Abstract
Treatment with vemurafenib, a potent and selective inhibitor of MAPK signaling downstream of the BRAFV600E oncogene, elicits dramatic clinical responses in patients with metastatic melanoma. Unfortunately, the clinical utility of this drug is limited by a high incidence of drug resistance. Thus, there is an unmet need for alternative therapeutic strategies to treat vemurafenib-resistant metastatic melanomas. We have conducted a high-throughput screening of two bioactive compound libraries (Siga and Spectrum libraries) against a metastatic melanoma cell line (A2058) and identified two structurally analogous compounds, deguelin and rotenone from a cell viability assay. Vemurafenib-resistant melanoma cell lines, A2058R and A375R (containing the BRAFV600E mutation), also showed reduced proliferation when treated with these two compounds. Deguelin, a mitochondrial complex I inhibitor, was noted to significantly inhibit oxygen consumption in cellular metabolism assays. Mechanistically, deguelin treatment rapidly activates AMPK signaling, which results in inhibition of mTORC1 signaling and differential phosphorylation of mTORC1’s downstream effectors, 4E-BP1 and p70S6 kinase. Deguelin also significantly inhibited ERK activation and Ki67 expression without altering Akt activation in the same timeframe in the vemurafenib-resistant melanoma cells. These data posit that treatment with metabolic regulators such as deguelin can lead to energy starvation thereby modulating the intracellular metabolic environment and reducing survival of drug-resistant melanomas harboring BRAFV600E mutations.
INTRODUCTION
Melanoma is one of the most aggressive and treatment-resistant human cancers. Besides early careful resection, no helpful methodology has been found to completely eliminate the possibility of recurrence. However, in recent years there has been a breakthrough in the understanding the molecular basis of the disease that has led to therapeutic strategies that produce dramatic clinical responses1. Mutations incurred by melanocytes during the process of malignant transformation result in hyperactivation of the mitogen-activated protein kinase (MAPK) and the PI3-K/Akt pathways1,2. Genetic alterations in BRAF are the most frequent activating mutations, accounting for 50–70% of oncogene mutations in melanoma tumors. The BRAFV600E mutation is the most prevalent alteration in malignant melanoma, which when combined with loss of the tumor suppressor PTEN causes tumor transformation and metastatic progression3,4.
Patients with metastatic melanoma harboring the BRAFV600E mutation became the focus for the development of BRAF inhibitors, including vemurafenib and dabrafenib. These drugs which have displayed significant efficacy especially when combined with the MEK½ inhibitor trametinib5-7. Despite the fact that initial responses and tumor control with BRAF inhibitors are impressive, a major challenge is limited durability of response due to development of resistance through multiple mechanisms. Tumor resistance inevitably leads to disease relapse within 6 to 8 months of the start of therapy8. Most patients who do respond initially, eventually develop acquired resistance to MAPK inhibitors through mechanisms including alterations in BRAF splicing, reactivation of the MAP kinase pathway, constitutive activation of receptor tyrosine kinase, overexpression of epidermal growth factor receptor as well as activation of the PI3K-Akt signaling8,9. Hence, there is a dire need for improved combinatorial and alternative treatment modalities that can be used to prevent disease progression and improve survival for patients who develop resistance to BRAF inhibition.
Altered metabolism is a hallmark of cancer cells which manifests as increased dependence on aerobic glycolysis, fatty acid and nucleotide synthesis, and glutaminolysis. This altered state is important for cancer maintenance and progression, and it is tightly linked to oncogenic signaling. Furthermore, melanocytes’ unique physiological role in melanin production ties together melanogenesis, cellular metabolism, and tumorigenesis. For example, it has been shown that induction of melanogenesis elevates hypoxia induced factor 1 (HIF-1) expression along with its target genes. These target genes are associated with angiogenesis, glucose metabolism, and stimulation of key glycolytic enzymes10. Metabolic alterations are therefore good targets for the design of novel therapeutic strategies for treatment of cancers.11,12. Resistance to BRAF inhibitors in melanoma has been shown to result in induction of oxidative phosphorylation (OXPHOS) and mitochondrial biogenesis13. Here, we have performed a high-throughput drug screen to evaluate the selectivity of drugs, that have an effect alone and in combination with vemurafenib, in the context of vemurafenib-resistant in BRAFV600E mutant melanoma cell lines. The MicroSource Spectrum library, comprised of 2,000 compounds including FDA approved drugs, natural products, and known bioactives, and a selection of 6,720 compounds from the Siga library were screened for reduced proliferation and induced cell death in vemurafenib-treated A2058 cells. Interestingly, we identified two mitochondrial complex I inhibitors, deguelin and rotenone, that inhibited the cellular proliferation of our resistant and metastatic melanoma cells by interfering with the altered metabolism observed in those vemurafenib resistant cell lines. Using phospho-proteomic array and subsequent western blot validation, we provide evidence that deguelin mechanistically acted by inhibiting oxygen consumption, which caused activation of AMPK signaling, resulting in inhibition of mTORC1 and alteration of its downstream effectors 4E-BP1 and p70S6 kinase.
MATERIALS AND METHODS
Chemicals and Reagents
Vemurafenib and compound C (Dorsomorphin) were purchased from Selleck Chemicals, Houston, TX, United States; rotenone from Tocris Bioscience, Bristol, United Kingdom; and deguelin (Item №: 10010706) from Cayman Chemical, Ann Arbor, MI, United States. A375 (CRL-1619) and A2058 (CRL-11147) cell lines were purchase from ATCC, Manassas, VA, United States. Normal human melanocytes were collected from tissue donated anonymously at the University of Utah. HCC1806 triple negative breast cancer cell lysates were a gift from Dr. Siva Kolluri at Oregon State University.
Chemical Libraries
The MicroSource Spectrum Collection is a library of 2,000 small molecules that include clinically approved drugs, natural produces, and compounds with known bioactivity. Compounds have shown biological potential, with some being fully characterized and some already available in the market. The OTRADI Siga Collection includes 240,000 compounds from the ChemBridge DIVERSet™-CL library, the ChemBridge CORE Library, and two focused antiviral libraries from Life Chemicals and ChemDiv. From this library, 6,720 compounds were selected to be in the primary screen. Compounds are at a concentration of 10 mM in 100% DMSO (Sigma-Aldrich, St. Louis, MO).
Cell Culture and Generation of Vemurafenib Resistance
Parental melanoma cell lines were cultured in complete growth media consisting of DMEM (Gibco, Waltham, MA) supplemented with 10% FBS (Atlanta Biologicals, Flowery Branch, GA) and 1% Penicillin-Streptomycin (Gibco). Cells were cultured in 75cm2 flasks (Greiner Bio-One, Monroe, NC) or 175cm2 flasks (Corning, Corning, NY). Normal human melanocytes were cultured in basal medium, MCDB153 (Sigma), supplemented with penicillin-streptomycin 50 U/mL (Gibco), Amphoteracin B 0.5 µg/mL (Gibco), Alpha-tocopherol 1 µg/mL (Sigma), sodium bicarbonate 14.05 mM (Fisher Scientific, Hampton, NH), FBS 4% (HyClone Laboratories, Inc., South Logan, UT), insulin 5 µg/mL (Sigma), bFGF 0.6 ng/mL (Sigma), and TPA 80 nM (Sigma) to make complete melanocyte medium. Normal human melanocytes were kept below 13 passages. All cell lines were incubated in a humidified environment at 37°C with 5% CO2. A375 cells are amelanotic and PTEN-wildtype while A2058 cells are melanotic and PTEN-null14,15.
Vemurafenib-resistant cell lines were developed by first treating cells with an initial dose of vemurafenib near each cell line’s IC50 value, then increasing the dose in a stepwise manner over a period of two weeks to a highpoint, and finally maintaining at a mid-range value. A375 was initially treated with 0.5 µM vemurafenib, then increased stepwise to 20 µM, and finally maintained in complete growth media supplemented with 2 µM vemurafenib. The initial vemurafenib dose for A2058 was 5 µM, then increased stepwise to 30 µM, and finally maintained in complete growth media supplemented with 10 µM vemurafenib. Resistant cell lines were considered those passaged 10 times at the maintenance dose prior to use in order to confirm resistance. For experiments using resistant cell lines, the cells were always cultured with the corresponding maintenance dose of vemurafenib. Parental and resistant A375 did not exceed 45 passages while parental and resistant A2058 did not exceed 60 passages.
High Throughput Screening
Primary:
Vemurafenib treated A2058 cells were then seeded at 2500 cells/well in 384 well microplates using a MultiFlo bulk reagent dispenser (BioTek, Winooski, VT) and incubated at 37°C with 5% CO2. After 24 hours, plates were delivered to the Oregon Translational Research and Drug Development Institute (OTRADI) satellite lab at Oregon State University for compound loading. Compound dilution plates were made in 96 well plates at 100 µM and 10% DMSO. Compounds were further diluted into assay plates at a final concentration 5 µM and 0.5% DMSO with the Hamilton Startlet (Hamilton, Reno, NV) liquid handler. Assay plates with compounds added were incubated for 48 hours. Control wells of cells with 0.5% DMSO were included on every plate. CellTiter-Glo (Promega, Madison, WI) cell viability reagent was added with the bulk dispenser. Luminescence was measured on the Synergy4 (Biotek, Winooski, VT) plate reader. Data from test wells were normalized to control wells. Hits were classified as compounds that reduced viability by more than 3.5 standard deviations from the mean of all the test compounds.
Secondary:
Assay plates were prepared similarly to the primary screen. Hit compounds were titrated into the assay plates with the D-300 (Hewlett Packard, Palo Alto, CA) digital dispenser and tested in an 8-point, half-log series in triplicate from 18 µM to 5.7 nM. Cell viability was assessed as before.
Cell Viability Assay
Cells were seeded at 7,000 cells/well into white, opaque 96 well plates (Greiner Bio-One). A375, A2058, and A375R cell lines were incubated 24 hours prior to compound addition while A2058R cells were allowed 48 hours. Compounds were then added to construct a 10-point dose-response curve in triplicate using a half-log serial dilution. The serial dilution for deguelin (Cayman Chemical) started at 200 µM while rotenone (Tocris Bioscience) started at 50 µM. For each compound serial dilution, a vehicle control serial dilution was also performed in triplicate. Cells were incubated with the compounds for 48 hours after which viability was determined using CellTiter-Glo assay. Percentage survival for each point in the compound serial dilution was relative to the equivalent point in the vehicle control serial dilution. Dose-response curves were fit to a 4-parmater IC50 model also using GraphPad Prism from which relative IC50 values were calculated.
Normal human melanocytes were plated in a 96 well TC treated plate (Greiner Bio-One) at 5000 cells/well in complete melanocyte Medium with 13 ng/mL bovine pituitary extract (ThermoFisher, Waltham, MA). The cells were allowed to rest for 48 hours then treated with either deguelin or rotenone at 0.18% of the total media volume. A DMSO control, no DMSO, and media only control was included for each treatment group. Cells were treated with the indicated concentration of compound, incubated for 48 hours then assayed for viability using CellTiter-Glo (Promega). There were 8 treatment doses ranging from 0.003mM to 10mM.
Live-Cell Metabolic Assay
The Seahorse XF24 assay was performed for live cell metabolism in accordance to the manufacturer’s instructions. Briefly, cells were seeded into Seahorse XF24 microplates (Agilent Technologies, Santa Clara, CA) at 15,000 cells/well A375, 17,500 cells/well A375R and A2058, and 20,000 cells/well A2058R in complete growth medium to achieve ~90% confluence at the start of the experiment. A2058R cells were seeded 48 hours prior to the start of the experiment while A375, A375R, and A2058 cells were seeded 24 hours before. On the day of the experiment cells were equilibrated and assayed in sterile filtered assay medium consisting of unbuffered low glucose DMEM (Gibco) supplemented with 640 mg/L glucose. Compounds were injected into the wells after 6 basal loops and the assay run for another 11 loops afterwards. Loops consisted of 3 minutes mixing, 2 minutes equilibration, and 3 minutes of measurement. Once the assay was complete, a CyQUANT® NF Cell Proliferation Assay (ThermoFisher Scientific) was performed in accordance with the manufacturer’s instructions to obtain cell counts for each well by extrapolation from a standard curve for normalization.
Western Blotting
Cells were grown in 10cm dishes (Falcon; Becton, Dickinson, and Company; Franklin Lakes; NJ) and deguelin added when 60% confluence was achieved. Cells were incubated for 1 or 12 hours and then harvested. To do so, cells were scraped into cold PBS, centrifuged, and protein lysates were obtained by homogenizing cell pellets in denaturing SDS lysis buffer (as described previously16) followed by sonication. Protein concentration was determined using the BCA assay (ThermoFisher). Protein lysates were resolved using 10% SDS-PAGE and transferred onto 0.45 µm nitrocellulose membrane (GE Healthcare) or 0.45 µm PVDF membrane (ThermoFisher) for LC3B. The blots were blocked for 1 hour at room temperature (RT) in 5% non-fat dry milk and incubated overnight (O/N) with specific antibodies diluted in 5% BSA (Jackson ImmunoResearch Laboratories, West Grove, PA). After a 1-hour incubation with secondary antibody, signals were detected with immunochemiluminescent reagents (GE Healthcare, Chicago, IL) using a MyECL Imager (ThermoFisher). The antibodies and dilutions used for immunoblotting are: P-AMPK (T172) 1:1000 (#2535, Cell Signaling, Danvers, MA), AMPK 1:1000 (#5832, Cell Signaling), P-p70S6K (T389) 1:1000 (#9205, Cell Signaling), p70S6K 1:1000 (#2708, Cell Signaling), 4E-BP1 1:1000 (Cell Signaling), P-ERK p44/42 (T202/Y204) 1:1000 (#9101, Cell Signaling), ERK p44/42 1:1000 (#4695, Cell Signaling), LC3B 1:1000 (#2775, Cell Signaling), cleaved caspase-3 0.2 µg/mL (#700182, ThermoFisher), Bax 1:200 (#SC-526, Santa Cruz Biotechnology, Dallas, TX), β-actin 1:2000 (#A300–491, Bethyl Laboratories, Inc., Montgomery, TX), and HRP-conjugated Goat anti-Rabbit IgG (H&L Chain) secondary antibody 1:2000 (#401315, Calbiochem, San Diego, CA).
Phospho-Kinase Array
The phosphorylation status of 43 kinase phosphorylation sites and 2 related proteins was done using the Human Phospho-Kinase Array Kit (R&D Systems, Inc., Minneapolis, MN). A375R cells were treated with either 24 µM deguelin or vehicle control for 30 minutes to 1 hour and then lysed in with Lysis Buffer 6 provided by the manufacturer. Membranes were blocked for 1 hour at RT prior to incubation with 300 µg protein O/N at 4°C. Membranes were then incubated with provided biotin-conjugated detection antibodies and Streptavidin-HRP according to the manufacturer’s instructions and signals detected as before using chemiluminescence reagents included in the kit.
Immunocytochemistry
ICC was performed as previously described17. Briefly, cells were seeded onto 0.1% gelatin-coated glass coverslips (Fisher Scientific) in 6 well plates (Falcon) at 200,000 cells/well. A375R cells were incubated 24 hours and A2058R cells were incubated 48 hours prior to deguelin treatment for 12 hours. Cells were then fixed in a 1:1 mixture of methanol and acetone at −20°C, dried, and frozen. Cell coated coverslips were then blocked for 1 hour at RT in 10% FBS in PBS and then incubated with Ki67 primary antibody 1:1000 (#15580, Abcam, Cambridge, UK) O/N at 4°C. The next day coverslips were incubated with Cy3-conjugated Donkey anti-Rabbit IgG (H&L Chain) secondary antibody 1:250 (#711–165-152, Calbiochem) for 1 hour at RT and 0.2 ng/mL DAPI for 10 minutes. Coverslips were then mounted onto slides using DPX mounting medium (VWR, Radnor, PA), dried O/N, and stored in the dark at 4°C. Slides were imaged using appropriate filters at 20x magnification with a Zeiss fluorescent microscope (ZEISS, Oberkochen, Germany). Percentage Ki67+ cells were then counted per field.
Crystal Violet Viability Assay
Cells were seeded 40,000 cells/well into 24 well plates (Greiner Bio-One) with A375R cells incubated 24 hours and A2058R cells incubated 48 hours prior to deguelin treatment for 24 hours. Crystal violet staining solution was prepared by dissolving crystal violet powder (Allied Chemical and Dye Corporation, Morristown, NJ) in DI H2O to final concentration of 0.5% (w/v) and subsequently adding methanol up to a 20% (v/v). Following deguelin treatment, cells were washed twice with PBS and incubated in 0.5% crystal violet staining solution for 20 minutes at RT. Wells were then washed four times with tap water and allowed to dry O/N. Crystal violet staining was quantified by adding 1 mL methanol to each well and incubating the plates for 20 minutes at RT. Plates were then read for absorbance at 570 nm on a Synergy4 (BioTek) plate reader.
Statistics and Software
Statistics were performed using GraphPad Prism 5 software (GraphPad Software, San Diego, CA). Results from the primary screen were analyzed and visualized using Vortex v.2018.03.7158-s (Dotmatics, Woburn, MA). Seahorse data were normalized using Wave v.2.4.0 (Agilent Technologies). Densiometric quantification of immunoblotting was performed using ImageJ software v.1.51j818. The heatmap for the phospho-proteomic array was constructed using the heatmap.2 function in RStudio v1.1.383 (RStudios, Inc., Boston, MA). Fluorescent microscopy images were prepared using Zeiss AxioVs40 software v.4.8.2.0 and then processed using Adobe Photoshop CC 2018 (Adobe, Inc., San Jose, CA) and ImageJ.
RESULTS
High-throughput Screening Identifies Metabolic Inhibitors as Effective Against Metastatic Melanoma
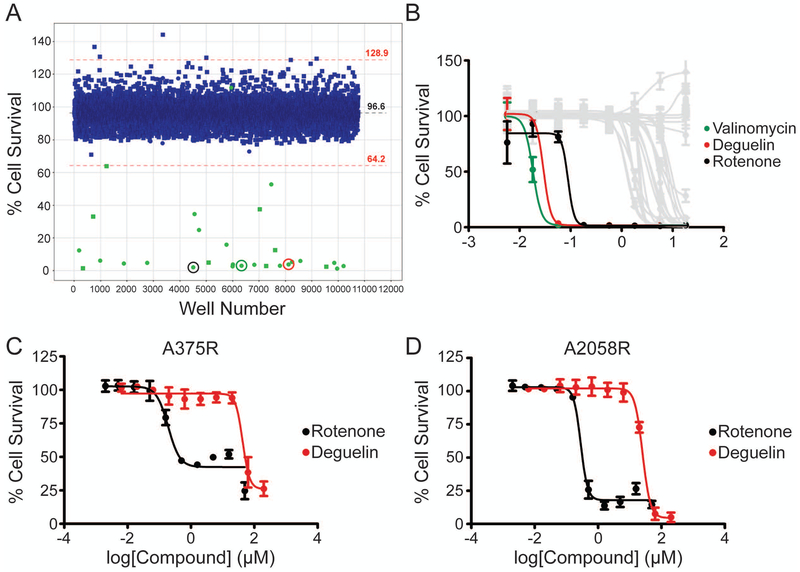
In order to identify novel compounds or targets for the treatment of BRAF inhibitor-resistant metastatic melanoma we screened a vemurafenib treated A2058 cell line (for details see methods), harboring the oncogenic BRAFV600E and PTEN-null mutations19, against two chemical libraries, the MicroSource Spectrum Collection of 2,000 known bioactive compounds and a selection of 6,720 compounds from the OTRADI Siga Collection. This cell line was selected due to its inherent resistance to vemurafenib treatment with an IC50 approximately10 µM20. From the primary screen a total of 32 hits were obtained, 21 from the Spectrum library and 11 from the Siga library (Figure 1A). These 32 hits underwent a dose-response secondary screen to obtain IC50 values. The three most potent compounds were valinomycin, deguelin, and rotenone with IC50 [95% (CI)] values of 0.0181 (0.0171 to 0.0193) µM, 0.0293 (0.0211 to 0.0406) µM, and 0.0885 (0.0240 to 0.3264) µM, respectively (Figure 1B). All three were part of the Spectrum library (detailed in methods section). Deguelin and rotenone were selected for further validation as they are structural analogs that may share similar mechanisms of action. Valinomycin was not further evaluated due to its known cytotoxicity21. To validate the results from the secondary screen and examine the potency of our identified compounds against BRAFV600E vemurafenib-resistant cell lines, A375R and A2058R cells were co-treated with vemurafenib and either deguelin or rotenone for 48 hours. The compounds were purchased from a vendor different from that of the libraries. The IC50 (95% CI) values for deguelin and rotenone against A375R cells were 42.84 (33.13 to 55.38) µM and 0.1912 (0.1283 to 0.2848) µM, respectively (Figure 1C). For A2058R cells the IC50 (95% CI) values for deguelin and rotenone were 25.34 (21.94 to 29.28) µM and 0.2839 (0.2345 to 0.3436) µM, respectively (Figure 1D). To determine if development of BRAF inhibitor resistance influenced the potency of either compound in a cell line specific manner we also tested parental A375 and A2058 cell lines against the two compounds. A similar trend was observed with nanomolar and micromolar IC50 values for rotenone and deguelin, respectively (Supplementary Figure S1A,B). The IC50 (95% CI) values for deguelin and rotenone in A375 cells were 26.71 (23.24 to 30.69) µM and 0.1325 (0.0342 to 0.5127) µM, respectively. For A2058 cells the IC50 (95% CI) values for deguelin and rotenone were 23.08 (18.96 to 28.10) µM and 0.2712 (0.2314 to 0.3179) µM, respectively. To determine whether our identified compounds exhibited selective toxicity against cancer cells, we examined the potency of both compounds against normal human melanocytes in vitro and no effect on viability was observed (Supplementary Figure S1C). In further experiments we chose to focus on the activity of deguelin as our model inhibitor of cancer cell metabolism due to rotenone’s known toxicity in vivo22,23.
Figure 1. High-throughput screening identifies Deguelin and Rotenone as effective against metastatic melanoma.
(A) Scatterplot depicting hits in green from the primary screen of A2058 cells using the Spectrum (circles) and Siga (squares) compound libraries. Hits were considered compounds that resulted in viabilities 3.5 standard deviations (red dotted lines) below the average of the screen (black dotted line) following 48 hours treatment. Circled are the lead compounds selected from the secondary screen in colors corresponding to panel B. 48-hour dose-response curves for the secondary screen (B) and validation in vemurafenib-resistant cell lines A375R (C) and A2058R (D). Data represent mean±SD from triplicate values (B) or from three independent experiments performed in triplicate (C,D).
Deguelin Inhibits Oxygen Consumption in Vemurafenib-Resistant Melanoma Cell Lines
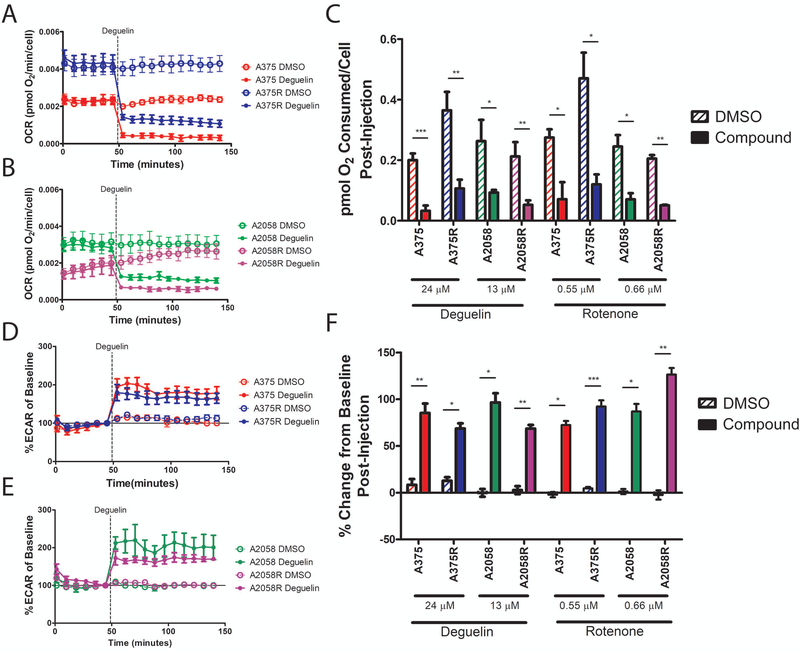
To determine if deguelin acts as a metabolic inhibitor in our cell lines, a Seahorse XF24 assay was performed to measure the effect of the compounds on oxygen consumption rate (OCR). A decrease in the amount of oxygen consumed suggests a reduction in the degree to which oxidative phosphorylation is occurring in cells. The effect of rotenone in the Seahorse XF system is well established as it is provided by the manufacturer as a positive control for inhibition of oxygen consumption in these assays and so it was used for the same purpose here. We observed a rapid decrease in OCR upon injection of deguelin for both parental and resistant A375 (Figure 2A) and A2058 (Figure 2B) cell lines as compared to vehicle control. We also determined that there was a significant decrease in the total pmoles O2 consumed for all cell lines post deguelin injection (Figure 2C). This effect was observed with rotenone treatment as well (Supplementary Figure S2A,B). To see if developing vemurafenib resistance induced changes in oxygen consumption, we compared the OCR values of deguelin vehicle control treated A375, A375R, A2058, and A2058R. There was a significant difference in pmoles O2 consumed between DMSO treated A375 and A375R (p = 0.0119, Student’s two-tailed t-test), while the difference between A2058 and A2058R was not significant (p = 0.3617, Student’s two-tailed t-test).
Figure 2. Deguelin and Rotenone significantly inhibit oxygen consumption in vemurafenib-resistant A375R and A2058R metastatic melanoma cell lines.
Seahorse XF24 rate graphs depicting OCR of A375 and A375R (A) or A2058 and A2058R (B) cell lines. Dotted line indicates the timepoint at which Deguelin was injected into the well. (C) Bar graph depicting the area under the curve of the rate graphs after compound injection. Baselined rate graphs depicting percentage change in ECAR of A375 and A375R (D) or A2058 and A2058R (E) cell lines. The baseline value for each treatment was considered the ECAR value immediately preceding compound injection into the well. Solid line indicates baseline value. (F) Bar graph depicting the average percentage change in ECAR from baseline for all the timepoints following compound injection. OCR and ECAR were normalized to cell counts. Data represent mean±SEM from three independent experiments (A,B,D,E) or mean±SD from three independent experiments for Deguelin and two independent experiments for Rotenone (C,F). All assays were performed in triplicates. Statistical significance was determined by two-tailed Student’s t-test (C) or by 2-way ANOVA (F). * p < 0.05, ** p < 0.01, *** p < 0.001.
In addition to the OCR, the extracellular acidification rate (ECAR) was concurrently measured. ECAR gives in indication of the glycolytic activity of the cells as the pH in the well microenvironment changes due to lactic acid production. It is expected that with inhibition of oxidative phosphorylation the cell will shift towards increased glycolysis to compensate for the loss of ATP in the cell. Our observations support this as we saw a simultaneous elevation in baselined ECAR upon injection of deguelin into the wells in both parental and resistant A375 (Figure 2D) and A2058 (Figure 2E) cell lines as compared to vehicle control. There was a significant difference between the curves for all compound treated wells as compared to the vehicle control wells for the time points following compound injection (Figure 2F). These data suggest that deguelin inhibits oxidative phosphorylation which results in a metabolic shift towards glycolysis in a manner similar to its analog rotenone (Supplementary Figure S2C,D).
Deguelin Treatment Induces Activation of Cellular Energy Stress Response
To get a global perspective in the signaling changes induced by deguelin a proteome profiling phospho-kinase array was performed to see the phosphorylation status of 43 kinase phosphorylation sites. In particular, we were interested in the early signaling events that would be directly caused by the rapid inhibition of oxygen consumption seen in our Seahorse XF assays. For this reason, cell lysates were collected at early time points and probed with the arrays. After 30 minutes of deguelin treatment there were 18 target phosphorylation sites with a significant change in signal intensity (Supplementary Figure S3D) and after an hour there were 11 targets with significant differences (Supplementary Figure S3E). From these normalized signal intensities, fold changes between deguelin and vehicle control treated membranes were calculated and a heatmap constructed to visualize the data (Supplementary S4). From this heatmap a number of trends were observed that were used to further examine deguelin’s activity. Of principle interest were pathways associated with metabolism and energy stress signaling. Firstly, at 30 minutes and 1 hour time points there was an increasing trend in AMPKα activation, which may be associated to elevated energy stress conditions in the cell. Furthermore, there was a trend in decreased activation of p70 S6 Kinase, suggesting a possible inhibition of upstream regulator mTORC1. We also observed a consistent activation of p27 which is involved in cell cycle regulation. We did not observe inhibition of Akt or ERK activation at this early timepoint.
Deguelin Treatment Leads to AMPK-Mediated Inhibition of mTORC1
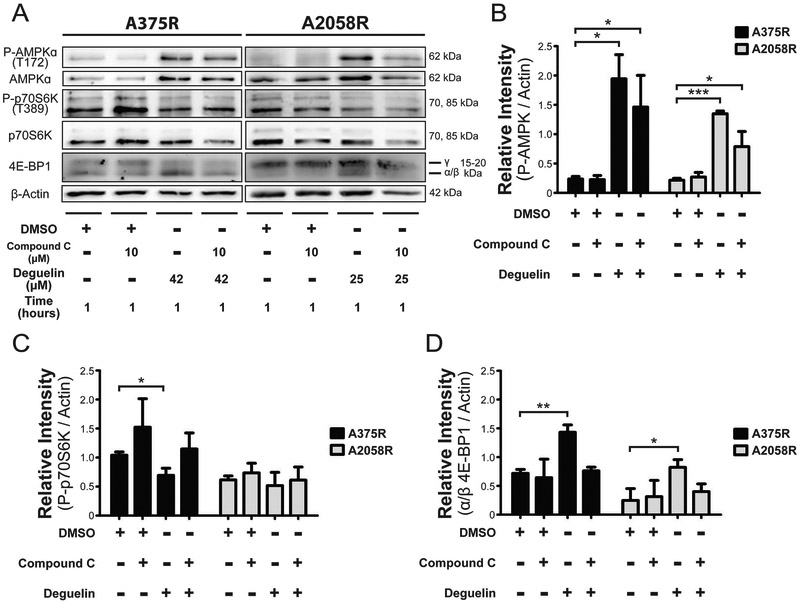
Given the potential activation of AMPKα and deactivation of p70 S6 Kinase observed with our micro array data, we then sought to determine if there was AMPK-mediated inhibition of mTORC1 activity in response to deguelin treatment in that timeframe. For that, we performed immunoblotting on protein lysates following 1 hour of treatment to probe for increased AMPKα phosphorylation, reduced p70 S6 Kinase phosphorylation, and shifted electrophoretic mobility of 4E-BP1 (Figure 3A). To confirm whether AMPK signaling was responsible for possible changes in P-p70 S6 Kinase and 4E-BP1 electrophoretic mobility, we included a known AMPK inhibitor, compound C. First, we observed a significant increase in AMPKα phosphorylation upon 1 hour treatment with deguelin consistent with our micro array observations (Figure 3B). Next, we observed that in A375R cells there was a significant decrease in phosphorylation of p70 S6 Kinase that was reversed upon treatment with compound C (Figure 3C). A similar trend was observed in A2058R cells, but the change in relative signal intensity was not significant. Lastly, we saw in both cell lines a significant shift in electrophoretic mobility in 4E-BP1 towards the α/β isoforms away from the γ isoform, which was reversed in presence of compound C (Figure 3D). This shift in mobility is associated with a loss of hyperphosphorylation caused by mTORC124,25 leading in turn to increased activity of 4E-BP1.
Figure 3. Deguelin inhibits AMPK-mediated MTORC1 activity as exemplified in the differential regulation of downstream effectors p70S6K and 4E-BP1.
(A) Representative western blotting to probe the phosphorylation status of AMPK and p70S6K as well as overall expression of 4E-BP1. Bar graphs depicting densiometric quantification of the preceding immunoblots relative to β-actin loading control for P-AMPK (B), P-p70S6K (C), and α/β isoforms of 4E-BP1 (D). Data represent the mean±SD from two independent experiments. Statistical significance was determined by one-tailed Student’s t-test. * p < 0.05, ** p < 0.01, *** p < 0.001.
Deguelin Treatment Does Not Inhibit Akt Activation nor Induce Apoptosis at Earlier Time Points
Our micro array data seemed to suggest that deguelin did not inhibit Akt activation at early time points following treatment. As was done for ERK phosphorylation, we probed 12-hour treated lysates for the phosphorylation status of Akt at residue S473 (Supplementary Figure S5A) and we observed a significant increase in phosphorylation for both cell lines (Supplementary Figure S5B). We also probed for the phosphorylation status of residue T308 (Supplementary Figure S5C) and observed no significant change (Supplementary Figure S5D). To confirm that there was a reduction in viable cells before the 48-hour time point used in the dose-response viability experiments, crystal violet staining was performed and a significant decrease in viable cells was observed (Supplementary Figure S5E,F). We did not see any evidence for an induction or inhibition of autophagy following 12 hours of deguelin treatment (Supplementary Figure S5G,H). Finally, we investigated whether apoptosis occurred at early time points post treatment. There was not an apparent increase in cleaved caspase-3 12 hours after deguelin treatment (Supplementary Figure S6A) nor was there an increase in expression of Bax at 1 hour or 12 hours post treatment (Supplementary Figure S6B).
Deguelin Treatment Leads to Inhibition of Cellular Proliferation
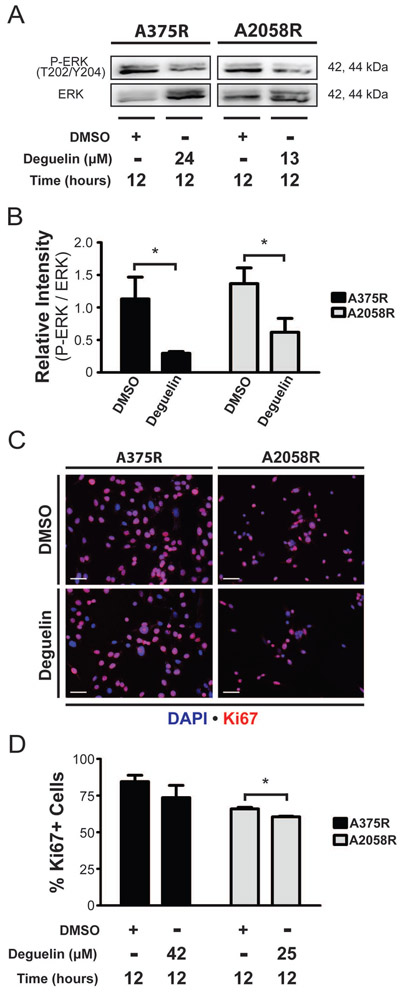
We were next interested in confirming whether the trend in ERK activation observed in the array was occurring at a later time point. For that, we did immunoblotting of lysates harvested following 12 hours of deguelin treatment for the phosphorylation status of ERK (Figure 4A), and we observed a significant decrease in P-ERK in both cell lines at this time point (Figure 4B). For further confirmation of inhibited cellular proliferation, the expression of proliferation marker Ki67 was examined at the same time point using immunocytochemistry (Figure 4C). We also found a significant decrease in expression of Ki67 in A2058R cells and decreasing trend A375R cells (Figure 4D)
Figure 4. Deguelin inhibits MAP kinase signaling and proliferation.
(A) Representative western blotting to probe for phosphorylation status of ERK. (B) Bar graph depicting densiometric quantification of the bands obtained from immunoblotting for relative intensity of phospho-protein over total protein for ERK. (C) Immunofluorescence to detect expression of the proliferation marker Ki67 following compound or DMSO treatment. (D) Counts of percentage Ki67+ cells obtained from immunofluorescence staining. Scale bar represents 50 µm. Data represent mean±SD from two independent experiments. Statistical significance was determined by one-tailed Student’s t-test. * p < 0.05.
DISCUSSION
The hallmark of metabolic reprogramming by cancer cells has long been held to be, under aerobic conditions, a shift towards glycolysis as the key energy production mechanism first observed by Dr. Otto Warburg in the mid twentieth century26. Interestingly, a consistent trend of elevated oxidative phosphorylation and mitochondrial biogenesis has been observed in BRAF inhibitor-resistant BRAFV600E melanomas as a consequence of the development of drug resistance 27,28. For that reason, the results of our high-throughput screening made sense as we identified the electron transport chain inhibitors rotenone and deguelin29 as two of the three most potent compounds against vemurafenib-treated BRAFV600E/PTEN-null A2058 melanoma cells. Interestingly, our results are consistent with another recent screen results that identified deguelin and rotenone as compounds effective against PTEN-null cells in the context of prostate cancer30, potentially suggesting additional reasons for the results we have observed.
Surprisingly, when we proceeded to validate those hits in vemurafenib-resistant cells lines A375R and A2058R we observed a shift in potency of deguelin from a nanomolar to micromolar IC50. To determine if that was due to the development of drug resistance we also went back and treated the parental cell lines with both compounds and saw a similar trend. It is possible that batch variation between the different deguelin source used in the primary/secondary screen and the subsequent validation could be responsible for the shift in drug potency. We also observed that in A2058 parental and resistant cells the IC50 values were equivalent, while in the A375 cell line the IC50 doubled in resistant cells as compared to parental. That could be related to the basal OCR values observed for each cell line. In A2058 cell lines the parental and resistant cells’ basal OCR values were similar while in the A375 cell lines the resistant A375R cells exhibited a significantly increased OCR compared to the parental. Our results suggest that a higher dose of deguelin would be needed in A375R cells to reduce the OCR to a similar level as in A375 cells. The lack of a difference in OCR between A2058 parental and resistant cells may be due to their PTEN-null status where they have an inherent resistance to vemurafenib treatment and may not undergo the same metabolic alterations as in a PTEN-WT context. As expected, treatment of primary normal human melanocytes exhibited no toxicity which has been observed in primary cells from other tissues as well31.
We then confirmed that deguelin rapidly (within 8 min of compound addition) inhibits oxygen consumption and found evidence that vemurafenib-resistant cells shift back to glycolysis following deguelin treatment. That observation made us interested in the signaling that occurred at early time points following treatment as those molecular changes/events could be directly tied to deguelin’s metabolic inhibitory activity in our context.
To probe early changes in signaling, we performed phospho-kinase arrays and observed a clear energy stress response with activation of AMPK signaling and deactivation of p70 S6 kinase suggesting mTORC1 inhibition within an hour of deguelin treatment. We also observed activation of ERK and Akt. Lastly, a consistent activation of p27 was observed, which agrees with deguelin’s known activity in promoting cell cycle arrest32 as phosphorylation of p27 at T198 has been associated with its stabilization33. However, further study is needed to determine if deguelin is acting through that mechanism in the current context.
We then confirmed deguelin’s early AMPK-mediated inhibition of mTORC1 through examination of the phosphorylation status of its downstream effectors p70 S6 kinase and 4E-BP1 one hour after treatment. As expected, AMPK was highly phosphorylated at T172 indicating a reduction in ATP production as a consequence of inhibiting OXPHOS. Phosphorylation of p70 S6 kinase was significantly reduced with deguelin treatment in A375R cells with a similar trend observed in A2058R cells. The lack of significance observed in A2058R cells could be due to delayed signaling kinetics particular to that cell line. Our data regarding electrophoretic mobility of 4E-BP1 was highly consistent between both cell lines with a significant increase of the hypo-phosphorylated isoforms present in both cases. Reversal of the phosphorylation status for both effectors with compound C treatment indicated that deguelin could be responsible for AMPK-mediated inhibition of mTORC1 (Figure 5).
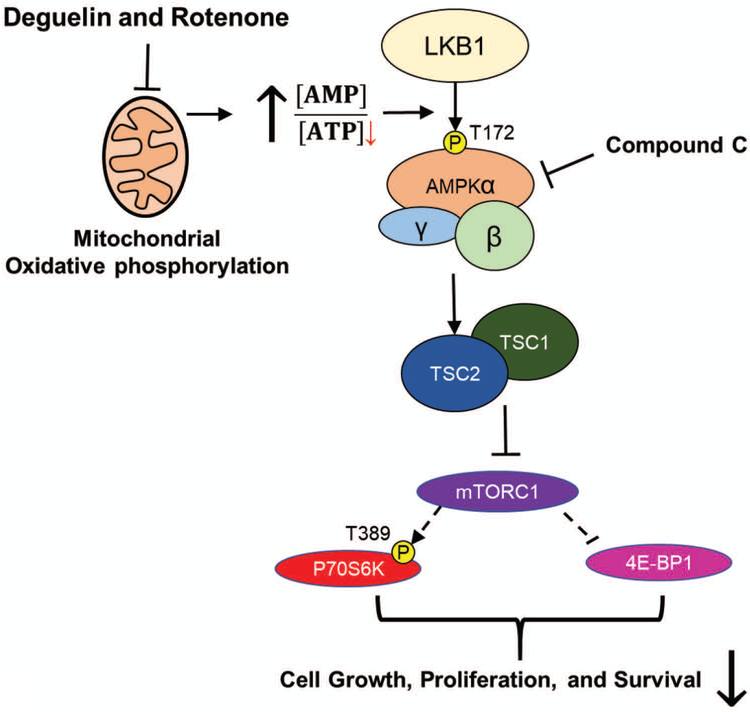
Figure 5. Mechanistic scheme portraying the partial activity of Deguelin and Rotenone in BRAF inhibitor-resistant metastatic melanoma.
Deguelin and Rotenone inhibit oxidative phosphorylation in the mitochondria which results in an elevation of the AMP to ATP ratio in the cell. This increase of AMP results in activation of AMPK signaling due to phosphorylation of the AMPKα2 subunit at T172 by LKB1. AMPK signaling activates the TSC complex which in turn inhibits MTORC1. Inhibition of MTORC1 results in a reduction in phosphorylation of p70S6 kinase, inhibiting its downstream activity. Additionally, the hyperphosphorylation of 4E-BP1 is inhibited resulting in its increased activity. The consequence of this is reduced cell survival and proliferation. Inhibition of MTORC1 can be reversed by addition of compound C (Dorsomorphin) which blocks AMPK signaling.
Inhibition of mTORC1 is associated with decreased cell growth and proliferation in cancer34,35 and so we were then interested in determining deguelin’s effect on proliferation given that the activation of ERK was observed in our study with phospho-proteomic array. The decrease in ERK activation and a general trend of reduced expression of the proliferation marker Ki67 observed after 12 hours support the notion that early inhibition of mTORC1 could lead to reduced cellular proliferation. The lack of significance seen in A375R cells could be due to deguelin’s lower potency in that cell line which may alter the signaling kinetics for proliferation.
We were surprised to see a trend in increased Akt activation in our phospho-array data as deguelin has been shown to inhibit Akt activation and signaling36,37. Additional validation by immunoblotting further confirmed the initial results from our phospho-array where we observed either an increase in Akt-phosphorylation at the residue S473 or no difference at residue T308. The contradictory results observed could be due to the early time points that were the focus of the present study as time kinetics for deguelin’s inhibition have been previously done in the context of H460 lung cancer cells and only show a decrease after 24 hours of treatment at both phosphorylation sites38. Those data suggest that the PI3K-Akt axis of deguelin’s activity may be only indirectly tied to its inhibition of OXPHOS requiring some time for the necessary signaling to occur. It follows that if PI3K-Akt signaling occurs at later time points then deguelin’s characteristic induction of apoptosis does as well37,38. Indeed, in the present study we observed no evidence for an induction of apoptosis at early time points. However, we did observe a significant decrease in viability as soon as 24 hours following treatment, which is around the time survival signaling that has been shown to be inhibited in other contexts.
Lastly, we were interested if there was an apparent induction or inhibition of autophagy following the activation of AMPK signaling observed. However, at a 12-hour time point there was not a consistent effect among our cell lines providing no evidence of autophagy being directly connected to deguelin treatment. Altogether, our results suggest that Deguelin treatment can lead to energy starvation thereby reducing survival of drug-resistant melanomas harboring BRAFV600E mutation.
Supplementary Material
Acknowledgments and Funding
We thank members of the Indra lab and the OSU College of Pharmacy for their continuous support and encouragement. We also thank Dr. Siva Kolluri at OSU for providing the materials for the apoptosis immunoblots. Research reported in this publication was supported in part by National Institute of Environmental Health Sciences (NIEHS) of the National Institutes of Health (NIH) under the award number 1R01ES016629-01A1 (PI :AI), National Cancer Institute of the NIH under award number T32CA106195 and the OSU/OHSU College of Pharmacy Pilot Project Grant to AI.
Footnotes
Availability of Data
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26(11):1131–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shain AH, Bastian BC. From melanocytes to melanomas. Nat Rev Cancer. 2016;16(6):345–358. [DOI] [PubMed] [Google Scholar]
- 3.Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41(5):544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demunter A, Stas M, Degreef H, De Wolf-Peeters C, van den Oord JJ. Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J Invest Dermatol. 2001;117(6):1483–1489. [DOI] [PubMed] [Google Scholar]
- 5.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467(7315):596–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manzano JL, Layos L, Buges C, et al. Resistant mechanisms to BRAF inhibitors in melanoma. Ann Transl Med. 2016;4(12):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benito-Jardon L, Diaz-Martinez M, Arellano-Sanchez N, Vaquero-Morales P, Esparis-Ogando A, Teixido J. Resistance to MAPK inhibitors in melanoma involves activation of the IGF-1R-MEK5-Erk5 pathway. Cancer Res. 2019. [DOI] [PubMed] [Google Scholar]
- 10.Baxter LL, Hou L, Loftus SK, Pavan WJ. Spotlight on spotted mice: a review of white spotting mouse mutants and associated human pigmentation disorders. Pigment Cell Res. 2004;17(3):215–224. [DOI] [PubMed] [Google Scholar]
- 11.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Voet A, Callewaert L, Ulens T, et al. Structure based discovery of small molecule suppressors targeting bacterial lysozyme inhibitors. Biochem Biophys Res Commun. 2011;405(4):527–532. [DOI] [PubMed] [Google Scholar]
- 13.Haq R, Fisher DE, Widlund HR. Molecular pathways: BRAF induces bioenergetic adaptation by attenuating oxidative phosphorylation. Clin Cancer Res. 2014;20(9):2257–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vitiello M, Tuccoli A, D’Aurizio R, et al. Context-dependent miR-204 and miR-211 affect the biological properties of amelanotic and melanotic melanoma cells. Oncotarget. 2017;8(15):25395–25417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutchison S, Rae C, Tesson M, Babich JW, Boyd M, Mairs RJ. Evaluation of Melanin-Targeted Radiotherapy in Combination with Radiosensitizing Drugs for the Treatment of Melanoma. Can and Oncol Res. 2014;2(4):58–67. [Google Scholar]
- 16.Carpenter EL, Le MN, Miranda CL, et al. Photoprotective Properties of Isothiocyanate and Nitrile Glucosinolate Derivatives From Meadowfoam (Limnanthes alba) Against UVB Irradiation in Human Skin Equivalent. Front Pharmacol. 2018;9:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guha G, Lu W, Li S, et al. Novel Pactamycin Analogs Induce p53 Dependent Cell-Cycle Arrest at S-Phase in Human Head and Neck Squamous Cell Carcinoma (HNSCC) Cells. PLoS One. 2015;10(5):e0125322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing F, Persaud Y, Pratilas CA, et al. Concurrent loss of the PTEN and RB1 tumor suppressors attenuates RAF dependence in melanomas harboring (V600E)BRAF. Oncogene. 2012;31(4):446–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao J, Egger ME, McMasters KM, Hao H. Differential expression of ABCB5 in BRAF inhibitor-resistant melanoma cell lines. BMC Cancer. 2018;18(1):675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroten MA, Bartoszewicz M, Swiecicka I. Cereulide and valinomycin, two important natural dodecadepsipeptides with ionophoretic activities. Pol J Microbiol. 2010;59(1):3–10. [PubMed] [Google Scholar]
- 22.Jiang XW, Qiao L, Feng XX, et al. Rotenone induces nephrotoxicity in rats: oxidative damage and apoptosis. Toxicol Mech Methods. 2017;27(7):528–536. [DOI] [PubMed] [Google Scholar]
- 23.Johnson ME, Bobrovskaya L. An update on the rotenone models of Parkinson’s disease: their ability to reproduce the features of clinical disease and model gene-environment interactions. Neurotoxicology. 2015;46:101–116. [DOI] [PubMed] [Google Scholar]
- 24.Gingras AC, Sonenberg N. Adenovirus infection inactivates the translational inhibitors 4E-BP1 and 4E-BP2. Virology. 1997;237(1):182–186. [DOI] [PubMed] [Google Scholar]
- 25.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc Natl Acad Sci U S A. 2008;105(45):17414–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Warburg O On the origin of cancer cells. Science. 1956;123(3191):309–314. [DOI] [PubMed] [Google Scholar]
- 27.Haq R, Shoag J, Andreu-Perez P, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer Cell. 2013;23(3):302–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corazao-Rozas P, Guerreschi P, Andre F, et al. Mitochondrial oxidative phosphorylation controls cancer cell’s life and death decisions upon exposure to MAPK inhibitors. Oncotarget. 2016;7(26):39473–39485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerhauser C, Lee SK, Kosmeder JW, et al. Regulation of ornithine decarboxylase induction by deguelin, a natural product cancer chemopreventive agent. Cancer Res. 1997;57(16):3429–3435. [PubMed] [Google Scholar]
- 30.Naguib A, Mathew G, Reczek CR, et al. Mitochondrial Complex I Inhibitors Expose a Vulnerability for Selective Killing of Pten-Null Cells. Cell Rep. 2018;23(1):58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun KH, Kosmeder JW 2nd, Sun S, et al. Effects of deguelin on the phosphatidylinositol 3-kinase/Akt pathway and apoptosis in premalignant human bronchial epithelial cells. J Natl Cancer Inst. 2003;95(4):291–302. [DOI] [PubMed] [Google Scholar]
- 32.Murillo G, Salti GI, Kosmeder JW 2nd, Pezzuto JM, Mehta RG. Deguelin inhibits the growth of colon cancer cells through the induction of apoptosis and cell cycle arrest. Eur J Cancer. 2002;38(18):2446–2454. [DOI] [PubMed] [Google Scholar]
- 33.Sun C, Wang G, Wrighton KH, et al. Regulation of p27(Kip1) phosphorylation and G1 cell cycle progression by protein phosphatase PPM1G. Am J Cancer Res 2016;6(10):2207–2220. [PMC free article] [PubMed] [Google Scholar]
- 34.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12(1):9–22. [DOI] [PubMed] [Google Scholar]
- 35.Dowling RJ, Topisirovic I, Alain T, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328(5982):1172–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rebolleda N, Losada-Fernandez I, Perez-Chacon G, et al. Synergistic Activity of Deguelin and Fludarabine in Cells from Chronic Lymphocytic Leukemia Patients and in the New Zealand Black Murine Model. PLoS One. 2016;11(4):e0154159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang YL, Ji C, Bi ZG, et al. Deguelin induces both apoptosis and autophagy in cultured head and neck squamous cell carcinoma cells. PLoS One. 2013;8(1):e54736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu YC, Chiang JH, Yu CS, et al. Antitumor effects of deguelin on H460 human lung cancer cells in vitro and in vivo: Roles of apoptotic cell death and H460 tumor xenografts model. Environ Toxicol. 2017;32(1):84–98. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.