Abstract
Purpose
The aim of this study was to investigate the relationship between CT imaging phenotypes and genetic and biological characteristics in pancreatic ductal adenocarcinoma (PDAC).
Methods
In this retrospective study, consecutive patients between April 2015 and June 2016 who underwent PDAC resection were included if previously consented to a targeted sequencing protocol. Mutation status of known PDAC driver genes (KRAS, TP53, CDKN2A, and SMAD4) in the primary tumor was determined by targeted DNA sequencing and results were validated by immunohistochemistry (IHC). Radiomic features of the tumor were extracted from the preoperative CT scan and used to predict genotype and stromal content.
Results
The cohort for analysis consisted of 35 patients. Genomic and IHC analysis revealed alterations in KRAS in 34 (97%) patients, and changes in expression of CDKN2A in 29 (83%), SMAD4 in 16 (46%), and in TP53 in 29 (83%) patients. Models created from radiomic features demonstrated associations with SMAD4 status and the number of genes altered. The number of genes altered was the only significant predictor of overall survival (p = 0.016). By linear regression analysis, a prediction model for stromal content achieved an R2 value of 0.731 with a root mean square error of 19.5.
Conclusions
In this study, we demonstrate that in PDAC SMAD4 status and tumor stromal content can be predicted using radiomic analysis of preoperative CT imaging. These data show an association between resectable PDAC imaging features and underlying tumor biology and their potential for future precision medicine.
Keywords: Pancreatic neoplasm, Computational biology, Survival, Radiogenomics, Genomics
Introduction
Pancreatic ductal adenocarcinoma (PDAC) has a five-year survival rate of approximately 7% [1] and is projected to become the second most common cause of cancer death within the next 10 years [2]. The majority of patients present with locally advanced or metastatic disease, and of the patients with resectable disease at diagnosis, most will recur locally or with distant metastasis. Therefore, deciphering the underlying biology of PDAC is critical to optimize patient selection for resection and to develop novel treatment strategies.
Large-scale studies of PDAC have demonstrated high-frequency alterations in the oncogenic genes KRAS, TP53, CDKN2A, and SMAD4 [3–5]. TP53 mutations are seen in approximately 75% of PDAC [6] and can be loss-of-function (LOF) or gain-of-function (GOF) as determined by immunohistochemistry (IHC). Mutations in SMAD4 are found in approximately 50% of PDAC tumors, and prior studies have demonstrated an association between SMAD4 mutation status in tumors and overall survival in PDAC patients [7–10].
The stromal component is known to be significant in PDAC and can interfere with acquisition of sufficient tumor tissue for analysis. Although the functional significance of stromal composition in pancreatic cancer is unclear, studies have shown decreased survival in PDAC patients whose tumors have lower stromal content [11, 12]. Ideally, by quantifying the amount of surrounding stroma, treatment modalities known to do better in certain low proportions of stroma may be prioritized, and similarly, patients would be spared systemic therapies that are ineffective in the setting of high stromal proportion.
In many cancer types, computational evaluation of tumor phenotype on diagnostic imaging (radiomics) has demonstrated the potential to describe underlying tumor biology [13, 14]. Radiomic assessment of tumor appearance can be performed using texture analysis, a technique that captures spatial variations in pixel intensities within a tumor. With this technique, an algorithm operating at the single-pixel level can detect microscopic changes otherwise invisible to the naked eye. In PDAC, radiomic features derived from CT imaging have been associated with overall survival in patients who underwent resection [15–18]. In other tumor types, radiomic features have been associated with tumor genotype [19, 20]; however, a similar radiogenomic correlation has not been established in PDAC.
The primary aim of this study was to determine whether radiomic analysis could accurately predict the genotype of PDAC driver genes. A secondary aim was to use radiomics to predict stromal content in these tumors.
Methods
Study population
Following Institutional Review Board approval at our institution, a waiver of informed consent was obtained to perform a retrospective study that selected patients who underwent resection for PDAC between April 2015 and June 2016, with available preoperative CT angiogram of the pancreas and targeted genomic sequencing. Consent for genomic sequencing was previously obtained prospectively to use tumor and matched normal tissue. Patients who received neoadjuvant chemotherapy or patients with pancreatitis on preoperative CT as determined by a radiologist (XX, 8 years of experience after fellowship) were excluded from analysis to account for variables that affect preoperative imaging. Demographic, clinical, and pathologic factors were collected from a prospectively maintained database and supplemented with retrospective review of the electronic medical record.
Targeted sequencing
We used a targeted sequencing panel that analyzes all exons and selected introns of 410 cancer-associated genes. An established pipeline was used for DNA extraction, sequencing, and analysis as previously described [21]. Data were analyzed through a custom bioinformatics pipeline. Point mutations were filtered for quality by the following criteria: (tumor variant allele frequency [VAF]/normal VAF) ≥ 5, tumor coverage ≥ 20, tumor VAF ≥ 0.02 (hotspot) or ≥ 0.05 (non-hotspot), tumor mutant reads ≥ 8 (hotspot) or ≥ 10 (non-hotspot). Finally, all filtered called mutations were manually reviewed by a bioinformatician to identify potential false positives. Copy number analysis was performed using FACETS, a software tool optimized for detecting copy number alterations (CNA) while incorporating variations in tumor purity, ploidy, and clonal heterogeneity [22].
Immunohistochemistry
To validate the sequencing results, IHC staining for known PDAC tumor suppressors (TP53, CDK2NA, and SMAD4) was performed on resected tumor specimens. All hematoxylin and eosin-stained slides of each case were examined by a gastrointestinal pathologist under low power (4 ×) objective to identify the best representative tumor section to perform IHC staining. Then unstained 5-μm slides were cut from paraffin-embedded tumor blocks and then deparaffinized by standard techniques. The slides were labeled with monoclonal antibodies to TP53 (DO-7, Agilent Technologies, Inc., Santa Clara, CA), CDKN2A (ink4a, Roche MTM Laboratories, Indianapolis, IN), and SMAD4 (B8, Santa Cruz Biotechnology, Dallas, TX). A pathologist, blinded to sequencing results and clinical outcomes, evaluated the IHC labeling of the tumor samples. TP53 slides were categorized as “normal,” “abnormal nuclear accumulation,” or “abnormal-homozygous deletion;” CDKN2A slides were categorized as “positive” (present) or “negative” (absent); and SMAD4 slides were categorized as “intact” or “lost.”
Mutation status determination
The point mutation, copy number, and IHC results were interpreted together to make a final determination of the allele status. For KRAS, the presence of a point mutation or CNA classified the sample as abnormal. For TP53, a point mutation, CNA, or abnormal IHC result classified the sample as abnormal. We further subdivided the TP53 genotypes into (1) wild type (WT) if it harbored no point mutations or CNA and stained normally for TP53 by IHC, (2) gain-of-function (GOF) for missense mutations with associated nuclear accumulation in > 30% of the cancer cells, and (3) loss-of-function (LOF) for nonsense or frameshift mutations with complete loss of protein expression compared to reactive fibroblasts or lymphocytes present in the same tissue section. Cases with a normal TP53 gene sequence, but with complete loss of protein expression, were presumed homozygous deletions and also categorized as LOF. For CDKN2A and SMAD4, samples were determined to be abnormal if the IHC was negative (lost) and normal if both the IHC was positive (intact) and no point mutations or CNA were present.
Tumor-stroma analysis
In slides with tumor identified by low power (4 ×) screening, percentages of epithelial and stromal components were assessed in a semiquantitative manner using the mean value of medium power fields in a 20 × objective on all tumor slides (range, 5 to 13 slides) of a given tumor.
CT image acquisition
Contrast-enhanced CT images were used for quantitative image analysis. Following administration of 150 mL of iodinated contrast (Omnipaque 300, GE Healthcare) at 4.0 mL/s, CT images were obtained using a multidetector CT (Light-speed 16 and VCT, GE Healthcare) during the pancreatic parenchymal phase (scan delay 40 s) and portal venous phase. The scan parameters for the portal venous phase were as follows: pitch/table speed 0.984–1.375/39.37–27.50 mm; autoMA 220–380; noise index 12.5–14; rotation time 0.7–0.8 ms; scan delay 80–85 s. Axial slices reconstructed at 2.5-mm intervals were used.
Quantitative CT image analysis
The tumor imaged in the portal venous phase was manually segmented over the entire tumor volume using Scout Liver Software (Pathfinder Technologies Inc., Analogic Corporation) by research study assistants with prior experience in tumor segmentation. A diagnostic radiologist specializing in pancreatic tumors (with 8 years of experience on the pancreas tumor board) reviewed the segmentations and adjusted tumor contours to ensure tumor region accuracy (Online Resource 1), using the pancreatic parenchymal phase as a guide when necessary. All were blinded to clinical and genetic variables. The decision to use the portal venous phase was due to the variability in use of dual-energy CT for the pancreatic parenchymal phase. 255 radiomic features describing image heterogeneity were extracted by computer scientists from the segmented volume as described previously [references blinded for review]. Briefly, the features were extracted using gray-level co-occurrence matrices (GLCM), run-length matrices (RLM), local binary patterns (LBP), fractal dimension (FD), intensity histogram (IH), and angle co-occurrence matrices (ACM) [23–27]. ACMs describe the directional edge patterns present in a tumor, whereas the other types quantify intensity patterns. A set of statistical features from each type are computed as follows: 19 statistical features from GLCM, 11 from RLM, 128 from LBP, 54 from FD, 5 from IH, and 38 from ACM. Radiomic features were extracted from each CT axial slice of the tumor region and averaged to a single value for the entire tumor volume. All image analysis was performed in MATLAB 2015a (MathWorks, Natick, MA).
Statistical analysis
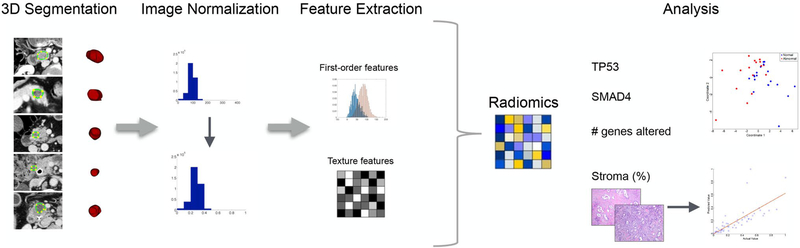
Radiomic features extracted from CT images were analyzed for significance in each of the four outcomes: SMAD4 status, TP53 status, number of genes altered, and stromal content. Neither KRAS nor CDKN2A were included in our analysis as KRAS is mutated in greater than 90% of PDACs [28, 29] and CDKN2A is inactivated in ~ 90% either through point mutations or epigenetic mechanisms [30, 31], and inclusion would have compromised the potential generation of a prediction model. SMAD4 status, TP53 status, and number of genes altered were analyzed as categorical variables, while stromal content was evaluated as a continuous variable. To identify the discriminatory features for each variable, multiple feature selection algorithms [i.e., univariate analysis and fuzzy minimum-redundancy-maximum-relevance (fMRMR)] were proposed, and the method with the best performance was selected. The workflow of the proposed method is outlined in Fig. 1.
Fig. 1.
CT imaging was analyzed and texture features extracted. The texture features were then used to predict TP53 status, SMAD4 status, number of genes altered, and stroma content of the primary PDAC tumor
The first predictive algorithm used radiomic feature analysis to predict SMAD4 status (normal vs. abnormal). Radiomic features found to be significant were selected using fMRMR in which the redundancy of a feature is calculated by the average fuzzy mutual information of the feature with the selected features, and relevancy is computed using fuzzy mutual information of the feature with the class levels. A multidimensional scaling (MDS) plot was created to visualize the level of similarity between groups for selected imaging features. In MDS plots, data with multiple dimensions are converted into a visually interpretable plot as to preserve the distance between objects while still allowing visualization of the difference between groups. In this study, a 2D MDS plot is used where the two coordinates represent the first two principal components. The second model aimed to stratify patients according to their TP53 protein effect (WT vs. GOF vs. LOF). Radiomic features were selected using the Kruskal–Wallis test (p < 0.05) and an MDS plot was created. The third model aimed to determine whether radiomic analysis could correctly classify a patient as harboring less than or equal to and greater than the median number of mutations. Radiomic features found to be significant in this analysis were selected using an fMRMR algorithm as described above, and subsequently, an MDS plot was created to illustrate the results. Lastly, we investigated whether radiomic analysis could correctly predict stromal content. Features associated with stromal content were first selected by univariate analysis with linear regression (p < 0.05); a multivariate analysis with the selected features was then performed using linear regression to observe their efficacy in predicting the percentage of stromal content.
Overall survival (OS) and recurrence-free survival (RFS) were analyzed for all study patients. OS was defined as the time interval between date of operation and date of death or date last known alive. RFS was defined as the time between the date of operation and the date of recurrence on imaging. For patients without recurrent disease, the most recent CT scan was recorded. OS and RFS were analyzed for each of the aforementioned variables.
Continuous variables are expressed as median and interquartile range (IQR), and categorical variables are expressed as number and percentages. Univariate survival analysis was performed using Cox proportional hazards regression models for both continuous and categorical variables. A p value of < 0.05 was considered statistically significant.
Results
Patient population
During the study period, 60 patients underwent a preoperative CT angiogram required for image analysis and had their tumors successfully sequenced after informed consent. We excluded 13 patients who received neoadjuvant therapy, five patients who had final pathological diagnoses other than PDAC, and three patients with pancreatitis on preoperative imaging. Additionally, four patients’ tumor blocks were unavailable for IHC staining. Therefore, our final cohort consisted of 35 patients. A summary of demographic and clinical data can be found in Table 1.
Table 1.
Cohort demographics
| All patients (n = 35)a | |
|---|---|
| Age | 67 (62–75) |
| Gender | |
| Male | 14 (40%) |
| Female | 21 (60%) |
| Procedure | |
| Whipple | 34 (97%) |
| Distal pancreatectomy | 1 (3%) |
| Pathology | |
| Tumor size | 2.6 (2.1–3.3) |
| Tumor grade | |
| Well | 2 (6%) |
| Moderate | 16 (46%) |
| Poor | 17 (48%) |
| Days between CT and operation | 8 (5–17) |
Numbers in this column represent mean (interquartile range) or N (%)
Radiogenomic analysis
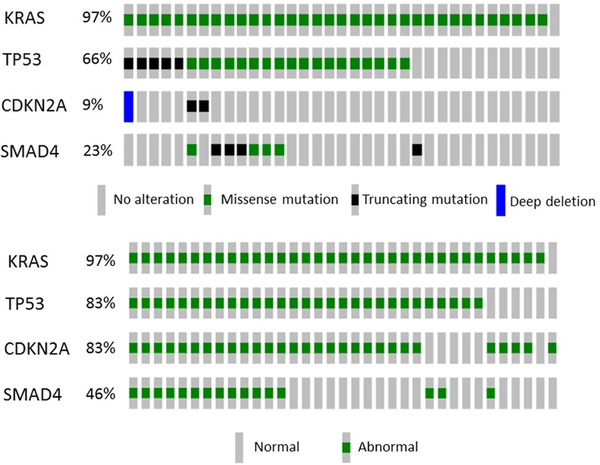
The results of the genetic and IHC analyses are presented in Fig. 2. Consistent with previous data, 97% (34/35) of tumor samples harbored mutations in KRAS, and 83% (29/35) of samples had alterations in CDKN2A. Given the high proportion of alterations in these genes, KRAS and CDKN2A were not pursued in further predictive analyses.
Fig. 2.
Top: Oncoprint showing genomic alterations. Bottom: Oncoprint showing status as determined by genomic alterations and IHC. Columns represent patients in the cohort (n = 35)
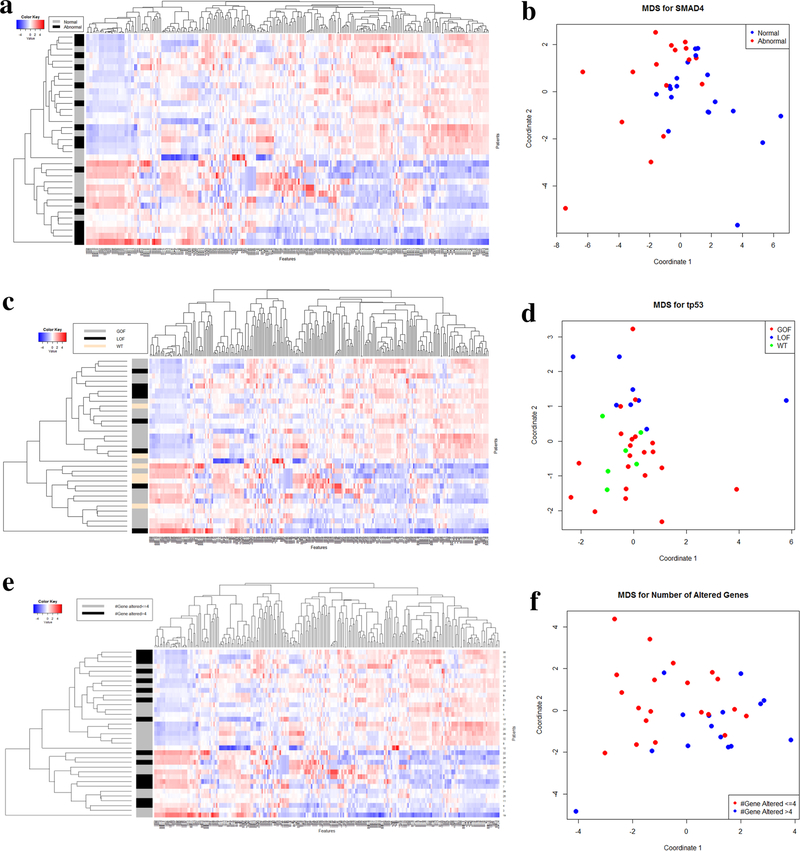
Of all patients included, 16 (46%) were found to have an alteration in SMAD4. 255 radiomic features were extracted from CT scans for analysis (Fig. 3a), and feature selection using fMRMR resulted in 28 significant features. All features were selected from LBP, with the exception of one RLM feature, one ACM-based feature, and one intensity-histogram-based skewness. An MDS plot (Fig. 3b) shows good discriminatory power between patients with normal and abnormal SMAD4 status.
Fig. 3.
Heatmap with classification of 255 texture features and 35 patients for SMAD4 (a), TP53 (c), and number of genes altered (e). Multidimensional scaling for SMAD4 using fMRMR for feature selection (b), TP53 using univariate analysis (p < 0.05) for feature selection (d), and number of genes altered separated by ≤ 4 and > 4 genes altered (f). GOF gain-of-function, LOF loss-of-function, WT wild type
In our cohort, 29 patients (83%) had alterations in TP53 expression (21 GOF; 8 LOF). The 255 texture features for TP53 presence/absence are shown in Fig. 3c. Following univariate analysis, 32 features were significant. The features were selected from all types of features except GLCM. An MDS plot (Fig. 3d) illustrates the model’s ability to discriminate between GOF and LOF. However, the model did not clearly isolate patients with WT status from those with an abnormal TP53 status.
The median number of altered genes per patient was four (IQR 3–6). Similar to Fig. 3c, features for patients with greater than 4 altered genes are shown in Fig. 3e. Using fMRMR, 14 significant features were identified. The features were selected from both LBP- and FD-based features. An MDS plot (Fig. 3f) was generated based on quantitative differences in radiomic features and shows patient discrimination between those with ≤ 4 and > 4 altered genes.
Stroma
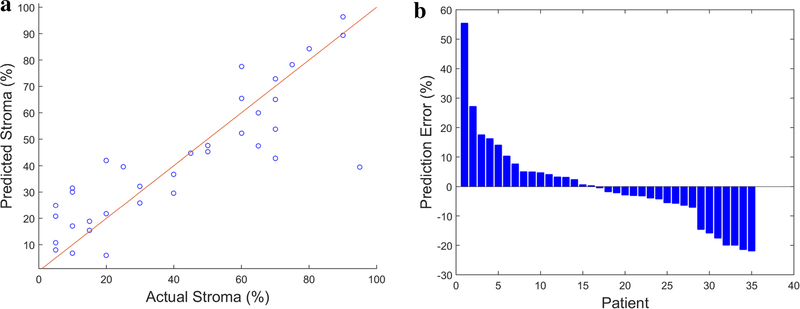
The median stromal content in the cohort was 40% (IQR 15–68%). There were 21 patients with ≤ 50% stroma and 14 patients with > 50% stroma in the tumor. Tumors with ≤ 50% stroma harbored significantly more altered genes than those with > 50% stroma (mean 5 vs. 3; p = 0.001). Tumor size was also significantly different between the two groups (mean 3.1 vs. 2.3 cm; p = 0.008), yet tumor grade was not significantly different (p = 0.774). Feature selection by univariate analysis led to 19 significant features. Energy and contrast features extracted from GLCM as well as 17 LBP-based features were selected. A multivariate analysis was then performed with the selected features using linear regression. The continuous prediction model resulted in an R-square value of 0.731 with a root mean square error of 19.5. The mean absolute prediction error was 10.15%. A calibration plot showing predicted versus actual stromal content is shown in Fig. 4.
Fig. 4.
Left: calibration plot showing predicted versus actual stromal content. Identity line (y = x) denotes perfect prediction. Right: histogram of prediction error percentage for all patients in the cohort
Survival and recurrence analysis
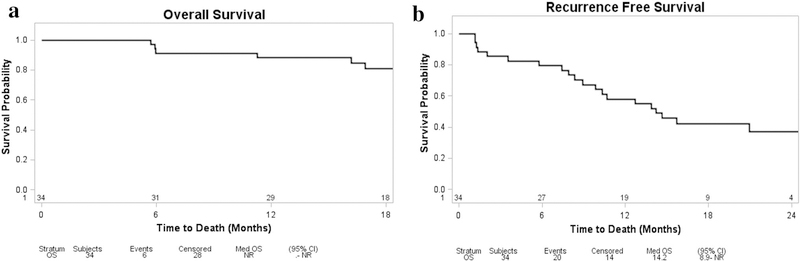
OS and RFS were analyzed in our cohort using a Cox regression model for each of the four variables: SMAD4 status, TP53 status, number of genes altered, and stromal content. One patient was excluded from analysis due to a lack of follow-up data, and therefore the final number of patients included in this analysis was 34. OS for the cohort is demonstrated in Fig. 5a. At the time of the analysis, 82% (28/34) of patients were alive with a median follow-up of 21.5 months. The number of genes altered was the only significant predictor of OS (p = 0.016). During the follow-up period, 20 patients (59%) were diagnosed with recurrent disease, and the median RFS was 14 months (Fig. 5b). Both the number of genes altered and percent stroma (p < 0.001 and p = 0.034, respectively) were predictive of RFS.
Fig. 5.
Kaplan–Meier curves for overall survival (a) and recurrence-free survival (b) of all patients in the cohort
Discussion
In this study, we identified radiomic features associated with PDAC genetic alterations and stromal content. These associations show the potential of using noninvasive imaging on pre-surgical pancreas cancer patients for precision medicine. Linking radiomic features to underlying tumor biology is an area of great interest, given the ubiquity of diagnostic imaging and the challenges and costs in performing molecular analyses. Recent DNA/RNA sequencing studies have provided in-depth insights into individual tumor’s genetic makeup and have demonstrated some prognostic power for survival that may guide personalized treatment. While biopsies can yield sufficient information for diagnosis, detecting characteristics specific to an individual tumor (such as with IHC) often requires more tissue. New computational approaches to extracting phenotypic information from CT scans could help optimize management for patients with PDAC, as targeted therapies become available.
While there have been prior radiomic studies predicting survival of pancreatic adenocarcinomas [15, 16, 18, 32, 33], there are no previous reports relating the underlying biology of PDAC to quantitative CT data. In the current study, we showed strong discrimination between tumors with and without SMAD4 alterations. SMAD4 expression was also associated with PDAC CT features in a previous study, although the features were assessed by radiologists visually [34]. The number of altered genes was also associated with radiomic features in our patient cohort, suggesting that the number of mutations is related to increased tumor heterogeneity in imaging. Visually, the MDS plot for the association of radiomics and SMAD4 status as well as number of genes altered demonstrated separation of distinct groups; in contrast, the MDS plot for TP53 status does not delineate the three groups as clearly, likely due to the limited number of patients in each group. Stromal content correlated to radiomic features (R2 value 0.731), suggesting that CT imaging also captures stromal information. Stromal composition could impact treatment decision making for PDAC. For one, the relative hypoxia inside the tumor mass from a thick stroma could impair drug delivery. Similarly, the immune system’s inability to engage the tumor may not be solely due to inhibitory signals from the tumor but also perhaps from a stromal barrier keeping crucial T lymphocytes at bay. The ability to quantify tumor stromal content using radiomics may serve as an important prognostic factor that can predict patients’ responsiveness to certain treatments.
Mutation burden has been shown to be prognostic in other cancer types, such as ovarian and lung [35, 36]. Conversely, a recent PDAC study found no association between number of mutations and survival [8]. Here, we found a significant difference in survival between patients who have mutations in four or less genes as compared to greater than four genes, and we show that this difference can be predicted on preoperative CT imaging. Further studies with larger cohorts are needed to validate our findings.
The strength of this study lies mainly in its use of a computational method to extract and process image data. Further, the sequencing results were manually reviewed to confirm the presence of called mutations, and we performed an orthogonal method of validation (IHC staining) to ensure accurate genotyping. The main weaknesses of the study are the small sample size and the lack of independent validation. As we accrue more patients and develop multi-institutional collaborations, we can increase our sample size and validate our models. Another limitation is that a majority of our tumors were found in the pancreatic head. Since neoadjuvant therapy is increasingly used for patients with resectable pancreatic adenocarcinomas, it may be challenging to validate our results going forward. Nevertheless, our study provides a potential biological rationale behind investigating texture analysis in this deadly tumor. We did not investigate if differences in degree of vascular contact in our resectable patient population had associations with tumor genetics or fibrosis; this question may be better investigated in a larger cohort of patients with and without neoadjuvant therapy. Finally, our study does not make use of CT scans from outside institutions which may utilize different imaging protocols limiting the widespread applicability. Further studies are needed to evaluate the reproducibility of radiomic analysis in PDAC with variable CT imaging protocols and to validate our results with a larger range of CT scan protocols.
In conclusion, we demonstrate that radiomic features extracted from clinical CT images are associated with genotype, the number of altered genes, and stromal content in PDAC. These associations may underlie the observation that PDAC imaging features are associated with survival. Further studies will be needed to increase sample size and perform external validation.
Supplementary Material
Acknowledgements
This research was funded in part through the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30 CA008748, the David M. Rubenstein Center for Pancreatic Research, and Cycle for Survival.
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Compliance with ethical standards
Ethical approval All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00261-019-02112-1) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66 (1):7–30. 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM (2014) Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research 74 (11):2913–2921 [DOI] [PubMed] [Google Scholar]
- 3.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, Bruxner TJ, Christ AN, Harliwong I, Idrisoglu S, Manning S, Nourse C, Nourbakhsh E, Wani S, Wilson PJ, Markham E, Cloonan N, Anderson MJ, Fink JL, Holmes O, Kazakoff SH, Leonard C, Newell F, Poudel B, Song S, Taylor D, Waddell N, Wood S, Xu Q, Wu J, Pinese M, Cowley MJ, Lee HC, Jones MD, Nagrial AM, Humphris J, Chantrill LA, Chin V, Steinmann AM, Mawson A, Humphrey ES, Colvin EK, Chou A, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Pettitt JA, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Jamieson NB, Graham JS, Niclou SP, Bjerkvig R, Grutzmann R, Aust D, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Falconi M, Zamboni G, Tortora G, Tempero MA, Australian Pancreatic Cancer Genome I, Gill AJ, Eshleman JR, Pilarsky C, Scarpa A, Musgrove EA, Pearson JV, Biankin AV, Grimmond SM (2015) Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518 (7540):495–501. 10.1038/nature14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, Chang DK, Cowley MJ, Gardiner BB, Song S, Harliwong I, Idrisoglu S, Nourse C, Nourbakhsh E, Manning S, Wani S, Gongora M, Pajic M, Scarlett CJ, Gill AJ, Pinho AV, Rooman I, Anderson M, Holmes O, Leonard C, Taylor D, Wood S, Xu Q, Nones K, Fink JL, Christ A, Bruxner T, Cloonan N, Kolle G, Newell F, Pinese M, Mead RS, Humphris JL, Kaplan W, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chou A, Chin VT, Chantrill LA, Mawson A, Samra JS, Kench JG, Lovell JA, Daly RJ, Merrett ND, Toon C, Epari K, Nguyen NQ, Barbour A, Zeps N, Australian Pancreatic Cancer Genome I, Kakkar N, Zhao F, Wu YQ, Wang M, Muzny DM, Fisher WE, Brunicardi FC, Hodges SE, Reid JG, Drummond J, Chang K, Han Y, Lewis LR, Dinh H, Buhay CJ, Beck T, Timms L, Sam M, Begley K, Brown A, Pai D, Panchal A, Buchner N, De Borja R, Denroche RE, Yung CK, Serra S, Onetto N, Mukhopadhyay D, Tsao MS, Shaw PA, Petersen GM, Gallinger S, Hruban RH, Maitra A, Iacobuzio-Donahue CA, Schulick RD, Wolfgang CL, Morgan RA, Lawlor RT, Capelli P, Corbo V, Scardoni M, Tortora G, Tempero MA, Mann KM, Jenkins NA, Perez-Mancera PA, Adams DJ, Largaespada DA, Wessels LF, Rust AG, Stein LD, Tuveson DA, Copeland NG, Musgrove EA, Scarpa A, Eshleman JR, Hudson TJ, Sutherland RL, Wheeler DA, Pearson JV, McPherson JD, Gibbs RA, Grimmond SM (2012) Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491 (7424):399–405. 10.1038/nature11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grutzmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM, Australian Pancreatic Cancer Genome I,, Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM (2016) Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531 (7592):47–52. 10.1038/nature16965 [DOI] [PubMed] [Google Scholar]
- 6.Redston MS, Caldas C, Seymour AB, Hruban RH, da Costa L, Yeo CJ, Kern SE (1994) p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer research 54 (11):3025–3033 [PubMed] [Google Scholar]
- 7.De Bosscher K, Hill CS, Nicolas FJ (2004) Molecular and functional consequences of Smad4 C-terminal missense mutations in colorectal tumour cells. Biochem J 379 (Pt 1):209–216. 10.1042/BJ20031886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackford A, Serrano OK, Wolfgang CL, Parmigiani G, Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Eshleman JR, Goggins M, Jaffee EM, Iacobuzio-Donahue CA, Maitra A, Cameron JL, Olino K, Schulick R, Winter J, Herman JM, Laheru D, Klein AP, Vogelstein B, Kinzler KW, Velculescu VE, Hruban RH (2009) SMAD4 gene mutations are associated with poor prognosis in pancreatic cancer. Clin Cancer Res 15 (14):4674–4679. 10.1158/1078-0432.CCR-09-0227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh P, Srinivasan R, Wig JD (2012) SMAD4 genetic alterations predict a worse prognosis in patients with pancreatic ductal adenocarcinoma. Pancreas 41 (4):541–546. 10.1097/MPA.0b013e318247d6af [DOI] [PubMed] [Google Scholar]
- 10.Iacobuzio-Donahue CA, Fu B, Yachida S, Luo M, Abe H, Henderson CM, Vilardell F, Wang Z, Keller JW, Banerjee P, Herman JM, Cameron JL, Yeo CJ, Halushka MK, Eshleman JR, Raben M, Klein AP, Hruban RH, Hidalgo M, Laheru D (2009) DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J Clin Oncol 27 (11):1806–1813. 10.1200/JCO.2008.17.7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neesse A, Algul H, Tuveson DA, Gress TM (2015) Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut 64 (9):1476–1484. 10.1136/gutjnl-2015-309304 [DOI] [PubMed] [Google Scholar]
- 12.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV, De Jesus-Acosta A, Sharma P, Heidari P, Mahmood U, Chin L, Moses HL, Weaver VM, Maitra A, Allison JP, LeBleu VS, Kalluri R (2014) Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer cell 25 (6):719–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.GGillies RJ, Kinahan PE, Hricak H (2016) Radiomics: Images Are More than Pictures, They Are Data. Radiology 278 (2):563–577. 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinker K, Shitano F, Sala E, Do RK, Young RJ, Wibmer AG, Hricak H, Sutton EJ, Morris EA (2018) Background, current role, and potential applications of radiogenomics. J Magn Reson Imaging 47 (3):604–620. 10.1002/jmri.25870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eilaghi A, Baig S, Zhang Y, Zhang J, Karanicolas P, Gallinger S, Khalvati F, Haider MA (2017) CT texture features are associated with overall survival in pancreatic ductal adenocarcinoma - a quantitative analysis. BMC Med Imaging 17 (1):38 10.1186/s12880-017-0209-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attiyeh MA, Chakraborty J, Doussot A, Langdon-Embry L, Mainarich S, Gonen M, Balachandran VP, D’Angelica MI, DeMatteo RP, Jarnagin WR, Kingham TP, Allen PJ, Simpson AL, Do RK (2018) Survival Prediction in Pancreatic Ductal Adenocarcinoma by Quantitative Computed Tomography Image Analysis. Ann Surg Oncol 25 (4):1034–1042. 10.1245/s10434-017-6323-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty J, Langdon-Embry L, Cunanan KM, Escalon JG, Allen PJ, Lowery MA, O’Reilly EM, Gonen M, Do RG, Simpson AL (2017) Preliminary study of tumor heterogeneity in imaging predicts two year survival in pancreatic cancer patients. PLoS One 12 (12):e0188022 10.1371/journal.pone.0188022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun G, Kim YH, Lee YJ, Kim B, Hwang JH, Choi DJ (2018) Tumor heterogeneity of pancreas head cancer assessed by CT texture analysis: association with survival outcomes after curative resection. Sci Rep 8 (1):7226 10.1038/s41598-018-25627-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinker K, Chin J, Melsaether AN, Morris EA, Moy L (2018) Precision Medicine and Radiogenomics in Breast Cancer: New Approaches toward Diagnosis and Treatment. Radiology 287 (3):732–747. 10.1148/radiol.2018172171 [DOI] [PubMed] [Google Scholar]
- 20.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross DS, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Ladanyi M, Berger MF (2015) Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 17 (3):251–264. 10.1016/j.jmoldx.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen R, Seshan VE (2016) FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44 (16):e131 10.1093/nar/gkw520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haralick RMSK, Dinstein I (1973) Textural Features for Image Classification. IEEE Trans Syst Man Cybern SMC-3 (6):610–621 [Google Scholar]
- 24.Tang X (1998) Texture information in run-length matrices. IEEE Trans Image Process 7 (11):1602–1609. 10.1109/83.725367 [DOI] [PubMed] [Google Scholar]
- 25.Buczkowski S, Hildgen P, Cartilier L (1998) Measurements of fractal dimension by box-counting: A critical analysis of data scatter. 252 (1–2):23–34. 10.1016/S0378-4371(97)00581-5 [DOI] [Google Scholar]
- 26.Chakraborty J, Rangayyan RM, Banik S, Mukhopadhyay S, Desautels JEL (2012) Statistical measures of orientation of texture for the detection of architectural distortion in prior mammograms of interval-cancer. J Electron Imaging 21 (3):12 [Google Scholar]
- 27.Ojala T, Pietikäinen M, Harwood D (1996) A Comparative Study of Texture Measures with Classification Based on Feature Distributions. 29 (1):51–59. 10.1016/0031-3203(95)00067-4 [DOI] [Google Scholar]
- 28.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M (1988) Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53 (4):549–554 [DOI] [PubMed] [Google Scholar]
- 29.Wood LD, Hruban RH (2012) Pathology and molecular genetics of pancreatic neoplasms. Cancer J 18 (6):492–501. 10.1097/PPO.0b013e31827459b6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caldas C, Hahn SA, da Costa LT, Redston MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ, Kern SE (1994) Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat Genet 8 (1):27–32. 10.1038/ng0994-27 [DOI] [PubMed] [Google Scholar]
- 31.Schutte M, Hruban RH, Geradts J, Maynard R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff I, Schmiegel W, Baylin SB, Kern SE, Herman JG (1997) Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer research 57 (15):3126–3130 [PubMed] [Google Scholar]
- 32.Sandrasegaran K, Lin Y, Asare-Sawiri M, Taiyini T, Tann M (2019) CT texture analysis of pancreatic cancer. Eur Radiol 29 (3):1067–1073. 10.1007/s00330-018-5662-1 [DOI] [PubMed] [Google Scholar]
- 33.Cozzi L, Comito T, Fogliata A, Franzese C, Franceschini D, Bonifacio C, Tozzi A, Di Brina L, Clerici E, Tomatis S, Reggiori G, Lobefalo F, Stravato A, Mancosu P, Zerbi A, Sollini M, Kirienko M, Chiti A, Scorsetti M (2019) Computed tomography based radiomic signature as predictive of survival and local control after stereotactic body radiation therapy in pancreatic carcinoma. PLoS One 14 (1):e0210758 10.1371/journal.pone.0210758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi SH, Kim HJ, Kim KW, An S, Hong SM, Kim SC, Kim MH (2017) DPC4 gene expression in primary pancreatic ductal adenocarcinoma: relationship with CT characteristics. Br J Radiol 90(1073):20160403 10.1259/bjr.20160403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birkbak NJ, Kochupurakkal B, Izarzugaza JM, Eklund AC, Li Y, Liu J, Szallasi Z, Matulonis UA, Richardson AL, Iglehart JD, Wang ZC (2013) Tumor mutation burden forecasts outcome in ovarian cancer with BRCA1 or BRCA2 mutations. PLoS One 8 (11):e80023 10.1371/journal.pone.0080023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park JH, Kim TM, Keam B, Jeon YK, Lee SH, Kim DW, Chung DH, Kim YT, Kim YW, Heo DS (2013) Tumor burden is predictive of survival in patients with non-small-cell lung cancer and with activating epidermal growth factor receptor mutations who receive gefitinib. Clin Lung Cancer 14 (4):383–389. 10.1016/j.cllc.2012.10.007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.