Abstract
Cancer is associated with a number of conditions such as hypoxia, nutrient deprivation, cellular redox, and pH changes that result in accumulation of unfolded or misfolded proteins in the endoplasmic reticulum (ER) and trigger a stress response known as the unfolded protein response (UPR). The UPR is a conserved cellular survival mechanism mediated by the ER transmembrane proteins activating transcription factor 6, protein kinase-like endoplasmic reticulum kinase, and inositol-requiring enzyme 1α (IRE1α) that act to resolve ER stress and promote cell survival. IRE1α is a kinase/endoribonuclease (RNase) with multiple activities including unconventional splicing of the messenger RNA (mRNA) for the transcription factor X-Box Binding Protein 1 (XBP1), degradation of other mRNAs in a process called regulated IRE1α-dependent decay (RIDD) and activation of a pathway leading to c-Jun N-terminal kinase phosphorylation. Each of these outputs plays a role in the adaptive and cell death responses to ER stress. Many studies indicate an important role for XBP1 and RIDD functions in cancer and new studies suggest that these two functions of the IRE1α RNase can have opposing functions in the early and later stages of cancer pathogenesis. Finally, as more is learned about the contextdependent role of IRE1α in cancer development, specific small molecule inhibitors and activators of IRE1α could play an important role in counteracting the protective shield provided by ER stress signaling in cancer cells.
Keywords: endoplasmic reticulum stress, inositol-requiring enzyme 1α, regulated IRE1α-dependent decay, unfolded protein response, X-Box Binding Protein 1
1 |. INTRODUCTION
The unfolded protein response (UPR) is a complex and wide-reaching adaptive mechanism that protects the cell from a variety of stresses such as an imbalance of endoplasmic reticulum (ER) calcium levels, hypoxia, redox imbalance, glucose depletion, or viral infection.1 These events disrupt the protein folding homeostasis of the ER and lead to a buildup of misfolded proteins that are damaging to the cell if not rectified. ER stress can be alleviated, and homeostasis restored by the signaling effects of three key ER transmembrane proteins: inositol-requiring enzyme 1 (IRE1α), activating transcription factor 6 (ATF6), and RNA activated protein kinase-like endoplasmic reticulum kinase (PERK). The abnormal conditions that contribute to ER stress are also found extensively in cancerous cells due to the intrinsic nature of tumors: rapid rate of proliferation, high consumption of glucose, and dense hypoxic environment. Therefore, the UPR is commonly activated in tumors and plays an important role in tumor development. In this review, we will discuss the role of the UPR in cancer focusing on IRE1α and the outputs of its RNase: X-Box Binding Protein 1 (XBP1) messenger RNA (mRNA) splicing and regulated IRE1α dependent decay (RIDD) of RNA.
2 |. OVERVIEW OF THE UPR
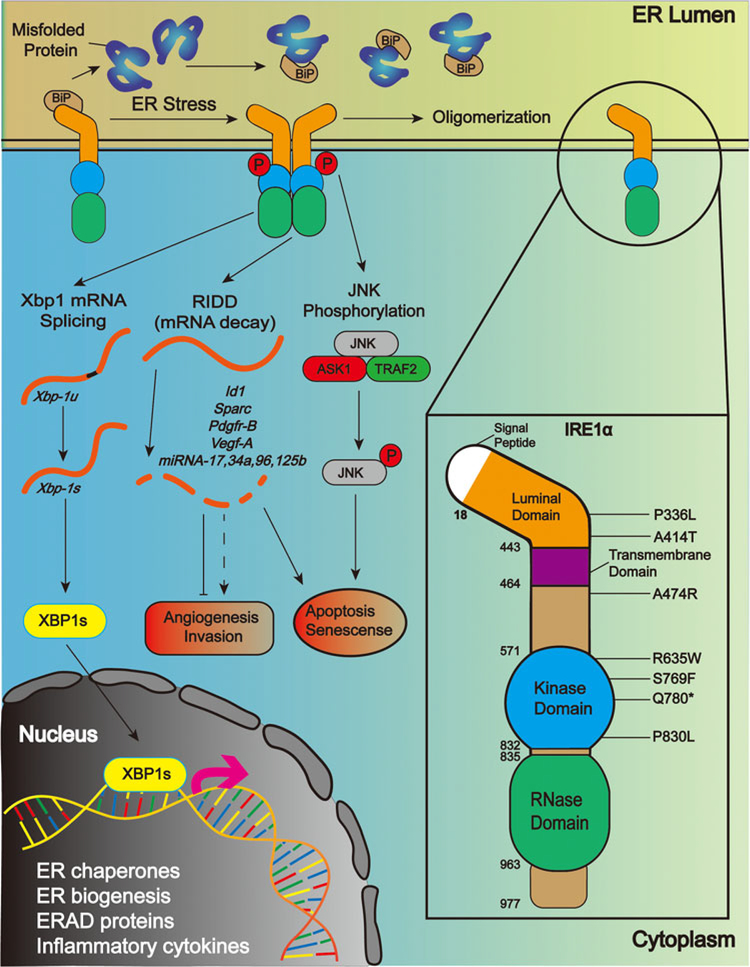
The ER transmembrane mediators of the UPR are normally held in an inactive conformation by the ER luminal stress sensor protein BiP, also known as GRP78. BiP has preferential binding for the hydrophobic residues found on misfolded proteins, and so in the presence of misfolded proteins, BiP detaches from the luminal domains of IRE1α, ATF6, and PERK and attaches instead to misfolded proteins, releasing these signaling proteins and triggering their activation.2,3 ATF6, a transcription factor, translocates to the Golgi apparatus where it is cleaved into the smaller, transcriptionally active form.4 Active ATF6 induces expression of genes involved in protein folding and the recycling of misfolded proteins, a process named endoplasmic reticulum-associated degradation (ERAD).5 PERK is a kinase that is activated by homodimerization and autophosphorylation.6 It functions to reduce strain on protein folding mechanisms by inhibiting global protein translation through phosphorylation of the translation initiation factor eIF2α, although selective mRNA translation such as that for the transcription factor ATF4 is retained because this is important in upregulation of ER chaperones.7 Activation of IRE1α also involves dimerization and trans-autophosphorylation, although it may form higher order oligomeric complexes with altered activity under the conditions of unmitigated ER stress.8 IRE1α contains both kinase and endoribonuclease domains, and activation of the kinase is required for RNase activity (Figure 1). The most highly conserved IRE1α RNase activity is the unconventional splicing of mRNA for the important UPR transcription factor XBP1. IRE1α removes a 26-nucleotide sequence from the XBP1 mRNA, causing a frameshift that allows translation of the mature transcription factor. The RNase domain of IRE1α can also cleave and degrade multiple mRNA and microRNA (miRNA) targets in a process called RIDD. IRE1α plays an important role in the adaptive and proapoptotic effects of UPR signaling. Although PERK is mainly responsible for inhibition of global protein translation, IRE1α RIDD activity plays a supporting role by selectively degrading mRNA transcripts encoding large, multidomain or transmembrane proteins as these complex proteins are the most taxing on the ER to fold and transport and so are terminated before translation to prevent further strain to the ER.9 In addition, a range of prosurvival effects are triggered by XBP1 to adapt the cell to unwanted stress conditions and rectify the underlying issues causing stress. XBP1 upregulates the protein folding capacity of the cell by increasing lipid production and the size of the ER10 to raise the capacity of ER luminal contents, including protein folding11 and ERAD machinery.5
FIGURE 1.
IRE1α structure, activation and potential roles in cancer. IRE1α is an ER transmembrane protein containing both protein kinase and RNase domains on its cytoplasmic side. Schematic of IRE1α activation in response to the accumulation of unfolded proteins in the ER by dimerization and trans/autophosphorylation (left). Signaling from IRE1α includes activation of the JNK pathway, splicing of the mRNA for XBP1 and targeted degradation of specific RNA in a process called regulated IRE1-dependent decay or RIDD. Schematic of IRE1α domain structure and the location of specific mutations that are found in human cancers and described in the manuscript text (right, not to scale). ER, endoplasmic reticulum; IRE1α, inositol-requiring enzyme 1α; JNK, c-Jun N-terminal kinase; mRNA, messenger RNA; RIDD, regulated IRE1-dependent decay; XPB1, X-Box Binding Protein 1 [Color figure can be viewed at wileyonlinelibrary.com]
The prosurvival steps carried out by XBP1 and other UPR transcription factors may be sufficient for misfolded proteins to be cleared and ER homeostasis to be restored. In this case, the UPR can be halted by the dephosphorylation12 and monomerization of IRE1α and reattachment of BiP to lock it in its monomeric form.13 The molecular details of the attenuation of ATF6 and PERK signaling are not fully understood. However, if misfolded proteins remain after the initial prosurvival phase of UPR signaling then an apoptotic phase of the UPR is triggered. This is mediated primarily by IRE1α through three distinct mechanisms. The first is a pathway leading to phosphorylation and activation of JNK and its downstream proapoptotic signaling.14 Second, RIDD is a driver of apoptosis through the degradation of miRNA downregulating caspase-2 and induction of apoptosis through the BAX/BAK-dependent pathway.15,16 Third, one of the downstream targets of XBP1 is the transcription factor C/EBP homologous protein (CHOP), a regulator of a wide range of apoptosis-related proteins. Amongst others, CHOP downregulates the antiapoptotic protein BCL-217 and upregulates proapoptotic proteins such as GADD3418 and BAX.17 CHOP expression is also increased by the PERK pathway via its primary downstream transcription factor ATF4, further enhancing the proapoptotic effect of late-stage UPR signaling.19
Unlike a conventional UPR where high unresolvable stress leads to cell death, in cancer cells the prosurvival effects of the UPR predominate to shield cells from tumor suppression mechanisms. If the apoptotic stage of the UPR is avoided by various mechanisms such as upregulation of the CHOP suppressor p58IPK,20 cancerous cells can exploit the prosurvival aspects of the UPR unhindered to resist the hostile environment inside a growing tumor. Upregulation of components of the UPR, such as BiP or the IRE1α-XBP1 pathway, occurs in many cancers and is linked to increased chemoresistance, aggressiveness, and poor outcomes for patients.21–24
More recent studies show that ER stress and the UPR are transmissible between cells via extracellular vesicles.25 This expands on the previous discovery of the transmission of hypoxia resistance between cancerous cells26 and the wider concept of extracellular vesicles as a method of communication within the tumor micro-environment.27 These vesicles have been shown to transmit mRNA, miRNA and functional proteins, as well as misfolded proteins that disrupt the ER homeostasis of the recipient cell. This discovery, therefore, highlights the crucial nature of the UPR in tumorigenesis of individual cells, as well as in field cancerization.
3 |. IRE1α EXPRESSION IN HUMAN CANCER
Changes in IRE1α expression have been observed in several cancer types but there is as yet no consistent pattern. Increased expression of IRE1α has been found in glioblastoma and is linked to reduced survival.28 In contrast, higher expression of IRE1α in surgically resected lung adenocarcinoma was associated with lower recurrence rates, lower clinical stage, and minimal vascular invasion.29 In CT26- BALB/c and B16-C57BL/6 tumor-bearing mice, a low-protein diet activated the IRE1α arm of the UPR leading to reduced tumor burden. Increased IRE1α activity and subsequent activation of RIDD, as measured by increased expression of RIG1, enhanced recruitment of CD8-positive cells through induction of IFNγ and CXCL10, thus initiating antitumor surveillance response in these xenograft models.30 Downregulation of IRE1α was also reported to drive tumorigenesis. Epigenetic inactivation of IRE1α by histone methyltransferase enhancer of zeste homolog 2 in germinal center B-cell-like diffuse large B-cell lymphoma subtype regulated the IRE1α-XBP1 axis to accelerate tumor growth in mouse xenografts. DNABJ9, a direct target of spliced XBP1, was implicated in arresting differentiation leading to cellular proliferation as well as tumor growth in the HBL1 mouse xenograft models.31 Analysis of The Cancer Genome Atlas (TCGA) using UALCAN, a portal that allows survival analysis by tumor subgroup gene expression32 found a correlation between elevated IRE1α expression as defined by the TPM values in the upper quartile in bladder urothelial carcinoma, brain lower grade glioma, cervical squamous cell carcinoma, renal clear cell carcinoma, and mesothelioma. However, analysis of the median IRE1α gene expression in TCGA using the analysis tool GEPIA,33 shows lower IRE1α expression across multiple cancer types compared with their paired normal samples with an exception of acute myeloid leukemia, esophageal cancer, cholangiocarcinoma, and glioblastoma (SM and ABG unpublished). The exact significance of higher or lower IRE1α expression levels in human cancer remains to be determined in part because IRE1α activation as measured by autophosphorylation may be independent of expression. Furthermore, the potential for opposing functions of XBP1 splicing and RIDD outputs of the IRE1α RNase in cancer pathogenesis make this analysis complex. This complexity is further increased by observations that targeted deletion of Xbp1 in mouse models leads to hyperactivation of IRE1α and RIDD34 suggesting that there may be interactions between these two outputs of the IRE1α RNase that are yet to be understood.
4 |. IRE1α OUTPUTS AND CANCER: XBP1
Given the role of XBP1 in the adaptive response to ER stress, it is not surprising that increased expression of spliced XBP1 mRNA is observed in several cancers such as breast cancer, hepatocellular carcinoma, lymphoma, and multiple myeloma (MM).35 Although the expression is upregulated in such a broad range of cancers, the overall outcome of XBP1 signaling can be either oncogenic or suppressive. The published mechanisms by which XBP1 is thought to affect the tumor microenvironment also vary greatly. For instance, in triple-negative breast cancer (TNBC) cell line, knockdown of XBP1 with siRNA decreased growth in vitro, and reduced invasiveness and relapse in xenografts due to lack of angiogenesis, rather than defects in proliferation and apoptosis.36 In these TNBC cells, an XBP1-hypoxia-inducible factor 1α (HIF1α) complex recruited RNA polymerase II to HIF1α target genes, critical for the response to hypoxic stress. Thus, in TNBC cells, overexpression of XBP1 promotes an adaptive response to both ER and hypoxic stress thereby enhancing survival of TNBC cells and decreasing patient survival.36,37 A similar link between HIF1α and XBP1 was shown with XBP1 knockdown in MEF and human fibrosarcoma cells which caused reduced blood vessel formation and tumor growth rate.38,39 In a different study of breast cancer, overexpression of lysyl oxidase-like 2 triggered epithelial-mesenchymal transition (EMT) in basal-like breast carcinoma cell lines through the activation of IRE1α and subsequent XBP1 splicing. In these cells, XBP1 was shown to directly induce several EMT-transcription factors such as SNAI1, ZEB2, and TCF3.40 In MM, however, increased levels of the IRE1α-XBP1 pathway are a survival mechanism that occurs in response to the stress of elevated antibody production and secretion. In several studies of MM patients, hyperactivation of IRE1α-XBP1 signaling, or a high ratio spliced to unspliced XBP1 mRNA was again linked to poor prognosis.41,42 Furthermore, in a mouse model of MM overexpression of XBP1 in B cells and plasma cells promoted B cell malignant transformation.43 Treatment with small molecules IRE1α RNase domain inhibitors, such as STF-083010 and 4μ8c, or inhibiting general RNA splicing with toyocamycin significantly impeded MM tumor progression.44,45
In contrast to these studies, Denoyelle and coworkers showed that in primary melanocytes mutant H-RasG12V induces the UPR and this activation lead to premature senescence.46 Knockdown of ATF4, ATF6, or XBP1 reduces premature senescence induced by oncogenic H-RasG12V suggesting that ER stress, including the IRE1α-XBP1 pathway, is a mechanism of tumor suppression in melanocytes.46 Furthermore, in the APCmin mouse model, mice with a deletion of XBP1 in intestinal epithelial cells are more susceptible to colorectal cancer through enhanced inflammation.47,48 This is reversed in compound XBP1/IRE1α null mice suggesting that a functional IRE1α is necessary for enhanced colon tumor formation caused by deletion of XBP1.47,48
5 |. IRE1α OUTPUTS AND CANCER: RIDD
While some consensus cleavage sites have been identified for mRNAs and miRNAs degraded by RIDD,49 the targets for this more promiscuous RNase activity may be cell and context specific as the effects will vary according to the nature of the transcript degraded. In in vitro organoid cultures as well as mouse xenograft models of colon cancer, knockdown of IRE1α reduces colon cancer by simultaneous suppression of β-catenin translation and RNase dependent activation of the PERK signaling.50 Similarly, expression of a dominant negative form of IRE1α in the U87 glioblastoma cell line reduced proliferation in vitro and tumor growth in xeno-grafts.51,52 This was associated with upregulation of extracellular matrix protein genes and enhanced expression of SPARC, an antiangiogenic protein and a target of RIDD, as well as down-regulation of proinflammatory and proangiogenic factors vascular endothelial growth factor A and interleukin-6 (IL-6). However, these cells also exhibited increased adhesion and migration on Matrigel through activation of RhoA.52 In TNBC cell lines IRE1α RNase activity is required for expression of protumorigenic factors such as IL-6, IL-8, CXCL1, and TGF-β, and contributes paclitaxel mediated expansion of tumor-initiating cells.53
Some of the tumor suppression activity of IRE1α may be mediated by its ability to trigger apoptotic signaling though RIDD regulation of specific microRNAs. For instance, sustained activation of IRE1α RIDD causes rapid degradation of several microRNAs (miR-17, miR-34a, miR-96, and miR-125b) that repress translation of caspases upstream of BAX/BAK-dependent apoptotic pathways. Degradation of the miRNAs induces expression of procaspase-2 and triggers caspase-2 activation and apoptosis.16 In the INS-1 insulinoma cell line, increased RIDD activity is associated with NLR-family pyrin domain containing 3 (NLRP3) inflammasome activation and inflammatory responses through degradation of miR-17 which normally blocks expression of the thioredoxin-interacting protein (TXNIP). Amplification or overexpression of IRE1α stabilizes TXNIP mRNA causing increased oxidative stress, NLRP3 inflammasome activation, and cell death via secretion of IL-1β and activation of the caspase-1 pathway.54
Recent studies examining both outputs of the IRE1α RNase suggest they could have distinct functions in cancer. We showed that in primary mouse keratinocytes oncogenic Ras causes transient activation of ER stress but this is followed by reduction of ER stress during Ras-driven senescence.55 Initially, oncogenic Ras caused increased expression and activation of IRE1α and XBP1 mRNA splicing that drove the Ras-induced proliferative response, whereas XBP1 knockdown accelerated senescence.Further analysis showed that IRE1α-RIDD function was critical for driving Ras-induced senescence and accelerated senescence in XBP1-deficient keratinocytes. We identified the prooncogenic transcription factor ID1 as a critical RIDD target whose degradation by IRE1α enhanced senescence in response to oncogenic Ras. Furthermore, under conditions of reduced ER stress and Xbp1 mRNA splicing, Id1 mRNA degradation is sustained.55 Thus, in this in vitro model of Ras-driven cancer the IRE1α-XBP1 splicing arm promotes while the IRE1α-RIDD arm suppresses survival and transformation of keratinocytes. Similarly, in a complex analysis of glioblastoma patients, Lhomond et al28 were able to classify cancers based on levels of XBP1 mRNA splicing and RIDD. Tumors with low XBP1 mRNA and high RIDD activity and had a neural phenotype and reduced tumor angiogenesis and invasiveness while tumors with high XBP1 mRNA splicing/low RIDD activity were more mesenchymal, aggressive and had enhanced tumor immune infiltration and angiogenesis.28 Survival was longer in patients with high RIDD and low XBP1. Taken together these studies provide strong evidence for divergent functions of the IRE1α RNase in cancer, although the specific context may be critical.
6 |. IRE1α MUTATIONS AND CANCER
The structure of human IRE1α (Figure 1) based on crystallographic analysis consists of a 367 amino acid N-terminal ER luminal domain organized into triangular β-sheet clusters.56 This region interacts with the ER stress sensor BiP and antiparallel β-sheet interactions between IRE1α luminal domains are required for dimerization and autophosphorylation after the release of BiP. The cytoplasmic region of IRE1α (aa465–977) contains a bi-lobal protein kinase fold (aa571–832) with the kinase activation segment from aa720–729 and a phosphorylation site for kinase activation at Ser724.57 The RNase domain is structurally continuous with the C-terminal lobe of the kinase domain followed by an extension of approximately 15 residues.57,58 During activation, face-to-face interaction of the cytoplasmic kinase domains allows for transphosphorylation and further stabilization of the active RNase domain.57
Analysis of TCGA data using cBioportal software59,60 reveals alterations in IRE1α in 3% of all human cancers (328 of 10950 samples) including amplification, deep deletions, truncating and missense mutations (SM ABG, unpublished). Somatic missense mutations are found throughout the luminal, kinase and RNase domains of IRE1α (Figure 1), without apparent clustering, but their significance and role in cancer development and progression are just beginning to be unraveled. A number of IRE1α human cancer-specific mutations such as A474R (near the transmembrane domain), and R635W, S769F, Q780Δ, and P830L (kinase domain) suppress chemically-induced ER stress-induced apoptosis when overexpressed in the INS-1 insulinoma cell line.61 The Q780Δ is a truncation mutant lacking the RNase domain and does not exhibit IRE1α phosphorylation with ER stress. The Q780Δ, S769F, and P830L mutants were also defective in IRE1α phosphorylation and XBP1 splicing while R635W and L474R retained these functions although the level of activation was attenuated when compared with wild-type IRE1α.61 In a glioblastoma xenograft model, the Q780Δ variant accelerated tumor development compared with controls,28 further supporting the idea that some component of the RNase can function to suppress tumor development. Two different missense mutants in the luminal domain, both resulting in increased IRE1α dimerization, oligomerization, hyperphosphorylation, and XBP1 mRNA splicing compared with WT-IRE1α, had opposite effects on glioblastoma xenografts. The P336L missense mutation completely prevented tumor formation while the A414T accelerated tumor growth and caused rapid death of tumor-bearing mice due to increased vascularization as well as low macrophage infiltration, unlike the S769F and Q780Δ mutants and a dominant negative IRE1α.28 Interestingly, the IRE1α P336L and the A414T mutants had distinct activity towards the known RIDD target miR-17, with the P336L mutant exhibiting significantly higher degradation of miR-17 expression relative to the A414T mutant.28 The underlying basis for these distinct downstream phenotypes of seemingly similar IRE1α hyperactivation levels is not known but suggests that different IRE1α mutations may alter RNase activity in a way to give very specific consequences for cancer development.
7 |. INHIBITION AND ACTIVATION OF IRE1α SIGNALING IN CANCER THERAPY
Given the importance of both XBP1 signaling and RIDD in tumor development, it can be surmised that small molecule inhibitors or activators of the IRE1α RNase domain have potential as a therapeutic strategy in combination with chemotherapy. For example, Logue et al53 showed that MKC8866, a small molecule IRE1α RNase inhibitor, strengthened the response to paclitaxel by downregulating the synthesis and secretion of protumorigenic cytokines in triple-negative MDA-MB-231 breast cancer cells. In vitro mammosphere formation was notably reduced in these cells, suggesting that this reduction in the number of tumor-initiating cells is a direct consequence of an IRE1α-dependent decrease in protumorigenic cytokines.53 Similarly, the small molecule IRE1α RNase inhibitor 8866 was shown to enhance the response to docetaxel in Myc-overexpressing breast tumors. This effect was proposed to be due to XBP1 acting as a synthetic lethal partner for myc.62 Another IRE1α RNase domain inhibitor, STF-083010, was used in the human colon cancer cell line HCT116 to manipulate a direct link between p53 and IRE1α, where p53 interacts with the E3 ubiquitin ligase HRD1 that reduces IRE1α levels by proteasomal degradation.63 Interestingly, this inhibitor suppressed the growth of HCT116 p53−/− cells to a much greater extent than for p53+/+ cells, as the lack of p53 contributed to an overabundance of IRE1α and dysregulation of the UPR. This connection between IRE1α and p53 opens a new therapeutic avenue for tumors with a p53 knockout, a common stumbling block in cancer therapy.
IRE1α inhibition has also been used to enhance the immune response to tumors. Aggressive ovarian cancer malignancies have been shown to cause ER stress in resident T cells by sabotaging the ability of the T cells to uptake glucose and perform N-linked protein glycosylation through inhibition of GLUT1 transporters and uridine diphosphate N-acetylglucosamine (UDP-GlcNAc).64 This stress triggers the IRE1α arm of the UPR, but under these circumstances, XBP1 exacerbates ER stress by limiting intracellular glutamine transport, which compromises mitochondrial respiration in the affected T cells. Inactivation of IRE1α, and thus of spliced XBP1, has been shown to enhance the antitumor activity of activated T cells in mouse models of advanced ovarian cancers. Indirect suppression of IRE1α has also been used in the treatment of breast cancers, where the estrogen receptor antagonist fulvestrant was shown to reduce proliferation and promote apoptosis by indirectly inhibiting the IRE1α-XBP1 axis in GH3 prolactinoma cells in culture.65
On the other hand, studies conducted using in vitro and in vivo models of prostate cancer showed that activation of IRE1α, along with other branches of the UPR, significantly reduced cell proliferation and tumor growth when treated with a combination of clofoctol and sorafenib.66 The IRE1α-JNK-JUN pathway has been shown to restore sensitivity to MEK inhibitors in colon cancer cell lines bearing mutations in KRAS.67 Similarly, the IRE1α-JNK-CHOP axis has been implicated in sensitizing gastric cancer cells to an HDAC inhibitor kaempferol to induce autophagy and cell death,68 and PPAR-γ antagonist fenofibrate induced autophagy by activating IRE1α and PERK in mouse models of pancreatic cancer.69 Taken together, these studies highlight the targetable potential along the IRE1α-XBP1 axis across a range of human cancers. However, whether suppression or activation of IRE1α is desired would depend on the context of the tumor and vary on a patient-by-patient basis.
8 |. CONCLUSIONS
Although the importance of ER stress and the UPR for cancer development is not a new concept, the emerging complexity of the IRE1α pathway indicates that much remains to be understood. The idea that IRE1α outputs can have distinct effects in cancer pathogenesis suggests further studies in different human cancers and cancer models are warranted. With improvements to personalized medicine and an increased understanding of the microenvironment context of each tumor these studies will illuminate potential avenues and pitfalls for targeting IRE1α in cancer therapy.
ACKNOWLEDGEMENTS
The authors would like to thank colleagues in the field for support and apologize for any unintentional oversight in citing their work due to constraints on manuscript length. This study is supported by R01 CA197942 to ABG, and by the USDA National Institute of Food and Agriculture and Hatch Appropriations under Project #4607, accession number 1009993.
Funding information
National Cancer Institute, Grant/Award Number: R01 CA197942; USDA National Institute of Food and Agriculture and Hatch Appropriations
Abbreviations
- APC
adenomatous polyposis coli
- EMT
epithelial-mesenchymal transition
- ERAD
endoplasmic reticulum-associated degradation
- MEF
mouse embryonic fibroblasts
- RIDD
regulated IRE1-dependent decay
- TCGA
The Cancer Genome Atlas
- JNK
c-Jun N-terminal kinase
Footnotes
This is the peer reviewed version of the following article: Chalmers F, Mogre S, Son J, Blazanin N, Glick AB. The multiple roles of the unfolded protein response regulator IRE1α in cancer. Mol Carcinog. 2019 which has been published in final form DOI:10.1002/mc.23031 This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Use of Self-Archived Versions.
REFERENCES
- 1.Kaufman RJ, Scheuner D, Schröder M, et al. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol. 2002;3:411–421. [DOI] [PubMed] [Google Scholar]
- 2.Bertolotti A, Zhang Y, Hendershot LM, et al. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. [DOI] [PubMed] [Google Scholar]
- 3.Oikawa D, Kimata Y, Kohno K, et al. Activation of mammalian IRE1alpha upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res. 2009;315:2496–2504. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Shen J, Prywes R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem. 2002;277:13045–13052. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto K, Sato T, Matsui T, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. [DOI] [PubMed] [Google Scholar]
- 6.Kebache S, Cardin E, Nguyen DT, et al. Nck-1 antagonizes the endoplasmic reticulum stress-induced inhibition of translation. J Biol Chem. 2004;279:9662–9671. [DOI] [PubMed] [Google Scholar]
- 7.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Promlek T, Ishiwata-Kimata Y, Shido M, et al. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol Biol Cell. 2011;22:3520–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollien J, Lin JH, Li H, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. [DOI] [PubMed] [Google Scholar]
- 11.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang TK, Lawrence DA, Lu M, et al. Coordination between two branches of the unfolded protein response determines apoptotic cell fate. Mol Cell. 2018;71:629–636. [DOI] [PubMed] [Google Scholar]
- 13.Amin-Wetzel N, Saunders RA, Kamphuis MJ, et al. A J-protein co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell. 2017;171:1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urano F, Wang X, Bertolotti A, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. [DOI] [PubMed] [Google Scholar]
- 15.Han D, Lerner AG, Vande WL, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Upton JP, Wang L, Han D, et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCullough KD, Martindale JL, Klotz LO, et al. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marciniak SJ, Yun CY, Oyadomari S, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. [DOI] [PubMed] [Google Scholar]
- 20.Huber AL, Lebeau J, Guillaumot P, et al. (IPK)-mediated attenuation of the proapoptotic PERK-CHOP pathway allows malignant progression upon low glucose. Mol Cell. 2013;49:1049–1059. p58 [DOI] [PubMed] [Google Scholar]
- 21.Huang LW, Lin CY, Lee CC, et al. Overexpression of GRP78 is associated with malignant transformation in epithelial ovarian tumors. Appl Immunohistochem Mol Morphol. 2012;20:381–385. [DOI] [PubMed] [Google Scholar]
- 22.Jin C, Jin Z, Chen NZ, et al. Activation of IRE1alpha-XBP1 pathway induces cell proliferation and invasion in colorectal carcinoma. Biochem Biophys Res Commun. 2016;470:75–81. [DOI] [PubMed] [Google Scholar]
- 23.Gifford JB, Huang W, Zeleniak AE, et al. Expression of GRP78, master regulator of the unfolded protein response, increases chemoresistance in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2016;15:1043–1052. [DOI] [PubMed] [Google Scholar]
- 24.Chen L, Li Q, She T, et al. IRE1alpha-XBP1 signaling pathway, a potential therapeutic target in multiple myeloma. Leuk Res. 2016;49:7–12. [DOI] [PubMed] [Google Scholar]
- 25.Wu CH, Silvers CR, Messing EM, et al. Bladder cancer extracellular vesicles drive tumorigenesis by inducing the unfolded protein response in endoplasmic reticulum of nonmalignant cells. J Biol Chem. 2019;294:3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zonneveld MI, Keulers TGH, Rouschop KMA. Extracellular vesicles as transmitters of hypoxia tolerance in solid cancers. Cancers. 2019;11:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker A, Thakur BK, Weiss JM, et al. Extracellular vesicles in cancer: cell-to-cell mediators of metastasis. Cancer Cell. 2016;30:836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lhomond S, Avril T, Dejeans N, et al. Dual IRE1 RNase functions dictate glioblastoma development. EMBO Mol Med. 2018;10:e7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakatani T, Maemura K, Hiyama N, et al. High expression of IRE1 in lung adenocarcinoma is associated with a lower rate of recurrence. Jpn J Clin Oncol. 2017;47:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubio-Patino C, Bossowski JP, De Donatis GM, et al. Low-protein diet induces IRE1alpha-dependent anticancer immunosurveillance. Cell Metab. 2018;27:828–842. [DOI] [PubMed] [Google Scholar]
- 31.Bujisic B, De GA, Tallant R, et al. Impairment of both IRE1 expression and XBP1 activation is a hallmark of GCB DLBCL and contributes to tumor growth. Blood. 2017;129:2420–2428. [DOI] [PubMed] [Google Scholar]
- 32.Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hur KY, So JS, Ruda V, et al. IRE1alpha activation protects mice against acetaminophen-induced hepatotoxicity. J Exp Med. 2012; 209:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tameire F, Verginadis II, Koumenis C. Cell intrinsic and extrinsic activators of the unfolded protein response in cancer: mechanisms and targets for therapy. Semin Cancer Biol. 2015;33:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, Iliopoulos D, Zhang Q, et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature. 2014;508:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koumenis C, Naczki C, Koritzinsky M, et al. Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol. 2002;22:7405–7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romero-Ramirez L, Cao H, Regalado MP, et al. X box-binding protein 1 regulates angiogenesis in human pancreatic adenocarcinomas. Transl Oncol. 2009;2:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drogat B, Auguste P, Nguyen DT, et al. IRE1 signaling is essential for ischemia-induced vascular endothelial growth factor-A expression and contributes to angiogenesis and tumor growth in vivo. Cancer Res. 2007;67:6700–6707. [DOI] [PubMed] [Google Scholar]
- 40.Cuevas EP, Eraso P, Mazon MJ, et al. LOXL2 drives epithelial-mesenchymal transition via activation of IRE1-XBP1 signalling pathway. Sci Rep. 2017;7:44988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura M, Gotoh T, Okuno Y, et al. Activation of the endoplasmic reticulum stress pathway is associated with survival of myeloma cells. Leuk Lymphoma. 2006;47:531–539. [DOI] [PubMed] [Google Scholar]
- 42.Bagratuni T, Wu P, Gonzalez de CD, et al. XBP1s levels are implicated in the biology and outcome of myeloma mediating different clinical outcomes to thalidomide-based treatments. Blood. 2010;116: 250–253. [DOI] [PubMed] [Google Scholar]
- 43.Carrasco DR, Sukhdeo K, Protopopova M, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papandreou I, Denko NC, Olson M, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mimura N, Fulciniti M, Gorgun G, et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denoyelle C, bou-Rjaily G, Bezrookove V, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–1063. [DOI] [PubMed] [Google Scholar]
- 47.Kaser A, Lee AH, Franke A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niederreiter L, Fritz TM, Adolph TE, et al. ER stress transcription factor Xbp1 suppresses intestinal tumorigenesis and directs intestinal stem cells. J Exp Med. 2013;210:2041–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bright MD, Itzhak DN, Wardell CP, et al. Cleavage of BLOC1S1 mRNA by IRE1 is sequence specific, temporally separate from XBP1 splicing, and dispensable for cell viability under acute endoplasmic reticulum stress. Mol Cell Biol. 2015;35:2186–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li XX, Zhang HS, Xu YM, et al. Knockdown of IRE1alpha inhibits colonic tumorigenesis through decreasing beta-catenin and IRE1alpha targeting suppresses colon cancer cells. Oncogene. 2017;36:6738– 6746. [DOI] [PubMed] [Google Scholar]
- 51.Dejeans N, Pluquet O, Lhomond S, et al. Autocrine control of glioma cells adhesion and migration through IRE1alpha-mediated cleavage of SPARC mRNA. J Cell Sci. 2012;125:4278–4287. [DOI] [PubMed] [Google Scholar]
- 52.Auf G, Jabouille A, Guerit S, et al. Inositol-requiring enzyme 1alpha is a key regulator of angiogenesis and invasion in malignant glioma. Proc Natl Acad Sci USA. 2010;107:15553–15558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logue SE, McGrath EP, Cleary P, et al. Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat Commun. 2018;9:3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lerner AG, Upton JP, Praveen PV, et al. IRE1alpha induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16:250–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blazanin N, Son J, Craig-Lucas AB, et al. ER stress and distinct outputs of the IRE1alpha RNase control proliferation and senescence in response to oncogenic Ras. Proc Natl Acad Sci USA. 2017;114:9900–9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J, Liu CY, Back SH, et al. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci USA. 2006;103:14343–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ali MM, Bagratuni T, Davenport EL, et al. Structure of the Ire1 autophosphorylation complex and implications for the unfolded protein response. EMBO J. 2011;30:894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee KP, Dey M, Neculai D, et al. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1–pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ghosh R, Wang L, Wang ES, et al. Allosteric inhibition of the IRE1alpha RNase preserves cell viability and function during endoplasmic reticulum stress. Cell. 2014;158:534–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao N, Cao J, Xu L, et al. Pharmacological targeting of MYC-regulated IRE1/XBP1 pathway suppresses MYC-driven breast cancer. J Clin Invest. 2018;128:1283–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Namba T, Chu K, Kodama R, et al. Loss of p53 enhances the function of the endoplasmic reticulum through activation of the IRE1alpha/XBP1 pathway. Oncotarget 2015;6:19990–20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song M, Sandoval TA, Chae CS, et al. IRE1alpha-XBP1 controls T cell function in ovarian cancer by regulating mitochondrial activity. Nature. 2018;562:423–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C, Bai M, Wang X, et al. Estrogen receptor antagonist fulvestrant inhibits proliferation and promotes apoptosis of prolactinoma cells by regulating the IRE1/XBP1 signaling pathway. Mol Med Rep. 2018;18:4037–4041. [DOI] [PubMed] [Google Scholar]
- 66.Fan L, He Z, Head SA, et al. Clofoctol and sorafenib inhibit prostate cancer growth via synergistic induction of endoplasmic reticulum stress and UPR pathways. Cancer Manag Res. 2018;10:4817–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sustic T, van WS, Bosdriesz E, et al. A role for the unfolded protein response stress sensor ERN1 in regulating the response to MEK inhibitors in KRAS mutant colon cancers. Genome Med. 2018;10:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim TW, Lee SY, Kim M, et al. Kaempferol induces autophagic cell death via IRE1-JNK-CHOP pathway and inhibition of G9a in gastric cancer cells. Cell Death Dis. 2018;9:875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tao T, Zhao F, Xuan Q, et al. Fenofibrate inhibits the growth of prostate cancer through regulating autophagy and endoplasmic reticulum stress. Biochem Biophys Res Commun. 2018;503:2685–2689. [DOI] [PubMed] [Google Scholar]