Abstract
Background
Laryngeal squamous cell carcinomas have a high risk of recurrence and poor prognosis. Patient derived cancer cell lines remain important preclinical models for advancement of new therapeutic strategies, and comprehensive characterization of these models is vital in the precision medicine era.
Methods
We performed exome and transcriptome sequencing as well as copy number analysis of a panel of LSCC-derived cell lines that were established at the University of Michigan and are used in laboratories worldwide.
Results
We observe a complex array of alterations consistent with those reported in the HNSCC TCGA project, including aberrations in PIK3CA, EGFR, CDKN2A, TP53, NOTCH family, and FAT1 genes. A detailed analysis of FAT family genes and associated pathways showed disruptions to these genes in most cell lines.
Conclusions
The molecular profiles we have generated indicate that as a whole, this panel recapitulates the molecular diversity observed in patients and will serve as useful guides in selecting cell lines for preclinical modeling.
Keywords: HNSCC, UM-SCC, cell lines, exome sequencing, laryngeal cancer, precision medicine
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common cancer worldwide and can arise in the oral cavity, oropharynx, hypopharynx, or larynx (1). The five-year survival rates for patients with HNSCC range from 40–80%, varying by anatomic site, tumor stage, and HPV status (2). Laryngeal squamous cell carcinomas (LSCCs), which comprise 20% of all HNSCCs, are typically HPV negative and have a five-year survival rate of 80–90% for stage I/II disease (3), but up to 50% of advanced LSCC patients experience recurrence following frontline therapy, at which point disease progression often occurs rapidly with significant regression in quality of life metrics (4). Thus, robust models of LSCC are important for identification of biomarkers distinguishing patients most likely to fail therapy, as well as to develop novel treatments for aggressive disease. We believe LSCC cell line models representing the range of cancer stages and genetic composition in both primary and recurrent/metastatic settings will aid in better understanding individual disease processes and responses to treatment, and in developing therapies that improve outcomes for LSCC.
From The Cancer Genome Atlas (TCGA) project, a molecular landscape of primary untreated LSCC is beginning to emerge (5). Alterations to TP53, NOTCH1, CDKN2A and PIK3CA are common, while the presence of HPV is relatively rare. Meanwhile, studies of recurrent and/or metastatic LSCC suggest that with progression, the molecular landscape shifts to contain more oncogenic lesions (6), though this relationship has not been confirmed in large cohorts of matched primary and metastatic tumors. Regardless, it is clear that distribution of genetic lesions varies among tumors, and as new questions emerge, it will be important to interrogate them using appropriate models within the context of genetic status. Tissue type and genetic background will likely impact the efficacy of targeted therapies, emphasizing the need for improved understanding of the unique complexity of individual cancers (7; 8).
Cell lines serve as valuable tools for assessing the impact of genetic alterations (9–12). The University of Michigan previously created a repository of HNSCC cell lines (UM-SCC) that were characterized by short tandem repeat typing (13), and while many of these have been utilized extensively throughout the world, thorough genetic characterization has not yet been performed for cell lines derived specifically from laryngeal carcinomas (8). This limitation prevents researchers from interpreting phenotypic and therapeutic results in the context of tumor genetics. Thus, we aimed to profile the genetic and transcriptomic landscape of laryngeal UM-SCC cell lines in order to provide a molecular basis for future studies that leverage this panel.
2. Materials and Methods
2.1. UM-SCC models.
LSCC cell lines were established and characterized in the Head and Neck Oncology laboratory at the University of Michigan with written informed consent from LSCC patient donors, who were treated for LSCC between 1980 and 2011. Cell lines were maintained in exponential growth phase in DMEM with 10% FBS, 5% penicillin/streptomycin, and 5mM Non-essential amino acids in a 5% CO2 incubator.
In all cases except UM-SCC-105, due to the age of the cell lines, donor tissue from either tumor or normal tissue was unavailable for further testing.
2.2. Exome Sequencing.
Exome capture library construction was performed using the Roche NimbleGen V2 (44.1 Mbp) Exome Enrichment Kit (Otogenetics) for UM-SCC-10A, 10B, 11A, 13, 17B, 23, 25, 28, 41, 46, 76, and 81B, and paired-end sequencing (2 × 100 bp) was performed on an Illumina Genome Analyzer IIx Platform, with an average coverage of 50×. Library construction for UM-SCC-12 and 105 was performed using the Roche NimbleGen V3 and paired-end sequencing (2 × 150 bp) was performed on an Illumina HiSeq 4000 platform with average coverage of 100×. All sequencing was carried out at the University of Michigan DNA sequencing core according to standard protocol. Whole exomes are available through the Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) accession # PRJNA525437.
2.3. Variant Calling.
Quality control checks were performed on the raw sequencing data using FastQC v.0.11.5 (14). Reads were aligned to hg19 reference genome using BWA v0.7.8 (15). Duplicates were marked using PicardTools v1.79 (Broad Institute). BAM files were created by following the GATK best practices workflow (16). Variants were called on each cell line using the HaplotypeCaller producing a VCF file for each sample. These VCFs were then combined using the GenotypeGVCFs tool and a single VCF file was obtained for all the samples. Variant Quality Score Recalibration was applied to this joint VCF file to filter out low quality variants. To annotate and filter the variants of interest, the commercially available tool Goldex Helix Varseq v1.4.0 (Golden Helix, Inc., Bozeman, MT) was used. Filters were set as previously described (17).
2.4. Variant pathogenicity analysis.
The Cancer-Related Analysis of Variants Toolkit (CRAVAT; http://www.cravat.us) was used to evaluate missense and indel mutations to predict pathogenicity via the Variant Effect Scoring Tool (VEST) and driver/passenger status via the Cancer-specific High-throughput Annotation of Somatic Mutations (CHASM) tool. Missense mutations were scored with both VEST and CHASM; Indels were scored with VEST only. The scores are used to generate p-values, and a cutoff of p<0.05 was used to designate highly pathogenic (VEST) or probable driver (CHASM) mutations.
2.5. Sanger Sequencing Validation.
Genomic DNA isolation was performed using the Gentra PureGene kit (Qiagen). DNA was then PCR amplified with Platinum Taq DNA Polymerase High Fidelity (Invitrogen) following manufacturer’s instructions. PCR products were cloned into the pCR8 TOPO vector (Invitrogen) and subjected to Sanger sequencing on a 3730XL DNA Sequencer (Applied Biosystems) at the University of Michigan DNA Sequencing Core. Sequence alignment was performed using the DNASTAR Lasergene software suite.
2.6. Copy Number Analysis.
The Affymetrix OncoScan Assay Kit was used to analyze copy number alterations in the cell lines. The CEL files produced by the kit were merged to produce OSCHP files using the OncoScan Console v1.3 software. These OSCHP files were then analyzed by applying the TuScan algorithm, which is a part of the Nexus Express for OncoScan software package. From our analysis, we found a disparity between the B-Allele Frequency plot and the copy number estimate made by the TuScan algorithm in case of some homozygous deletion calls (Copy Number = 0). To improve the accuracy of copy number calls in these cases, we used the presence or absence of exome sequencing reads to confirm complete loss of the gene locus. Thus, we corrected copy number calls that were assigned a copy number call of zero by the TuScan software, but had exome sequencing reads, in Table S1 to indicate a single copy of the gene. Each of these corrections was annotated with an asterisk to denote the change. Copy number data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO; http://www.ncbi.nlm.nih.gov/geo/) and are available through GEO Series accession #GSE127231.
2.7. Transcriptome Analysis.
RNA sequencing was performed for UM-SCC-10A, 10B, 12, 17B, 23, 25, 28, 46, 81A, 81B, and 105 using Illumina stranded transcriptome library preparation kits with 75 nucleotide paired end sequencing to >100x depth on an Illumina HiSEQ4000. FPKM were calculated as previously described (17) and values for specific genes are listed in Table S2. Gene expression data from RNA-seq experiments have been deposited in the NCBI GEO and are available through GEO Series accession # GSE126975.
3. Results:
We performed exome sequencing and high-density arrays on a panel of UM-SCC cell lines generated from LSCC patients. We analyzed 16 cell lines total, generated from 14 patients. Our panel represented a range of disease states (stage I through stage IV LSCC) and included cell lines from 8 primary untreated, 3 recurrent, and 4 metastatic LSCCs (Table 1). Smoking/alcohol use was reported in all except the patient from whom UM-SCC-105 was derived, who was HPV-18 positive (18). Two matched pairs of cell lines were included in certain analyses: UM-SCC-10A and 10B, derived from primary tumor and lymph node metastasis, respectively, and UM-SCC-81A and 81B, derived from two masses resected in separate procedures.
Table 1.
Clinical characteristics of LSCC patients from whom UM-SCC cell lines were derived. UM-SCC-10A and 10B were derived from samples taken from the same patient at different times and sites. UM-SCC-81A and 81B were derived from two vocal cord masses resected in separate procedures. HPV-18 was detected in UM-SCC-105 by HPV-PCR mass array. HPV was not detected in other cell lines. Categories for which no data was available are marked with a dash; M, male; F, female; P, primary; R, recurrence; Met, metastasis; ICX, induction chemotherapy; S, surgery; RT, radiation therapy.
| Cell Line | Age | Sex | Clinical TNM | Stage | Specimen site | Type of Lesion | Previous Therapy | HPV Status | Smoking (pack-years) | Alcohol |
|---|---|---|---|---|---|---|---|---|---|---|
| UMSCC-10A | 57 | M | T3N0M0 | III | True Cord | P | None | 20 | Heavy | |
| UMSCC-10B | 58 | M | rT3N1M0 | III | Lymph Node | Met | S | 20 | Heavy | |
| UMSCC-11A | 65 | M | T2N2aM0 | IV | Epiglottis | P | ICX | 35 | Heavy | |
| UMSCC-12 | 71 | M | T2N1M0 | III | Larynx | R | S, RT | - | - | |
| UMSCC-13 | 60 | M | T3N0M0 | III | Stoma | R | RT,S | 90 | Heavy | |
| UMSCC-17B | 47 | F | T1N1M0 | III | Soft tissue-neck | Met | RT | 40 | None | |
| UMSCC-23 | 36 | F | T2N0M0 | II | Supraglottis | P | None | 48 | Heavy | |
| UMSCC-25 | 50 | M | rT3N2bM0 | III | Lymph Node | Met | RT,S,S | Heavy | Heavy | |
| UMSCC-28 | 61 | F | T1N0M0 | I | True Cord | P | None | 40 | - | |
| UMSCC-41 | 78 | M | T2N1M0 | III | Arytenoid | P | None | 50 | Moderate | |
| UMSCC-46 | 58 | F | T2N2M0 | IV | Supraglottis | R | RT,S | 60 | Heavy | |
| UMSCC-57 | 69 | M | - | - | Supraglottis | - | - | - | - | |
| UMSCC-76 | 66 | M | T3N2cM0 | IV | Lymph Node | Met | ICX | 40 | Heavy | |
| UMSCC-81A | 53 | M | T2N0M0 | II | Anterior Commisure | P | None | 100 | Heavy | |
| UMSCC-81B | 58 | M | T2N0M0 | II | True Cord | P | None | 100 | Heavy | |
| UMSCC-105 | 51 | M | T4N0M0 | IV | True Cord | P | None | HPV-18 | 0 | None |
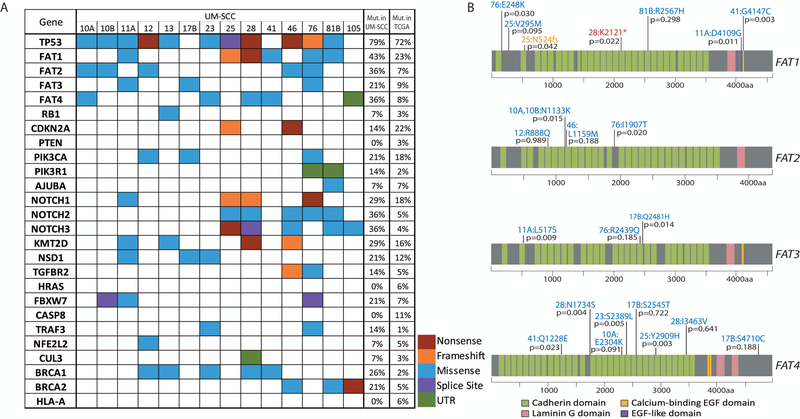
Comprehensive capture based exome sequencing was performed on 14 cell lines. Our analysis showed a large mutational load, with ~30–50 nonsynonymous mutations identified per Mb (Figure S1). To annotate the cell line panel, we assessed common genetic aberrations previously reported by the HNSCC TCGA consortium (5). We identified non-synonymous mutations affecting several of these genes, including TP53 in 11/14 (79%) and FAT atypical cadherin 1 (FAT1) in 6/14 (43%) of cell lines (Figure 1A). Table S3 lists the specific mutations observed. Mutation rates for each gene are provided as compared to TCGA HNSCC data (5; 19). Importantly, our study lacks matched normal samples, and therefore cannot account for germline variants, although most genes were mutated with similar frequencies in the LSCC cell line panel as in the TCGA tumors. Notable exceptions included FAT and NOTCH family genes and BRCA1/2, which are mutated at higher rates in our models than in TCGA specimens.
Figure 1. Single nucleotide variants identified in the laryngeal UM-SCC cell line panel.
(A) Non-synonymous mutations in laryngeal UM-SCC cell lines as called using Nimblegen capture-based exome sequencing were color coded by mutation type as indicated. Genes selected for this analysis were previously found to be mutated in the HNSCC TCGA project (5), or were otherwise of interest as common cancer-related genes (FAT, NOTCH, and BRCA families). SNPs occurring in introns were excluded with the exception of those affecting splice sites. Also excluded were SNPs occurring at a frequency >0.01 according to the 1000 Genomes Project. The percentage of cell lines in the LSCC panel harboring a mutation in each gene is indicated immediately to the right. The percentage of TCGA HNSCC samples (TCGA provisional dataset) harboring a point mutation in each gene is noted in the rightmost column (http://cancergenome.nih.gov/). (B) Schematic diagrams of FAT1–4 proteins showing distribution of mutations found in the FAT1 gene in UM-SCC laryngeal cell lines. Resulting amino acid changes are noted for each mutation next to the cell line in which it was identified. VEST-4 pathogenicity score is indicated as a p-value for each mutation.
The FAT family mutations identified in our LSCC cell line panel are depicted in Figure 1B and were validated by Sanger sequencing (Figure S2). For each FAT mutation identified in our panel, we used the Variant Effect Scoring Tool (VEST4) (20–22) to predict pathogenic impact. Variant score p-values are reported for each mutation in Figure 1B and support a pathogenic impact on FAT1 function of 5/7 of the identified alterations (p<0.05). Interestingly, FAT4 mutations were also especially prevalent, and VEST4 scores predicted a pathogenic impact in 5/8 cases. For missense mutations, the Cancer-specific High-throughput Annotation of Somatic Mutations (CHASM-3.1) tool was used to predict driver mutations (Table S4). Similar analysis was completed for the NOTCH family genes, and BRCA1/2 alterations identified in our panel as these genes were altered at slightly higher than expected rates. While VEST pathogenicity scores for BRCA1/2 were not significant, the CHASM predictor of tumorigenic impact classified the mutations reported in both genes as likely driver mutations (Table S4).
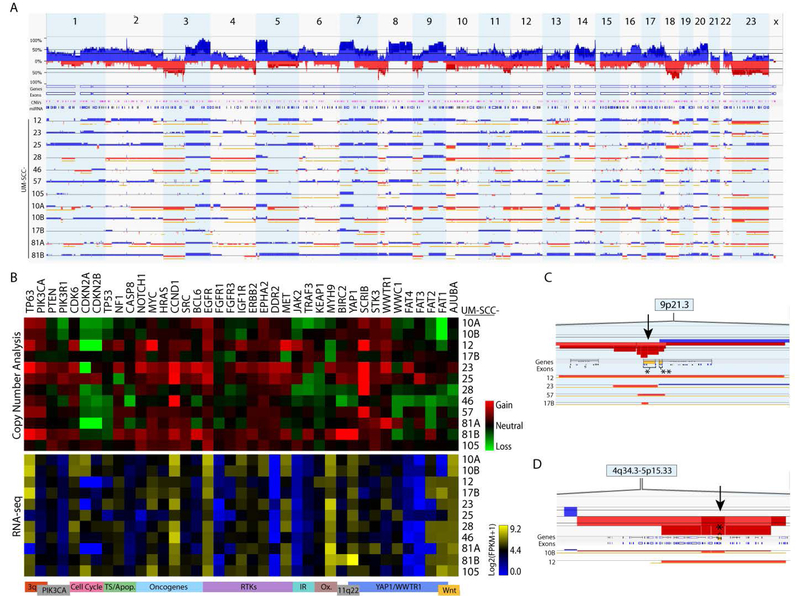
After annotating molecular alterations found in the panel, we assessed copy number alterations in 12 LSCC cell lines via high-density SNP arrays to provide additional molecular detail. We first performed a combined analysis of all 12 lines by summing copy number alterations at each probe site (Figure 2A). Our analysis revealed common copy number alterations in the cell line panel consistent with those reported in previous HNSCC studies, including broad amplifications of chromosome 3q, 5p, 7p, 8q, and 20q arms, and deep deletions in the chromosome 3p, 8p, 9p, 11q, and 18q arms (5; 23–25). Importantly, the 3q amplicon includes transcription factors TP63 and SOX2, as well as the oncogene PIK3CA. As 35% of HNSCCs in the TCGA study harbor an alteration in PIK3CA, it is widely considered a potential therapeutic target, with several clinical trials investigating PI3K inhibitors in HNSCC patients (26). Additionally, both broad and focal deletions were observed in the 4q35 region containing the FAT1 gene. Thus, our analysis suggests that this panel as a whole recapitulates the landscape of major chromosomal aberrations found in HNSCC tissues.
Figure 2. Genetic characterization of laryngeal UM-SCC cell lines by copy number analysis.
Genomic DNA was harvested from low passage UM-SCC cell lines and analyzed using high density SNP arrays (Affymetrix OncoScan Assay) and compared to a commercially available pooled control. Affymetrix software was used to call copy number alterations. (A) Copy number alterations were summed across UM-SCC-10A, 10B, 12, 17B, 23, 25, 28, 46, 57, 76, 81A, 81B, and 105. Alterations for individual cell lines are shown below with gains indicated in blue and losses indicated in red. (B) Heat maps displaying median copy numbers (upper panel) and RNA expression (lower panel) for selected genes. Key functions and relevant chromosomal regions are noted below each column. (C) Focal deletions (arrow) at the CDKN2A-CDKN2B (9p21) locus occurring in UM-SCC 12, 23, 57, and 17B. The CDKN2A gene is indicated by a bracket in the row labeled “Genes.” CDKN2A,* CDKN2B**. (D) Focal deletions (arrow) at the FAT1 locus (4q35) occurring in UM-SCC-10B and 12. The FAT1 locus is indicated by a bracket and an asterisk in the row labeled “Genes.”
Next, we further interrogated our panel to characterize key genes and pathways. Genes chosen for analysis were previously identified as commonly altered in the TCGA HNSCC cohort (5), are otherwise implicated in HNSCC pathogenesis (SRC, BCL6, JAK2), or are reportedly linked to FAT1 signaling (SCRIB, STK3, WWTR1, WWC1, MTNR1A, FAT3). Copy number calls are reported in Table S1. Median copy numbers are depicted in a heat map (Figure 2B, upper panel), with numerical values provided in Table S5. We refer to median values ≥0.5 as amplifications and values ≤ −0.5 as copy losses. Consistent with TCGA findings, we observed amplifications of EGFR in 8/12 cell lines, and amplifications of PIK3CA in 5/12. Copy losses at the CDKN2A-CDKN2B locus were especially prevalent (10/12 cell lines). We observed broad 9p deletions in 6/12 cell lines, with an additional 4 cell lines exhibiting focal deletions at the CDKN2A-CDKN2B locus (Figure 2C). We also performed RNAseq for a subset of UM-SCC larynx cell lines (Figure 2B, lower panel). As expected, EGFR was highly expressed in all cell lines. In many cases, copy number alterations corresponded with variations in gene expression (Figure S3). For example, we report deletion and low expression of CDKN2A/B in UM-SCC-12 and 81A, as well as amplification and high expression of YAP1 in UM-SCC-81B. We also asked whether cell lines harboring nonsense mutations likely to confer loss of function might exhibit altered gene expression, but in this small sample size we observed no trends with regard to RNA expression and mutation status (Figure S4).
FAT1 copy loss was observed in 4/12 cell lines in this analysis, with focal deletions in UM-SCC-10B and 12 (Figure 2D). Notably, FAT1 was also a commonly mutated gene in our panel (Figure 1A). Interestingly, of those cell lines that lacked a point mutation, some (UM-SCC 10A, 10B, and 46) did exhibit FAT1 deletions, for a total of 9 cell lines with potential loss of FAT1 function. Some cell lines exhibited loss of multiple FAT family genes: FAT1 and FAT2 losses were both observed in the UM-SCC-10A/B pair, and losses of all 4 FAT genes were observed in UM-SCC-46 (Figure 2B). However, RNA-seq indicated high expression of FAT1 in most cell lines, with the exception of UM-SCC-10A/B.
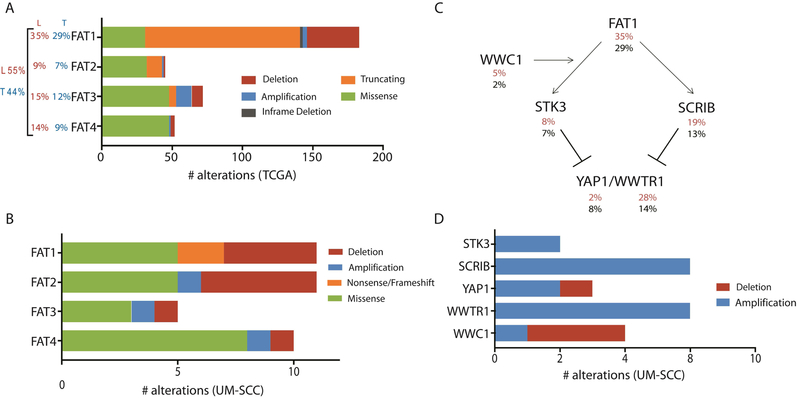
Given the high rate of FAT1 alterations with predicted functional impact in our panel, we sought to summarize alterations in FAT family genes. FAT1 alterations occurred in 35% of the 110 LSCCs in the TCGA cohort and 29% of the overall cohort (Figure 3A) (19), and of the 9 UM-SCC cell lines with both copy number and single nucleotide variant data available, 7 exhibited loss of function alterations in FAT1, consistent with its purported role as a tumor suppressor. 55% of LSCCs and 44% of all HNSCCs in the TCGA study harbored at least one FAT family gene alteration. Among these samples, the majority 110/143 (77%) of FAT1 mutations are reported as truncating mutations (Figure 3A) which was consistent with our observation of missense mutations, a frameshift, and a stopgain among UM-SCC lines (Figure 1). Figure 3B summarizes FAT family alterations observed in the cell line panel. While most FAT1 mutations were truncating or deep deletions, mutations in other FAT family members were predominantly missense mutations in TCGA samples and UM-SCC cell lines.
Figure 3. Summary of aberrations in FAT related genes in TCGA tumors and laryngeal UM-SCC cell lines.
(A) Alterations reported in TCGA provisional dataset (http://cbioportal.org; http://cancergenome.nih.gov/) for FAT1–4. Total numbers observed for each category of mutation or copy number variation are displayed. For each gene, the percentage of 510 tumors in the total HNSCC dataset with an alteration is reported to the left in black (T). The percentage of the 110 laryngeal primary tumors in this dataset harboring an alteration in each gene is reported in red (L). The percentage of all HNSCC (black) and laryngeal SCC (30) harboring one or more mutations in any FAT gene are reported in the far left. (B) Alterations identified in the laryngeal UM-SCC cell line panel for FAT1–4. This analysis considers available data for all 16 cell lines in this study, although exome sequencing was not performed for UM-SCC-57 or 81A, and copy number data is not available for UM-SCC-11A, 13, 41, or 76. (C) Schematic diagram describing proposed signaling interactions involving the FAT1 protein. Percentages of HNSCC tumors bearing alterations (amplifications, deletions, and mutations) in each gene are displayed below the gene name for LSCC only (30) and the overall cohort (black). (D) Alterations identified in the laryngeal UM-SCC cell line panel for Hippo/YAP1 pathway genes.
Our data support a model where functionally recurrent alterations to multiple genes within a pathway contribute to overall pathway disruption. To further understand functional recurrence of alterations to FAT signaling, we next examined alterations to genes linked to FAT1 signaling in both the TCGA dataset and our cell line panel. FAT1 has been shown to inhibit Hippo/YAP1 pathway-induced proliferation and survival through its interactions with Scribble (SCRIB) and Serine/Threonine Kinase 3 (STK3) (27–29). Figure 3C summarizes the prevalence of genetic alterations in these genes identified in TCGA primary larynx tumors (30) and all TCGA HNSCC tumors (black). From this summary, it appears that alterations to FAT signaling are more common in LSCC than other HNSCC disease sites; in particular, we noted that alterations affecting WWTR1, a YAP1 paralog, were especially prevalent in larynx tumors, with 31/110 (28%) larynx tumors harboring a WWTR1 amplification, compared with 39/394 (10%) at other subsites. Unfortunately, the relatively low number of tumors from each subsite limits the ability to test this association statistically.
In the UM-SCC panel, we observed broad copy gains to 11q22, which contains the YAP1 gene, in 2 /12 cell lines, further implicating Hippo/YAP1 activation in promoting growth and survival in these models (Figure 2B). WWTR1 is also frequently amplified, with copy gains occurring in 6/12 cell lines. This is consistent with frequent WWTR1 amplifications observed in the TCGA HNSCC dataset. Figure 3D summarizes alterations to Hippo/YAP1 pathway genes in UM-SCC cell lines. Interestingly, contrary to their documented tumor suppressive functions, amplifications of both STK3 and SCRIB were observed in 2/14 and 6/14 cell lines, respectively (Figure 2B), with modest copy gains observed in several additional cell lines. This is consistent with the broad 8q copy gains observed in the UM-SCC panel and TCGA data (5).
Also linked to this pathway is the KIBRA protein, encoded by WWC1, which is thought to promote the phosphorylation and inhibition of YAP1 and WWTR1 (31). We observed WWC1 loss in 3/12 cell lines, consistent with a role in dampening Hippo/YAP1 signaling, although UM-SCC-81A exhibits a modest copy gain. Furthermore, NOTCH3, mutated in 5/14 cell lines in our mutation analysis, was recently shown to act as a tumor suppressor in breast cancer cells by inducing KIBRA upregulation (32). Overall, the prevalence of alterations in FAT1 related genes supports a role for Hippo/YAP1 and FAT family signaling in these models, warranting further investigation of this network in LSCC.
4. Discussion
As precision medicine protocols are developed, comprehensive genetic stratification of tumors becomes increasingly crucial to correlate with disease prognosis and to target known driver mutations (33). Large-scale, integrated analyses have recently provided unparalleled molecular detail toward stratification of tumors, paving the way for precision medicine protocols based upon comprehensive molecular profiles (5; 25). To advance novel targets and combinations, an array of well-characterized models representative of the diversity of disease observed in the clinic must also develop in tandem, as preliminary studies using these tools are critical to predicting therapeutic response. The UM-SCC cell line panel is widely used to model HNSCC, but genetic characterization of these cell lines has been limited. Furthermore, LSCC is a challenging clinical entity, with limited response to current treatment modalities and poor survival rates, especially in recurrent disease (34). We therefore sought to create a profile of UM-SCC cell lines derived from LSCC patients to better understand these models and determine how accurately they reflect genetic characteristics of patients.
Overall, our analysis indicates that many aberrations recurrently identified in the HNSCC TCGA study are well represented in the UM-SCC larynx cell line panel. EGFR, PIK3CA, and CCND1 copy gain, CDKN2A copy loss, and TP53 and FAT1 mutation are among the most common aberrations observed in our panel, consistent with reports of clinical specimens. Thus, this panel appears to adequately represent many well-studied, targetable alterations in HNSCC, and should serve as an important tool in advancing combination therapies targeting these pathways. Importantly, there were some genes (FBXW7, BRCA1, NOTCH2, NOTCH3) for which our analysis indicated considerably higher mutation rates than those observed in the TCGA dataset. Such discrepancies may be attributed to our small sample size, differences in variant calling pipelines, or to the fact that the TCGA report includes only somatic mutations, while our analysis cannot distinguish somatic mutations from germline. It is also possible that certain alterations have been selected for during cell line derivation and culture.
Notably, significant variation in molecular profiles exists within tumor sets and across the UM-SCC panel. For example, UM-SCC-17B is strikingly devoid of point mutations and copy number alterations, but harbors a PIK3CA hotspot mutation at the 3:178938934 position (Table S3) (35). Likewise, UM-SCC-105, an HPV-18 positive cell line, appears similarly genomically stable, but harbors a pathogenic nonsense mutation in BRCA2. Identification and stratification of such molecular subsets will benefit research applications and could aid in selection of appropriate models based on patient characteristics.
Our data show an unexpectedly high prevalence of FAT1 inactivating mutations or genetic deletions and support a deeper analysis of the pathway. While FAT1 alterations are well documented in HNSCC, few reports address FAT2, FAT3 and FAT4, which have lower alteration rates than FAT1 in the TCGA dataset (Figure 3A). In our cell line panel, we found that FAT family alterations were prevalent, and UM-SCC-10A, 10B, and 46 harbored alterations in multiple FAT genes. We observed FAT1 and FAT2 copy loss in the UM-SCC-10A/10B pair, along with both copy loss and mutation in FAT4 in UM-SCC-10A. Furthermore, a FAT2 mutation with high predicted pathogenic impact was identified in both cell lines, further supporting a prominent pathogenic role for FAT genes in these particular models.
Although FAT3 is considered paralogous to FAT1 and exhibits similar functions (29), fewer FAT3 alterations were discovered in the TCGA cohort, and these appeared less likely to confer loss of function, consisting of a mix of amplifications, deletions, missense mutations, and truncating mutations (Figure 3A). Similarly, in the 12 cell lines subjected to copy number analysis, we observed one loss and one gain in FAT3 (in UM-SCC-46 and −25, respectively) (Figure 2B, 3B). Only three FAT3 mutations were observed in our cell line panel, and all were missense mutations (Figure 1). However, UM-SCC-11A and 17B both harbored mutations classified as pathogenic by the VEST tool, and the mutation in 11A is a predicted driver according to the CHASM score. Interestingly, expression of FAT3 was very low in most cell lines in our panel (Figure 2B). This suggests that further dissection of FAT3 genetic alterations may be required to understand how each type of alteration affects FAT3 pathway activity.
When we expanded our analysis to additional genes linked to FAT signaling, we noted frequent copy number alterations consistent with dysregulated Hippo/YAP1 signaling, both in our cell lines and in the TCGA dataset, particularly affecting WWTR1. While amplifications of purported tumor suppressors STK3 and SCRIB were also observed, this may be due to the fact that both genes are located in a broadly amplified region of 8q. Although a mechanistic role for STK3 and SCRIB in promoting tumorigenesis has not been clearly defined, the seemingly paradoxical overexpression of these proteins is commonly reported in human cancers (36).
FAT1 is in the cadherin class of membrane-bound proteins, with functions that remain to be fully characterized. Notably, FAT1 mutations may have context-dependent effects depending on the tissue source. In HNSCC and esophageal squamous cell carcinomas, it appears to act as a tumor suppressor gene, inhibiting epithelial-mesenchymal transition and cell proliferation (37), whereas in other tumors, it may have oncogenic function (38; 39). There is limited understanding of the role of FAT1 in HNSCC in general, apart from the high mutational rate reported in the recent TCGA study. Interestingly, FAT1 mutant HNSCCs may have better overall survival (40), suggesting that it may portend a better prognosis for which clinical treatment modification may be investigated. A prognostic role for FAT1 will need to be investigated in confirmatory cohorts, and further characterization of tumors harboring FAT1 alterations will be necessary. Recently, Martin et al showed that FAT1 participates in assembly of a Hippo signaling complex responsible for negatively regulating YAP1 in HNSCC cell lines, thus its loss may result in unrestrained YAP1 activity (30). Reintroduction of FAT1 intracellular domain into FAT1 deficient cell lines resulted in decreased YAP1 activity, reduced proliferation, and abrogated tumorigenesis in vivo. These effects were rescued by YAP1 overexpression. A 2017 study by Pan et al assessed YAP1 protein by immunohistochemistry in 121 LSCC tumor samples, and found positive YAP1 expression to be associated with clinical stage, TNM classification, lymph node metastasis, and poor overall survival (41). Taken together, these studies support YAP1 as a promising therapeutic target in the context of genetic alterations in FAT1 and the Hippo signaling pathway.
Another recent study identified a potential interaction between FAT1 and CASP8 in oral cavity SCC (42), showing increased growth and migration in cell lines with FAT1 loss of function, further corroborating the functional role of FAT1 as a tumor suppressor gene. FAT1 may function as a tumor suppressor by binding to β-catenin and blocking its nuclear translocation, thereby inhibiting Wnt signaling pathways fundamental to growth and proliferation (38). As discussed above, FAT1 may inhibit the YAP1 pathway, which is critical for cell growth and survival. Thus, with FAT1 loss of function in LSCC, there may be unchecked tumor cell growth and proliferation through both Wnt/β-catenin and YAP1 pathways. In selecting targeted therapies in LSCC, consideration of FAT1 status may be beneficial, as agents inhibiting the Wnt/β-catenin pathway, many of which are in development, may be particularly efficacious.
The UM-SCC larynx cell line panel has been in use in laboratories throughout the world for the past several decades. We now provide a comprehensive genetic characterization of these models that can be used to contextualize past and future studies in terms of the genetic diversity seen in patients. However, in utilizing cell lines as model systems it is important to note the likelihood of variations between stocks of the same cell line. In support of the concept that genetic diversity exists between models cultured separately, cytogenetic analysis was originally performed for UM-SCC-17A/B at several different passages by Carey et al in 1989 (43). There were no karyotypic differences between UM-SCC-17A cells analyzed at passages 8, 23, 28, and 37, nor between UM-SCC-17B cells at passages 13, 17, and 52. However, a UM-SCC-17A subline was also discovered, differing from the UM-SCC-17A stemline both at the cytogenetic level and by expression of various surface antigens, including the E7 and A9 antigens. This subline was believed to represent a distinct population present in the primary tumor, indicating that multiple heterogeneous populations existed initially. Furthermore, Ludwig et al performed comprehensive profiling of the UM-SCC oral cavity cell line panel, and provided evidence of multiple clones through copy number analysis and fluorescence in situ hybridization (17).
The concept of cell line evolution in culture was highlighted more recently in a comprehensive characterization of 27 MCF7 strains in which the authors observed considerable variations in genetics, gene expression programs, morphology, and drug response (44). Many of the cell lines discussed here have been distributed to laboratories throughout the world, and genetic drift and divergence among lineages cultured in different laboratories is highly likely. As the purpose of the present study is to offer a baseline profile of the LSCC cell lines, a direct comparison between the genetics of our cell lines and lineages propagated in other laboratories is beyond the scope of this report. However, we do describe and reference many of the same genetic alterations reported by other laboratories. For example, a 2018 study by Cheng et al assessed mutations and copy number variations in a panel of 26 HNSCC cell lines, notably including UM-SCC-46 and UM-SC-105 (45). We recapitulate many of their findings in UM-SCC-46, such as 3q copy gain, YAP1/BIRC2 deletion, a TP53 nonsense mutation, and KMT2D frameshift. Cheng et al also report a 3q gain in UM-SCC-105, as well as CASP8 deletion, etc., which our study did not detect further supporting the concept of potential genetic drift between models cultured in different labs over time. Additionally, while Cheng et al report FAT1 copy gain in nearly all cell lines, including UM-SCC-46 and 105, our data show copy loss in UM-SCC-46 and no alteration in UM-SCC-105.
Further, Nisa et al analyzed alterations in several UM-SCC cell line pairs in 2018, including the UM-SCC-10A/B, 17A/B, and 81A/B pairs (46). They report several differences between the 10A (primary tumor) and 10B (lymph node metastasis) lines, including as a FAT4 mutation only in 10A, an observation recapitulated in our study. We also reproduce their findings of TP53 and FAT2 mutations in both lines, but interestingly, Nisa et al also report FAT1, 2, and 4 mutations in UM-SCC-17B, as well as a PTEN mutation in UM-SCC-81B, which we did not observe.
The present study emphasizes the utility of continuing to expand the available array of well-characterized HNSCC cell lines. Importantly, this report also highlights an underappreciated but broad range of molecular alterations to multiple genes associated with FAT signaling and supports a need to deeply dissect the function of this pathway in HNSCC pathogenesis. As we refine our understanding of molecular complexity and heterogeneity in HNSCC, our study provides a foundation for modeling therapeutic responses and advancing personalized medicine protocols.
Supplementary Material
Grant Support:
J.C.B. and T.E.C. received funding from NIH Grants U01-DE025184, P30-CA046592 and R01-CA194536. M.L. was funded by F31-CA206341 and T32-DC005356. J.E.M. was funded by F31-DE027600 and 5T32CA140044–07. H.J. received funding from NIH Grant U01-DE025184 and P30-CA046592.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. 2011. Global cancer statistics. CA Cancer J Clin 61:69–90 [DOI] [PubMed] [Google Scholar]
- 2.Gillison ML, D’Souza G, Westra W, Sugar E, Xiao W, et al. 2008. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. Journal of the National Cancer Institute 100:407–20 [DOI] [PubMed] [Google Scholar]
- 3.Mendenhall WM, Werning JW, Hinerman RW, Amdur RJ, Villaret DB. 2004. Management of T1-T2 glottic carcinomas. Cancer 100:1786–92 [DOI] [PubMed] [Google Scholar]
- 4.Forastiere AA, Weber RS, Trotti A. 2015. Organ Preservation for Advanced Larynx Cancer: Issues and Outcomes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 33:3262–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas N. 2015. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517:576–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris LG, Chandramohan R, West L, Zehir A, Chakravarty D, et al. 2016. The Molecular Landscape of Recurrent and Metastatic Head and Neck Cancers: Insights From a Precision Oncology Sequencing Platform. JAMA oncology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birkeland AC, Ludwig ML, Meraj TS, Brenner JC, Prince ME. 2015. The Tip of the Iceberg: Clinical Implications of Genomic Sequencing Projects in Head and Neck Cancer. Cancers 7:2094–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ludwig ML, Birkeland AC, Hoesli R, Swiecicki P, Spector ME, Brenner JC. 2016. Changing the paradigm: the potential for targeted therapy in laryngeal squamous cell carcinoma. Cancer biology & medicine 13:87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grenman R, Carey TE, McClatchey KD, Wagner JG, Pekkola-Heino K, et al. 1991. In vitro radiation resistance among cell lines established from patients with squamous cell carcinoma of the head and neck. Cancer 67:2741–7 [DOI] [PubMed] [Google Scholar]
- 10.Bradford CR, Zhu S, Ogawa H, Ogawa T, Ubell M, et al. 2003. P53 mutation correlates with cisplatin sensitivity in head and neck squamous cell carcinoma lines. Head & neck 25:654–61 [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Pan S, Hsieh MH, Ng N, Sun F, et al. 2013. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proceedings of the National Academy of Sciences of the United States of America 110:20224–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akervall J, Guo X, Qian CN, Schoumans J, Leeser B, et al. 2004. Genetic and expression profiles of squamous cell carcinoma of the head and neck correlate with cisplatin sensitivity and resistance in cell lines and patients. Clin Cancer Res 10:8204–13 [DOI] [PubMed] [Google Scholar]
- 13.Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, et al. 2010. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head & neck 32:417–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews S FastQC A Quality Control tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 15.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics (Oxford, England) 25:1754–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics 43:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludwig ML, Kulkarni A, Birkeland AC, Michmerhuizen NL, Foltin SK, et al. 2018. The genomic landscape of UM-SCC oral cavity squamous cell carcinoma cell lines. Oral oncology 87:144–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walline HM, Goudsmit CM, McHugh JB, Tang AL, Owen JH, et al. 2017. Integration of high-risk human papillomavirus into cellular cancer-related genes in head and neck cancer cell lines. Head & neck 39:840–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. 2012. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery 2:401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. 2013. Identifying Mendelian disease genes with the variant effect scoring tool. BMC genomics 14 Suppl 3:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douville C, Carter H, Kim R, Niknafs N, Diekhans M, et al. 2013. CRAVAT: cancer-related analysis of variants toolkit. Bioinformatics (Oxford, England) 29:647–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douville C, Masica DL, Stenson PD, Cooper DN, Gygax DM, et al. 2016. Assessing the Pathogenicity of Insertion and Deletion Variants with the Variant Effect Scoring Tool (VEST-Indel). Human mutation 37:28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liehr T, Ries J, Wolff E, Fiedler W, Dahse R, et al. 1998. Gain of DNA copy number on chromosomes 3q26-qter and 5p14-pter is a frequent finding in head and neck squamous cell carcinomas. International journal of molecular medicine 2:173–9 [DOI] [PubMed] [Google Scholar]
- 24.Carey TE, Frank CJ, Raval JR, Jones JW, McClatchey KD, et al. 1997. Identifying genetic changes associated with tumor progression in squamous cell carcinoma. Acta oto-laryngologica. Supplementum 529:229–32 [DOI] [PubMed] [Google Scholar]
- 25.Campbell JD, Yau C, Bowlby R, Liu Y, Brennan K, et al. 2018. Genomic, Pathway Network, and Immunologic Features Distinguishing Squamous Carcinomas. Cell reports 23:194–212.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai Y, Dodhia S, Su GH. 2017. Dysregulations in the PI3K pathway and targeted therapies for head and neck squamous cell carcinoma. Oncotarget 8:22203–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed AF, de Bock CE, Lincz LF, Pundavela J, Zouikr I, et al. 2015. FAT1 cadherin acts upstream of Hippo signalling through TAZ to regulate neuronal differentiation. Cellular and Molecular Life Sciences 72:4653–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao B, Li L, Lei Q, Guan KL. 2010. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes & development 24:862–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katoh M 2012. Function and cancer genomics of FAT family genes. International Journal of Oncology 41:1913–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin D, Degese MS, Vitale-Cross L, Iglesias-Bartolome R, Valera JLC, et al. 2018. Assembly and activation of the Hippo signalome by FAT1 tumor suppressor. Nature Communications 9:2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hill VK, Dunwell T, Catchpoole D, Krex D, Brini AT, et al. 2011. Frequent epigenetic inactivation of KIBRA, an upstream member of the Salvador/Warts/Hippo (SWH) tumor suppressor network, is associated with specific genetic event in B-cell acute lymphocytic leukemia. Epigenetics 6:326–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Liu X, Luo J, Xiao W, Ye X, et al. 2016. Notch3 inhibits epithelial-mesenchymal transition by activating Kibra-mediated Hippo/YAP signaling in breast cancer epithelial cells. In Oncogenesis, p. e269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giefing M, Wierzbicka M, Szyfter K, Brenner JC, Braakhuis BJ, et al. 2016. Moving towards personalised therapy in head and neck squamous cell carcinoma through analysis of next generation sequencing data. European journal of cancer 55:147–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Birkeland AC, Beesley L, Bellile E, Rosko AJ, Hoesli R, et al. 2017. Predictors of survival after total laryngectomy for recurrent/persistent laryngeal squamous cell carcinoma. Head & neck 39:2512–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, et al. 2016. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nature biotechnology 34:155–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaira V, Faversani A, Dohi T, Maggioni M, Nosotti M, et al. 2011. Aberrant Overexpression of the Cell Polarity Module Scribble in Human Cancer. The American Journal of Pathology 178:2478–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu X, Zhai Y, Kong P, Cui H, Yan T, et al. 2017. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer letters 397:83–93 [DOI] [PubMed] [Google Scholar]
- 38.Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, et al. 2013. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nature genetics 45:253–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valletta D, Czech B, Spruss T, Ikenberg K, Wild P, et al. 2014. Regulation and function of the atypical cadherin FAT1 in hepatocellular carcinoma. Carcinogenesis 35:1407–15 [DOI] [PubMed] [Google Scholar]
- 40.Kim KT, Kim BS, Kim JH. 2016. Association between FAT1 mutation and overall survival in patients with human papillomavirus-negative head and neck squamous cell carcinoma. Head & neck 38 Suppl 1:E2021–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan C, Du Z, Cai Z, Liu Y, Sun Y, et al. 2017. Elevated expression of yesassociated protein is associated with the malignant status and prognosis of laryngeal squamous cell carcinoma. Molecular medicine reports 16:4934–40 [DOI] [PubMed] [Google Scholar]
- 42.Hayes TF, Benaich N, Goldie SJ, Sipila K, Ames-Draycott A, et al. 2016. Integrative genomic and functional analysis of human oral squamous cell carcinoma cell lines reveals synergistic effects of FAT1 and CASP8 inactivation. Cancer letters 383:106–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carey TE, Van Dyke DL, Worsham MJ, Bradford CR, Babu VR, et al. 1989. Characterization of Human Laryngeal Primary and Metastatic Squamous Cell Carcinoma Cell Lines UM-SCC-17A and UM-SCC-17B. Cancer Research 49:6098. [PubMed] [Google Scholar]
- 44.Ben-David U, Siranosian B, Ha G, Tang H, Oren Y, et al. 2018. Genetic and transcriptional evolution alters cancer cell line drug response. Nature 560:325–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng H, Yang X, Si H, Saleh AD, Xiao W, et al. 2018. Genomic and Transcriptomic Characterization Links Cell Lines with Aggressive Head and Neck Cancers. Cell reports 25:1332–45.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nisa L, Barras D, Medová M, Aebersold DM, Medo M, et al. 2018. Comprehensive Genomic Profiling of Patient-matched Head and Neck Cancer Cells: A Preclinical Pipeline for Metastatic and Recurrent Disease. Molecular Cancer Research 16:1912. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.