Abstract
Voltage-gated ion channels (VGICs) contain positively-charged residues within the S4 helix of the voltage-sensing domain (VSD) that are displaced in response to changes in transmembrane voltage, promoting conformational changes that open the pore. Pacemaker HCN channels are unique among VGICs because their open probability is increased by membrane hyperpolarization rather than depolarization. Here we measured the precise movement of the S4 helix of a sea urchin HCN channel using transition metal ion fluorescence resonance energy transfer (tmFRET). We show that the S4 undergoes a significant (~10 Å) downward movement in response to membrane hyperpolarization. Furthermore, by applying distance constraints determined from tmFRET experiments to Rosetta modeling, we reveal that the C-terminal part of the S4 helix exhibits an unexpected tilting motion during hyperpolarization activation. These data provide a long-sought glimpse of the hyperpolarized state of a functioning VSD and also a framework for understanding the dynamics of reverse gating in HCN channels.
INTRODUCTION
Voltage-gated ion channels (VGICs) underlie almost all electrical signaling in the body1. They change their open probability in response to changes in transmembrane voltage, allowing permeant ions to flow across the cell membrane. To sense the transmembrane voltage, these channels evolved a voltage-sensing domain (VSD) consisting of four transmembrane helices (S1-S4) on the periphery of the channel2,3. The S4 helix contains a series of positively-charged arginines that are thought to sense the transmembrane voltage and move towards the inside of the cell (downwards) during membrane hyperpolarization4–8. Clusters of negatively-charged amino acids in the S1-S3 helices stabilize the S4 charges in the resting and activated states3,9. A hydrophobic constriction site and a charge transfer center (CTC), near the middle of the membrane, are thought to focus the membrane electric field and facilitate the conformational rearrangement of the VSD3,10,11. Although many X-ray crystal and cryo-EM structures have been solved for the depolarized state (“up” state) of the VSD, no structures have yet been solved at hyperpolarized voltages. Instead structures of the hyperpolarized state have had to be inferred from structures obtained by trapping the VSD in the “down” state using various gating modifiers12–14. Therefore, to fully understand the molecular mechanisms underlying voltage-dependent activation of VGICs, we need to be able to measure the structure and dynamics of the VSD at hyperpolarized voltages in a membrane environment.
Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels underlie the If and Ih currents in the heart and brain, and play a key role in pacemaking activity, resting membrane potential, and synaptic integration15. These channels are particularly enigmatic in the VGIC family because they are activated by membrane hyperpolarization rather than depolarization, referred to as reverse gating16–18. Each HCN subunit contains an amino-terminal HCN domain (HCND), a VSD (S1-S4 helices), a pore domain (S5-S6 helices) and a carboxyl-terminal C-linker/cyclic nucleotide-binding domain (C-linker/CNBD) (Fig. 1a)19,20. HCN channels assemble in a non-domain-swapped subunit arrangement, i.e. the VSD is in proximity to the pore domain of the same subunit, which is distinct from the domain-swapped arrangement in most VGICs19,20. Although the mechanism for reverse gating is unknown, the S4 has been suggested to move in a similar direction with voltage compared to depolarization-activated VGICs, but the coupling between the VSD and the pore domain is thought to be reversed8,21. Because of close contact between the S4 and S5 helices and the lack of a requirement for a contiguous S4-S5 linker, the S4 helix may impinge on the pore domain via a direct interaction with the S5 helix19,22,23. A high-resolution cryo-EM structure is available for the depolarized state of the human HCN1 channel19, but little information exists about the hyperpolarized state or conformational dynamics associated with HCN channel activation.
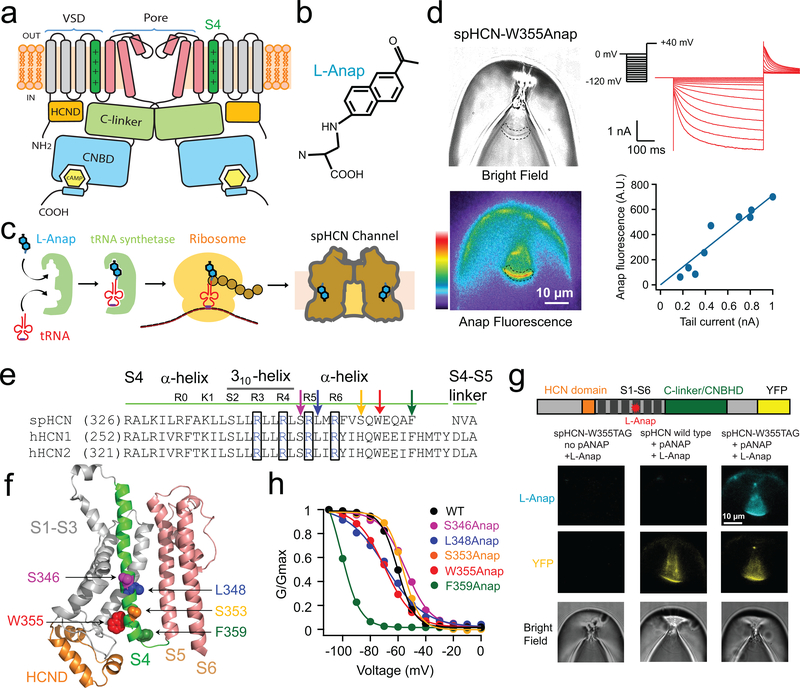
Figure 1 |. Patch-clamp fluorometry (PCF) simultaneously measures current and Anap fluorescence in HCN channels.
a, Cartoon showing the architecture of an HCN channel subunit (only two subunits are shown). Each subunit contains a cyclic nucleotide-binding domain (CNBD) in the carboxy-terminal region that is connected to the pore region via a gating ring formed by the C-linker of each subunit. b, Structure of L-Anap. c, Cartoon illustrating amber stop-codon suppression strategy for incorporating noncanonical amino acids. d, Representative PCF images showing a giant inside-out patch expressing spHCN-W355Anap channels in bright field (top left) and epifluorescence (heat map, bottom left) from the same patch; the dashed area in both images is an exemplar region of interest used for quantifying the patch fluorescence. Top right, channel current elicited by a series of hyperpolarizing voltages from 0 mV to −120 mV with 10 mV steps (Bottom right, the intensity of Anap fluorescence is proportional to the magnitude of the peak tail current (bottom right). e, Sequence alignment of the S4 helix of spHCN, human HCN1, and HCN2 channels. L-Anap sites are indicated with arrows, positively-charged arginines on the S4 close to the L-Anap sites are highlighted in black boxes, and the 310-helical segment of the S4 is indicated with a black line. f, Positions of Anap sites within S4 shown in the homology model of spHCN based on human HCN1 (PDB 5U6O). g, Top, map of spHCN constructs showing eYFP fused to the C-terminal end (size of eYFP is not to scale). Bottom, exemplar PCF images showing the specific incorporation of L-Anap at the spHCN-W355 site. h, G-V relationships of spHCN constructs in the presence of 1 mM cAMP with L-Anap incorporated into different sites in the S4, compared to wild-type spHCN (WT) (also see Supplementary Table 1). Current traces are in Supplementary Fig. 1.
Here, using transition metal ion fluorescence resonance energy transfer (tmFRET) combined with patch-clamp fluorometry (PCF) and Rosetta modeling, we determined the structure of the hyperpolarized state of the VSD of a functioning HCN channel. In this study we found that the S4 helix of the voltage sensor exhibits a large downward movement and a significant tilting motion during hyperpolarization. This new model of the VSD movement in HCN channels may help elucidate the reverse gating mechanism of HCN channels. In addition, our methods can be broadly applied to probe short-distance rearrangements in other ion channels and membrane proteins.
RESULTS
spHCN as a model system for studying VSD structure and dynamics
For these studies, we used a sea urchin HCN channel (spHCN, from Strongylocentrotus purpuratus) that exhibits significant sequence similarity to mammalian HCN channels (78% sequence similarity in the VSD) and is robustly expressed in heterologous expression systems24,25. Like many mammalian HCN channels, spHCN channels are weakly K+ selective and potentiated by the direct binding of cyclic nucleotides to the intracellular CNBD24. In the absence of cyclic nucleotides, spHCN channels rapidly enter a non-conducting or inactivated state upon membrane hyperpolarization24,26 (Supplementary Fig. 1a). In the presence of cAMP, spHCN channels exhibit similar hyperpolarization dependence to mammalian HCN channels. Therefore, in this paper, all of the experiments were done in the presence of cAMP. Previous scanning cysteine accessibility mutagenesis studies have suggested that the conformational change of the VSD during channel activation is conserved between spHCN and mouse HCN1 channels8,21.
To measure the structure and dynamics of the VSD during channel activation, we used tmFRET on intact spHCN channels in the plasma membrane27–32. tmFRET measures the distance between a donor fluorophore and an acceptor non-fluorescent transition metal ion, such as Ni2+, Co2+, and Cu2+. Minimal transition metal ion binding sites were introduced by site-directed mutagenesis. Efficient and specific fluorophore labeling was achieved using amber stop-codon (TAG) suppression to introduce the fluorescent, noncanonical amino acid L-Anap (Fig. 1b, c)33,34. Anap is a small environmentally sensitive fluorophore with a short linker to the protein backbone, and is well suited as a tmFRET donor for distance measurements29. We also employed PCF to allow simultaneous measurement of the structure (with tmFRET) and function (with electrophysiology) of intact spHCN channels28,35, while controlling membrane voltage and rapidly applying intracellular ligands (e.g. cAMP and transition metals) (Fig. 1d). By combining these methods, we established a powerful way to measure the conformational dynamics associated with the gating of ion channels in a membrane environment.
PCF measurements of spHCN channels during activation by hyperpolarization
We introduced amber stop codons individually at five positions along the S4 transmembrane helix of a spHCN channel with a carboxyl-terminal Yellow Fluorescent Protein (YFP) fusion (Fig. 1e and f). YFP was used to confirm the expression of the full-length channel, correlate with Anap incorporation, and calculate the relative quantum yield of L-Anap for tmFRET (Supplementary Fig. 2). The five different spHCN constructs were expressed in Xenopus oocytes which were also injected with L-Anap and an L-Anap-specific aminoacyl tRNA synthetase/tRNA plasmid (pANAP)33. Inside-out patches were excised from the oocyte plasma membrane, and fluorescence and current were recorded simultaneously using PCF (Fig. 1d). Substantial Anap fluorescence, YFP fluorescence, and K+ current were observed from all five engineered spHCN channels. Anap fluorescence was directly proportional to ionic current and YFP fluorescence (Fig. 1d and Supplementary Fig. 2). In addition, little or no Anap fluorescence was observed in the absence of pANAP or in the absence of the introduced amber stop codon (Fig. 1g). These results indicate that the fluorescent labeling was efficient and specific for each of the five L-Anap sites introduced into the channels.
The functional properties of the engineered channels were measured using a family of hyperpolarizing voltage pulses from 0 mV to −110 mV in the presence of 1 mM cAMP. All channels exhibited appreciable hyperpolarization activation (Supplementary Fig. 1). The conductance-voltage (G-V) relationships were generally similar to that of wild-type spHCN channels in four of the channels (Fig. 1h, Supplementary Fig. 1b and Supplementary Table 1). However, the F359Anap site at the end of S4 resulted in activation kinetics that were substantially slower, and a G-V curve that was shifted by more than 30 mV in the hyperpolarizing direction, in comparison to wild-type spHCN (Fig. 1h, Supplementary Fig. 1b and Supplementary Table 1). These results suggest that, for all of our sites except F359Anap, introduction of L-Anap into the S4 helix did not substantially perturb the hyperpolarization-dependent gating of these channels.
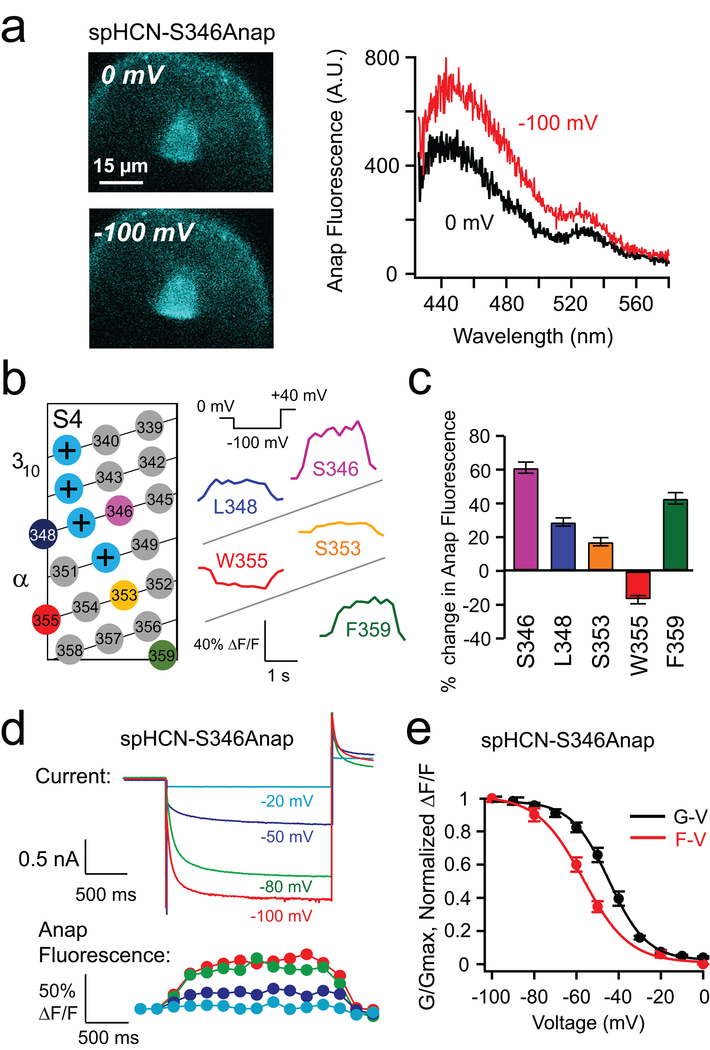
The environmental sensitivity of Anap fluorescence allowed us to test for any changes to the environment of the S4 residues during voltage-dependent activation of spHCN channels. For spHCN-S346Anap, there was a substantial increase in fluorescence during hyperpolarizing voltage pulses to −100 mV (61.2 ± 3.3%, n = 6 patches) (Fig. 2a and Supplementary Video 1). This increase was accompanied by a shift in the peak emission spectrum to longer wavelengths (8.3 ± 1.5 nm, n = 3 patches) (Fig 2a and Supplementary Fig. 3a, f), suggesting that S346Anap enters a more hydrophilic environment at −100 mV. An increase in fluorescence was also observed for L348Anap, S353Anap, and F359Anap, and a decrease was observed for W355Anap (Supplementary Video 2), with or without a shift in the peak emission (Fig. 2b, c and Supplementary Fig. 3). For S346Anap, the time course and voltage dependence of these fluorescence increases matched the time course and voltage dependence of the ionic currents (Fig. 2d, e) The F-V for S346Anap is shifted to negative voltages compared to the G-V (Fig. 2e), similar to the way the gating charge-voltage (Q-V) relationship of related HCN channels is shifted compared to the G-V, suggesting some charge movement after opening36. In contrast, an Anap site in the S1 transmembrane segment (W218Anap) exhibited little or no fluorescence change when hyperpolarized to −100 mV (Supplementary Fig. 4b and Supplementary Fig.3e). These results suggest that the fluorescence changes at the S4 Anap sites are reporting, at least in part, the rearrangement of the VSD during channel activation.
Figure 2 |. Hyperpolarization-dependent change in Anap fluorescence of spHCN channels with L-Anap incorporated into the S4 voltage sensor.
a, Representative epifluorescence images and spectra of the S346Anap fluorescence at 0 mV and −100 mV. b, Two-dimensional S4 topology and the Anap fluorescence changes at different S4 sites in response to a −100 mV pulse in the presence of 1 mM cAMP. c, Summary of the percent change of the Anap fluorescence for different sites within the S4 helix due to a −100 mV pulse. Data shown are mean ± s.e.m., n = 6 patches for S346, n = 4 for L348 and S353, n = 7 for W355 and n = 4 for F359. d, Simultaneous current and fluorescence measurements from spHCN-S346Anap channels in response to a family of hyperpolarizing voltage pulses in 1 mM cAMP. e, Fluorescence change-voltage (F-V) and conductance-voltage (G-V) relationships for the spHCN-S346Anap channels. Data shown are mean ± s.e.m., n = 3 patches.
Hyperpolarization-induced movement of S4 measured by tmFRET
In order to measure the S4 movement directly, we performed tmFRET experiments. Transition metals such as Ni2+, Co2+, and Cu2+ have absorption spectra that overlap with the emission spectrum of L-Anap (Fig. 3a)27 and hence can serve as nonfluorescent FRET acceptors that quench the donor’s fluorescence in a highly distance-dependent manner. Because the absorption of most transition metals is low, with multiple transition dipoles, tmFRET can measure short distances (10–25 Å) with little or no orientation dependence27,37,38. Various metals can be reversibly attached to engineered metal binding sites on the protein, extending the useful distance range for tmFRET27. Finally, by measuring the degree of metal quenching for each state of the channel, the FRET efficiency can be accurately determined for each state even if there is an environmental change in fluorescence32.
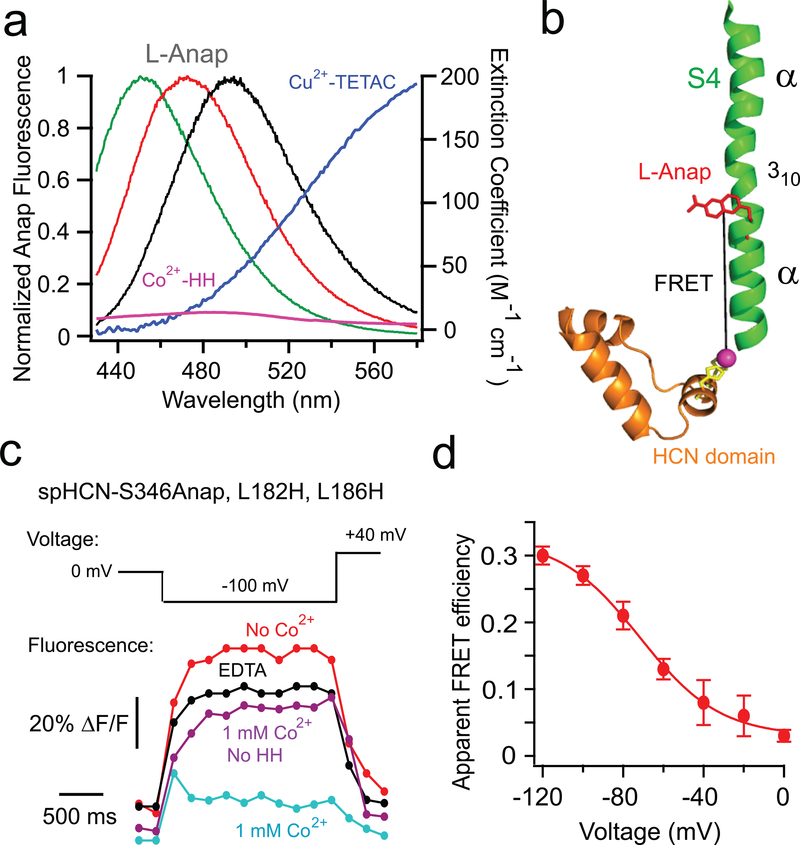
Figure 3 |. tmFRET detects a hyperpolarization-dependent downward movement of S4 in spHCN-S346Anap channels.
a, Spectral properties of free L-Anap emission and transition metal ion absorption. Emission spectra from L-Anap in different solvents (black, SBT buffer; red, EtOH; green, DMSO) are overlaid on the absorption spectra of Cu2+-TETAC (dark blue ) and Co2+-HH (magenta) b, Cartoon showing FRET between the S346Anap site and the diHis site in the HCN domain (L182H, L186H). c, Time course of Anap fluorescence in spHCN-S346Anap, L182H, L186H channels in response to a −100 mV pulse in the presence of cAMP before and after Co2+ application and EDTA to sequester Co2+. The same experiment for spHCN-S346Anap channels without the introduced diHis in the presence of Co2+ is also shown as a control. d, Voltage dependence of the apparent FRET efficiency for the spHCN-S346Anap, L182H-L186H channels in the presence of 1 mM cAMP. Data shown are mean ± s.e.m, n = 3 patches.
A transition metal ion binding site was introduced into the spHCN-S346Anap channel as a dihistidine (diHis) motif (L182H, L186H) on the second α helix in the HCN domain, directly below S4 (Fig. 3b). Upon application of 1 mM Co2+, there was substantial quenching of Anap fluorescence which was reversible by the addition of a metal chelator EDTA (Fig. 3c). The quenching at −100 mV was substantially more in spHCN-S346Anap, L182H, L186H channels (35.3 ± 1%) than in spHCN-S346Anap channels lacking the diHis site (14.5 ± 1%) (n = 3 patches). These results indicate that the quenching arose primarily from FRET between S346Anap and Co2+ bound to the diHis site in the HCN domain, though some solution quenching by 1 mM Co2+ was also present.
Remarkably, Co2+ quenching of Anap in spHCN-S346Anap, L182H, L186H channels was much greater at hyperpolarized voltages than at depolarized voltages (Fig. 3c). We quantified the apparent FRET efficiency by calculating the fractional decrease in Anap fluorescence in 1 mM Co2+ and correcting for the solution quenching in spHCN-S346Anap lacking the diHis site (see methods). The FRET efficiency increased by ten-fold, from 0.03 ± 0.01 at 0 mV to 0.30 ± 0.01 at −120 mV (Fig. 3d) with a voltage-dependence that is similar to that of previously measured gating currents from spHCN36,39. These results suggest that the S346 position in the S4 helix moves downward – closer to the HCN domain – during hyperpolarization activation of HCN channels.
To measure the movement of the S4 helix more precisely we switched to a new method for tmFRET, called ACCuRET (Anap Cyclen-Cu2+ resonance energy transfer)40. Instead of a diHis motif, ACCuRET introduces a higher affinity transition metal ion-binding site using TETAC (1-(2-pyridin-2-yldisulfanyl)ethyl)-1,4,7,10-tetraazacyclododecane) (Fig. 4a)40. TETAC is a cysteine-reactive compound with a short linker to a cyclen ring that binds transition metal ions with subnanomolar affinity41. Single cysteines introduced into the protein react with TETAC to create a high-affinity metal binding site for tmFRET. This binding site has two major advantages over diHis: 1) it avoids using high (millimolar) concentrations of transition metals in solution, which cause solution quenching and could bind to other endogenous sites or have unwanted effects on the protein; 2) TETAC increases and shifts the absorbance of Cu2+ to create greater overlap with the emission of Anap, extending the range of distances that can be measured with tmFRET40 (Fig. 3a). These advantages recently enabled the accurate measurement of absolute distances and distance changes in in soluble and membrane proteins40.
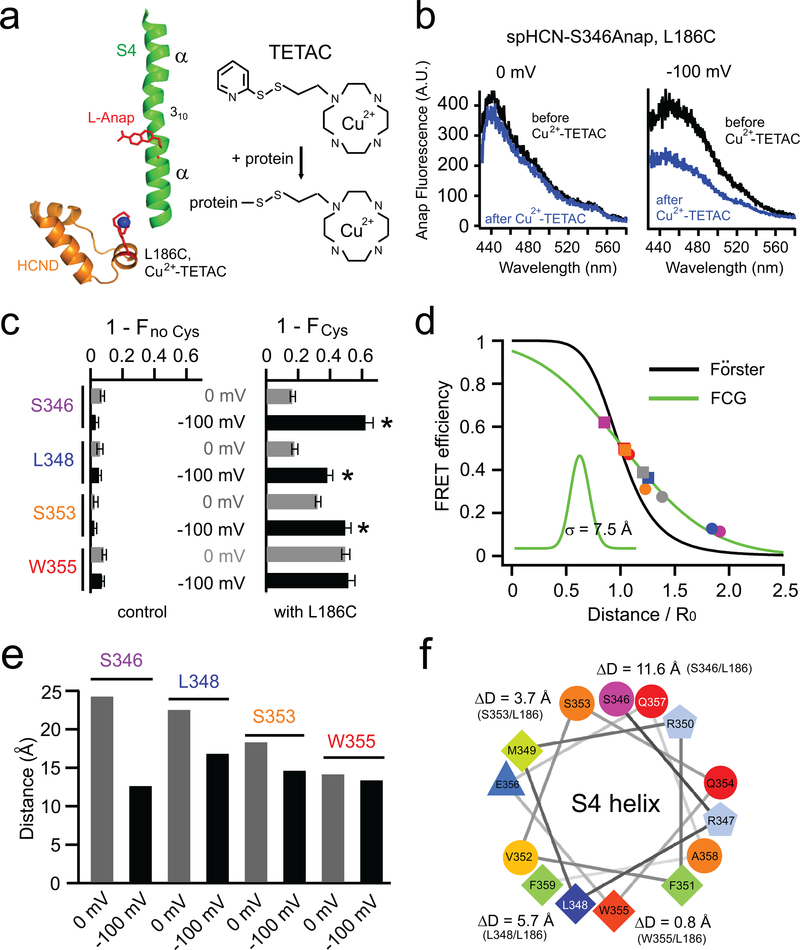
Figure 4 |. The distance change measured by ACCuRET decreases as Anap is positioned closer to the C-terminal end of S4.
a, Cartoon showing ACCuRET between S346Anap and Cu2+-TETAC attached to L186C in the HCN domain. Also shown is the reaction of Cu2+-TETAC with a cysteine in a protein. b, Representative spectra of the Anap emission at 0 mV and −100 mV, before and after quenching by Cu2+-TETAC, for spHCN-S346Anap, L186C channels. c, Summary of the fraction of Anap fluorescence quenched by Cu2+-TETAC for the four Anap sites in S4, without (left, n = 4 for S346, n = 3 for L348, S353, and W355) and with (right, n = 5 for S346, n = 4 for L348 and S353, n = 7 for W355) the introduced cysteine L186C. Data shown are mean ± s.e.m., n is number of patches, * p < 0.05 by two-tailed Student’s t-test. d, Distance dependence of the measured FRET efficiency at 0 mV for four L-Anap sites (circle markers) in S4 and the W218Anap site in S1 versus the β-carbon distances between each site and L186C (based on the spHCN homology model). Also shown are the predicted distance dependencies of the Förster equation (black) and the Förster Convolved with Gaussian (FCG) equation (green), using a Gaussian distribution with a standard deviation σ of 7.5 Å (inset). The distances of these 5 pairs at −100 mV estimated using the FCG equation are also shown with square markers. e, Summary of the distances of each FRET pair at 0 mV and −100 mV, calculated from the FCG equation in panel d, using the FRET efficiencies in Supplementary Fig. 5a. f. Helical-wheel display of the C-terminal α-helical part of S4 (S346 to F359) viewed from the extracellular side, highlighting the magnitude of the hyperpolarization-induced distance changes for the four Anap sites in the S4.
We employed ACCuRET to measure the size of the movement of the S4 helix during hyperpolarization activation of spHCN. In combination with the Anap sites, we introduced a single cysteine (L186C) into the second helix of the HCN domain of cysteine-depleted spHCN constructs. Neither the cysteine mutations nor Cu2+-TETAC labeling of L186C had an appreciable effect on hyperpolarization-dependent gating (Supplementary Fig. 5). Similar to the effect of Co2+ on the diHis motif, the modification by 10 μM Cu2+-TETAC caused a significant reduction in Anap fluorescence for spHCN-S346Anap, L186C, particularly at −100 mV (Fig. 4b, c). This Cu2+-TETAC quenching had no effect on the shape of the Anap emission spectrum and was negligible in the absence of the introduced cysteine, indicating that it arose from FRET between S346Anap and Cu2+-TETAC bound to L186C on the HCN domain (Fig. 4b, c). Like the effect of Co2+ on the diHis motif, the quenching was much greater at −100 mV (62.5 ± 3.6 %) than at 0 mV (16.7 ± 0.8 %) (n = 5 patches) (Fig. 4c).
We performed similar ACCuRET experiments for the three other Anap positions closer to the C-terminal end of the S4 (L348, S353, and W355). For each position, there was substantial quenching by Cu2+-TETAC modification of L186C (Fig. 4c). At 0 mV the quenching increased steadily as the Anap position moved towards the C-terminal end of the S4 helix, as expected from the decreasing FRET distance in the cryo-EM structure of hHCN119. Unexpectedly, however, the increase in quenching at −100 mV compared to that at 0 mV progressively diminished as Anap sites became closer to the C-terminal end of S4 helix (Fig. 4c). These data imply that S4 is not simply translating as a rigid body directly towards the HCN domain during hyperpolarization activation.
Distance changes between the Anap sites and HCN domain by hyperpolarization
We used our tmFRET data to calculate the distances between each Anap site and L186 at 0 mV and −100 mV. FRET efficiencies were calculated from the fractional decrease in Anap fluorescence after application of Cu2+-TETAC and corrected for the small Cu2+-TETAC-dependent quenching in Anap constructs lacking L186C (Supplementary Fig. 6 and see online Methods). We also determined the R0 values (the distance predicted to produce 50% FRET efficiency) for each site at both 0 mV and −100 mV, using emission spectra and fluorescence intensity data (Supplementary Fig. 2 and 3, and see online Methods). The measured FRET efficiencies for each site at 0 mV were plotted against the β-carbon distances between each FRET pair (in distance/R0 units) determined from an spHCN homology model built from the cryo-EM structure of hHCN1 (also at 0 mV) (Fig. 4d). The FRET efficiencies decreased with distance, and this relationship provides an empirical measurement of the distance dependence of tmFRET that can be used to measure the distances at −100 mV.
The distance dependence measured for tmFRET at 0 mV had the expected R0 value but was shallower than predicted by the Förster equation (Fig. 4d). Previously this shallower distance dependence has been attributed, in part, to heterogeneity of the interatomic distances in proteins and could be accounted for by convolving the Förster equation with a Guassian distribution of distances40. We found a Gaussian distribution of distances with a standard deviation of 7.5 Å could explain the distance dependence of the FRET efficiency we observed at 0 mV (Fig. 4d). Such wide distance distributions have been directly observed using double-electron-electron resonance spectroscopy (DEER) in other proteins and arise from rotameric and conformational heterogeneity42,43. The wide distribution observed in the distances between sites in the S4 and L186 in the HCN domain may be partially due to flexibility of the HCN domain. Note, however, that our mean distances estimated from the average tmFRET measurements of many channels are more accurate than the standard deviation of the individual distances38,40. The distances calculated from the FRET efficiencies at 0 mV using the Förster Convolved Gaussian (FCG) fit closely matched those in our homology model based on the known hHCN1 structure (RMSD = 1 Å) (Fig. 4d). This is similar to our previous estimates of the accuracy of tmFRET using FCG theory which have ranged from 1.5 to 2.9 Å40. Therefore, FCG theory provides an effective way to accurately determine the atomic distances based on the FRET efficiencies.
Using our calibrated distance dependence at 0 mV and our measured FRET efficiencies and R0 values determined at −100 mV (see online Methods), we calculated the distances for each FRET pair at −100 mV. While this estimate assumes the same Gaussian distribution of distances at 0 mV and −100 mV, this assumption had only a small effect on the estimated distances at −100 mV because they were all very near R0 (Fig. 4d, e). The distances at −100 mV deviated significantly from the distances at 0 mV. The change in distance between 0 and −100 mV decreased steadily along the S4 helix from 11.6 Å at S346 to only 0.8 Å at W355. These results support our proposal that the movement of S4 is not simply a rigid body movement directly towards L186 in the HCN domain. Furthermore, since our sites on the same side of the helix (S346 and S353, as well as L348 and W355) reported very different distance changes and residues on different sides of the helix (L348 and S353) reported more similar distance changes (Fig. 4f), the majority of the distance changes cannot result from a rotation. Instead, our results suggest that the S4 helix moves downward and tilts during hyperpolarization activation.
The interpretation above assumes that L186 in the HCN domain does not move substantially during activation. To test for movement of the HCN domain, we measured tmFRET between L186C and W218Anap on the S1 helix, and calculated distances at 0 mV and −100 mV as above (Supplementary Fig. 4d, e). Interestingly, W218Anap moved a little closer to the HCN domain at −100 mV (ΔD = 2.7 Å). This could reflect a small upward movement of the HCN domain, or a subtle downward movement of S1, or a combination of the two. In any case, this small movement was included in our structural modeling below, and did not appreciably affect our measurements of the much larger movements of some sites on the S4 segment.
Structural model for HCN S4 movement by Rosetta modeling
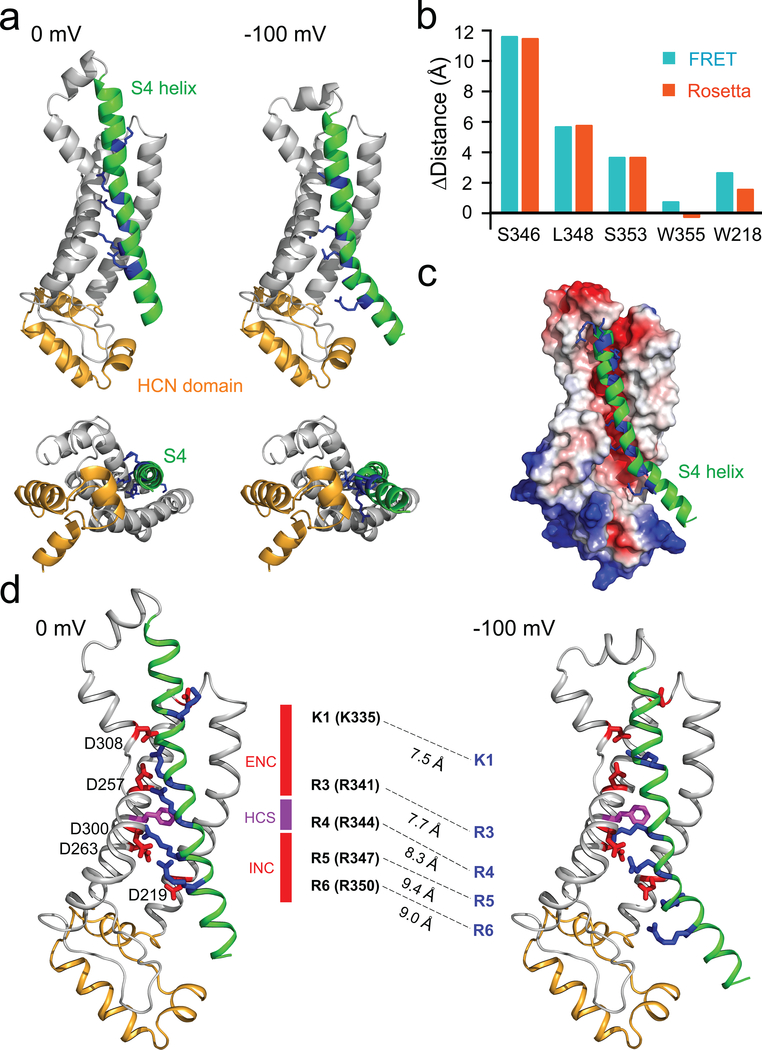
To estimate the actual movement of the S4 helix, we used our distances as constraints in Rosetta-based structural modeling of the conformational change of the VSD (Fig. 5a, Supplementary Video 3). The models at 0 mV and −100 mV accurately reproduced the distances and distance changes measured from our ACCuRET experiments (Fig. 5b and Supplementary Video 4). At 0 mV, the model was very similar to the spHCN homology model based on the HCN1 cryo-EM structure at 0 mV19 (Fig. 5a). However, with the −100 mV constraints, the model revealed a large downward movement of S4 of about two turns of the helix (Fig. 5a, c and Supplementary Video 3). In addition, the C-terminal end of the S4 helix tilted away from the HCN domain, producing a kink in the helix near position R344. Little or no rotation of S4 was observed in the model (indeed, rotation caused a poorer fit to our measured distances), and the HCN domain underwent only a small upward movement. Applying different constraining weights for the experimentally-determined distances at −100 mV generated models with very similar movement, reflecting the robustness of our modeling at the hyperpolarized voltage (Supplementary Fig. 7).
Figure 5 |. Rosetta model of S4 movement in HCN channels based on experimentally-determined distance constraints.
a, Model structures of the voltage-sensor domain of spHCN at 0 mV and −100 mV. Top: side view parallel to the membrane. Bottom: view from the intracellular side. b, Comparison of the measured distance changes using tmFRET from Fig. 4 and the distance changes in the Rosetta models. c, A vacuum electrostatic surface illustration of the S1-S3 helices and the HCN domain, showing a negative charged surface (red) facing the S4 helix. d, Structural diagrams showing the ion pair partners between arginines or lysine (blue) within S4 and aspartic acids (red) within S1-S3 at 0 mV and −100 mV in the Rosetta models. The “phenylalanine cap” F260 in the hydrophobic constriction site (HCS) of the charge transfer center is highlighted in magenta. The α-carbon distance changes between K1, R3-R6 residues at 0 mV and −100 mV are illustrated.
While our tmFRET method does not directly measure the positions of side chains, extensive salt bridges between S4 arginines and S1-S3 aspartic acids were present for the models at both voltages (Fig. 5d). In addition, the S1-S3 form a negatively-charged electrostatic surface which interfaces with S4 positive charges (Fig. 5c and Supplementary Video 3). At 0 mV, both R3 (R341) and R4 (R344) were above the “phenylalanine cap” F260 in the hydrophobic constriction site (HCS) and interacting with the extracellular negative cluster (ENC: D308, D257). R5 (R347) and R6 (R350) formed salt bridges with the intracellular negative cluster (INC: D263, D300). At −100 mV, R3 and R4 moved down below the HCS and were stabilized by interacting with the INC while the K1 (K335) formed a salt bridge with D308 of the ENC. The 310-helical nature of S4 in the gating charge transfer center (indicated in Fig. 1e)10,44 positions the voltage-sensing arginines R341 and R344 in line on the same side of S4 at both voltages. Therefore, this region moves almost vertically to exchange their ion-pair partners without much rotation (Fig 5d and Supplementary Video 3). Together, our experiments provide an explicit model of the conformational dynamics of the spHCN VSD during hyperpolarization activation, based on experimentally-determined distance measurements under physiological conditions.
DISCUSSION
The new model for the movement of the S4 helix presented here is consistent with the gating characteristics of HCN channels. A downward movement corresponding to two turns of the S4 helix could produce an equivalent charge movement of up to two electronic charges per subunit (eight charges per channel) during voltage-dependent activation of spHCN. This is more than enough charge movement to reproduce the steepness of the G-V, Q-V, and F-V curves for these channels8,39. Furthermore, the large downward translation of S4 strongly supports a previous model based on scanning cysteine accessibility mutagenesis experiments in mouse HCN1 channels21. In addition, the similar results from the cysteine accessibility studies for spHCN and mouse HCN1 channels confirm that the spHCN channel shares very similar VSD movement with its mammalian orthologs8,21. The tilting motion of the S4 that we have observed has not been previously described in any other VGIC, but is compatible with the short S4-S5 linker seen in these non-domain-swapped channels.
The mechanism for coupling this S4 movement to pore opening in HCN channels remains a mystery. Unlike the previously proposed mechanisms in domain-swapped channels, the coupling in non-domain-swapped HCN channels does not require an intact S4-S5 linker23. The S4-S5 linker is also not required for gating in the related depolarization-activated EAG and hERG channels which are also not domain swapped45,46. Considering the tight packing of S4 and S5 helices in HCN channels19, the downward and tilting movement of the S4 helix could bypass the short S4-S5 linker to influence the S5 helix directly, thus altering channel opening22,23. Potentially, the tilting motion of the C-terminal end of the S4 could move the N-terminal end of the S5 helix to allow the S6 bundle-crossing gate to open when the S4 is in the “down” position at hyperpolarized voltages. Our model of the S4 movement provides a framework to decipher this coupling and the reverse gating mechanism of HCN channels.
Despite decades of research on the VSD of VGICs, it remains controversial how the S4 voltage sensor moves during voltage-dependent activation. Using a combination of tmFRET, PCF, and Rosetta modeling, we have provided a first glimpse into the movement of the S4 at hyperpolarized voltages under physiological conditions that does not rely on gating modifiers to trap a potentially down state of the VSD. Our model is incompatible with other models proposing that the S4 helix has relatively little “up/down” movement based on cryo-EM, scanning cysteine mutagenesis, and FRET47–52. On the other hand, our model is similar to a recently proposed movement of the S4 in VSD2 of the depolarization-activated two-pore channel 1 (TPC1) inferred from structures of mouse TPC1 (“up” state) and Ba2+-inhibited Arabidopsis TPC1 (“down” state) at 0 mV14,53 (Supplementary Video 5). Furthermore, a similar movement of S4 was recently proposed for voltage-gated sodium channels based on the “down” state of VSD2 trapped by spider toxins as well as that of VSD4 trapped by α-scorpion toxins12,13 (Supplementary Video 5). The similarity between our model, obtained from tmFRET experiments and using a functioning VSD at hyperpolarized voltages, and the models based on cryo-EM structures with cation- or toxin-trapped states provides further support for the assumption that these high-resolution “down” states of the VSD may represent physiological conformations of the S4 at negative voltages. Our methods further provide an avenue to measure not just a snapshot of the “down” state structure, but also the conformational dynamics of the VSDs as they move to promote channel opening.
METHODS
Molecular Biology
The full-length spHCN cDNA (a gift from U. B. Kaupp, Molecular Sensory Systems, Center of Advanced European Studies and Research, Bonn, Germany; GenBank: Y16880) was subcloned into a modified pcDNA3.1 vector (Invitrogen, Carlsbad, CA) that contained a C-terminal eYFP, a T7 promoter, and 5’ and 3’ untranslated regions of a Xenopus β-globin gene. For the ACCuRET experiments, a cysteine-depleted spHCN construct was used with the following mutations: C211A, C224A, C369A, C373A. Point mutations were made using Quickchange II XL Site-Directed Mutagenesis kit (Agilent technologies, Santa Clara, CA). The sequences of the DNA constructs were confirmed by fluorescence-based DNA sequencing (Genewiz LLC, Seattle, WA). The mRNA was synthesized in vitro using the HiScribe T7 ARCA mRNA Kit (New England Biolabs, Ipswich, MA) or the mMESSAGE mMACHINE T7 ULTRA Transcription Kit (ThermoFisher, Waltham, MA) from the linearized plasmid.
Oocyte Expression and Electrophysiology
The animal-use protocols were consistent with the recommendations of the American Veterinary Medical Association and were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Washington. Xenopus oocytes were prepared as previously described54. L-Anap (free-acid form, AsisChem, Waltham, MA) was made as a 1 mM stock in water at a high pH by adding NaOH and stored at −20°C. The pANAP plasmid (purchased from Addgene, Cambridge, MA) contained the orthogonal tRNA/aminoacyl-tRNA synthetase specific to L-Anap33. The pANAP plasmid (~40 nL of 200 ng/μl) was injected into the Xenopus oocyte nucleus. Then, L-Anap (~50 nL of 1 mM) and channel mRNA were injected separately into the oocyte cytosol.
Channel currents were recorded from the oocytes 2 to 4 days after injection using the inside-out configuration of the patch-clamp technique with an EPC-10 (HEKA Elektronik, Germany) or Axopatch 200B (Axon Instruments, Union City, CA) patch-clamp amplifier and PATCHMASTER software (HEKA Elektronik). Inside-out patch-clamp recordings were made approximately 5 mins after patch excision, when the run-down of HCN channels due to PI(4,5)P2 depletion was almost complete55. Borosilicate patch electrodes were made using a P97 micropipette puller (Sutter Instrument, Novato, CA). The initial pipette resistances were 0.3–0.7 MΩ. Recordings were made at 22° C to 24° C. A μFlow microvolume perfusion system (ALA Scientific Instruments, Farmingdale, NY) was used to change solutions during the experiment. All recordings were made in the presence of 1 mM cAMP on the cytosolic side of the patch. For oocyte patch-clamp recording and Co2+-HH tmFRET experiments, the standard bath and pipette saline solutions contained 130 mM KCl, 10 mM HEPES, 0.2 mM EDTA, pH 7.2. 1 mM CoSO4 was added to the perfusion solution with EDTA eliminated. For ACCuRET experiments, stabilization buffer (SBT) (130 mM KCl; 30 mM TRIS; 0.2 mM EDTA, pH 7.4) was used for the pipette and the bath solution. The Cu2+-TETAC was prepared as previously described40. TETAC (Toronto Research Chemicals, Toronto, Canada) was made as a 100 mM stock in DMSO and stored at −20°C. On the day of the experiment, 1 μL each of 100 mM TETAC stock and 110 mM CuSO4 stock were mixed together and incubated for one minute until the solution turned to a deeper shade of blue, indicating binding of Cu2+ to the TETAC. To this mixture, 98 μL of SBT was added, giving a solution of 1.1 mM Cu2+ and 1 mM TETAC. The 10% over-abundance of Cu2+ ensured that all the TETAC was bound with Cu2+. This solution was then diluted 1:100 in SBT buffer, giving a final concentration of 10 μM Cu2+-TETAC, and perfused on to the patch for 1 minute before fluorescence measurements.
Fluorescence Measurements
Patch-clamp fluorometry (PCF) simultaneously records fluorescence and current signals in inside-out patches. Our PCF experiments were performed using a Nikon Eclipse TE2000-E inverted microscope with a 60X 1.2 NA water immersion objective. Epifluorescence recording of L-Anap was performed with wide-field excitation using a Lambda LS Xenon Arc lamp (Sutter Instruments) and a filter cube containing a 376/30 nm excitation filter and a 460/50 nm emission filter. A 425 nm long-pass emission filter was used for the spectral measurement of L-Anap. YFP was measured with a filter cube containing a 490/10 nm excitation filter and a 535/30 nm emission filter. Images were collected with a 200 ms exposure using an Evolve 512 EMCCD camera (Photometrics, Tucson, AZ) and MetaMorph software (Molecular Devices, Sunnyvale, CA). For the ACCuRET experiments, the steady-state Anap fluorescence at −100 mV was captured a few seconds after switching the voltage from 0 mV to −100 mV. For comparison of the Anap and YFP fluorescence intensities, the settings of the EMCCD camera were kept the same.
For spectral measurements, images were collected by a spectrograph (Model: 2150i, 300 g/mm grating, blaze = 500 nm; Acton research, Acton, MA) mounted between the output port of the microscope and the Evolve 512 EMCCD camera. The membrane patch was positioned in the entrance slit of the spectrograph with the pipette parallel to the slit. The slit width was made slightly smaller than the width of the patch to block light from regions of the patch attached to the glass. The slit width we used had little or no effect on the shape of the emission spectrum (Supplementary Fig. 8b). The spectrograph produced an image on the camera where the vertical dimension was positioned along the axis of the pipette, and the horizontal dimension was wavelength. The wavelength scale was calibrated with known filters and laser lines. Anap spectra were recorded by measuring line-scans through the patch area, background subtracted using line scans through the non-fluorescent region in the pipette outside of the patch, and corrected for the filter characteristics of the spectrograph.
Fluorometry experiments on free L-Anap in solvents were performed using a Jobin Yvon Horiba FluoroMax-3 spectrofluorometer (Edison, NJ). Absorption spectra were recorded with a DU 800 spectrophotometer (Beckman Coulter, Brea, CA). Sub-micro fluorometer cuvettes (100 μL Starna, Atascadero, CA) were used for both fluorometry and spectrophotometry. For emission spectra of free L-Anap-ME (AsisChem), an excitation wavelength of 350 nm and 1 nm slits for excitation and emission were used.
Rosetta Modeling
A homology model of the spHCN channel based on the human HCN1 structure19 (PDB 5U6O) was created using RosettaCM without any experimental constraints56,57. The spHCN homology model and hHCN1 structure were very similar (RMSD of α carbons for the HCN domain and VSD was 0.8 Å). To obtain models of the spHCN channel at 0 mV and −100 mV, the structures were refined using RosettaCM with three types of constraints applied to the homology model: (1) Coordinate constraints (tethering the Cartesian coordinates of an atom) were applied on every non-hydrogen atom except for those in the S4 helix. These were “top-out” constraints, which are roughly harmonic up to 1 Å from the minimum, and flat beyond; (2) harmonic pairwise distance restraints were applied on all backbone hydrogen bonding groups; and (3) experimentally-derived distances at 0 mV (State 1) and −100 mV (State 2) based on our tmFRET measurements were applied as “flat-bottom” harmonic constraints, where the constraint energies were zero in a 1 Å window centered on the measured distance, and harmonic beyond this. This constraint roughly corresponds to the estimated resolution of our tmFRET measurements. For setting the constraint weights for a satisfactory model, we performed a grid search of the weights on each of the three constraint types. The weakest overall weights that satisfied the experimental constraints in both states were used: a weight of 15 kcal/mol/Å2 on the experimental data, and a weight of 2 kcal/mol/Å2 on the hydrogen-bond and coordinate constraints. To better establish the robustness of our modeling procedure, we varied the weight on the experimental data at −100 mV in 11 increments ranging from 7.5 to 30 kcal/mol/Å2 and show that the final model is relatively insensitive to the experimental weights in this range (see Supplementary Fig. 7). All modeling was carried out starting with the complete spHCN tetrameric homology model with C4 symmetry imposed. Since only FRET constraints between the HCN domain and S4 helix were determined experimentally, we only expect to accurately measure the rearrangements in those regions. Therefore, we imposed coordinate constraints outside the S4 helix to resist movements and, hence, did not observe significant movements in other regions. This yielded a voltage-sensor activated but closed-pore conformation, which is compatible with the weak coupling observed in these channels36. No manual adjustment or refinement was used. The model at 0 mV (State 1) was highly similar to the homology model, while the model at −100 mV (State 2) showed a large downward and tilting rearrangement of the S4 (Fig. 5). Rosetta scripts are available in Supplementary Data Set 1. The coordinates for the models were deposited at PDB-dev with accession code: PDBDEV_00000032. A structural morph was created between the State 1 and State 2 models (Supplementary Video 3, 4 and 5), highlighting the hyperpolarization-dependent movement of the S4. Structural representations and morphs were created using the PyMOL software (https://pymol.org).
Data Analysis
Data were analyzed using IgorPro (Wavemetrics) and Image J (NIH).
The conductance-voltage (G-V) relationships were measured from the instantaneous tail currents at +40 mV following voltage pulses from 0 mV to between 0 and −110 mV. The leak tail currents following pulses to +40 mV were subtracted, and the currents were normalized to the maximum tail current following voltage pulses to −110 mV (G/Gmax). The relative conductances were plotted as a function of the voltage of the main pulse and fitted with a Boltzmann equation:
where V is the membrane potential, V1/2 is the potential for half-maximal activation, and Vs is the slope factor.
Fluorescence images from the membrane patches were imported into ImageJ58 for analysis. Regions of interest were drawn by hand around the dome of the patch, excluding any regions that appeared attached to the glass (c.f. Fig. 1d). For each patch, a background region was selected in the pipette outside the area of the patch. The mean gray value of the background region of interest was subtracted from the mean gray value of the region of interest of the patch to give the mean fluorescence intensity (fl).
The FRET efficiency was calculated using the following equations (E1) as previously described32:
| E1 |
Where FHH and Fno HH are the fractions of fluorescence that are unquenched by Co2+ in channels with and without HH sites, respectively, e.g. . Similarly, FCys and Fno Cys are the fractions of fluorescence that are unquenched by Cu2+-TETAC in channels with and without cysteines respectively. These equations for calculating FRET efficiency assume that the decrease in Fno HH is due to nonspecific energy transfer such as solution quenching or FRET to a metal ion bound to an endogenous metal binding site.
Alternatively, the FRET efficiency was calculated using the following equations (E2), which only account for nonspecific decreases in fluorescence (e.g. photobleaching) but not energy transfer.
| E2 |
The FRET efficiencies produced by E1 and E2 were similar because of the low degree of background quenching32,40 (Supplementary Fig. 6). For this paper we primarily used E1 to calculate the FRET efficiencies. To calculate the mean and standard error of the mean for the FRET efficiency, we used the mean and standard error of the mean for our fractional quenching measurements (i.e. FHH, Fno HH, FCys, and Fno Cys) in Monte Carlo resampling (1 × 106 cycles; NIST Uncertainty Machine v1.3.4; https://uncertainty.nist.gov).
The R0 values, the distances that predict 50% energy transfer, were calculated using the established equation as previously published40.
Where C is the scaling factor, J is the overlap integral of the donor emission spectrum and the acceptor absorption spectrum, Q is the quantum yield of the donor, η is the index of refraction, and κ2 is the orientation factor. η was assumed to be 1.33, and κ2 was assumed to be 2/3, a reasonable assumption for an isotropic metal ion37,38,59. Assuming κ2 = 2/3 in the case of an isotropic acceptor limits the worst-case error in R0 to −11% to +12%37,59. The J values for each L-Anap site at 0 mV and −100 mV were calculated using the emission spectrum of each site at each voltage (Supplementary Fig. 3) and the absorption spectrum of Cu2+-TETAC in solution. Since the shape of the Anap emission spectrum does not change appreciably in different solvents (Supplementary Fig. 8a inset), we shifted the free L-Anap spectrum measured in the cuvette to match the peak of the emission spectra measured on the microscope for each Anap site at 0 mV and −100 mV (Supplementary Fig. 3). This strategy removes the contamination of the Anap emission spectrum by the YFP emission peaking at around 530 nm. The YFP emission is most likely due to the direct excitation of YFP by the light used to excite Anap, although the possibility of a small amount of FRET between Anap and YFP cannot be completely excluded. Finally, we estimated the quantum yield Q of L-Anap at each site at 0 mV or −100 mV. The relative quantum yields of L-Anap at each site at 0 mV and −100 mV were estimated from the slopes of the plots of Anap intensity vs. YFP intensity at 0 mV (Supplementary Fig. 2) and the relative brightness at −100 mV vs. 0 mV (flAnap, −100 mV / flAnap, 0 mV) (Fig. 2c). For each site, Q0mV = 0.3 * Slope (Anap vs YFP) (Supplementary Fig. 2 a–e) and Q-100mV = Q0mV * (flAnap, −100 mV / flAnap, 0 mV). A scaling factor of 0.3 was used for converting the slopes to quantum yields to make the quantum yields fall in the range of the quantum yields for Anap in different solvents (0.2–0.5)29,40. This estimate of quantum yield assumes that the change in brightness of Anap is primarily due to a change in quantum yield and not extinction coefficient, as appears to be true for the solvents that we tested (Supplementary Fig. 8c). R0 values calculated using these estimated spectral overlaps and quantum yields were as follows: 13.1 Å (0 mV) and 14.9 Å (−100 mV) for S346Anap; 12.6 Å (0 mV) and 13.5 Å (−100 mV) for L348Anap; 13.8 Å (0 mV) and 14.1 Å (−100 mV) for S353Anap; 13.3 Å (0 mV) and 13.0 Å (−100 mV) for W355Anap; and 13.9 Å (0 mV) and 13.9 Å (−100 mV) for W218Anap.
Statistics and Reproducibility
Data parameters were expressed as mean ± s.e.m. of n independent patches. Statistical significance (*p < 0.05) was determined by using a two-tailed Student’s t-test.
REPORTING SUMMARY STATEMENT
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
CODE AVAILABILITY STATEMENT
Rosetta scripts used for the modeling are in Supplementary Data Set 1.
DATA AVAILABILITY STATEMENT
The coordinate files of the State 1 and State 2 models were deposited in PDB-dev (https://pdb-dev.wwpdb.org/) with accession codePDBDEV_00000032. Source data for Fig. 3, 4 and Supplementary Fig. 6 are available online. Other data are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ximena Optiz-Araya and Sarie Haraguchi for animal care and surgical support; Sharona E. Gordon, Lucie Delemotte, and all members of the Zagotta laboratory for their helpful advice and support, Dan Farrell for help with the PDB-dev submission, and Lesley Anson for comments on the manuscript. This work was funded by NIH Grants R01EY010329, R01MH102378, R01GM125351, and American Heart Association Award 14CSA20380095 (to W.N.Z.) and NIH Grant F32NS077622 (to T. K. A.).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Hille B Ion Channels of Excitable Membranes 3rd edn, (Sinauer Associates, 2001). [Google Scholar]
- 2.Armstrong CM & Hille B Voltage-gated ion channels and electrical excitability. Neuron 20, 371–380 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Long SB, Campbell EB & Mackinnon R Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science 309, 903–908, doi: 10.1126/science.1116270 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Stühmer W et al. Structural parts involved in activation and inactivation of the sodium channel. Nature 339, 597–603, doi: 10.1038/339597a0 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Papazian DM, Timpe LC, Jan YN & Jan LY Alteration of voltage-dependence of Shaker potassium channel by mutations in the S4 sequence. Nature 349, 305–310, doi: 10.1038/349305a0 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Yang N & Horn R Evidence for voltage-dependent S4 movement in sodium channels. Neuron 15, 213–218 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Larsson HP, Baker OS, Dhillon DS & Isacoff EY Transmembrane movement of the shaker K+ channel S4. Neuron 16, 387–397 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Mannikko R, Elinder F & Larsson HP Voltage-sensing mechanism is conserved among ion channels gated by opposite voltages. Nature 419, 837–841, doi: 10.1038/nature01038 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Catterall WA, Wisedchaisri G & Zheng N The chemical basis for electrical signaling. Nat Chem Biol 13, 455–463, doi: 10.1038/nchembio.2353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tao X, Lee A, Limapichat W, Dougherty DA & MacKinnon R A gating charge transfer center in voltage sensors. Science 328, 67–73, doi: 10.1126/science.1185954 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long SB, Tao X, Campbell EB & MacKinnon R Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382, doi: 10.1038/nature06265 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Clairfeuille T et al. Structural basis of alpha-scorpion toxin action on Nav channels. Science 363, doi: 10.1126/science.aav8573 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Xu H et al. Structural Basis of Nav1.7 Inhibition by a Gating-Modifier Spider Toxin. Cell 176, 1238–1239, doi: 10.1016/j.cell.2019.01.047 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Guo J et al. Structure of the voltage-gated two-pore channel TPC1 from Arabidopsis thaliana. Nature 531, 196–201, doi: 10.1038/nature16446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biel M, Wahl-Schott C, Michalakis S & Zong X Hyperpolarization-activated cation channels: from genes to function. Physiol Rev 89, 847–885, doi: 10.1152/physrev.00029.2008 (2009). [DOI] [PubMed] [Google Scholar]
- 16.DiFrancesco D Characterization of single pacemaker channels in cardiac sino-atrial node cells. Nature 324, 470–473, doi: 10.1038/324470a0 (1986). [DOI] [PubMed] [Google Scholar]
- 17.Santoro B et al. Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell 93, 717–729 (1998). [DOI] [PubMed] [Google Scholar]
- 18.Ludwig A, Zong X, Jeglitsch M, Hofmann F & Biel M A family of hyperpolarization-activated mammalian cation channels. Nature 393, 587–591, doi: 10.1038/31255 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Lee CH & MacKinnon R Structures of the Human HCN1 Hyperpolarization-Activated Channel. Cell 168, 111–120 e111, doi: 10.1016/j.cell.2016.12.023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James ZM & Zagotta WN Structural insights into the mechanisms of CNBD channel function. J Gen Physiol 150, 225–244, doi: 10.1085/jgp.201711898 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vemana S, Pandey S & Larsson HP S4 movement in a mammalian HCN channel. J Gen Physiol 123, 21–32, doi: 10.1085/jgp.200308916 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cowgill J et al. Bipolar switching by HCN voltage sensor underlies hyperpolarization activation. Proc Natl Acad Sci U S A, doi: 10.1073/pnas.1816724116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flynn GE & Zagotta WN Insights into the molecular mechanism for hyperpolarization-dependent activation of HCN channels. Proc Natl Acad Sci U S A 115, E8086–E8095, doi: 10.1073/pnas.1805596115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gauss R, Seifert R & Kaupp UB Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature 393, 583–587, doi: 10.1038/31248 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Flynn GE, Black KD, Islas LD, Sankaran B & Zagotta WN Structure and rearrangements in the carboxy-terminal region of SpIH channels. Structure 15, 671–682, doi: 10.1016/j.str.2007.04.008 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin KS, Maertens C, Proenza C, Rothberg BS & Yellen G Inactivation in HCN channels results from reclosure of the activation gate: desensitization to voltage. Neuron 41, 737–744 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Taraska JW, Puljung MC, Olivier NB, Flynn GE & Zagotta WN Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat Methods 6, 532–537, doi: 10.1038/nmeth.1341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordon SE, Senning EN, Aman TK & Zagotta WN Transition metal ion FRET to measure short-range distances at the intracellular surface of the plasma membrane. J Gen Physiol 147, 189–200, doi: 10.1085/jgp.201511530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zagotta WN, Gordon MT, Senning EN, Munari MA & Gordon SE Measuring distances between TRPV1 and the plasma membrane using a noncanonical amino acid and transition metal ion FRET. J Gen Physiol 147, 201–216, doi: 10.1085/jgp.201511531 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aman TK, Gordon SE & Zagotta WN Regulation of CNGA1 Channel Gating by Interactions with the Membrane. J Biol Chem 291, 9939–9947, doi: 10.1074/jbc.M116.723932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai G, James ZM & Zagotta WN Dynamic rearrangement of the intrinsic ligand regulates KCNH potassium channels. J Gen Physiol 150, 625–635, doi: 10.1085/jgp.201711989 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai G & Zagotta WN Molecular mechanism of voltage-dependent potentiation of KCNH potassium channels. Elife 6, doi: 10.7554/eLife.26355 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee A, Guo J, Lee HS & Schultz PG A genetically encoded fluorescent probe in mammalian cells. J Am Chem Soc 135, 12540–12543, doi: 10.1021/ja4059553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalstrup T & Blunck R Dynamics of internal pore opening in Kv channels probed by a fluorescent unnatural amino acid. Proc Natl Acad Sci U S A 110, 8272–8277, doi: 10.1073/pnas.1220398110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J & Zagotta WN Patch-clamp fluorometry recording of conformational rearrangements of ion channels. Sci STKE 2003, PL7, doi: 10.1126/stke.2003.176.pl7 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Ryu S & Yellen G Charge movement in gating-locked HCN channels reveals weak coupling of voltage sensors and gate. J Gen Physiol 140, 469–479, doi: 10.1085/jgp.201210850 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stryer L Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem 47, 819–846, doi: 10.1146/annurev.bi.47.070178.004131 (1978). [DOI] [PubMed] [Google Scholar]
- 38.Taraska JW, Puljung MC & Zagotta WN Short-distance probes for protein backbone structure based on energy transfer between bimane and transition metal ions. Proc Natl Acad Sci U S A 106, 16227–16232, doi: 10.1073/pnas.0905207106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruening-Wright A, Elinder F & Larsson HP Kinetic relationship between the voltage sensor and the activation gate in spHCN channels. J Gen Physiol 130, 71–81, doi: 10.1085/jgp.200709769 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gordon SE, Munari M & Zagotta WN Visualizing conformational dynamics of proteins in solution and at the cell membrane. Elife 7, doi: 10.7554/eLife.37248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kodama M & Kimura E Equilibria and kinetics of complex formation between zinc(II), lead(II), and cadmium(II), and 12-, 13-, 14-, and 15-membered macrocyclic tetra-amines. Journal of the Chemical Society, Dalton Transactions, 2269–2276, doi: 10.1039/DT9770002269 (1977). [DOI] [Google Scholar]
- 42.Polyhach Y & Jeschke G Prediction of favourable sites for spin labelling of proteins. Spectrosc-Int J 24, 651–659, doi: 10.3233/Spe-2010-0490 (2010). [DOI] [Google Scholar]
- 43.Jeschke G DEER distance measurements on proteins. Annu Rev Phys Chem 63, 419–446, doi: 10.1146/annurev-physchem-032511-143716 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Vieira-Pires RS & Morais-Cabral JH 310 helices in channels and other membrane proteins. J Gen Physiol 136, 585–592, doi: 10.1085/jgp.201010508 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorinczi E et al. Voltage-dependent gating of KCNH potassium channels lacking a covalent link between voltage-sensing and pore domains. Nat Commun 6, 6672, doi: 10.1038/ncomms7672 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomczak AP et al. A new mechanism of voltage-dependent gating exposed by KV10.1 channels interrupted between voltage sensor and pore. J Gen Physiol 149, 577–593, doi: 10.1085/jgp.201611742 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cha A, Snyder GE, Selvin PR & Bezanilla F Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature 402, 809–813, doi: 10.1038/45552 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Glauner KS, Mannuzzu LM, Gandhi CS & Isacoff EY Spectroscopic mapping of voltage sensor movement in the Shaker potassium channel. Nature 402, 813–817, doi: 10.1038/45561 (1999). [DOI] [PubMed] [Google Scholar]
- 49.Bell DC, Yao H, Saenger RC, Riley JH & Siegelbaum SA Changes in local S4 environment provide a voltage-sensing mechanism for mammalian hyperpolarization-activated HCN channels. J Gen Physiol 123, 5–19, doi: 10.1085/jgp.200308918 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Posson DJ, Ge P, Miller C, Bezanilla F & Selvin PR Small vertical movement of a K+ channel voltage sensor measured with luminescence energy transfer. Nature 436, 848–851, doi: 10.1038/nature03819 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chanda B, Asamoah OK, Blunck R, Roux B & Bezanilla F Gating charge displacement in voltage-gated ion channels involves limited transmembrane movement. Nature 436, 852–856, doi: 10.1038/nature03888 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Kintzer AF et al. Structural basis for activation of voltage sensor domains in an ion channel TPC1. Proc Natl Acad Sci U S A 115, E9095–E9104, doi: 10.1073/pnas.1805651115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.She J et al. Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature 556, 130–134, doi: 10.1038/nature26139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS-ONLY REFERENCES
- 54.Varnum MD, Black KD & Zagotta WN Molecular mechanism for ligand discrimination of cyclic nucleotide-gated channels. Neuron 15, 619–625 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Pian P, Bucchi A, Decostanzo A, Robinson RB & Siegelbaum SA Modulation of cyclic nucleotide-regulated HCN channels by PIP2 and receptors coupled to phospholipase C. Pflugers Arch 455, 125–145, doi: 10.1007/s00424-007-0295-2 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Rohl CA, Strauss CE, Misura KM & Baker D Protein structure prediction using Rosetta. Methods Enzymol 383, 66–93, doi: 10.1016/S0076-6879(04)83004-0 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Song Y et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742, doi: 10.1016/j.str.2013.08.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Selvin PR Principles and biophysical applications of lanthanide-based probes. Annu Rev Biophys Biomol Struct 31, 275–302, doi: 10.1146/annurev.biophys.31.101101.140927 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinate files of the State 1 and State 2 models were deposited in PDB-dev (https://pdb-dev.wwpdb.org/) with accession codePDBDEV_00000032. Source data for Fig. 3, 4 and Supplementary Fig. 6 are available online. Other data are available from the corresponding author upon reasonable request.