Abstract
Aptamers that bind live bacterial cells have been widely investigated, but their potential to inhibit Candida albicans biofilm formation needs to be further explored. The aims of this study were to evaluate the binding of C. albicans to RNA aptamers and to examine the potential of aptamers to inhibit C. albicans biofilm formation in vitro. In this study, RNA aptamers selected against yeast cells of C. albicans ATCC 10231 were developed using the systematic evolution of ligands by exponential enrichment (SELEX) technique. The binding affinity of the resulting aptamers was then determined by an aptamer‐linked immobilized sorbent assay (ALISA), and a colorimetric (MTT) assay was used to measure the metabolic activity of Candida biofilms. After 11 rounds of SELEX, two candidate aptamers, Ca‐apt‐1 and Ca‐apt‐12, were identified. The Ca‐apt‐1 aptamer also recognized C. albicans isolated from clinical specimens but did not recognize other oral microorganisms (i.e., Streptococcus mutans and Saccharomyces cerevisiae). The ALISA results showed that the binding affinity of these aptamers was comparable to that of an anti‐C. albicans monoclonal antibody. In addition, Ca‐apt‐1 could inhibit biofilm and hyphal formation of C. albicans in vitro, as demonstrated using biofilm assays. This study shows that RNA aptamers could potentially be used in diagnostic and therapeutic applications for C. albicans‐related disease in the future.
Keywords: ALISA, aptamer, biofilms, C. albicans, SELEX
1. INTRODUCTION
Candida albicans is a component of normal human flora and an opportunistic pathogen (Akpan & Morgan, 2002; Soll, 2002). Compared to other fungal pathogens that exist primarily in either yeast or hyphal forms, C. albicans shows phenotypic plasticity because it has the ability to switch between different morphological forms in response to environmental cues (Whiteway & Bachewich, 2007). This morphogenic switching from yeast to hyphal form contributes to the overall virulence of C. albicans (Bastidas, Heitman, & Cardenas, 2009). Because of this characteristic associated with its virulent character, developing antifungals that target the early phase of C. albicans biofilm formation is challenging.
Aptamers are short, single‐stranded oligonucleotides that have emerged as a new class of small molecule ligands that can recognize and bind specific target molecules with high affinity and specificity (Jayasena, 1999). Aptamers bind target molecules with the affinity and specificity equal to or greater than those of antibodies (Tang et al., 2007). Aptamers are selected by an in vitro selection process called SELEX (systematic evolution of ligands by exponential enrichment) (Tuerk & MacDougal‐Waugh, 1993). Although numerous reports have detailed the selection of aptamers against different bacterial species (Cao et al., 2009; Chen, Zhou, Luo, Mohammed, & Zhang, 2007; Hamula, Le, & Li, 2011), few studies screening aptamers for their clinical value and potential use to inhibit the morphology switching that occurs during C. albicans‐related infections have been reported.
In this study, we used the SELEX technique to select RNA aptamers with high affinity and specificity for C. albicans yeast cells. Furthermore, we used an aptamer‐linked immobilized sorbent assay (ALISA) to demonstrate the potential use of high‐affinity aptamers in quantitative determination of C. albicans. Finally, we investigated the ability of the aptamers to inhibit C. albicans growth in vitro.
2. MATERIALS AND METHODS
2.1. Preparation of cells for aptamer selection
Candida albicans (ATCC 10231) was used as a targeted aptamer ligand, while Saccharomyces cerevisiae (ATCC 9763) was used for counter selection. For the binding specificity test, we used Streptococcus mutans (Xc), a stock culture of C. albicans that was previously isolated in our dental hospital (Universitas Indonesia) using Chromogenic Candida Agar (CCA; Oxoid, Basingstoke, UK) (Ghelardi et al., 2008), and S. cerevisiae. All microorganisms were maintained and propagated as described elsewhere (Bachtiar et al., 2014; Shibata, Kawada, Nakano, Toyoshima, & Yamashita, 2005; Vazquez‐Reyna, Balcazar‐Orozco, & Flores‐Carreon, 1993). To obtain budding yeast cells of C. albicans and S. cerevisiae, we used YPD (1% yeast extract, 2% peptone, 2% glucose) broth shaking at 30°C that was inoculated with C. albicans or S. cerevisiae that had been grown overnight on a YPD agar plate under aerobic conditions. The resulting budding yeasts were washed twice in phosphate‐buffered saline (PBS, Oxoid Ltd, Basingstoke, UK) and resuspended in RPMI 1640 supplemented with L‐glutamine and buffered with MOPS (Sigma, St Louis, MO, USA). The yeast density was measured by using a hemocytometer and adjusted for the SELEX procedure, while the bacterial number was counted by the plating method.
2.2. In vitro selection of RNA aptamers for C. albicans ATCC 10231
A library of RNAs containing 40‐nt randomized central sequences flanked by defined primer binding sites with the sequence of 5′‐GGGAGUCGACCGACCAGAA [N40] UAUGUGCGUCUACAUCUAGACUCAU‐3′ (84 nt) was generated as previously described (Srisawat & Engelke, 2001) with a calculated library complexity of 1 × 1013 different RNA sequences. The selection conditions of the SELEX process are shown in Table 1. The specified amounts of RNA and yeast were mixed in a 0.45‐μm spin column (Millipore), and the binding reaction was performed in a total volume of 50 μl in the binding buffer (50 mmol/L HEPES pH 7.4, 10 mmol/L MgCl2, 100 mmol/L NaCl) with 5 μg of baker's yeast tRNA. The reaction was incubated at room temperature for 45 min in rounds 1–5 and for 30 min from round 6 onwards with gentle rotation. The cells were then washed with the binding buffer, and the bound RNAs were eluted with 500 μl of the elution buffer (8 mol/L urea, 5 mmol/L EDTA, pH 8.0). The eluted RNAs were then recovered by ethanol precipitation, amplified by quantitative real time‐PCR (q‐PCR), and transcribed in vitro to generate RNAs for the next round of selection (Srisawat & Engelke, 2001). To enrich the aptamers specific to C. albicans, a counterselection step was included at rounds 3, 5, and 10 using 5 × 107 S. cerevisiae cells. The RNAs unbound after S. cerevisiae binding were used in the binding reaction with C. albicans as described above.
Table 1.
Candida albicans‐specific aptamer selection protocol*
| Round | Input RNA (pmol) | C. albicans (cells) | Washing |
|---|---|---|---|
| 1 | 100 | 5 × 109 | 100 μl 5 times, 1 min each |
| 2 | 25 | 5 × 108 | 100 μl 5 times, 3 min each |
| 3 | 25 | 5 × 108 | 100 μl 5 times, 3 min each |
| 4 | 12.5 | 5 × 107 | 100 μl 5 times, 5 min each |
| 5 | 12.5 | 5 × 107 | 100 μl 5 times, 5 min each |
| 6 | 6.25 | 5 × 106 | 100 μl 5 times, 10 min each |
| 7 | 6.25 | 5 × 106 | 100 μl 5 times, 10 min each |
| 8 | 3.13 | 5 × 105 | 100 μl 5 times, 15 min each |
| 9 | 3.13 | 5 × 105 | 100 μl 5 times, 15 min each |
| 10 | 1.56 | 5 × 104 | 100 μl 5 times, 15 min each |
| 11 | 1.56 | 5 × 104 | 100 μl 5 times, 20 min each |
Saccharomyces cerevisiae was used in a subtraction step at rounds 3, 5, and 10.
After 11 rounds of selection, the RNAs were cloned into a plasmid using a TOPO TA Cloning Kit, and the ligated plasmids were transformed into One Shot® Top 10 Escherichia coli (Invitrogen, Carlsbad, CA). The plasmids containing the aptamers were purified using a QIAprep Miniprep Kit (Qiagen, Hilden, Germany), and the aptamer sequences were determined by First BASE Laboratories Sdn Bhd (Malaysia). The obtained RNA sequences were further evaluated for binding affinity and specificity as follows: approximately 106 tested cells were mixed with 100 pmol of either the control or C. albicans‐specific aptamers in the presence of baker's yeast tRNA. After the binding reaction, washing, and elution steps described above, the amount of input and bound RNAs were quantified using q‐PCR, and the binding percentage was calculated as bound RNAs × 100/input RNAs. Predicted secondary structures were generated using RNAstructure 5.3 (Reuter & Mathews, 2010).
2.3. Aptamer‐linked immobilized sorbent assay
The binding affinity of the selected aptamers was measured using an ALISA. To do this, 96‐well microtiter plates (Iwaki, Tokyo, Japan) were coated with 50 μl of mouse anti‐human C. albicans monoclonal antibody (U.S. Biological, Swampscott, Mass) diluted to 1 μg/ml in coating buffer (50 mmol/L Na2CO3, pH 9.6) and incubated overnight at 4°C. The plates were washed twice with PBST (50 mmol/L phosphate‐buffered saline, pH 7.2 containing 0.05% Tween 20) and blocked with 100 ml of 1% BSA (Sigma, MA, USA) in PBST for 90 min at room temperature (RT). After washing, various concentrations of the C. albicans preparation in PBST (100 μl, triplicate), ranging from 50 to 5,000 cells/ml, were incubated for 1 hr at RT by gentle shaking in 100 μl of binding buffer. After the designated time, the unbound target was removed, and the plates were washed twice with PBST containing 0.1% Tween 20. Subsequently, 100 μl (100 μg/ml) of the biotinylated aptamer that was prepared as described elsewhere (Tsuji et al., 2009) was added into each well, and binding was allowed to proceed protected from light for 1 hr at RT.
The unbound materials were removed by washing with washing buffer (three times). Finally, 100 μl of a 1:1,000 dilution of a solution of streptavidin conjugated to horseradish peroxidase (HRP) was added to the individual wells. Following a 30‐min incubation on a shaking platform at RT, wells were washed twice with PBST and developed using ABTS as a substrate (Sigma, MA, USA). The reaction was stopped with 100 ml of 0.25 M H2SO4, absorbance was measured at 450 nm using a microplate reader (BioRad, USA), and washing buffer was used as a background control. The same procedure was used for enzyme‐linked immunosorbent assays (ELISA). However, the biotinylated aptamer used in the ALISA was replaced by biotinylated polyclonal anti‐C. albicans antibodies in the ELISA.
2.4. The effect of RNA aptamers on biofilm formation
Biofilm formation assays were performed as previously described (Bachtiar et al., 2014). Briefly, 100 μl containing 1.8 × 105 yeast cells of C. albicans from overnight culture at 35°C was aliquoted into microtiter plates. Three different concentrations of each tested aptamer in buffer (1 ng/μl, 10 ng/μl, and 10 ng/μl) were then added into separate wells, and the plates were incubated at 37°C in 5% CO2 in air for 90 min with gentle shaking. To promote biofilm formation, the wells were treated with 150 μl of fresh yeast nitrogen base (Sigma‐Aldrich) medium without aptamer, and the culture period was lengthened to 24 hr. Candida albicans growth was determined by evaluating the metabolic activity of growing C. albicans using MTT (3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide) reagent (Sigma‐Aldrich). Candida albicans biofilms with PBS (pH 7.2) added instead of aptamer were included to serve as negative controls. Statistical significance for all experiments was determined by pairwise comparison by t test using GraphPad Prism 7.0 software. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. In vitro selection of RNA aptamers against yeast cells of C. albicans
In this study, q‐PCR was used to monitor the enrichment progress of RNA aptamers. To decrease nonspecific binding of RNA, the experimental conditions in each selection round (Table 1) included 5 μg of baker's yeast tRNA as a binding competitor in a volume of 50 μl. The obtained RNA pool was further utilized to select for aptamers that specifically bind to live C. albicans yeast cells.
From the first to fifth rounds of the SELEX process, the components of the library that were enriched included specific and nonspecific binders. However, when the counterselection step was performed at the fifth round, the retention rate decreased gradually from the sixth to 10th rounds. We found that at the 11th round, the enrichment of the RNAs capable of binding to the target C. albicans increased significantly compared with the fifth round RNAs and the original library RNAs (data not shown). Therefore, the PCR products from round 11 were cloned to characterize each aptamer, and the randomized regions of the aptamers are shown in Figure 1a. They do not exhibit any consensus sequences and are unique except for Ca‐apt‐1 and Ca‐apt‐12, which were identified four and two times during the screening, respectively. Their preferential retention during selection suggests that they might have high affinity toward the target cells. The predicted secondary structures of both aptamers are shown in Figure 1b. Interestingly, the length of the randomized region of Ca‐apt‐1 [41 nucleotide (nt)] was longer than that of the original 40‐nt pool RNAs. Such changes may occur either during reverse transcription or PCR steps in multiple rounds of SELEX (Doudna, Cech, & Sullenger, 1995; Takemura et al., 2006; Ye et al., 2014).
Figure 1.
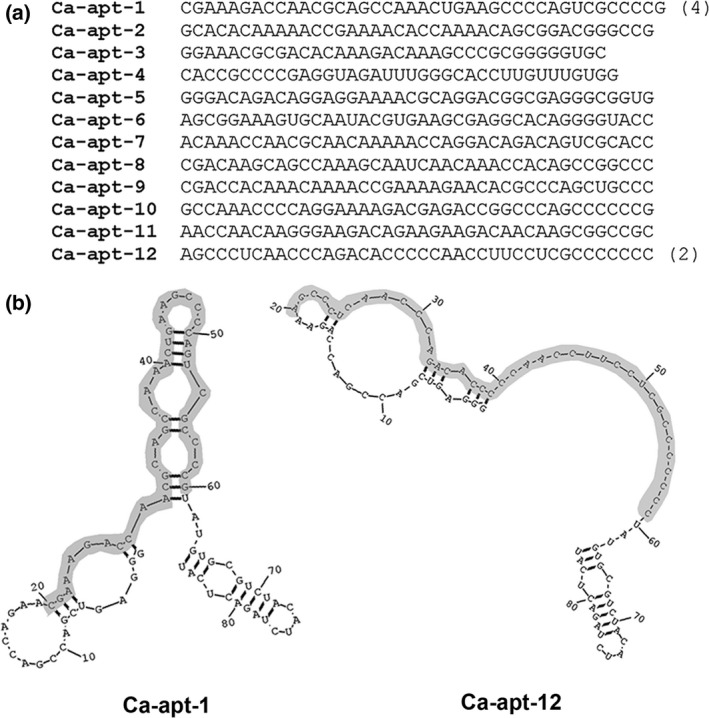
The sequences and predicted secondary structure of the Candida albicans‐specific aptamers (Ca‐apt). (a) The randomized region of the aptamers is shown. The number in parentheses represents the number of clones identified during the aptamer screening. (b) The predicted secondary structures of Ca‐apt‐1 and Ca‐apt‐12 with the lowest folding energy are shown. The nucleotides in the shaded area correspond to the randomized region of the aptamer
Next, the aptamers were screened for their binding to the target C. albicans. As expected, only the Ca‐apt‐1 and Ca‐apt‐12 aptamers demonstrated significantly higher binding percentages than the negative control RNA, which is a clone randomly chosen from the original RNA library (Figure 2a). Therefore, these aptamers were chosen for subsequent characterization.
Figure 2.
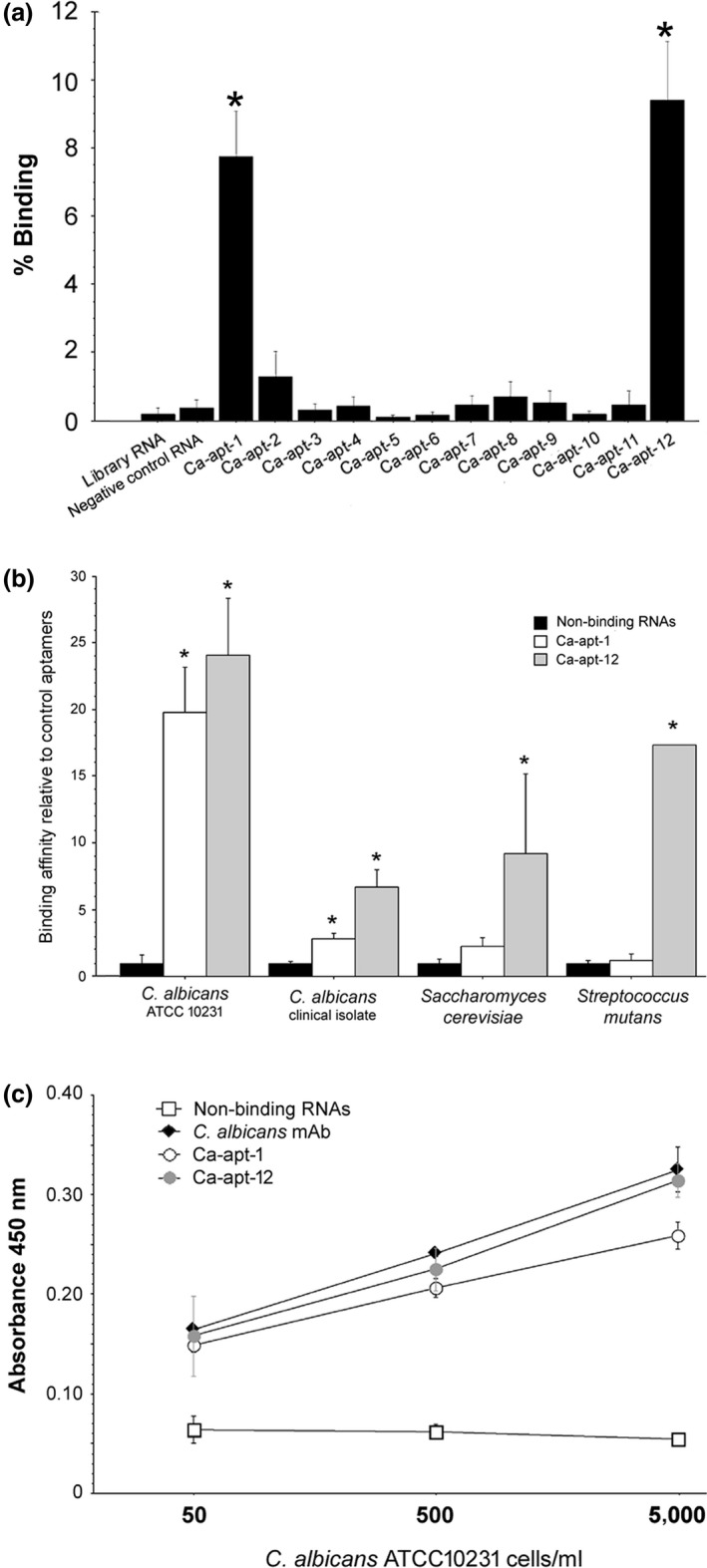
The binding properties of the Candida albicans‐specific aptamers. (a) The aptamers were screened for their binding to the C. albicans ATCC 10231 strain, which was used as the selection target. The plot shows the binding percentage (bound RNA × 100/input RNA). Only Ca‐apt‐1 and Ca‐apt‐12 demonstrated significantly higher binding than the negative control RNA. (b) The specificity of Ca‐apt‐1 and Ca‐apt‐12 was tested using either the target C. albicans strain or a clinical strain isolated from the oral cavity, a related yeast strain Saccharomyces cerevisiae, and Streptococcus mutans. The aptamer‐linked immobilized sorbent assay (ALISA) using both aptamers is shown and compared with an ELISA using antibodies against C. albicans. The plots show the mean values, and an error bar represents the standard error of the mean (SEM). An asterisk indicates a statistically significant difference compared with the nonbinding RNAs using an unpaired t test (p < 0.05)
3.2. Binding specificity test
To test the specificity of the aptamers and their binding to various targets, that is, C. albicans strains, either from the ATCC or a clinical specimen, S. cerevisiae, and S. mutans, was evaluated. As shown in Figure 2b, Ca‐apt‐1 and Ca‐apt‐12 aptamers show significantly higher binding affinities to C. albicans ATCC strain compared with those of the nonbinding RNAs, which are the aptamer clones showing no target binding. Moreover, both aptamers can also recognize C. albicans from a clinical specimen albeit at somewhat lower binding percentages (Figure 2b).
To confirm the results and to test whether the aptamers can be used in diagnostics for C. albicans, we evaluated the aptamers ability to detect C. albicans using the ALISA method. The results showed that both Ca‐apt‐1 and Ca‐apt‐12 can detect C. albicans at concentrations ranging from 50 to 5,000 cells/ml, and the ALISA performance is comparable to that of the ELISA method using C. albicans‐specific antibodies (Figure 2c).
3.3. Biofilm inhibition assay
The RNAs from the original library, round 11 RNAs, nonbinding RNAs, Ca‐apt‐1, and Ca‐apt‐12 were preincubated with C. albicans yeast cells before the biofilm was allowed to form. As shown in Figure 3a, the MTT assay results after 24 hr of biofilm growth demonstrate that the presence of the round 11 RNA and the Ca‐apt‐1 aptamer can cause a significant reduction in cell viability compared with that of the untreated group (p < 0.05). Moreover, there was a decrease in hyphal formation from yeast cells treated with the Ca‐apt‐1 aptamer compared with the untreated or library RNA‐treated group, as observed under a light microscope (Figure 3b). Interestingly, the Ca‐apt‐12 aptamer did not seem to affect biofilm formation, although it demonstrates binding affinity to C. albicans, as shown by ALISA (Figure 2c). Moreover, incubation of C. albicans during the early stage of biofilm formation with Ca‐apt‐1 at concentrations as low as 1 ng/μl was sufficient to inhibit viability and hyphal formation.
Figure 3.
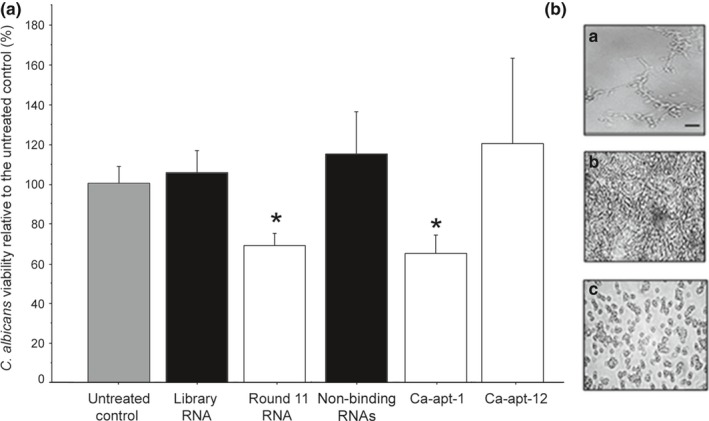
The effect of the Candida albicans‐specific aptamers on C. albicans biofilm formation. (a) C. albicans viability was determined after 24 hr of growth using the MTT assay in the presence of the RNAs from the original library, round 11, nonbinding RNAs, Ca‐apt‐1, and Ca‐apt‐12 at a concentration of 1 ng/μl. The plots show the mean values of the relative viability compared to the untreated control, and an error bar represents the standard error of the mean. An asterisk indicates a statistically significant difference compared with the untreated control using an unpaired t test (p < 0.05). (b) Representative light microscopy images of C. albicans in the untreated control (top) or the cells treated with either the original library RNA (middle) or Ca‐apt‐1 (bottom) are shown. The images from the light microscope (400× magnification) were edited for brightness and contrast, and the bars represent 20 μm for all images. Decreased hyphal formation is observed in the presence of the aptamer
4. DISCUSSION
Candida albicans is the most common fungus causing oral candidiasis and is involved in the pathogenesis of early childhood caries (Falsetta et al., 2014). However, no vaccine against this oral opportunistic pathogen is currently available. Thus, effective strategies for detecting and inhibiting Candida infection are needed. Aptamers have been reviewed in detail as highly sensitive and specific ligands to detect pathogens (Torres‐Chavolla & Alocilja, 2009). In this study, whole yeast cells of C. albicans strain ATCC 10231 were used as the target of RNA aptamer selection, as this may generate multiple targets in parallel (Shangguan et al., 2008) toward C. albicans cell surface molecules. Using whole‐cell targets in the SELEX process can be faster, easier, and more reproducible than using other targets (Guo, Lin, Zhang, Simon, & Kushner, 2009). In addition, when using the whole‐cell SELEX method, it is not necessary to isolate and purify a single target protein, which might change the presentation of the target molecule compared to an intact, live, bacterial cell (Dwivedi, Smiley, & Jaykus, 2010). Thus, this study showed that using the whole cell as a targeted ligand in the SELEX procedure has the potential to result in aptamers with binding affinity for targets on the cell in their native conformations.
In the current study, we used q‐PCR to monitor library enrichment after each selection cycle, showing relatively increasing amounts of RNA binding that reached a maximum in the 11th round. To increase aptamer binding, we used a relatively low number of C. albicans cells, as studies have shown that compared with using a large amount of target, using small amounts of target increases the success of the selection and often results in higher‐affinity aptamers (Chen et al., 2007).
When the specificities of our samples were compared, our data showed that the Ca‐apt‐1 aptamer bound to only C. albicans, whereas the Ca‐apt‐12 aptamer can also bind to the related yeast species S. cerevisiae and the oral bacterium S. mutans, which has a symbiotic relationship with C. albicans (Falsetta et al., 2014). As this study cannot determine the target molecule of the aptamer, we speculated that some of the potential targets of Ca‐apt‐12 are mannoproteins, immunodominant outer cell wall components of C. albicans (Lopez‐Ribot, Casanova, Murgui, & Martinez, 2004). These glycoproteins mediate C. albicans–S. mutans interplay in plaque biofilm (Bachtiar & Bachtiar, 2018; Hwang et al., 2017), and they have a critical role in the pathogenesis of early childhood caries (Kim et al., 2017). However, additional studies are required to identify the possible targets of both aptamers.
The aptamers were further tested for their effects on C. albicans biofilm formation, which is considered to contribute to the virulence characteristics of C. albicans (Chandra et al., 2001).We found that the Ca‐apt‐1 has the ability to interfere with C. albicans biofilm formation at the intermediate stage (24 hr) (Bachtiar, Dewiyani, Akbar, & Bachtiar, 2016) because the aptamer must act at the earliest stage of biofilm formation, which was set at 90 min in our experiment. We hypothesized that the effects of the Ca‐apt‐1 aptamer on biofilm formation might be physicochemical in nature or due to direct contact between the aptamer and the fungus, as shown in this study by a binding affinity test using an ALISA. However, in this study, we cannot explain the mechanisms by which the aptamer interferes with biofilm formation. Thus, further studies are necessary.
In conclusion, in this study, two candidate RNA aptamers, Ca‐apt‐1 and Ca‐apt‐12, were obtained through the SELEX method. The Ca‐apt‐1 aptamer binds specifically to C. albicans and possesses the ability to inhibit the fungus as it develops as a biofilm, whereas Ca‐apt‐12 shows cross‐binding with S. cerevisiae and S. mutans and does not affect biofilm formation. Additional studies are necessary to identify the aptamer targets and to explore the potential applications of these aptamers in clinical settings.
CONFLICT OF INTEREST
The author(s) declare no financial or commercial conflicts of interest.
AUTHORS CONTRIBUTION
BMB and CS designed, coordinated, and performed the experiments, as well as drafted the manuscript. EWB and CS analysed the data. All authors read and approved the manuscript.
ETHICS STATEMENT
None declared.
ACKNOWLEDGEMENTS
This study was supported by a grant (PUPT) provided by Kemenristekdikti RI to BMB. CS was supported by Chalermphrakiat grant, the Faculty of Medicine Siriraj Hospital, Mahidol University, and the National Nanotechnology Center (NANOTEC), NSTDA, Ministry of Science and Technology, Thailand through its Center of Excellence Network program. We thank Prof. Yamashita, Kyushu University, Japan, for providing the S. mutans Xc. We thank TWAS for providing a grant (RGA No: 11‐104 RG/Bio/As G‐UNESCO FR: 340262673) for some equipment and reagents. The authors thank Maysaroh Shiddiq for her skillful technical assistance.
Bachtiar BM, Srisawat C, Bachtiar EW. RNA aptamers selected against yeast cells inhibit Candida albicans biofilm formation in vitro. MicrobiologyOpen. 2019;8:e812 10.1002/mbo3.812
DATA ACCESSIBILITY
All data are included in the manuscript. Raw data are available on request.
REFERENCES
- Akpan, A. , & Morgan, R. (2002). Oral candidiasis. Postgraduate Medical Journal, 78(922), 455–459. 10.1136/pmj.78.922.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtiar, E. W. , & Bachtiar, B. M. (2018). Relationship between Candida albicans and Streptococcus mutans in early childhood caries, evaluated by quantitative PCR. F1000Research, 7, 1645 10.12688/f1000research.16275.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtiar, E. W. , Bachtiar, B. M. , Jarosz, L. M. , Amir, L. R. , Sunarto, H. , Ganin, H. , … Krom, B. P. (2014). AI‐2 of aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Frontiers in Cellular and Infection Microbiology, 4, 94 10.3389/fcimb.2014.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtiar, E. W. , Dewiyani, S. , Akbar, S. M. S. , & Bachtiar, B. M. (2016). Inhibition of Candida albicans biofilm development by unencapsulated Enterococcus faecalis cps2. Journal of Dental Sciences, 11(3), 1–8. 10.1016/j.jds.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas, R. J. , Heitman, J. , & Cardenas, M. E. (2009). The protein kinase Tor1 regulates adhesin gene expression in Candida albicans . PLoS Pathogens, 5(2), e1000294 10.1371/journal.ppat.1000294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X. , Li, S. , Chen, L. , Ding, H. , Xu, H. , Huang, Y. , … Shao, N. (2009). Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus . Nucleic Acids Research, 37(14), 4621–4628. 10.1093/nar/gkp489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra, J. , Kuhn, D. M. , Mukherjee, P. K. , Hoyer, L. L. , McCormick, T. , & Ghannoum, M. A. (2001). Biofilm formation by the fungal pathogen Candida albicans: Development, architecture, and drug resistance. Journal of Bacteriology, 183(18), 5385–5394. 10.1128/JB.183.18.5385-5394.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F. , Zhou, J. , Luo, F. , Mohammed, A. B. , & Zhang, X. L. (2007). Aptamer from whole‐bacterium SELEX as new therapeutic reagent against virulent Mycobacterium tuberculosis . Biochemical and Biophysical Research Communications, 357(3), 743–748. 10.1016/j.bbrc.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Doudna, J. A. , Cech, T. R. , & Sullenger, B. A. (1995). Selection of an RNA molecule that mimics a major autoantigenic epitope of human insulin receptor. Proceedings of the National Academy of Sciences of the United States of America, 92(6), 2355–2359. 10.1073/pnas.92.6.2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi, H. P. , Smiley, R. D. , & Jaykus, L. A. (2010). Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole‐cell SELEX. Applied Microbiology and Biotechnology, 87(6), 2323–2334. 10.1007/s00253-010-2728-7 [DOI] [PubMed] [Google Scholar]
- Falsetta, M. L. , Klein, M. I. , Colonne, P. M. , Scott‐Anne, K. , Gregoire, S. , Pai, C. H. , … Koo, H. (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo . Infection and Immunity, 82(5), 1968–1981. 10.1128/IAI.00087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelardi, E. , Pichierri, G. , Castagna, B. , Barnini, S. , Tavanti, A. , & Campa, M. (2008). Efficacy of chromogenic Candida Agar for isolation and presumptive identification of pathogenic yeast species. Clinical Microbiology and Infection, 14(2), 141–147. 10.1111/j.1469-0691.2007.01872.x [DOI] [PubMed] [Google Scholar]
- Guo, R. , Lin, W. , Zhang, J. , Simon, A. E. , & Kushner, D. B. (2009). Structural plasticity and rapid evolution in a viral RNA revealed by in vivo genetic selection. Journal of Virology, 83(2), 927–939. 10.1128/JVI.02060-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamula, C. L. , Le, X. C. , & Li, X. F. (2011). DNA aptamers binding to multiple prevalent M‐types of Streptococcus pyogenes . Analytical Chemistry, 83(10), 3640–3647. 10.1021/ac200575e [DOI] [PubMed] [Google Scholar]
- Hwang, G. , Liu, Y. , Kim, D. , Li, Y. , Krysan, D. J. , & Koo, H. (2017). Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross‐kingdom biofilm development in vivo . PLoS Pathogens, 13(6), e1006407 10.1371/journal.ppat.1006407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayasena, S. D. (1999). Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clinical Chemistry, 45(9), 1628–1650. [PubMed] [Google Scholar]
- Kim, D. , Sengupta, A. , Niepa, T. H. , Lee, B. H. , Weljie, A. , Freitas‐Blanco, V. S. , … Koo, H. (2017). Candida albicans stimulates Streptococcus mutans microcolony development via cross‐kingdom biofilm‐derived metabolites. Scientific Reports, 7, 41332 10.1038/srep41332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Ribot, J. L. , Casanova, M. , Murgui, A. , & Martinez, J. P. (2004). Antibody response to Candida albicans cell wall antigens. FEMS Immunology & Medical Microbiology, 41(3), 187–196. 10.1016/j.femsim.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Reuter, J. S. , & Mathews, D. H. (2010). RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics, 11, 129 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangguan, D. , Meng, L. , Cao, Z. C. , Xiao, Z. , Fang, X. , Li, Y. , … Tan, W. (2008). Identification of liver cancer‐specific aptamers using whole live cells. Analytical Chemistry, 80(3), 721–728. 10.1021/ac701962v [DOI] [PubMed] [Google Scholar]
- Shibata, Y. , Kawada, M. , Nakano, Y. , Toyoshima, K. , & Yamashita, Y. (2005). Identification and characterization of an autolysin‐encoding gene of Streptococcus mutans . Infection and Immunity, 73(6), 3512–3520. 10.1128/IAI.73.6.3512-3520.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soll, D. R. (2002). Candida commensalism and virulence: The evolution of phenotypic plasticity. Acta Tropica, 81(2), 101–110. 10.1016/S0001-706X(01)00200-5 [DOI] [PubMed] [Google Scholar]
- Srisawat, C. , & Engelke, D. R. (2001). Streptavidin aptamers: Affinity tags for the study of RNAs and ribonucleoproteins. RNA, 7(4), 632–641. 10.1017/S135583820100245X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura, K. , Wang, P. , Vorberg, I. , Surewicz, W. , Priola, S. A. , Kanthasamy, A. , … Sreevatsan, S. (2006). DNA aptamers that bind to PrP(C) and not PrP(Sc) show sequence and structure specificity. Experimental Biology and Medicine (Maywood, N.J.), 231(2), 204–214. 10.1177/153537020623100211 [DOI] [PubMed] [Google Scholar]
- Tang, J. , Yu, T. , Guo, L. , Xie, J. , Shao, N. , & He, Z. (2007). In vitro selection of DNA aptamer against abrin toxin and aptamer‐based abrin direct detection. Biosensors & Bioelectronics, 22(11), 2456–2463. 10.1016/j.bios.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Torres‐Chavolla, E. , & Alocilja, E. C. (2009). Aptasensors for detection of microbial and viral pathogens. Biosensors & Bioelectronics, 24(11), 3175–3182. 10.1016/j.bios.2008.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, S. , Tanaka, T. , Hirabayashi, N. , Kato, S. , Akitomi, J. , Egashira, H. , … Ohtsu, T. (2009). RNA aptamer binding to polyhistidine‐tag. Biochemical and Biophysical Research Communications, 386(1), 227–231. 10.1016/j.bbrc.2009.06.014 [DOI] [PubMed] [Google Scholar]
- Tuerk, C. , & MacDougal‐Waugh, S. (1993). In vitro evolution of functional nucleic acids: High‐affinity RNA ligands of HIV‐1 proteins. Gene, 137(1), 33–39. 10.1016/0378-1119(93)90248-2 [DOI] [PubMed] [Google Scholar]
- Vazquez‐Reyna, A. B. , Balcazar‐Orozco, R. , & Flores‐Carreon, A. (1993). Biosynthesis of glycoproteins in Candida albicans: Biochemical characterization of a soluble alpha‐mannosidase. FEMS Microbiology Letters, 106(3), 321–325. 10.1111/j.1574-6968.1993.tb05983.x [DOI] [PubMed] [Google Scholar]
- Whiteway, M. , & Bachewich, C. (2007). Morphogenesis in Candida albicans . Annual Review of Microbiology, 61, 529–553. 10.1146/annurev.micro.61.080706.093341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, F. , Zheng, Y. , Wang, X. , Tan, X. , Zhang, T. , Xin, W. , … Wang, J. (2014). Recognition of Bungarus multicinctus venom by a DNA aptamer against beta‐bungarotoxin. PLoS ONE, 9(8), e105404 10.1371/journal.pone.0105404 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are included in the manuscript. Raw data are available on request.


