues.
INTRODUCTION
Cyclic AMP (cAMP) stimulates cellular gene expression via the protein kinase A (PKA)-mediated phosphorylation of CREB and dephosphorylation of the cAMP-responsive transcriptional coactivators (CRTCs) (1). CREB phosphorylation at Ser133 stimulates its association with the histone acetyltransferase (HAT) coactivators CBP/p300, which are thought to enhance target gene expression by acetylating resident nucleosomes over relevant target genes (2).
In contrast, CRTCs are phosphorylated by salt-inducible kinases (SIKs) and sequestered in the cytoplasm under basal conditions through interactions with 14-3-3 proteins (1). Exposure to cAMP triggers the PKA-mediated phosphorylation and inactivation of the SIKs, leading to the dephosphorylation and nuclear entry of the CRTCs, which bind to CREB over relevant promoters.
Genome-wide studies suggest that the mammalian genome contains about 5,000 CREB target genes (3, 4). Only a small fraction of these targets appear to be activated by cAMP in any one tissue.
CREB stimulates the expression of ubiquitous core target genes via recruitment of CBP/p300 and CRTC to promoter-proximal CREB binding sites (1). Many of these targets are genes for transcription factors that induce a secondary wave of gene transcription (e.g., FOS, FOSB, and NR4A1, NR4A2, and NR4A3) as well as genes for signaling molecules that feed back to attenuate upstream signaling pathways (e.g., RGS2, ICER, and PDE4B) (5). CRTC activity appears critical for induction of most core targets, but the importance of CBP/p300 in this setting is less clear (6, 7). Indeed, a number of core genes remain cAMP inducible following targeted disruption of both CBP and p300, suggesting that these coactivators are dispensable for cAMP-dependent transcription, at least in this context.
In addition to its effects on ubiquitous targets, CREB also stimulates the expression of cell-type-restricted genes (3, 4). Cell-specific genomic responses to extracellular stimuli are thought to proceed via the recruitment of lineage-determining transcription factors (LDTFs) to distal enhancers that also contain binding sites for stimulus-induced transcription factors (8). Indeed, signal-dependent factors such as NF-κB, Lxr, Smad3, and AP-1 have been shown to establish enhancer activity in a variety of cell contexts (9–13).
Superenhancers also contribute importantly to cell-type-restricted gene expression; these loci are composed of extended genomic regions that are heavily occupied by LDTFs, transcriptional coactivators (e.g., CBP/p300, BRD4, and mediator), RNA polymerase II (Pol II), and activating chromatin modifications, such as H3AcK27, over long genomic regions (>10 kb) (14–16). Notably, superenhancers are acutely sensitive to inhibitors of the BET/bromodomain protein BRD4, a chromatin reader that links histone acetylation to the release of paused Pol II and to its productive elongation over the gene body (17).
Although it appears to function in a wide variety of settings, CREB has been shown to mediate effects of hormonal and nutrient signals on glucose homeostasis in metabolic tissues such as liver and pancreatic islets (1). Activation of CREB and CRTC2 in response to the incretin hormone glucagon-like peptide 1 (GLP1) during feeding stimulates beta cell genes that promote insulin secretion and islet viability (18). Depletion of CREB or CRTC2 activity in pancreatic beta cells leads to hyperglycemia due in part to decreases in insulin secretion (18–20).
Conversely, CREB also promotes increases in hepatic glucose production in response to circulating pancreatic glucagon during fasting by stimulating expression of gluconeogenic genes (21). Depleting CRTC2 in hepatocytes lowers gluconeogenic gene expression and blood glucose concentrations in the fasted state and in response to insulin resistance.
Based on the importance of CREB in mediating effects of two hormones that both stimulate cAMP signaling, we explored the mechanism by which this activator promotes the expression of distinct genetic programs in response to the same signal. We found that cAMP triggers the expression of core CREB targets in hepatocytes and beta cells by binding to promoter-proximal cAMP response elements (CREs), whereas CREB stimulates pancreatic beta cell-specific genes by binding to CREs on distal enhancers and superenhancers.
In contrast with its transient effects on promoter-proximal target genes, CREB stimulated beta cell-specific transcription in a more sustained manner, in part by cooperating with the LDTF Neurod1. Reducing Neurod1 function, by RNA interference (RNAi)-mediated depletion or by deletion of enhancer binding sites for Neurod1 and CREB, abolishes cAMP-inducible expression of neighboring genes. Neurod1 was found to promote cAMP-dependent transcription by creating nucleosome-free regions that enhance occupancy of CREB and its coactivators over relevant binding sites. Collectively, these results point to an important role of Neurod1 and perhaps other LDTFs in cooperating with signal-dependent activators to stimulate the expression of tissue-restricted gene programs in response to hormonal stimuli.
RESULTS
CREB mediates a broad transcriptional response to cAMP in pancreatic beta cells.
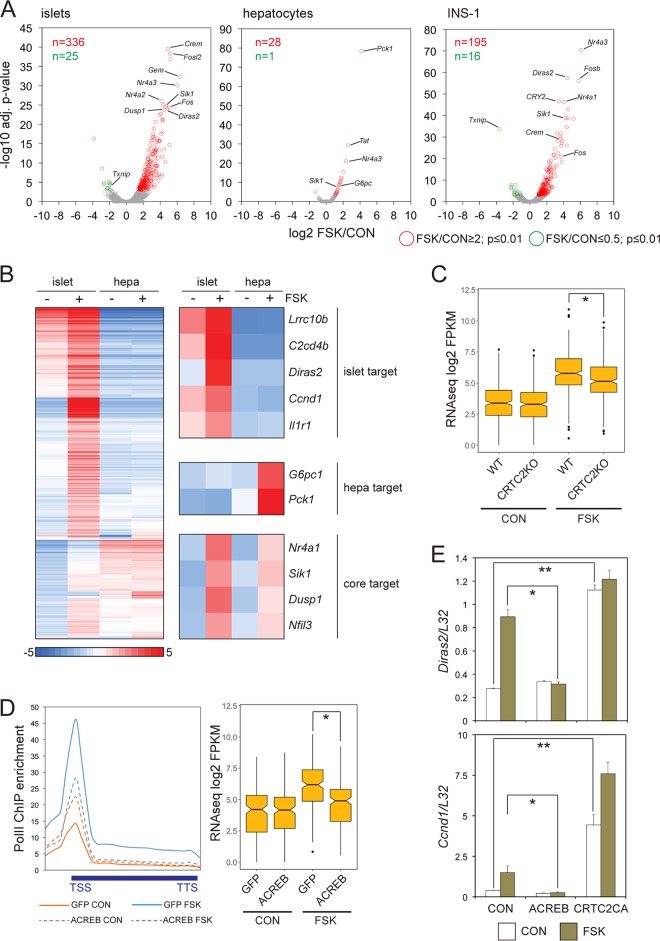
We compared the transcriptional responses of cultured mouse pancreatic islets with INS-1 insulinoma cells and primary hepatocytes to cAMP signaling (Fig. 1A and B). The effect of forskolin (FSK) on gene expression was overwhelmingly stimulatory. Exposure to FSK upregulated a similar set of core CREB target genes in each cell type (e.g., Nr4a2, Crem, and Sik1), but FSK had more robust effects on gene expression in islets and INS-1 insulinoma cells, triggering the expression of nearly 10-fold more genes than hepatocytes; these included genes involved in calcium signaling, insulin secretion, and circadian cycling (e.g., Ncald, Syt4, Sstr1, Per1, and Vgf) as well as a number of genes with unknown function (e.g., C2cd4a, C2cd4b, Fndc4, Lrrc10b, and Diras2) (Fig. 1B) (20).
FIG 1.
CREB mediates a broad transcriptional response to cAMP in pancreatic beta cells. (A) Volcano plots of RNA-seq data comparing genomic responses to cAMP in cultured primary mouse islets, primary hepatocytes (hepa), and INS-1 insulinoma cells (n = 2 independent experiments for each; 2 h of FSK exposure). adj., adjusted. (B) Heat map comparing the magnitude of gene induction in response to cAMP in cultured mouse islets relative to primary hepatocytes. Genes upregulated 2-fold or better in primary mouse islets or hepatocytes selected from RNA-seq experiments conducted independently from replicates shown in panel A. Tissue-specific and ubiquitously induced (core) CREB target genes are highlighted. (C) Box plot showing relative effect of FSK on gene expression in cultured pancreatic islets from wild-type and CRTC2 whole-body knockout littermates (n = 882; *, P < 1 × 10−8). (D, left) Metagene analysis of RNA Pol II occupancy over cAMP-induced genes in INS-1 cells infected with control adenovirus (GFP) or adenovirus expressing dominant-negative ACREB. (Right) Effect of ACREB expression on cAMP-induced gene expression relative to that of control (GFP) INS-1 cells. *, P < 1 × 10−11. (E) Effect of adenovirally encoded ACREB inhibitor or phosphorylation-defective, constitutively active CRTC2 [CRTC2CA(S171A)] on expression of beta cell-specific CREB target genes. mRNA amounts for Diras2 and Ccnd1 are shown. *, P < 0.05; **, P < 0.01.
We investigated the role of the CREB/CRTC pathway in promoting cAMP-dependent gene expression in beta cells. Cultured pancreatic islets from whole-body Crtc2−/− mice were less responsive to FSK than islets from wild-type littermates (genes induced 2-fold or greater following exposure to FSK [FSK/CON], ≥2; fragments per kilobase million [FPKM], ≥8) by RNA sequencing (RNA-seq) analysis (Fig. 1C). Loss of CRTC2 only partially disrupted target gene expression, however, likely reflecting compensatory effects of other CRTC family members (CRTC1 and CRTC3) in this setting.
Similar to CRTC2 depletion, adenoviral expression of the dominant-negative CREB inhibitor ACREB (22), which heterodimerizes with and blocks binding of all three CREB family members (CREB1, ATF1, and CREM) to DNA, disrupted genome-wide cAMP-inducible gene expression to a greater degree in INS-1 cells (Fig. 1D). Consistent with these effects, ACREB expression decreased FSK-induced Pol II occupancy over the transcription start site (TSS) as well as elongation over the gene body (Fig. 1D). Amounts of paused Pol II at the promoter were unexpectedly increased in ACREB-expressing cells under basal conditions, suggesting that CREB enhances Pol II elongation under these conditions.
In keeping with the inhibitory effects of CRTC2 or CREB disruption, adenoviral expression of phosphorylation-defective constitutively active CRTC2 [CRTC2(S171A)] upregulated the expression of CREB target genes, particularly under basal conditions, when endogenous CRTC2 is normally phosphorylated and sequestered in the cytoplasm (Fig. 1E). Taken together, these results indicate that cAMP exerts extensive genome-wide effects on beta cell gene expression, stimulating both core and beta cell-specific gene expression through induction of the CREB-CRTC2 pathway.
CREB triggers cell-specific gene expression through distal enhancer activation.
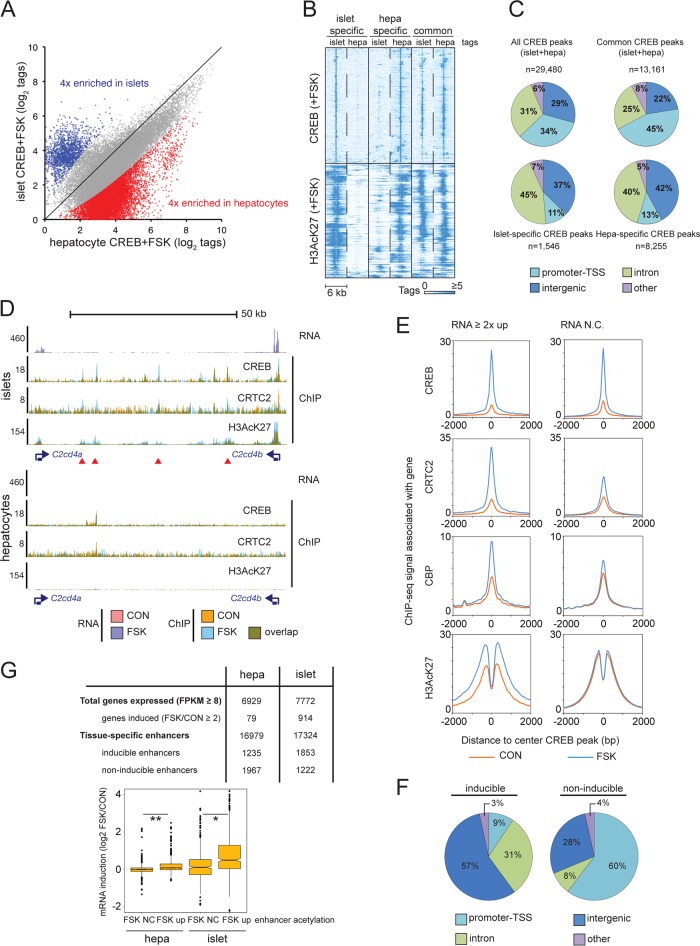
To determine the mechanism by which CREB and its coactivators promote cell-type-specific gene expression, we compared genome-wide occupancy patterns for CREB and CRTC2 in primary mouse hepatocytes and pancreatic islets (Fig. 2A). In chromatin immunoprecipitation sequencing (ChIP-seq) studies, we detected significantly fewer CREB- and CRTC2-bound regions in islets than hepatocytes, likely due to the relatively harsher genomic DNA shearing conditions required to generate ChIP-seq libraries.
FIG 2.
CREB triggers cell-specific gene expression through distal enhancer activation. (A) Scatter plot comparing tag enrichment in CREB ChIP-seq experiments of cultured primary mouse islets and hepatocytes (1 h of FSK exposure). Tissue-specific enrichment (≥4-fold) of CREB binding in islets and hepatocytes is highlighted. (B) Heat map depicting CREB occupancy restricted to activated genomic regions (H3AcK27-decorated promoters and enhancers) that are tissue specific or shared between islets and hepatocytes. (C) Pie charts showing genomic distribution of common and cell-restricted CREB peaks. The majority of tissue-restricted CREB occupancy occurs in TSS-distal genomic loci (promoter-TSS, −1,000/+100 bp from TSS). (D) Browser plot of a genomic region containing two beta cell-restricted CREB target genes, C2cd4a and C2cd4b, and intervening superenhancer. RNA-seq and ChIP-seq data collected under basal conditions (orange) and upon FSK exposure (blue) in primary islets and hepatocytes (2 h for RNA-seq, 1 h for ChIP-seq). (E) Histograms showing genome-wide occupancy profiles for CREB, CRTC2, CBP, and H3AcK27 across CREB binding sites annotated to cAMP-inducible or noninducible (RNA no change [RNA N.C.]) genes. Exposure to FSK is indicated. (F) Pie charts showing relative genomic distribution of CREB occupancy for cAMP-inducible and noninducible enhancers in INS-1 cells. (G) Enhancer activation (H3AcK27 signal FSK/CON, ≥2) correlates with cAMP-inducible gene expression in primary mouse hepatocytes and islet tissue. Relative numbers of inducible enhancers and target genes are indicated. Box plot shows correspondence between enhancer H3AcK27 inducibility in response to FSK and target gene induction. RNA-seq data were taken from experiment shown in Fig. 1B. *, P < 1 × 10−13; **, P < 1 × 10−15.
We compared cohorts of tissue-specific CREB-occupied regions that were preferentially enriched in islets or hepatocytes (n = 1,546 loci in which islet tags were enriched ≥4 above the level for hepatocyte tags; n = 8,255 loci in which hepatocyte tags were enriched ≥4 above the level for islet tags) (Fig. 2A). In both cases, roughly 90% of cell-type-specific CREB occupancy was confined to activated TSS-distal enhancer regions that are enriched in H3AcK27 by ChIP assay (Fig. 2B and C).
The importance of distal enhancer regions for activation of cell-type-specific CREB targets is illustrated by the C2cd4a/b locus. C2cd4a and C2cd4b are cAMP-inducible CREB target genes in mouse pancreatic islets and INS-1 cells; they are not expressed detectably in hepatocytes (Fig. 2D). The shared ∼69-kb genomic region between C2cd4a and C2cd4b genes corresponds to a conserved islet-restricted superenhancer, which contains multiple type 2 diabetes-associated single-nucleotide polymorphisms (23–25). Within this superenhancer, we identified four CREB/CRTC2-bound loci that are absent from hepatocytes (Fig. 2D).
We compared CREB occupancy profiles over loci, which were annotated to genes that were upregulated 2-fold or better by FSK, and we compared these to loci annotated to genes that are unresponsive to FSK in INS-1 cells. Although exposure to FSK increased CREB binding comparably for both groups, it selectively enhanced CBP and CRTC2 occupancy as well as H3AcK27 enrichment for inducible targets (Fig. 2E). These results indicate that cAMP-inducible genes are distinguishable from noninducible genes in their ability to recruit CRTC2 and CBP/p300 to CREB binding sites in response to cAMP.
Having seen effects of cAMP on CREB and coactivator occupancy for CREB binding loci annotated to cAMP-inducible genes, we tested the importance of CREB occupancy for enhancer activation by H3K27 acetylation in beta cells. To that end, we assembled a list of high-confidence CREB-bound regions from four independent CREB ChIP-seq experiments representing 6,247 CREB peaks in INS-1 cells. We extracted a subset of CREB-inducible enhancers from this list by quantifying effects of FSK on H3AcK27 amounts over a 4-kb region centered on high-confidence CREB-bound loci (n = 741; log2 H3AcK27 tag [FSK/CON], ≥0.8; adjusted P value of ≤0.04).
ACREB expression in INS-1 cells decreased amounts of CREB, CRTC2, CBP, and H3AcK27 compared to those of inducible enhancers under basal conditions and following exposure to FSK. Notably, most (88%) of these inducible loci occur within promoter-distal regions, whereas only 9% of regions with cAMP-inducible CREB binding and H3K27 acetylation are located in gene promoters (Fig. 2F). Conversely, the majority (60%) of noninducible H3AcK27 enrichment at CREB-occupied loci is located at promoter-proximal sites (Fig. 2F). These results support the idea that stimulus-induced binding of CREB to distal enhancers triggers coactivator recruitment and H3K27 acetylation, leading to target gene activation.
In keeping with the relatively robust transcriptional response to cAMP signaling in islets, FSK-induced enhancers (H3AcK27 tag FSK/CON, ≥2) were increased in pancreatic islets compared to those in hepatocytes (Fig. 2G, top). Target genes annotated to inducible enhancers were also more responsive to FSK than genes annotated to noninducible enhancers (Fig. 2G, bottom). Taken together, these results point to an important role for CREB in stimulating target gene expression through cAMP-dependent increases in distal enhancer acetylation.
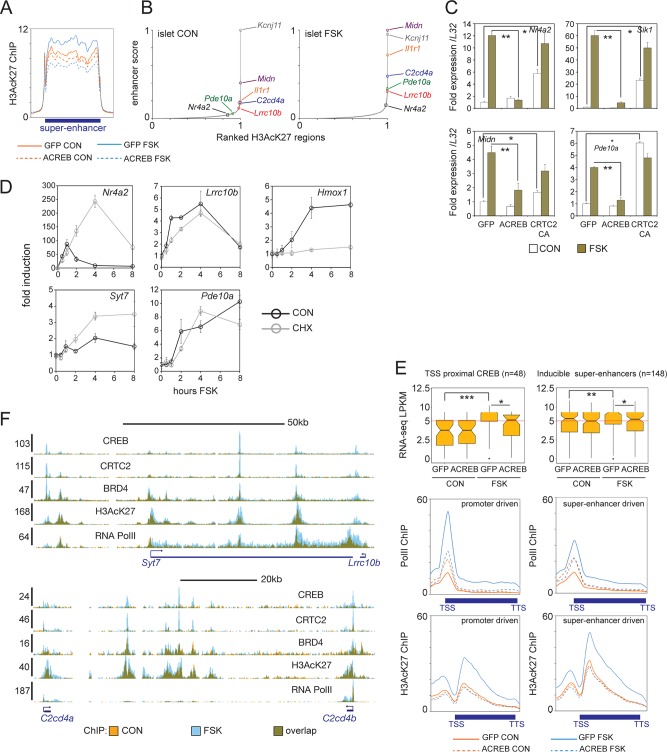
CREB promotes superenhancer activity.
Genes critical for cell identity and function are often controlled by superenhancer regions that are enriched in coactivator occupancy and activated chromatin modifications along extended genomic regions (14–16). Using H3AcK27 amounts as an index of enhancer strength, we noticed that exposure to FSK promoted activation of superenhancer regions in INS-1 cells and islets (Fig. 3A and B). In beta cells, genes located in the same stimulus-induced superenhancer region are coregulated by cAMP (C2cd4a/C2cd4b and Midn/Atp5d/Dos). Similar to proximal promoter-driven targets (Nr4a2 and Sik1), superenhancer-dependent genes are also repressed by ACREB and induced by CRTC2(S171A) overexpression in INS-1 cells (Fig. 3C).
FIG 3.
CREB promotes gene expression from superenhancers. (A) Metagene analysis of H3AcK27 occupancy over superenhancers in INS-1 cells (n = 606). Effects of FSK treatment and ACREB expression are shown. (B) Enhancer ranking based on H3AcK27 ChIP-seq signal in primary islet tissue under basal and FSK-treated conditions. Genes located inside enhancers are indicated. (C) Relative effect of FSK on core (Nr4a2 and Sik1) and beta cell-restricted CREB target genes located in superenhancers (Midn and Pde10a) in INS-1 cells. Cells infected with control (GFP), ACREB, or CRTC2(S171A) adenovirus are indicated. qPCR data are presented as expression over basal levels in control GFP adenovirus-infected cells. *, P < 0.007; **, P < 0.003. (D) Time course analysis of mRNA levels in INS-1 cells exposed to FSK. mRNA profiles of core (Nr4a2) and beta cell-restricted (Lrrc10b, Syt7, and Pde10a) CREB target genes are shown. Effect of cycloheximide (CHX) on CREB or HIF target (Hmox1) gene expression are indicated. (E, top) Box plots showing FPKM values from RNA-seq studies of promoter- or superenhancer-driven CREB target genes in INS-1 cells maintained under basal conditions and following exposure to FSK. Cells expressing control (GFP) or ACREB are indicated. (Bottom) Metagene analysis of RNA Pol II and H3AcK27 enrichment over promoter- and enhancer-driven CREB targets. *, P < 0.004; **, P < 0.002; ***, P < 1 × 10−6. TTS, transcription termination site. (F) Browser plots of two conserved beta cell-specific superenhancer regions in INS-1 cells. Effect of FSK on CREB, CRTC2, BRD4, and H3AcK27 occupancy on Pol II elongation is shown. y axis values indicate normalized tag enrichment.
We compared the activation profiles of genes that are controlled by TSS-proximal recruitment of CREB (n = 48) or in the context of an inducible superenhancer (n = 148). Consistent with their location in activated chromatin regions, superenhancer-regulated genes are expressed at higher levels under basal conditions, and they exhibit a more modest transcriptional response to FSK than core CREB targets (Fig. 3D and E). Although prolonged exposure to cAMP agonist has been shown to promote islet gene expression via protein synthesis-dependent increases in the hypoxia-inducible factor HIF1a (18), the induction of superenhancer-driven CREB targets by cAMP appears to be direct; indeed, exposure to cycloheximide (CHX) did not reduce their expression in response to FSK compared to that of the HIF1a target gene Hmox1 (Fig. 3D) (18, 26).
Enhancer clusters near the gene promoter have been shown to increase transcription by triggering release of paused Pol II (27). The bromodomain-containing protein BRD4 appears to promote transcription via its association with the pause release complex pTEFb, which mediates release of paused Pol II to productive elongation in part by phosphorylating the C-terminal domain of Pol II (17, 28). Following its activation, CREB appears to induce strong Pol II recruitment to the TSS as well as elongation over the gene body (Fig. 3E). Disrupting CREB occupancy with ACREB decreased Pol II elongation in cells exposed to FSK, but ACREB actually increased Pol II density over the TSS for CREB targets in the basal state, suggesting again that CREB activity is required for Pol II release from target promoters under these conditions.
Triggering of the cAMP pathway in INS-1 cells increased Pol II recruitment to superenhancer-driven target genes more modestly than promoter-driven genes (Fig. 3E), but FSK treatment increased H3AcK27 amounts to a greater degree over superenhancer-driven targets (Fig. 3E). Correspondingly, exposure to FSK increased BRD4 recruitment to CREB binding sites within superenhancers (Fig. 3F). Taken together, these experiments indicate that CREB maintains dynamic expression of tissue-specific genes through chromatin activation and recruitment of BRD4 in response to increases in H3K27 acetylation.
BRD4 and CBP/p300 are required for induction of enhancer-driven CREB target genes by cAMP.
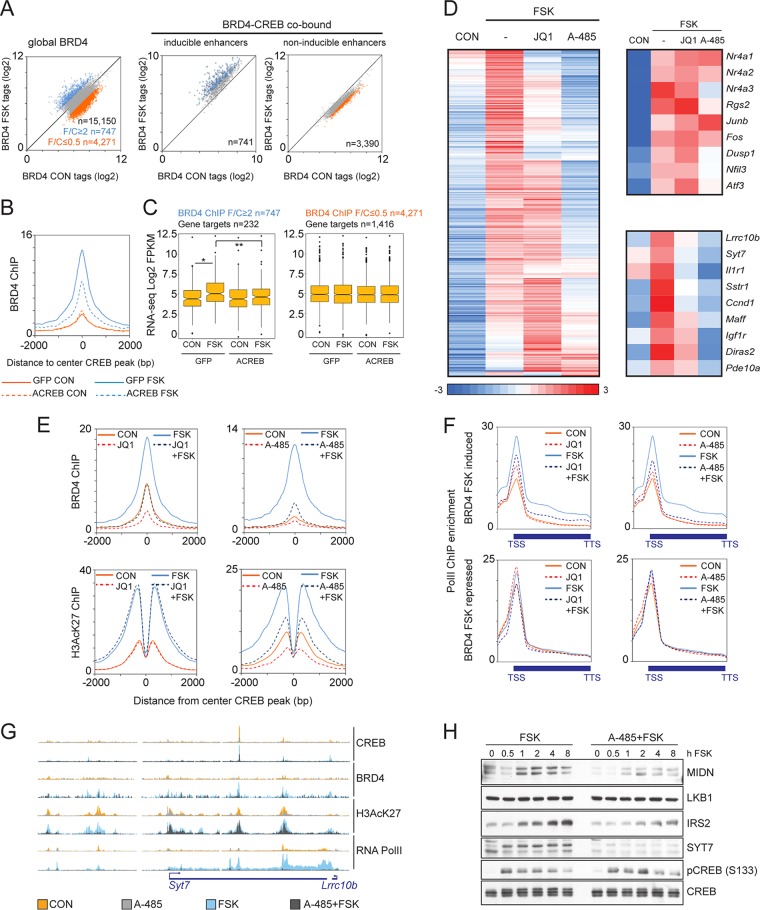
Realizing that BRD4 links enhancer acetylation to target gene expression (28), and knowing that BRD4 is recruited to CREB-bound superenhancers, we investigated the genome-wide effects of FSK on BRD4 occupancy and gene expression.
Exposure of INS-1 cells to FSK promoted the recruitment of BRD4 to CREB-occupied cAMP-inducible enhancers but not to noninducible enhancers (Fig. 4A). Disrupting CREB activity by overexpression of dominant-negative ACREB blocked BRD4 binding to cAMP-inducible enhancers (Fig. 4B). Consistent with the notion that BRD4 is recruited to chromatin by binding to acetylated lysine residues in histone tails, we noted that BRD4 occupancy over inducible enhancers extends across the nucleosome-free region and into H3AcK27-decorated flanking regions (Fig. 4B).
FIG 4.
Role of BRD4 and CBP/p300 in CREB target gene induction. (A) Scatter plot showing BRD4 ChIP-seq tag counts in INS-1 cells under basal or FSK-induced conditions. Twofold tag enrichment (blue) or depletion (orange) upon stimulation is indicated. Total BRD4 (left) and BRD4 occupancy over CREB-bound loci in inducible and noninducible enhancers (right) is shown. (B) Histogram showing effect of FSK on BRD4 occupancy over CREB-bound loci in inducible enhancers. Effect of ACREB is indicated. (C) Box plots showing relative mRNA levels (FPKM) for genes associated with FSK-inducible BRD4 peaks (left) or FSK-inhibited BRD4 peaks (right) in INS-1 cells. Effect of ACREB is indicated. *, P < 3 × 10−7; **, P < 4 × 10−4. (D) Heat map showing effect of BRD4 inhibitor JQ1 (1 μM) or the CBP/p300 HAT inhibitor A-485 (10 μM; 2 h) on CREB target gene expression in INS-1 cells exposed to FSK (10 μM). Genes induced by FSK (FSK/CON, ≥2) are shown. A comparison of core promoter-driven (top) and enhancer-driven (bottom) beta cell-specific targets is shown on the right. (E) Histograms showing effect of BRD4 inhibitor JQ1 (1 μM) (left) or CBP/p300 HAT inhibitor (A-485; 10 μM) (right) on occupancy profiles for BRD4 (top) and H3AcK27 (bottom) across CREB-bound, inducible enhancers in INS-1 cells exposed to FSK for 1 h. (F) Metagene analysis of RNA Pol II ChIP enrichment over genes associated with BRD4 peaks that are either induced (top) or repressed (bottom) by FSK. INS-1 cells were preexposed to the BRD4 inhibitor JQ1 (1 μM) (left) or the CBP/p300 HAT inhibitor A-485 (10 μM) (right) for 2 h and then stimulated with FSK for 1 h. (G) Browser plot showing effect of CBP/p300 HAT inhibitor A-485 on occupancy of CREB, BRD4, H3AcK27, and RNA Pol II over the Syt7/Lrrc10b locus in INS-1 cells. Effect of FSK is shown. (H) Immunoblot showing effect of A-485 inhibitor on protein amounts for enhancer-driven endogenous CREB target gene products (MIDN, IRS2, and SYT7). Effect of A-485 on LKB1, expressed from a gene adjacent to the Midn,Atp5d,Dos enhancer is also shown. Effects of A-485 on total and phospho-CREB amounts are indicated.
Because the transcriptional response to FSK exposure is overwhelmingly stimulatory, we examined whether BRD4 occupancy correlates with stimulus-induced gene expression. As expected, genes associated with BRD4 recruitment were upregulated in response to FSK, but genes associated with BRD4-depleted loci were not repressed (Fig. 4C). These results are consistent with the largely positive effect of FSK on cellular gene expression in hepatocytes and pancreatic islets (Fig. 1A).
Genes under the control of superenhancers appear to be highly sensitive to small molecules that inhibit epigenetic cofactors (13, 29, 30). Having seen that CREB activates genes controlled by superenhancers, we wondered whether small-molecule inhibitors that target either the bromodomain of BRD4 (JQ1) (31) or CBP/p300 histone acetyltransferase (HAT) activity (A-485) (32) correspondingly attenuate CREB target gene expression.
In line with this idea, we found clusters of beta cell-specific genes in INS-1 cells that are sensitive to small-molecule inhibition of CBP/p300 HAT activity or BRD4 recruitment by RNA-seq analysis (Fig. 4D). Indeed, a majority of inducible CREB targets appeared sensitive to CBP/p300 HAT inhibition (n = 284 out of 484 induced genes; FSK/A-485 plus FSK, ≥2). Notably, core proximal promoter-driven target genes were comparatively insensitive to A-485 (Nr4a1, Nr4a2, Junb, and Fos). Significantly, the cAMP-dependent induction of these core target genes also appears to be unaffected by targeted genetic disruption of both p300 and CBP (7).
Similar to effects of CBP/p300 HAT inhibitor, exposure to the BRD4 inhibitor JQ1 (33) also decreased FSK-induced gene expression from enhancer-driven CREB targets (Fig. 4D). In keeping with the importance of CBP/p300-induced histone acetylation for BRD4 recruitment, 110 out of 120 JQ1-repressed genes were also inhibited by exposure to A-485. Notably, genes with enhancers that are active in both islets and hepatocytes (e.g., Irs2 and Midn/Atp5d/Dos) appear to be resistant to JQ1, while genes controlled selectively by beta cell-specific enhancers (e.g., C2cd4a/b, Lrrc10b/Syt7, Ncald, and Il1r1) are sensitive to JQ1 (Fig. 4D).
Based on the ability for JQ1 and A-485 to block beta cell-specific CREB target gene expression, we tested effects of these inhibitors on BRD4 recruitment and H3K27 acetylation for CREB target genes. Exposure of INS-1 cells to JQ1 inhibitor blocked BRD4 recruitment and reduced the subsequent elongation of Pol II over target gene bodies without disrupting H3AcK27 induction in response to FSK (Fig. 4E and F). In contrast, exposure to the CBP/p300 inhibitor A-485 disrupted both H3K27 acetylation and BRD4 recruitment (Fig. 4E), indicating that CBP and p300 act upstream of BRD4. Exposure to FSK increased recruitment of Pol II to gene bodies near BRD4-induced enhancers; these effects were repressed in cells treated with JQ1 or A-485 (Fig. 4F and G). Consistent with the role of BRD4 in promoting pause release, exposure to JQ1 increased amounts of paused Pol II at the TSS. This increase in paused Pol II appeared similar to effects of ACREB expression noted earlier.
Pol II occupancy for genes with BRD4-depleted enhancers was unaffected by FSK and did not decrease in response to BRD4 or CBP/p300 inhibitor, indicating that BRD4 depletion in response to FSK occurs near genes that are not dependent on CBP/p300 HAT activity or BRD4 recruitment for their expression (Fig. 4F).
Having seen that CREB target gene expression is linked to genomic loci that are sensitive to CBP/p300 HAT inhibition, we compared effects of A-485 on genes controlled by CREB-regulated enhancers. Exposure to A-485 had no effect on CREB protein amounts or phosphorylation in response to FSK (Fig. 4H). Confirming its effects on transcript abundance for these enhancer-driven genes, exposure to A-485 inhibitor also decreased basal and FSK-induced protein amounts for Midn, Irs2, and Syt7 but not for Lkb1, a cellular gene that is directly adjacent to the Atp5d/Midn/Cirbp superenhancer. Taken together, these results show that cAMP/CREB-dependent recruitment of CBP/p300 and subsequent targeting of BRD4 to cell-defining enhancers are key events in maintaining tissue-restricted gene expression in beta cells.
Neurod1 promotes cAMP-inducible gene expression in beta cells.
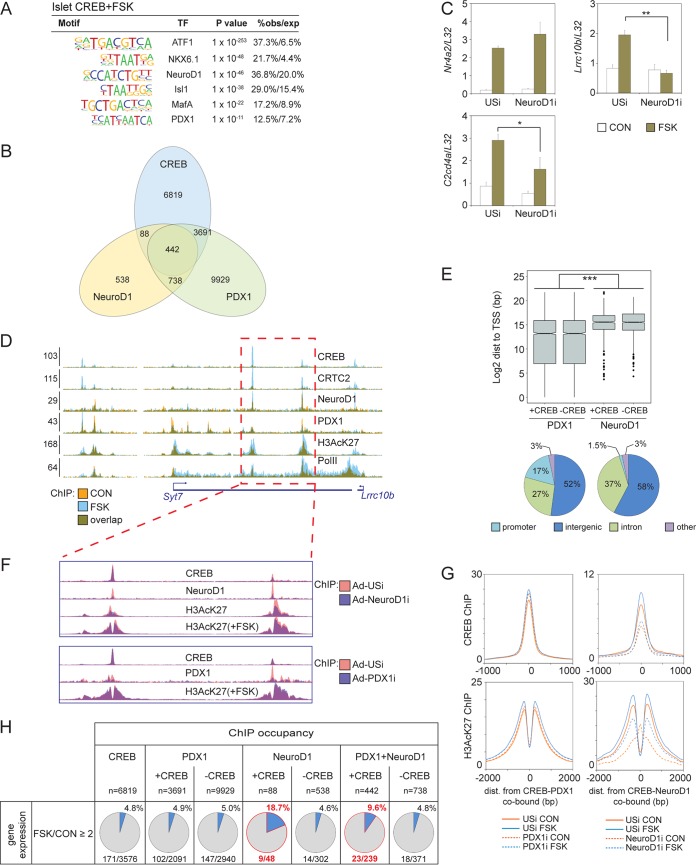
Based on its ability to stimulate beta cell-specific gene expression, we considered that CREB cooperate with lineage-dependent transcription factors (LDTFs) to activate distal enhancers in relevant tissues. To test this idea, we searched for transcription factor binding motif enrichment in tissue-specific CREB-bound genomic loci. This analysis revealed potential cooccupancy of CREB and LDTFs such as the homeobox factor Pdx1 and E-box binding factor Neurod1 across the genome in islet tissue (Fig. 5A).
FIG 5.
Neurod1 promotes cAMP-inducible gene expression in beta cells. (A) Motif analysis of tissue-specific CREB binding loci in mouse islets showing enrichment of lineage-defining transcription factor (TF) binding motifs. obs/exp, observed/expressed. (B) Overlap of CREB, NeuroD1, and PDX1 genomic occupancy in INS-1 cells. (C) Effect of RNAi-mediated depletion of Neurod1 on induction of core (Nr4a2) or cell-restricted (Lrrc10b and C2cd4a) CREB target genes by FSK in INS-1 cells. *, P < 0.04; **, P < 7 × 10−4. (D) Browser plot depicting binding of Neurod1 and Pdx1 in the Syt7/Lrrc10b-inducible superenhancer in INS-1 cells. Colocalization of Neurod1 and Pdx1 with CREB over inducible enhancers is indicated. Effect of FSK is shown. y axis values indicate normalized tag enrichment. (E, top) Box plot showing genomic distribution (dist) of Pdx1- and Neurod1-bound loci relative to TSS for genes with (+CREB) or without (−CREB) cobound CREB peaks in INS-1 cells. ***, P = 0. (Bottom) Pie charts showing relative genomic distribution of Pdx1 and Neurod1 peaks in INS-1 cells. (F) Effect of Neurod1 or Pdx1 depletion on enhancer activity and CREB occupancy on the Syt7/Lrrc10b enhancer. (G) Histograms showing relative effect of Pdx1 or Neurod1 knockdown on CREB and H3AcK27 occupancy over CREB-Pdx1-cobound (left) or CREB-Neurod1-cobound loci (right) in INS-1 cells. Exposure to FSK is shown. (H) Correspondence between CREB-Neurod1 or CREB-Pdx1 cobinding relative to CREB, Neurod1, and Pdx1 alone on cAMP inducibility of proximal genes in INS-1 cells.
To test the potential role of Neurod1 in beta cell-specific CREB target gene induction, we generated adenovirus expressing a short hairpin RNA (shRNA) against this LDTF. Depletion of Neurod1 in INS-1 cells disrupted the cAMP-dependent induction of beta cell-specific genes (Lrrc10b and C2cd4a), having no effect on the expression of a core target gene (NRr4a2) (Fig. 5C). Neurod1 is expressed in mature beta cells, where it plays a key role in islet cell survival and insulin gene expression (34, 35). Neurod1 has also been shown to stimulate the expression of genes that govern neuronal specification through chromatin derepression and enhancer activation (36).
We examined whether, similar to its effects in neurons, Neurod1 also contributes to enhancer activation in beta cells. Using ChIP-seq studies of INS-1 cells, we detected both Neurod1 and Pdx1 binding to beta cell-specific enhancer regions that were also occupied by CREB (Fig. 5A and B). The number of Pdx1-bound loci was nearly 10-fold higher than the number of Neurod1-bound loci (n = 14,800 and 1,806 for Pdx1 and Neurod1, respectively), and most Neurod1-bound loci (n = 1,180) were also cooccupied by Pdx1 (Fig. 5B). Pointing to a potential role for this factor in the activation of beta cell-specific enhancers, Neurod1 was recruited primarily to TSS-distal genomic loci but Pdx1 was not (Fig. 5D and E).
We tested the relative importance of Neurod1 and Pdx1 for CREB occupancy and enhancer activation in INS-1 cells. In keeping with its role in chromatin activation, depletion of Neurod1 but not Pdx1 reduced H3AcK27 protein amounts across CREB-cooccupied enhancers (Fig. 5F and G). Notably, knockdown of Neurod1 also decreased binding of CREB to nucleosome-free regions (NFRs) over CREB-Neurod1 cooccupied enhancers (Fig. 5F and G). Depletion of Neurod1 also triggered a significant increase in H3AcK27 binding within the NFRs of Neurod1-bound enhancers (Fig. 5G).
Based on the ability of Neurod1 to function as a pioneering factor that associates with the SWI/SNF chromatin remodeling complex (37), loss of Neurod1 may lead to nucleosome infiltration within enhancer regions. Taken together, these results suggest that Neurod1 enhances cAMP-dependent transcription by promoting CREB occupancy at beta cell-specific enhancers that contain E-box motifs adjacent to CREs.
We considered that cooccupancy of CREB and Neurod1 may be predictive of cAMP inducibility for nearby genes. Less than 5% of genes annotated to CREB or CREB-Pdx1-cobound loci were upregulated 2-fold or better in response to FSK. In contrast, 18.7% of genes annotated to Neurod1-CREB-cobound loci and 9.6% of genes near Neurod1-Pdx1-CREB-cobound loci were upregulated by FSK (Fig. 5H). Taken together, these results indicate that CREB stimulates beta cell-specific gene expression cooperatively with Neurod1 and that, surprisingly, cooccupancy between CREB and a lineage-determining transcription factor can be more predictive of target gene induction than occupancy of CREB alone.
Neurod1 promotes CREB occupancy at binding sites within beta cell-specific enhancers.
The GLP1 agonist exendin-4 has been shown to promote the conversion of progenitor cells into hormone-expressing beta cell-like cells (38, 39). Based on the ability of GLP1 to stimulate cAMP signaling, we considered that Neurod1 and CREB may promote beta cell identity through their cooperative effects on superenhancer activation.
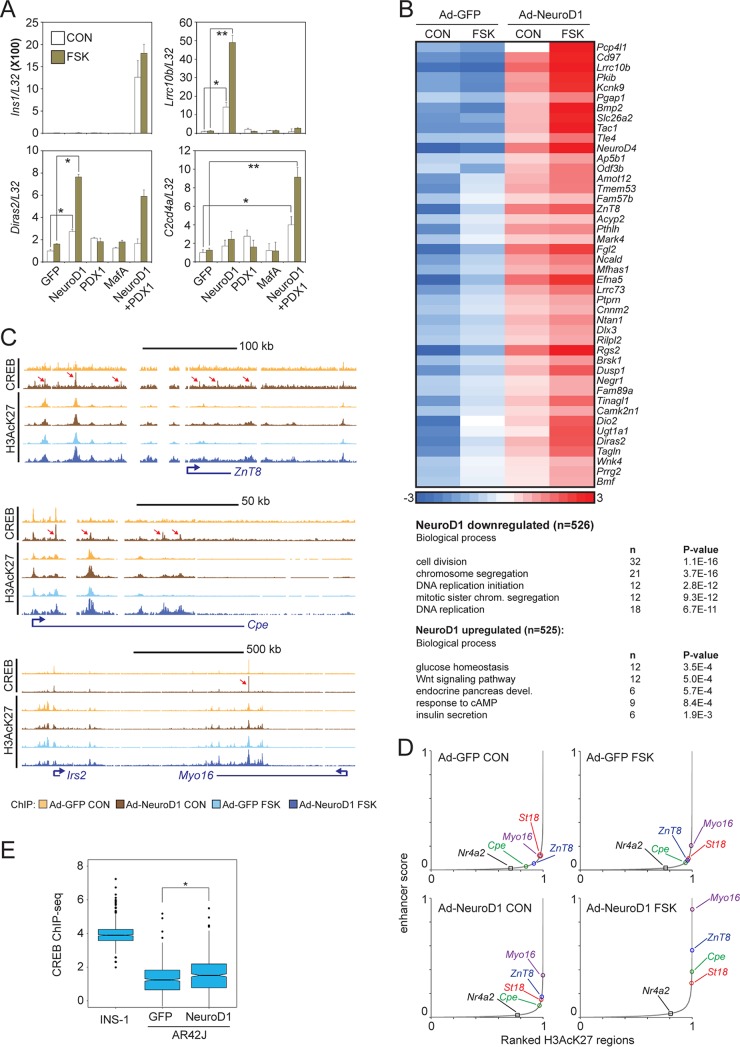
The pancreatic acinar cell line AR42J has been shown to transdifferentiate into beta-like cells following coexpression of Neurod1, Pdx1, and MafA (40). Indeed, coexpression of adenovirally encoded Neurod1 and Pdx1 triggered expression of the insulin 1 (Ins1) gene (Fig. 6A). In the absence of beta cell factors, exposure of AR42J cells to FSK did not increase mRNA amounts for the beta cell-specific CREB targets Lrrc10b and Diras2 (Fig. 6A). Overexpression of Neurod1 was sufficient to upregulate both genes in response to FSK (Fig. 6A). Overexpression of both Pdx1 and Neurod1 appeared necessary for the induction of the CREB target gene C2cd4a in response to FSK (Fig. 6A). Taken together, these results indicate that Neurod1 plays an important role in beta cell-restricted CREB target gene expression. The coordinated expression of multiple LDTFs may be required to establish active, signal-responsive enhancers in mature tissue.
FIG 6.
Neurod1 promotes CREB occupancy at binding sites within beta cell-specific enhancers. (A) Effect of beta cell transcription factor (Pdx1, Neurod1, and MafA) overexpression on mRNA amounts for beta cell-restricted CREB targets Lrrc10b, Diras2, and C2cd4a in AR42J pancreatic exocrine cells. *, P < 0.02; **, P < 0.005. Ins1 expression is shown. Exposure to FSK (green) or vehicle (white) is indicated. (B) Heat map showing effect of adenoviral Neurod1 on cAMP-inducible gene expression in pancreatic exocrine AR42J cells. (Top) AR42J cells were treated with FSK for 2 h. Heat map depicts RNA-seq data of a cluster of Neurod1- and FSK-induced genes. (Bottom) Enrichment in biological process of NeuroD1-induced and repressed genes. (C, top tracks) Browser plots showing effect of adenoviral NeuroD1 expression on CREB occupancy over beta cell-specific loci. (Bottom tracks) Effect of FSK on enhancer activation (H3AcK27 occupancy) over beta cell-specific gene loci. AR42J cells were treated with FSK for 1 h. Arrows point to induced CREB-bound loci. (D) Effect of Neurod1 expression and FSK treatment alone or together on beta cell-specific enhancer strength. AR42J cells infected with control (GFP) or Neurod1-expressing adenovirus and treated with FSK for 1 h. (E) Box plot showing genome-wide enrichment of CREB binding over INS-1-specific CREB-bound loci following adenoviral Neurod1 expression in AR42J cells. INS-1-specific CREB binding loci are defined as regions with a ≥4× enrichment of CREB tags in INS-1 cells relative to that in AR42J cells. *, P < 2 × 10−9.
We evaluated the genome-wide effects of Neurod1 on the induction of beta cell-specific CREB target genes by cAMP. In keeping with its ability to promote the expression of beta cell-specific genes, Neurod1 overexpression in AR42J cells stimulated the expression of genes involved in glucose homeostasis, insulin secretion, and exocytosis while decreasing the expression of genes that promote cell proliferation (Fig. 6B).
To test for the potential cooperativity between Neurod1 and CREB at the level of enhancer activation, we measured effects of virally encoded Neurod1 on H3AcK27 enrichment over these regions. Neurod1 expression alone activated beta cell-specific enhancers (Fig. 6C and D) and triggered recruitment of endogenous CREB to these regions (Fig. 6C). Although FSK had only modest effects alone, it strongly potentiated effects of Neurod1 on beta cell-specific enhancer strength (Fig. 6D).
To further evaluate the role of Neurod1 in targeting CREB to beta cell-specific loci, we isolated INS-1-specific CREB-occupied loci (INS-1 cells showed ≥4× enrichment compared to that in AR42J cells) and measured CREB occupancy over these regions under basal conditions and following overexpression of Neurod1 in AR42J cells. Consistent with the eviction of CREB from beta cell-specific enhancers that we observed following depletion of Neurod1 from INS-1 cells, overexpression of Neurod1 in AR42J cells increased CREB occupancy over the same INS-1-specific loci (Fig. 6E).
A conserved CREB-Neurod1-regulated superenhancer is required for beta cell function.
Having seen effects of CREB and Neurod1 on enhancer activity, we tested the role of CREB and Neurod1 binding sites in regulating beta cell function. The cAMP-responsive genes Lrrc10b and Syt7 are located within a conserved inducible superenhancer. Lrrc10b is a protein of unknown function, whereas Syt7 is a well-characterized member of the synaptotagmin protein family that promotes glucose-stimulated insulin secretion (GSIS). Indeed, Syt7 activity is also modulated through PKA-mediated phosphorylation in response to GLP1 signaling (41, 42).
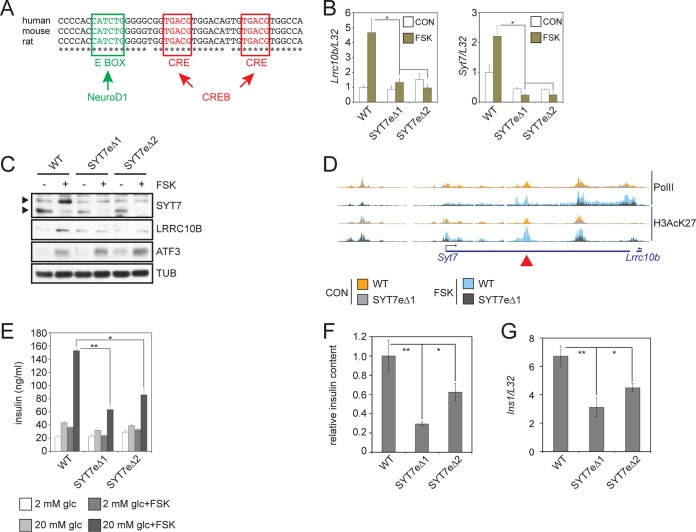
Lrrc10b and Syt7 expression appears to be controlled by a conserved CREB/Neurod1-bound enhancer in the second intron of Syt7, ≈27.8 kb from the Syt7 TSS and ≈39 kb from the Lrrc10b TSS; it contains two conserved cAMP response elements (CREs) and a conserved E-box motif (Fig. 7A). We used the Crispr/Cas9 system and two sets of guide pairs to generate two mutant INS-1 cell lines carrying homozygous deletions of the entire enhancer region (Syt7eΔ1, rn5 chromosome 1 [Chr1] 233,410,068–778Δ; Syt7eΔ2, rn5 Chr1 233,410,103–721Δ). Relative to those of wild-type (WT) cells, mRNA and protein amounts for Lrrc10b and Syt7 in both mutant lines are decreased, and their responsiveness to FSK is eliminated (Fig. 7B and C).
FIG 7.
Conserved CREB-Neurod1-regulated superenhancer is required for insulin secretion. (A) Conservation of E-box and CRE sites within the Syt7 enhancer in human, mouse, and rat. (B) Relative effect of FSK on mRNA amounts for Lrrc10b and Syt7 in wild-type and enhancer-deleted SYT7eΔ lines. *, P < 0.02. Two immunoreactive Syt7 bands represent phospho (top) and dephospho (bottom) forms of Syt7. (C) Immunoblot showing effect of FSK on protein amounts for Lrrc10B and Syt7 in wild-type and SYT7eΔ lines. Cells were exposed to 10 μM FSK for 8 h. ATF3 included as a control for CREB target protein induction. Exposure to FSK reduces Syt7 mobility due to PKA-mediated phosphorylation. Arrows indicate phospho- and unphospho-Syt7. (D) Browser plot showing relative Pol II and H3AcK27 occupancy over the Lrrc10b/Syt7 enhancer in wild-type and SYT7eΔ1 mutant cells. Exposure to FSK (10 μM, 1 h) is shown. The location of deleted enhancer is indicated with an arrowhead (top). (E) Signal-induced insulin secretion assay in wild-type and SYT7eΔ lines. Cells were exposed for 2 h to glucose (glc) and FSK as indicated. Insulin secretion measured in KRBH buffer by radioimmunoassay. *, P < 0.003; **, P < 2 × 10−4. (F) Relative content of mature insulin in extract from wild-type and SYT7eΔ lines measured by radioimmunoassay. *, P < 0.04; **, P < 0.02. (G) Ins1 mRNA levels in wild-type and SYT7eΔ lines. *, P < 0.005. **, P < 0.0004.
Consistent with the loss of Lrrc10b and Syt7 expression in mutant cells, Pol II occupancy is disrupted across the deleted enhancer and over the Lrrc10b and Syt7 gene bodies, particularly following exposure to FSK by ChIPseq analysis (Fig. 7D). H3AcK27 protein amounts are also reduced across the deleted enhancer and for the promoters of both Lrrc10b and Syt7 (Fig. 7D). Thus, a single TSS-distal CREB-Neurod1-cooccupied enhancer appears to induce coordinated expression of two genes within a superenhancer (43, 44).
We evaluated the functional significance of the Syt7-Lrrc10b enhancer in INS-1 cells by measuring insulin secretion from Syt7eΔ1 and Syt7eΔ2 cells in response to glucose or cAMP. Exposure to high glucose (20 mM) and, to a greater extent, FSK plus glucose increased insulin secretion from wild-type cells. These effects were diminished by half in the mutant lines relative to those in control cells (Fig. 7E). We next measured Ins1 mRNA expression levels and intracellular insulin content in WT, Syt7eΔ1, and Syt7eΔ2 lines and found that both were also decreased in mutant cells (Fig. 7F and G). These results demonstrate that the phenotypic effects of this deletion extend beyond simply loss of Syt7 expression and point to potential effects of Lrrc10b in this setting. The Syt7/Lrrc10b superenhancer and the Ins1 gene are syntenic in rat (Chr1, ≈46.8 Mbp apart), mouse (Chr19, ≈41.8 Mbp), and human (INS) (Chr11, ≈59.1 Mbp). While gene regulation by enhancers in cis has not been described over such large genomic distances, our results suggest that the Syt7/Lrrc10b enhancer affects beta cell function and identity beyond controlling Syt7 gene expression and concurrent insulin secretion defects. Collectively, these results demonstrate the importance of signal-dependent activators and LDTFs in promoting pancreatic islet function through the cooperative activation of beta cell-restricted superenhancers in response to extracellular signals.
DISCUSSION
In most cell types, the second messenger cAMP promotes cell cycle exit and growth arrest. However, in a subset of endocrine cells, most notably beta cells of the pancreatic islets, cAMP functions as a potent growth factor signal that also enhances insulin secretion and beta cell survival (45). The incretin hormone GLP1 and its agonists have been found to improve glucose homeostasis in individuals with type II diabetes in part by triggering the cAMP pathway. cAMP in turn appears to stimulate beta cell function at least in part through activation of CREB. Depletion of CREB or its coactivator, CRTC2, in beta cells disrupts glucose homeostasis by reducing insulin secretion and beta cell viability (18, 19). Our results show that activation of the cAMP pathway in pancreatic islets triggers a robust transcriptional response that far exceeds its effects on cultured hepatocytes. In addition to epigenetic contributions noted in our study, CREB activity may also be preferentially upregulated in beta cells due to upstream differences in cAMP signaling that enhance CREB phosphorylation and CRTC2 dephosphorylation.
Genome-wide studies indicate that CREB regulates up to 5,000 genes in the mammalian genome (3, 4). Although CREB undergoes phosphorylation at a majority of these sites in response to cAMP signaling, only a small fraction (2 to 4%) of these putative target genes is upregulated in any one cell type. The discordance between CREB phosphorylation and transcriptional induction has argued for the presence of additional cAMP regulators that might function with CREB to promote gene expression (46). Supporting this idea, the CRTC coactivators have been shown to promote induction of CREB targets following their dephosphorylation and nuclear entry in response to cAMP, where they bind to CREB over relevant sites (47). Indeed, recruitment of CRTC2 to CREB binding sites was predictive of cAMP inducibility.
To date, most mechanistic studies of CREB-dependent transcription have focused on cAMP-inducible genes that contain promoter-proximal CREB binding sites and that are ubiquitously activated by cAMP (6, 7). Many of these are unaffected by loss of CBP and P300 (7). In contrast with these core target genes, which contain promoter-proximal CREB binding sites, we found that cell-restricted targets are controlled by CREB binding sites on distal enhancers. CREB appears to stimulate enhancer activity by associating with both CRTC2 and CBP/p300, leading to the acetylation of resident nucleosomes. In turn, the enrichment of H3AcK27 appears to mediate recruitment of the chromatin reader BRD4 to such enhancers and to trigger the pTEFb-dependent phosphorylation and release of RNA polymerase II from paused promoters. In contrast with its dispensable role in the regulation of genes with promoter-proximal CREB binding sites, CBP/p300 HAT activity appears critical for the expression of beta cell-specific CREB target genes.
Compared to core targets, cell-restricted CREB target genes are expressed at higher levels under basal conditions, apparently reflecting constitutive increases in H3K27 acetylation at relevant enhancers. The effects of CREB in this setting appear to be largely epigenetic, as small-molecule inhibitors that disrupt CBP-HAT activity or BRD4 occupancy decrease cell-specific but not core CREB target gene expression. Overexpression of the CREB inhibitor ACREB actually increased amounts of paused Pol II over that of the TSS, apparently by decreasing Pol II elongation. Notably, exposure to the BRD4 inhibitor JQ1 also increased amounts of paused Pol II, suggesting that CREB regulates Pol II elongation in the basal state via an association with BRD4.
Our data indicate that the CRTCs contribute importantly to induction of both core and cell-type-restricted target genes in beta cells. Based on the ability of CRTCs to coactivate genes involved in insulin secretion, small molecules that enhance CRTC activity may provide therapeutic benefit to individuals with type II diabetes. Indeed, selective small-molecule inhibitors against the salt-inducible kinases (SIKs), which otherwise phosphorylate and sequester the CRTCs, have been shown to trigger the expression of CREB target genes in different cell types (48, 49). These would be expected to have salutary effects on insulin secretion and glucose homeostasis by potentiating effects of GLP1 on CREB target gene expression. Complicating this picture, SIK2 has also been found to promote insulin secretion in response to glucose stimulation (50) by phosphorylating the CDK5 activator p35 and thereby promoting its degradation; loss of SIK2 disrupts insulin secretion. Future studies should address whether the effects of SIK2 on insulin secretion and CREB activity can be distinguished by different small-molecule inhibitors.
We found that CREB stimulates beta cell-specific gene expression in cooperation with the beta cell transcription factor Neurod1. Indeed, depletion of Neurod1 reduced CREB-dependent transcription, while Neurod1 overexpression increased it. Based on the increased density of histone H3 within the nucleosome-free region of Neurod1-depleted cells, our results suggest that Neurod1 reconfigures the architecture of CREB binding sites to permit coactivator recruitment in response to cAMP. Indeed, CREB-Neurod1-cobound loci are substantially more responsive to cAMP than CREB- or CREB-Pdx1-bound loci. Future studies should reveal the extent to which such combinatorial effects between CREB and other LDTFs contribute to cAMP-dependent transcription in other tissues.
CREB and Neurod1 appear critical for Syt7/Lrrc10b enhancer activation and for increases in beta cell function beyond Syt7 gene expression and insulin secretion. Deletion of CREB and Neurod1 binding sites in this superenhancer reduced Ins1 mRNA and protein levels in two independent mutant INS-1 clones, suggesting a larger role for this locus in regulating beta cell function. Notably, the Syt7/Lrrc10b superenhancer is syntenic with the Ins1 gene in rat (Chr1, ≈46.8 Mbp apart), mouse (Chr19, ≈41.8 Mbp), and human (INS) (Chr11, ≈59.1 Mbp). Although transcriptional effects of enhancers in cis have not been reported over such large genomic distances, future studies should provide insight into the mechanisms underlying this process.
MATERIALS AND METHODS
Contact for reagent and resource sharing.
Requests for further information and for reagents should be directed to Marc Montminy (montminy@salk.edu).
Experimental model and subject details.
Primary tissues were isolated from male wild-type C57BL/6J mice obtained from The Jackson Laboratory. To test the role of CRTC2 in forskolin-induced gene expression, we used whole-body CRTC2 knockout mice (60).
Mice were housed in a temperature-controlled environment under 12-h light/dark cycle conditions with free access to water and standard chow diet.
INS-1 insulinoma cells (51) were cultured in RPMI 1640 with 10% fetal bovine serum, 2 mM glutamine (Mediatech), 1 mM sodium pyruvate, 100 μg/ml penicillin-streptomycin, and 0.05 mM β-mercaptoethanol. AR42J cells (CRL-1492; ATCC) were cultured in F-12K with 20% fetal bovine serum and 100 μg/ml penicillin-streptomycin.
Recombinant adenovirus was grown in HEK293 cells cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 100 μg/ml penicillin-streptomycin.
Adenovirus production.
Adenovirus was generated as described earlier. Briefly, clones were ligated in the AdTrack(+CMV) vector. PmeI-digested AdTrack with insert was recombined with AdEasy-1 by cotransformation in BJ5183 Escherichia coli cells. PacI-linearized recombined vector was transfected in 293 cells (61). Recombinant adenovirus was amplified in 293 cells and purified by ultracentrifugation over CsCl gradients. For shRNA-expressing adenovirus, oligonucleotides containing hairpin sequences (see Table S2 in the supplemental material) were cloned in pBS/U6 and subcloned in AdTrack(−CMV) vector using NotI and XbaI restriction endonucleases.
Preparation of primary mouse islets.
Islet tissue was isolated from wild-type C57BL/6J mice (000664; The Jackson Laboratory) as described earlier (18). Primary islets were cultured in complete RPMI medium for 1 to 2 days before analysis.
Preparation of primary mouse hepatocytes.
Hepatocytes were isolated from wild-type C57BL/6J mice (000664; The Jackson Laboratory) as described earlier (52).
Reporter assays.
Enhancer regions were cloned from mouse genomic DNA and ligated in the pGL3 promoter vector (E1751; Promega). Reporter constructs and β-galactosidase (β-Gal) under the control of the respiratory syncytial virus promoter were cotransfected in INS-1 cells using Lipofectamine 2000 (11668019; ThermoFisher) according to the manufacturer’s instructions. Cells were exposed to FSK for 6 h and lysed in luciferase extraction buffer (25 mM Gly-Gly, 15 mM MgSO4, 4 mM EGTA, 1 mM dithiothreitol [DTT], 1% Triton X-100). For luciferase activity assay, 20 μl of extract was added to assay buffer (25 mM Gly-Gly, 15 mM MgSO4, 4 mM EGTA, 15 mM K2HPO4, pH 7.8, 2 mM DTT, 2.5 mM ATP). Fifty microliters of 0.1 mM d-luciferin K+ salt was added just before measuring luminescence. For β-Gal activity assay, 20 μl of extract was added to 50 μl of β-Gal assay buffer (1.33 mg/ml ortho-nitrophenyl-β-galactoside, 100 mM β-mercaptoethanol, 2 mM MgCl2, 200 mM Na2HPO4) and incubated at 37°C until the reaction looked faint yellow. Absorbance was measured at 420 nm.
Western blotting.
Immunoblots were performed as described previously (18). Densitometry for Western blot quantification was performed with ImageJ, v. 1.51s (53).
qPCR.
mRNA amounts for these studies were determined by reverse transcription-quantitative PCR (qPCR) analysis. RNA was extracted from cultured cells or tissue with TRIzol (15596026; Invitrogen)-chloroform. cDNA was synthesized from 1 μg total RNA using a Transcriptor first-strand cDNA synthesis kit (04897030001; Roche) according to the manufacturer’s instructions. Quantitative PCR was performed using LightCycler 480 SYBR green I master mix (04887352001; Roche) in a LightCycler 480 II (Roche) with oligonucleotides listed in Table S1.
Insulin secretion.
A total of 0.5 × 106 INS-1 cells were seeded in a 24-well plate. After 2 days, cells were washed once and incubated in 0.5 ml KRBH buffer (10 mM HEPES [pH 7.4], 5 mM NaHCO3, 129 mM NaCl, 4.8 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 1 mM CaCl2) for 2 h to starve cells. Glucose and FSK were added at the indicated concentrations. Buffer was collected and insulin measured with sensitive rat insulin radioimmunoassay (SRI-13K; EMD Millipore) according to the manufacturer’s instructions.
CRISPR/CAS9-mediated deletion of Syt7/Lrrc10b enhancer.
Two unique guide pairs flanking the Syt7/Lrrc10b enhancer region were designed using the guide design tool on crispr.mit.edu (now defunct) (Table S3) and were cloned into the lentiCRISPRv2 vector as described previously (62). Three-microgram aliquots of each LentiCRISPRv2 plasmid containing enhancer flanking guides were cotransfected into 5 million INS-1 cells using the Amaxa cell line nucleofection kit T (Lonza) and program T-20 according to the manufacturer’s instructions. After transfection, cells were selected in 2 μg/ml puromycin for 4 days. Clones were isolated by single-cell sorting in a 96-well plate. Homozygous enhancer deletion mutants were screened by PCR on genomic DNA using oligonucleotides listed in Table S3, cloned in pUC19, and sequenced.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described earlier (54). Briefly, cells were fixed in 0.75% formaldehyde for 10 min and quenched with 125 mM glycine for 5 min. Cells were washed and scraped in ice-cold phosphate-buffered saline (PBS). Cell pellets were resuspended in buffer LB3 (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, 0.5% N-laurylsarcosine, protease inhibitor cocktail; P9599; Sigma) and sonicated (Active Motif EpiShear probe sonicator) (Table 1). After sonication, Triton X-100 was added to the extract (1% final) and chromatin was cleared by centrifugation at 17,000 × g for 10 min.
TABLE 1.
Sonicator settings
| Cell or tissue | Output (%) | Pulse time (s) | No. of cycles |
|---|---|---|---|
| Islets | 30 | 10 | 28 |
| 70 | 10 | 12 | |
| Hepatocytes | 25 | 10 | 30 |
| INS-1/AR42J | 25 | 10 | 10 |
Thirty-five microliters of protein A-agarose beads (20333; Thermo Scientific) was washed twice with PBS plus 0.1% bovine serum albumin (BSA) and coupled with antibody for 4 h in PBS plus 0.1% BSA at 4°C with rotation. For ChIP, 500 μl extract was added to antibody-coupled beads and incubated overnight at 4°C with rotation.
Beads were washed three times in 500 μl wash buffer 1 (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100) and three times in wash buffer 2 (20 mM Tris-HCl [pH 7.4], 250 mM LiCl, 1 mM EDTA, 1% Triton X-100, 0.7% Na-deoxycholate). Elution was then performed by incubating beads in 50 μl elution buffer 1 (Tris-EDTA [TE], 1% SDS) for 15 min at 50°C and 50 μl elution buffer 2 (TE, 1% SDS, 300 mM NaCl) for 15 min at 50°C with shaking. Both elutions were combined and incubated with proteinase K and RNase A for 2 h at 37°C. DNA-protein complexes were de-cross-linked at 65°C with shaking overnight. ChIP DNA was purified using Agencourt AMPure XP beads (A63881; Beckman Coulter).
RNA-seq analysis.
FASTQ files were aligned against the mouse mm10 or rat rn6 genome builds using Tophat v2.0.9 or STAR (55). For the differential expression analysis shown in Fig. 1A, raw read count tables of two biological replicates from control and FSK-treated material were generated using Homer (54) (analyzeRepeats.pl with option –raw). Differential expression was calculated with Homer (getDiffExpression.pl using DESeq2 [56]). Cluster analysis was performed on log2-transformed normalized FPKM data generated with Cufflinks v2.2.1. RNA-seq data were filtered for expression (FPKM under at least one condition of ≥8). Genes induced by FSK were defined as an FSK/CON of ≥2. Hierarchical clustering was performed with Cluster 3.0 (57) and visualized with Java TreeView (version 1.1.6r4) (58). RNA-seq data for Fig. 1D, 3E, and 4C were taken from GEO accession number GSE60158 (20).
ChIP-seq analysis.
Reads were aligned to the mouse (mm10) or rat (rn5 and rn6) genomes using bowtie2 (59) or STAR (55). Tag directories of uniquely mapped reads were generated with HOMER. Binding peaks were called using HOMER (findPeaks in “factor” mode for CREB, CRTC2, BRD4, PDX1, and NeuroD1 and in “histone” mode for H3AcK27).
We identified high-confidence CREB peaks in INS-1 cells from four independent CREB ChIP-seq experiments (+FSK, 1 h) using HOMER (getDifferentialPeaksReplicates.pl, default parameters). We extracted FSK-inducible enhancers by quantifying H3AcK27 signal over a 4-kb region centered on high-confidence CREB-bound loci (log2 H3AcK27 tag [FSK/CON], ≥0.8; adjusted P value of ≤0.04).
Histograms of ChIP-seq data were generated with using HOMER (annotatePeaks.pl <peakfile/bed> <genome> -size –hist 10 –d <Tagdirectory1, Tagdirectory2,…>).
ChIP-seq binding data were correlated with RNA-seq expression data by annotation of ChIP-seq peaks to nearby gene loci using HOMER (annotatePeaks.pl, default parameters). annotatePeaks.pl was also used to measure genomic distance of ChIP-seq binding data to TSS of the nearest genes.
Sequencing-depth-normalized BigWig files were generated with HOMER (makeMultiWigHub.pl, default parameters) and visualized in the UCSC Genome Browser (http://genome.ucsc.edu).
Tissue-enriched ChIP-seq peaks were identified with HOMER (getDifferentialPeaks with ≥4× read coverage).
Superenhancers were identified based on H3AcK27 occupancy with HOMER (findPeaks –style super –L 0; maximum distance to stitch peaks together, 12.5 kb).
Motif analysis was performed using HOMER (findMotifsGenome.pl –size 200 –len 5,6,8,10,12).
Motif enrichment near ChIP-seq peaks was performed with HOMER using motif files created with findMotifsGenome or custom motifs created using seq2profile.pl. Motif enrichment was plotted using the annotatePeaks.pl command (annotatePeaks.pl <peakfile> <genome> -size 1000 –hist 5 –m <motiffile>).
CREB, NeuroD1, and PDX1 binding cooccupancy was analyzed with HOMER using the mergePeaks command. First, peak files from untreated and FSK-treated ChIP-seq experiments were merged for each transcription factor. Peak files for overlapping and unique binding sites then were generated for each transcription factor using the mergePeaks command using the merged peak files with the –prefix option.
To analyze Pol II occupancy profiles over a selection of gene bodies, we first generated lists of RefSeq gene coordinates in rn5 using the filterListBy.pl program in HOMER. Using the gene coordinates as a peak file and Pol II or H3AcK27 tag directories, we built occupancy histograms with the makeMetaGeneProfile.pl program.
Reagents.
Reagents used included rabbit anti-CREB serum (244; in-house), rabbit anti-CRTC2 serum (6865 in-house), rabbit anti-CBP serum (5613; in-house), rabbit anti-PDX1 serum (5538; in-house), rabbit anti-H3AcK27 (ab4729; Abcam), goat anti-NeuroD1 (Sc-1086X; Santa Cruz), rabbit anti-RPB1 (RNA Pol II) (2629; Cell Signaling), rabbit anti-Lrrc10B (TA337416; OriGene), mouse anti-SYT7 (OASE00272; Aviva), rabbit anti-MIDN (18939-1-AP; Proteintech), rabbit anti-ATF-3 (Sc-188 X; Santa Cruz), mouse antitubulin (05-829; EMD Millipore), mouse anti-IRS2 (06-506; Upstate), rabbit anti-BRD4 (A301-985A; Bethyl), rabbit anti-LKB1 (3047; Cell Signaling), forskolin (F6886; Sigma), and (+)-JQ1 (11187; Cayman Chemical).
Quantification and statistical analysis.
Significance was calculated with two-tailed t test.
Data availability.
RNA-seq and ChIP data sets are available under GEO accession number GSE126558.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grants R01 DK083834 (M.M.) and GM62437 (P.C.), the FAMRI Foundation (P.C.), the Leona M. and Harry B. Helmsley Charitable Trust (M.M.), the Clayton Foundation for Medical Research (M.M.), and the Kieckhefer Foundation (M.M.).
S.V.D.V. designed and performed experiments, analyzed data, and wrote the manuscript. E.W. performed RNA-seq experiments and analyzed the data. M.T. performed experiments. Y.H. and P.A.C. provided reagents and expertise. M.M. analyzed the data and wrote the manuscript.
We have no competing interests to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00200-19.
REFERENCES
- 1.Altarejos JY, Montminy M. 2011. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol 12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vo NK, Goodman RH. 2001. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem 276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- 3.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. 2005. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A 102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. 2004. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 5.Mayr B, Montminy M. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol 2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 6.Ravnskjaer K, Kester H, Liu Y, Zhang X, Lee D, Yates JR III, Montminy M. 2007. Cooperative interactions between CBP and TORC2 confer selectivity to CREB target gene expression. EMBO J 26:2880–2889. doi: 10.1038/sj.emboj.7601715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasper LH, Lerach S, Wang J, Wu S, Jeevan T, Brindle PK. 2010. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J 29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan AL, Benner C, Heinz S, Huang W, Xie L, Miano JM, Glass CK. 2011. Serum response factor utilizes distinct promoter- and enhancer-based mechanisms to regulate cytoskeletal gene expression in macrophages. Mol Cell Biol 31:861–875. doi: 10.1128/MCB.00836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinz S, Romanoski CE, Benner C, Allison KA, Kaikkonen MU, Orozco LD, Glass CK. 2013. Effect of natural genetic variation on enhancer selection and function. Nature 503:487–492. doi: 10.1038/nature12615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinz S, Romanoski CE, Benner C, Glass CK. 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16:144–154. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. 2011. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malik AN, Vierbuchen T, Hemberg M, Rubin AA, Ling E, Couch CH, Stroud H, Spiegel I, Farh KK, Harmin DA, Greenberg ME. 2014. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci 17:1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown JD, Feldman ZB, Doherty SP, Reyes JM, Rahl PB, Lin CY, Sheng Q, Duan Q, Federation AJ, Kung AL, Haldar SM, Young RA, Plutzky J, Bradner JE. 2018. BET bromodomain proteins regulate enhancer function during adipogenesis. Proc Natl Acad Sci U S A 115:2144–2149. doi: 10.1073/pnas.1711155115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, NISC Comparative Sequencing Program, Black BL, Visel A, Pennacchio LA, Collins FS. 2013. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A 110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, Howley PM. 2011. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol 31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van de Velde S, Hogan MF, Montminy M. 2011. mTOR links incretin signaling to HIF induction in pancreatic beta cells. Proc Natl Acad Sci U S A 108:16876–16882. doi: 10.1073/pnas.1114228108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jhala US, Canettieri G, Screaton RA, Kulkarni RN, Krajewski S, Reed J, Walker J, Lin X, White M, Montminy M. 2003. cAMP promotes pancreatic beta-cell survival via CREB-mediated induction of IRS2. Genes Dev 17:1575–1580. doi: 10.1101/gad.1097103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanchet E, Van de Velde S, Matsumura S, Hao E, LeLay J, Kaestner K, Montminy M. 2015. Feedback inhibition of CREB signaling promotes beta cell dysfunction in insulin resistance. Cell Rep 10:1149–1157. doi: 10.1016/j.celrep.2015.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M. 2005. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 22.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty D, Vinson C. 1998. A dominant negative inhibitor of CREB reveals that it is a general mediator stimulus-dependent transcription of c-fos. Mol Cell Biol 18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamauchi T, Hara K, Maeda S, Yasuda K, Takahashi A, Horikoshi M, Nakamura M, Fujita H, Grarup N, Cauchi S, Ng DPK, Ma RCW, Tsunoda T, Kubo M, Watada H, Maegawa H, Okada-Iwabu M, Iwabu M, Shojima N, Shin HD, Andersen G, Witte DR, Jørgensen T, Lauritzen T, Sandbæk A, Hansen T, Ohshige T, Omori S, Saito I, Kaku K, Hirose H, So W-Y, Beury D, Chan JCN, Park KS, Tai ES, Ito C, Tanaka Y, Kashiwagi A, Kawamori R, Kasuga M, Froguel P, Pedersen O, Kamatani N, Nakamura Y, Kadowaki T. 2010. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet 42:864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 24.Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, Rybin D, Petrie JR, Travers ME, Bouatia-Naji N, Dimas AS, Nica A, Wheeler E, Chen H, Voight BF, Taneera J, Kanoni S, Peden JF, Turrini F, Gustafsson S, Zabena C, Almgren P, Barker DJP, Barnes D, Dennison EM, Eriksson JG, Eriksson P, Eury E, Folkersen L, Fox CS, Frayling TM, Goel A, Gu HF, Horikoshi M, Isomaa B, Jackson AU, Jameson KA, Kajantie E, Kerr-Conte J, Kuulasmaa T, Kuusisto J, Loos RJF, Luan J, Makrilakis K, Manning AK, Martínez-Larrad MT, Narisu N, Nastase Mannila M, Ohrvik J, Osmond C, Pascoe L, Payne F, Sayer AA, Sennblad B, Silveira A, Stancáková A, Stirrups K, Swift AJ, Syvänen A-C, Tuomi T, van 't Hooft FM, Walker M, Weedon MN, Xie W, Zethelius B, Ongen H, Mälarstig A, Hopewell JC, Saleheen D, Chambers J, Parish S, Danesh J, Kooner J, Ostenson C-G, Lind L, Cooper CC, Serrano-Ríos M, Ferrannini E, Forsen TJ, Clarke R, Franzosi MG, Seedorf U, Watkins H, Froguel P, Johnson P, Deloukas P, Collins FS, Laakso M, Dermitzakis ET, Boehnke M, McCarthy MI, Wareham NJ, Groop L, Pattou F, Gloyn AL, Dedoussis GV, Lyssenko V, Meigs JB, Barroso I, Watanabe RM, Ingelsson E, Langenberg C, Hamsten A, Florez JC. 2011. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 60:2624–2634. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kycia I, Wolford BN, Huyghe JR, Fuchsberger C, Vadlamudi S, Kursawe R, Welch RP, Albanus RD, Uyar A, Khetan S, Lawlor N, Bolisetty M, Mathur A, Kuusisto J, Laakso M, Ucar D, Mohlke KL, Boehnke M, Collins FS, Parker SCJ, Stitzel ML. 2018. A common type 2 diabetes risk variant potentiates activity of an evolutionarily conserved islet stretch enhancer and increases C2CD4A and C2CD4B expression. Am J Hum Genet 102:620–635. doi: 10.1016/j.ajhg.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlessi R, Chen Y, Rowlands J, Cruzat VF, Keane KN, Egan L, Mamotte C, Stokes R, Gunton JE, Bittencourt PIH, Newsholme P. 2017. GLP-1 receptor signalling promotes beta-cell glucose metabolism via mTOR-dependent HIF-1alpha activation. Sci Rep 7:2661. doi: 10.1038/s41598-017-02838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henriques T, Scruggs BS, Inouye MO, Muse GW, Williams LH, Burkholder AB, Lavender CA, Fargo DC, Adelman K. 2018. Widespread transcriptional pausing and elongation control at enhancers. Genes Dev 32:26–41. doi: 10.1101/gad.309351.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. 2013. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol 33:2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chipumuro E, Marco E, Christensen CL, Kwiatkowski N, Zhang T, Hatheway CM, Abraham BJ, Sharma B, Yeung C, Altabef A, Perez-Atayde A, Wong KK, Yuan GC, Gray NS, Young RA, George RE. 2014. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell 159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. 2010. Selective inhibition of BET bromodomains. Nature 468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lasko LM, Jakob CG, Edalji RP, Qiu W, Montgomery D, Digiammarino EL, Hansen TM, Risi RM, Frey R, Manaves V, Shaw B, Algire M, Hessler P, Lam LT, Uziel T, Faivre E, Ferguson D, Buchanan FG, Martin RL, Torrent M, Chiang GG, Karukurichi K, Langston JW, Weinert BT, Choudhary C, de Vries P, Van Drie JH, McElligott D, Kesicki E, Marmorstein R, Sun C, Cole PA, Rosenberg SH, Michaelides MR, Lai A, Bromberg KD. 2017. Discovery of a selective catalytic p300/CBP inhibitor that targets lineage-specific tumours. Nature 550:128–132. doi: 10.1038/nature24028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi X, Liu C, Liu B, Chen J, Wu X, Gong W. 2018. JQ1: a novel potential therapeutic target. Pharmazie 73:491–493. doi: 10.1691/ph.2018.8480. [DOI] [PubMed] [Google Scholar]
- 34.Sharma A, Moore M, Marcora E, Lee JE, Qiu Y, Samaras S, Stein R. 1999. The NeuroD1/BETA2 sequences essential for insulin gene transcription colocalize with those necessary for neurogenesis and p300/CREB binding protein binding. Mol Cell Biol 19:704–713. doi: 10.1128/MCB.19.1.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao CS, Loomis ZL, Lee JE, Sussel L. 2007. Genetic identification of a novel NeuroD1 function in the early differentiation of islet alpha, PP and epsilon cells. Dev Biol 312:523–532. doi: 10.1016/j.ydbio.2007.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pataskar A, Jung J, Smialowski P, Noack F, Calegari F, Straub T, Tiwari VK. 2016. NeuroD1 reprograms chromatin and transcription factor landscapes to induce the neuronal program. EMBO J 35:24–45. doi: 10.15252/embj.201591206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seo S, Richardson GA, Kroll KL. 2005. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development 132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 38.D'Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, Baetge EE. 2006. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 24:1392–1401. doi: 10.1038/nbt1259. [DOI] [PubMed] [Google Scholar]
- 39.Zhu S, Russ HA, Wang X, Zhang M, Ma T, Xu T, Tang S, Hebrok M, Ding S. 2016. Human pancreatic beta-like cells converted from fibroblasts. Nat Commun 7:10080. doi: 10.1038/ncomms10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lima MJ, Docherty HM, Chen Y, Docherty K. 2012. Efficient differentiation of AR42J cells towards insulin-producing cells using pancreatic transcription factors in combination with growth factors. Mol Cell Endocrinol 358:69–80. doi: 10.1016/j.mce.2012.02.024. [DOI] [PubMed] [Google Scholar]
- 41.Gustavsson N, Lao Y, Maximov A, Chuang JC, Kostromina E, Repa JJ, Li C, Radda GK, Sudhof TC, Han W. 2008. Impaired insulin secretion and glucose intolerance in synaptotagmin-7 null mutant mice. Proc Natl Acad Sci U S A 105:3992–3997. doi: 10.1073/pnas.0711700105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu B, Wei S, Petersen N, Ali Y, Wang X, Bacaj T, Rorsman P, Hong W, Sudhof TC, Han W. 2015. Synaptotagmin-7 phosphorylation mediates GLP-1-dependent potentiation of insulin secretion from beta-cells. Proc Natl Acad Sci U S A 112:9996–10001. doi: 10.1073/pnas.1513004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ptashne M. 1986. Gene regulation by proteins acting nearby and at a distance. Nature 322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- 44.Blackwood EM, Kadonaga JT. 1998. Going the distance: a current view of enhancer action. Science 281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 45.Drucker DJ. 2006. The biology of incretin hormones. Cell Metab 3:153–165. doi: 10.1016/j.cmet.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Brindle P, Nakajima T, Montminy M. 1995. Multiple protein kinase A-regulated events are required for transcriptional induction by cAMP. Proc Natl Acad Sci U S A 92:10521–10525. doi: 10.1073/pnas.92.23.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR III, Takemori H, Okamoto M, Montminy M. 2004. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell 119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 48.Kumagai A, Horike N, Satoh Y, Uebi T, Sasaki T, Itoh Y, Hirata Y, Uchio-Yamada K, Kitagawa K, Uesato S, Kawahara H, Takemori H, Nagaoka Y. 2011. A potent inhibitor of SIK2, 3, 3', 7-trihydroxy-4'-methoxyflavon (4'-O-methylfisetin), promotes melanogenesis in B16F10 melanoma cells. PLoS One 6:e26148. doi: 10.1371/journal.pone.0026148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel K, Foretz M, Marion A, Campbell DG, Gourlay R, Boudaba N, Tournier E, Titchenell P, Peggie M, Deak M, Wan M, Kaestner KH, Goransson O, Viollet B, Gray NS, Birnbaum MJ, Sutherland C, Sakamoto K. 2014. The LKB1-salt-inducible kinase pathway functions as a key gluconeogenic suppressor in the liver. Nat Commun 5:4535. doi: 10.1038/ncomms5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakamaki J, Fu A, Reeks C, Baird S, Depatie C, Al Azzabi M, Bardeesy N, Gingras AC, Yee SP, Screaton RA. 2014. Role of the SIK2-p35-PJA2 complex in pancreatic beta-cell functional compensation. Nat Cell Biol 16:234–244. doi: 10.1038/ncb2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. 1992. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology 130:167–178. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
- 52.Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J III, Montminy M. 2007. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 53.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saldanha AJ. 2004. Java Treeview–extensible visualization of microarray data. Bioinformatics 20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 59.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Inoue H, Ravnskjaer K, Viste K, Miller N, Liu Y, Hedrick S, Vera L, Montminy M. 2010. Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proc Natl Acad Sci U S A 107:3087–3092. doi: 10.1073/pnas.0914897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He T, Zhou S, Costa LD, Yu J, Kinzler K, Vogelstein B. 1998. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A 95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ChIP data sets are available under GEO accession number GSE126558.