Abstract
Cancer cells have the unique ability to overcome natural defense mechanisms, undergo unchecked proliferation and evade apoptosis. While chemotherapeutic drugs address this, they are plagued by a long list of side effects and have a poor success rate. This has spurred researchers to identify safer bioactive compounds that possess chemopreventive and therapeutic properties. A wide range of experimental as well as epidemiological data encourage the use of dietary agents to impede or delay different stages of cancer. In the present study, we have examined the anti-ancer property of ubiquitous phytochemical quercetin by using cell viability assay, flow cytometry, nuclear morphology, colony formation, scratch wound assay, DNA fragmentation and comet assay. Further, qPCR analysis of various genes involved in apoptosis, cell cycle regulation, metastasis and different signal transduction pathways was performed. Proteome profiler was used to quantitate the expression of several of these proteins. We find that quercetin decreases cell viability, reduces colony formation, promotes G2-M cell cycle arrest, induces DNA damage and encourages apoptosis. Quercetin induces apoptosis via activating both apoptotic pathways with a stronger effect of the extrinsic pathway relying on the combined power of TRAIL, FASL and TNF with up-regulation of caspases and pro-apoptotic genes. Quercetin could inhibit anti-apoptotic proteins by docking studies. Further, quercetin blocks PI3K, MAPK and WNT pathways. Anticancer effect of quercetin observed in cell-based assays were corroborated by molecular biology studies and yielded valuable mechanistic information. Quercetin appears to be a promising candidate with chemopreventive and chemotherapeutic potential and warrants further research.
Keywords: apoptosis, cervical cancer, chemoprevention, extrinsic pathway, Quercetin
Introduction
Cancer is one of the foremost causes of mortality across the world. Conventional cancer therapies lead to severe side effects, resulting in poor quality of life for the patient [1]. Therefore, there is a gradual shift toward a more targeted mechanism-based chemopreventive approach in lieu of conventional cytotoxic chemotherapeutics. Extensive epidemiological evidences suggest that a diet of fruit and vegetables can prevent a range of human cancers and are associated with a decreased risk of cancer-related mortality [2–7].
Plant polyphenols are structural group of naturally occurring organic chemicals distinguished by the existence of multiple phenolic functional units present in commonly consumed foods. Dietary polyphenols exhibit anti-inflammatory, immunomodulatory, antioxidant and pro-apoptotic properties and modulate cell signaling pathways that effectively suppress various stages of carcinogenesis [8–11]. Earlier in vitro studies demonstrate anticancer effect of phytochemicals derived from fruits and vegetables like genistein, EGCG, capsaicin, curcumin, sulforaphane, 6-gingerol and eugenol [12–17]. The modulation of cell signaling pathways, activation of cell death signals and induction of apoptosis in precancerous or malignant cells make phytochemicals a promising strategy in the management of malignancies [18–22].
Quercetin, a flavonoid (a subclass of polyphenolic compounds) is ubiquitously available in several vegetables and fruits. This compound has antioxidant, prooxidant, antivirus, anti-allergic and analgesic properties along with a variety of pharmacological effects [23]. Previous in vivo and in vitro experiments have demonstrated that quercetin impedes the growth of several tumors including breast, colon, ovary and stomach by inhibiting the cell cycle and cell signaling pathways (PI3K and MAPK pathways), regulating growth factors and apoptosis induction [24,25]. The prevention of colon and lung carcinogenesis by diet-derived quercetin has been demonstrated in the recent past [26,27].
The present study investigates the anti-proliferative and anti-apoptotic potential of quercetin on HeLa cells. Although, anti-proliferative potential of quercetin is known, there is no conclusive evidence available regarding its mechanistic action. In the present study, we have undertaken a comprehensive analysis of quercetin-induced apoptosis in cervical cancer cells and its effect on genes involved in apoptosis and tumorigenesis.
Materials and methods
Cell line and cell culture
Human cervical carcinoma HeLa cells were gifted by Dr. Tahir A. Rizvi, UAE University, Al-Ain, UAE. The cell line was maintained in Dulbecco’s modified Eagle’s medium (Sigma, St. Louis, MO) and supplemented with 10% fetal bovine serum (Sigma) and 100X Pen-strep (Sigma) in a humidified atmosphere of 5% CO2 in air at 37°C.
Preparation of quercetin
Quercetin (Sigma, U.S.A.) was prepared in 66.17 mM stock using DMSO and stored at −20°C. The working concentration of 1 mM quercetin was made in a complete medium. A range of 1–150 µM quercetin was tested in MTT assay followed by utilization of sublethal doses of 25 and 50 µM quercetin for all the assays.
Viability assay of HeLa cells and lymphocytes
Approximately 10000 HeLa cells/well were plated in 96-well plate and incubated for 24 h. After attachment, the cells were treated with different concentrations of quercetin ranging from 1 to 150 µM for 24 and 48 h. Similarly, cells were treated with vehicle control using DMSO. Morphological changes in HeLa cells were recorded using an inverted microscope (Labomed, U.S.A.). Following the treatment, MTT (Sigma–Aldrich) at final concentration of 0.5 mg/ml was added and incubated at 37°C for 2 h. The formazan crystals were solubilized with 100 µl of DMSO with 20-min incubation at 37°C (Sigma–Aldrich). Absorbance Microplate Reader (BioTek, U.S.A) was used to record the absorbance at 570 nm and calculate the viability of the cells. The experiments were repeated thrice and expressed as an average. The cell viability was calculated following the below-mentioned formula:
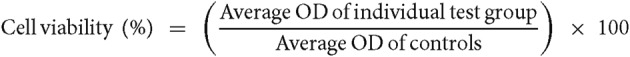 |
Lymphocytes were isolated from fresh blood using HiSep Media (HiMedia, India) following the manufacturer’s instructions. They were then resuspended in RPMI media and plated in 96-well microplates at approximately 10,000 cells/well and treated with quercetin as stated above. MTT assay was performed after 24 h exposure.
Colony formation assay
Approximately 25 x 104 cells were plated in six-well plates and treated the following day with 25 and 50 μM (24 and 48 h) quercetin. The cells were harvested post treatment, counted and plated at approximately 500 cells/well. After 2 weeks, the cells were fixed in 100% methanol, stained with 0.5% Crystal Violet and colonies were counted [28,29].
Nuclear morphology analysis with propidium iodide staining
Nuclear morphology analysis using propidium iodide (PI) stain was employed to analyze whether quercetin enables apoptotic cell death in HeLa cells. Briefly, the cells (approximately 25 × 104 cells/ml) were seeded on glass coverslips and left overnight to attach in a complete medium at 37°C, followed by the treatment with 25 and 50 μM quercetin (24 and 48 h). After the treatment, the cells were fixed in a mixture of acetone:methanol (1:1) at −20°C for 10 min and washed with PBS (pH 7.4) twice, and further stained with PI (10 μg/ml) for 30 s in dark at room temperature. The coverslips were then rinsed with PBS and mounted on a slide and observed at 515 nm under the Progress Fluorescent Microscope (Olympus, U.S.A.).
Cell cycle analysis with PI staining using flow cytometry
To determine the effect of quercetin on the cell cycle, approximately 2 × 106 cells were treated with 25 and 50 μM quercetin (24 and 48 h). The treated cells were fixed with 70% ethanol and incubated at −20°C. The fixed cells were washed with PBS, stained with PI (10 mg/ml PI; 0.5% Tween-20; 0.1% RNase in 0.01 M phosphate buffered saline) and processed using fluorescence-activated cell sorter (FACS; BD flow cytometer). The data were analyzed using FlowJo® software program. Untreated cells were used as control.
DNA ladder assay
DNA fragmentation kit (Abcam, U.S.A.) was used to extract the nucleosomal fraction and to analyze the fragmentation of DNA. Briefly, approximately 1 × 106 cells were plated and incubated at 37°C. The cells were treated with 25 and 50 μM quercetin for 24 h and harvested. The untreated cells were used as control. Nucleasomal DNA was extracted using the manufacturer’s protocol and electrophoresed at 80 V for 1 h on 1.2% agarose gel alongside a 100-bp DNA ladder. The samples were visualized in a gel documentation system and images were recorded.
Single cell gel electrophoresis assay
The single cell gel electrophoresis assay or comet assay was used to detect DNA damage following quercetin treatment if any. Approximately 25 × 104 cells were plated in six-well plates and treated with 25 and 50 μM quercetin (24 and 48 h). The cells were harvested after the treatment and Alkaline CometAssay® (Trevigen, U.S.A.) was performed according to the manufacturer’s protocol to detect single- and double-stranded breaks. The samples were stained with 20 µg/ml PI in PBS and then visualized using fluorescent microscope. The images were scored using the OpenComet plugin with Image Lab (www.rsbweb.nih.gov/ij/) [30]. The samples were scored on the basis of their tail length.
Caspase 3 activity assay
Sigma’s Caspase 3 Colorimetric Assay Kit was used to detect caspase 3 activity in treated and untreated cell lysates. Approximately 1 × 106 cells were plated and incubated at 37°C. The cells were treated with 25 and 50 μM quercetin (24 and 48 h) and then harvested. Untreated cells were used as control. The cells were lysed, and the assay was set up according to the manufacturer’s protocol. After overnight incubation, the plate was read at 562 nm. OD562 readings corresponded to caspase 3 activity and fold change with respect to the control was calculated and expressed as a graph.
Scratch wound assay
The effect of various dosages of quercetin on tumor cell migration was examined by performing the cell migration assay as described previously [31]. The cells were seeded in six-well plates at a density of approximately 5 × 106 cells per well and cultured until completely confluent. A yellow tip was used to score a constant diameter wound or cell free line. The cells were treated with 25 and 50 μM quercetin. The untreated cells were used as control. The migration of the cells across the cell free line was monitored microscopically at 0, 24, 48 and 72 h and images were obtained every 24 h. Monitoring was continued until the cell free line in the control wells reached complete closure. The wound width was measured and the percentage of wound closure was calculated and represented as a graph.
Expression analysis of genes involved in apoptosis, tumorigenesis and cancer-related pathways using qPCR
Total RNA isolation was carried out by using Gen Elute Mammalian Genomic Total RNA Kit (Sigma) from untreated and quercetin-treated HeLa cells (25 and 50 µM for 48 h) and cDNA was synthesized (ABI RT-PCR Kit). The synthesized cDNA was then used as a template for TaqMan® Human Apoptosis Array (Thermo Fisher), which has a range of different apoptosis regulators from both intrinsic and extrinsic pathways. A TaqMan-based custom array was designed consisting of several tumor suppressor genes and regulatory genes from various signal transduction pathways. PCR array was run on QuantStudio3 and analyzed by the ΔΔCT method using DataAssist™ software from Thermo Fisher. The data were normalized using 18s rRNA expression (apoptosis array) and global normalization (custom array). RQ indicates the fold change in gene expression against untreated control after normalization with the selected endogenous gene.
Docking of anti-apoptotic proteins with quercetin
Anti-apoptotic proteins BCL2 (PDB ID:2o22), BCL-xl (PDB ID: 2YXJ) and MCL1 (PDB ID: 6O6F) with co-crystallized inhibitors were retrieved [32–34]. Quercetin was retrieved from zinc database in mol2 format. The protein chains were dockprepped and docked with quercetin using SwissDock docking server [35]. The docked poses where chosen on the basis of the least energy (lowest fullfitness) values and compared with the docked pose of the co-crystallized known inhibitor bound to the protein using the visualizing software, UCSF Chimera [36].
Quantitative analysis of proteins involved in apoptosis and regulatory pathways using proteome profiler
The Human Apoptosis Array (R & D systems) was used to detect the expression of apoptosis-related proteins. Briefly, 1 × 107 cells were plated and treated with 25 and 50 quercetin for 48 h. After treatment, cells were harvested, and lysates were extracted according to the manufacturer’s instructions. The total protein quantity in the lysates was estimated by Pierce BCA assay according to the manufacturer’s protocol. The amount of lysate equal to 400 μg of protein was used for the proteome array. Antibody-spotted nitrocellulose membrane was incubated overnight with the cell lysates and the array was processed according to manufacturer’s protocol to enable chemiluminescent detection of proteins. Likewise, the Human Oncogene Array was processed following the manufacturer’s protocol. The image of the blot was captured by chemiluminescent gel doc system (Bio-Rad, U.S.A.), and analyzed using ImageLab software. The intensity of the blot corresponds to the expression of the protein.
Statistical analysis
All the data are expressed as means ± SD of at least three experiments. One-way ANOVA followed by t test was adopted for the statistical evaluation of the results. Significant differences were established at P≤0.05.
Results
Differential reduction in viability of Hela cells in a time- and dose-dependent manner
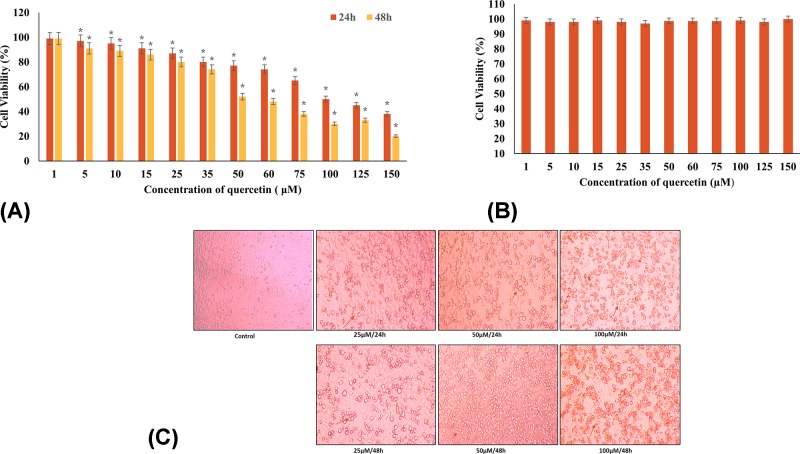
The effect of quercetin on the viability of HeLa cells was determined by treating the cells with different concentrations of quercetin and evaluating the cell viability using MTT assay. HeLa cells when treated with increasing concentrations of quercetin (1–150 µM) for 24 and 48 h showed significant growth inhibition in a dose- and time-dependent manner. The concentration at which quercetin inhibited the viability by 50% (EC50) was 100 µM after 24 h treatment (Figure 1A). The results indicate that quercetin has a significant inhibitory effect on the growth of HeLa cells in comparison with the untreated controls. Quercetin (25 µM) treatment reduced viability by 13 and 20% after 24 and 48 h, respectively. Whereas, 50 µM quercetin brought approximately 23% and 48% reduction in viability after 24 and 48 h exposure, respectively. The lymphocytes showed no adverse response against quercetin treatment (1–150 µM for 24 h) and did not inhibit their growth at the concentrations tested (Figure 1B). The results indicate that quercetin is safe on non-tumor cells and appears to specifically target cancerous cells.
Figure 1. Quercetin induces differential cell viability.
(A) Cell viability assay using the MTT assay: dose- and time-dependent cytotoxicity of quercetin (1–150 µM) treatment on HeLa cells for 24 (red) and 48 h (blue). The EC50 of quercetin was found to be 100 µΜ at 24 h (*P≤0.05). (B) Cell viability assay: dose-dependent viability of quercetin (1–150 µM) treatment on lymphocytes. Quercetin was found to have no effect on cell viability. (C) Morphological changes in HeLa cells at varying concentrations of quercetin. Microscopic features of HeLa cells treated with different concentrations (25, 50 µM for 24 and 48 h) of quercetin (magnification 20×). Arrows indicate the rounding of cells (indicative of death) with increasing concentrations.
The microscopic examination of the cells treated with various concentrations of quercetin for 24 and 48 h in comparison with untreated control showed the characteristic rounding off of the dying cells (Figure 1C).
Quercetin restricts colony formation in HeLa cells
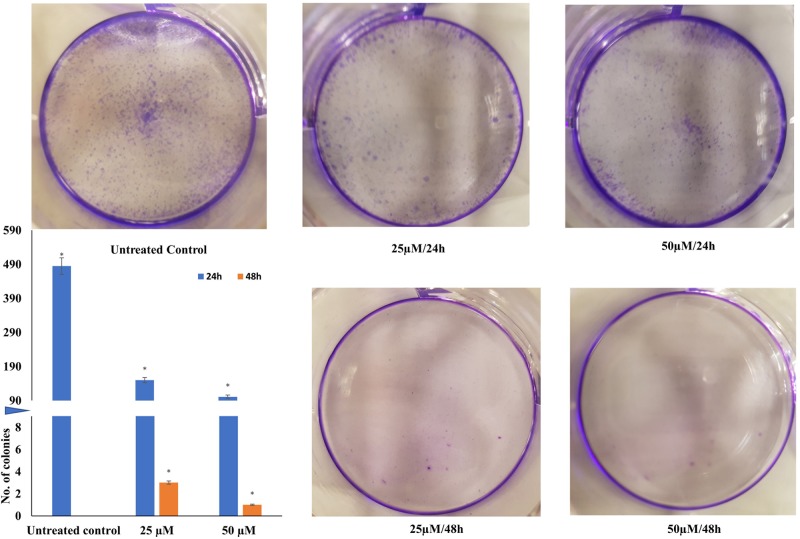
In order to examine the impact of quercetin on colony formation capacity of HeLa cells, 500 treated cells were seeded on to six-well plates. The untreated cells were used as control and to calculate the plating efficiency. While untreated cells formed colonies bearing approximately 50 cells each within 10 days, quercetin was observed to have impacted the ability of the cells to form colonies. The cells treated with 25 and 50 µM quercetin for 24 h formed 150 and 100 colonies, respectively. When treatment was extended to 48 h, no colonies were observed. The results indicate that quercetin is anti-proliferative and cytostatic (Figure 2).
Figure 2. Colony formation assay: HeLa cells treated with different concentration (25, 50 µM for 24 and 48 h), counted (500 cells) and plated.
Colonies were monitored microscopically and photographed after 2 weeks. Colonies formed after 2 weeks were counted and the mean represented as a graph, which is inset. A split Y-axis graph has been used to clearly indicate all the values. Axis is split at value 8 and restarts at value 90 (*P≤0.05).
Quercetin induces nuclear morphology changes in HeLa cells
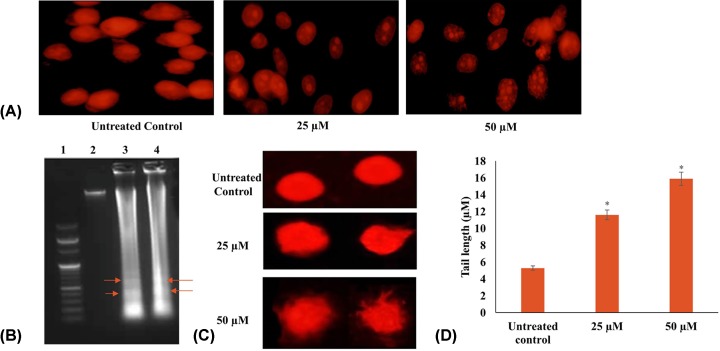
Following PI staining, fluorescent microscopy was used to observe apoptotic changes in more detail. Quercetin (25 and 50 µM treatment for 24 and 48 h) produced a substantial increase in nuclear condensation, nuclear fragmentation and apoptotic bodies in a dose- and time-dependent manner (Figure 3A).
Figure 3. Quercetin induces apoptosis in HeLa cells.
(A) Nuclear morphological features of HeLa cells treated with different concentrations (25, 50 µM for 24 and 48 h) of quercetin (magnification 100×). Figures indicate nuclear condensation, fragmentation and formation of apoptotic bodies indicative of apoptosis. (B) DNA ladder assay: HeLa cells treated with different concentrations (25, 50 µM for 24 h) of quercetin were found to produce a DNA laddering pattern consistent with apoptosis. (C) Single cell gel electrophoresis assay: HeLa cells treated with different concentrations (25, 50 µM for 24 h) of quercetin induce DNA damage. (D) The tail length of the comets are represented as a graph and indicate extent of damage (*P≤0.05).
Quercetin induces DNA fragmentation in HeLa cells
Apoptotic cells demonstrate ladder formation due to the fragmentation of DNA. Quercetin (25 and 50 µM for 24 h) was found to cause DNA fragmentation in HeLa cells in comparison with untreated control confirming the induction of apoptosis (Figure 3B).
Quercetin induces comet formation in HeLa cells
In order to understand the impact of quercetin on DNA damage, alkaline comet assay was performed on HeLa cells treated with 25 and 50 µM of quercetin for 24 h. The untreated cells yielded negligible DNA damage in the cells; whereas quercetin induced single- and double-stranded breaks in the DNA in a dose-dependent manner as observed in Figure 3C. The comet tail length was calculated and represented as a graph (Figure 3D).
Quercetin induces cell cycle arrest in HeLa cells in a dose- and time-dependent manner
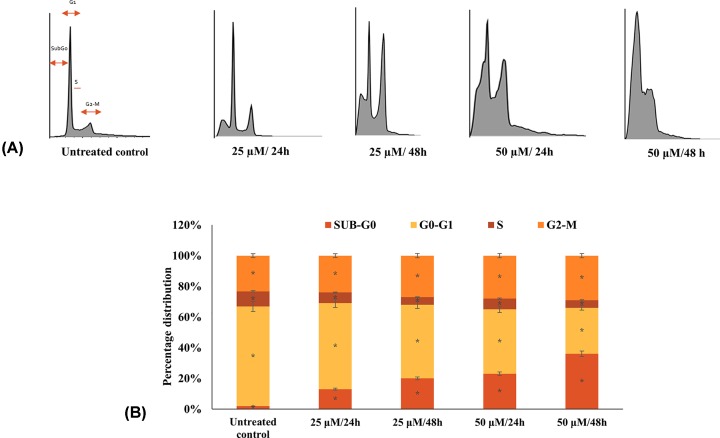
In order to understand the influence of quercetin on the cell cycle, HeLa cells were treated with quercetin (25 and 50 µM treatment for 24 and 48 h) and analyzed by flow cytometry. The results indicate that quercetin induces cell cycle arrest in G2-M phase with the G2-M population increasing proportional to the dose and duration of treatment (Figure 4). Further, the sub-G0 population, which is representative of apoptotic cells was also found to increase in a dose- and time-dependent manner (13% and 23% with 25 and 50 µM quercetin treatment for 24 h).
Figure 4. Flow cytometry: cell cycle of HeLa cells treated with different concentrations (25, 50 µM for 24 and 48 h) of quercetin was analyzed after staining with PI.
(A) Quercetin induces G2/M arrest with increase in sub-G0 apoptotic population. (B) The distribution of cells across the cell cycle is represented as a graph.
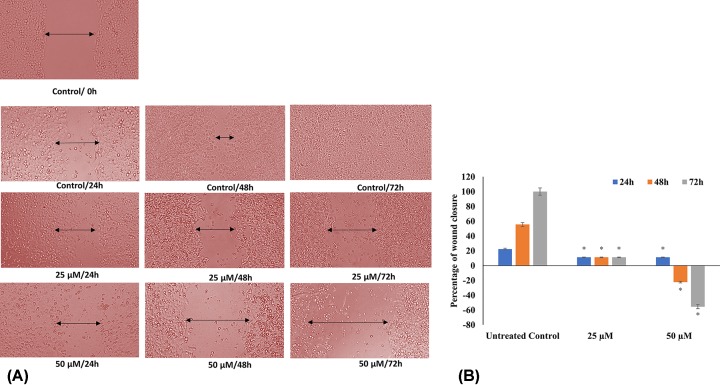
Quercetin mitigates HeLa cell migration as evidenced in scratch wound assay
In the metastatic cascade, the migration of tumor cells is a significant occurrence. The scratch-wound assay demonstrated a significant reduction in the migration capacity of HeLa cells treated with quercetin compared with the controls (Figure 5). While, the untreated HeLa cells showed complete wound closure by 72 h; the cell-free line remained clear at concentrations above 25 µM. In 50 μM quercetin treated well, the acellular line was persistent, showing that there was no migration even after 72 h.
Figure 5. Scratch wound assay: HeLa cells treated with different concentrations (25, 50 µM) of quercetin and the migration of the cells across the cell free line was monitored microscopically.
(A) Images of the wound were obtained at 0, 24, 48 and 72 h. (B) The wound width was measured, and the percentage of wound closure was calculated and represented as a graph (*P≤0.05).
Quercetin increases caspase 3 activity in a dose- and time-dependent manner
In order to examine the ability of quercetin in affecting the activity of the central executioner caspase, caspase 3 was assessed by ELISA-based activity assay. Quercetin was found to increase the activity of caspase in a dose- and time-dependent manner. Fold change was calculated in comparison with the untreated controls. A significant fold change, 8- and 12-folds was observed against 25 and 50 µM quercetin treatment of 48 h, respectively (Figure 6).
Figure 6. Caspase 3 activity: HeLa cells treated with different concentrations (25, 50 µM for 24 and 48 h) of quercetin increase the activity of caspase 3.
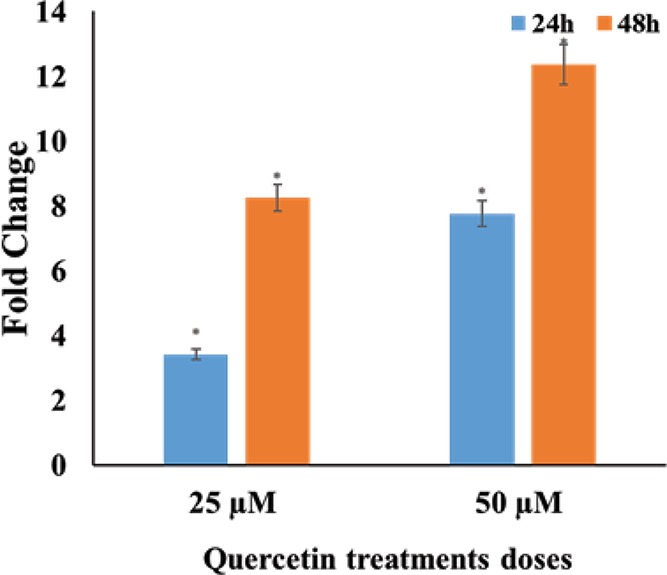
The fold change with respect to untreated control is represented as a fold change (*P≤0.05).
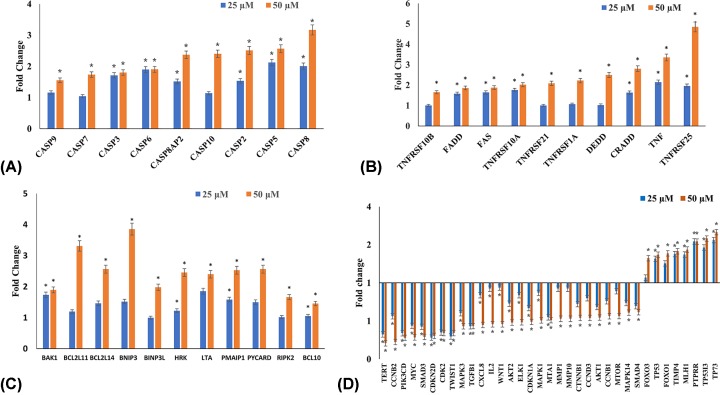
Quercetin induces apoptosis in HeLa cells mainly via extrinsic pathway
In order to ascertain the mechanism by which quercetin induces apoptosis, the expression of various genes involved in apoptosis was studied by TaqMan-based Real time qPCR array. Genes having RQ greater than 1.5 were considered as up-regulated; and those with RQ lower than 0.5 were considered as down-regulated.
The results (Figure 7) indicate that the genes involved in the extrinsic pathway of apoptosis are up-regulated and therefore this could be the mechanism through which quercetin induces apoptosis. The expression of TRAIL, FASL, TNF and their receptors (FAS, TNFSF10, TNFRSF10A, TNFRSF10B, TNFRSF1A, TNFRSF1B, TNFRSF21, TNFRSF25) increased and mediated the extrinsic pathway. TRADD, CRADD, DEDD were also found to be elevated. Further increase in expression of caspase 8, 10, 3 and 7 indicate the role of the extrinsic pathway. Caspase 8 and 10 expression via the normal course of the extrinsic pathway should lead to the activation of caspase 3 and 7, which are the effector caspases. The expression of caspase 9, 2, 14, and 6 were also found to be increased. Additionally, several genes from BCL2 family, which are involved in the pro-apoptotic action were also found to be elevated such as BAK1, HRK, NOXA, BIM, BCL10 and BCL2L14. Other pro-apoptotic responders such as BNIP3, BNIP3L, LTA, PYCARD and RIPK2 were found to increase in response to quercetin. The expression of several tumor suppressor genes involved in anti-proliferative and apoptotic response such as FOXO1, FOXO3, TP53, TP5313, TP73 were found to be increased.
Figure 7. Expression analysis of genes involved in apoptosis and cell signaling after treatment with 25, 50 µM of quercetin for 48 h.
(A) RQ plot of caspases. (B) RQ plot of extrinsic receptors and ligands. (C) RQ plot of pro-apoptotic genes. (D) RQ plot of cell cycle regulators, tumor suppressors and genes involved in PI3K, MAPK and WNT signaling (*P≤0.05).
Quercetin modulates expression of genes involved in cell cycle regulation
Quercetin down-regulates genes involved in G2-M stage of the cell cycle, viz. CCNB1, CCNB2 and CDK2, which is consistent with the observed G2-M arrest. Further it down-regulates CCND3 and CDKN1A but does not impact other genes involved in G1 stage. Telomerase reverse transcriptase (TERT) expression was also significantly down-regulated (Figure 7D).
Quercetin modulates expression of genes involved in migration
Quercetin down-regulates genes involved in migration and invasion, viz. MMP14, MMP9 and MTA1; while up-regulating TIMP4. TWIST1, an inhibitor of e-cadherin, was also steeply down-regulated (Figure 7D).
Quercetin suppresses MAPK, PI3K and WNT pathways to bring out its anti-proliferative and anti-migratory effect
ELK1 and MEKK/MAP3K5 gene expressions are down-regulated along with an increase in PTPRR, which is an inhibitor of MAP pathway. A significant decrease in AKT1, AKT2, MTOR, PI3KCTB and PI3KCD was observed, which suggests an inhibition of the PI3K pathway. WNT pathway is also suppressed by quercetin as evidenced by the decreased expression of CTNNB1 and WNT2. Gene expression of SMAD2, SMAD3, SMAD4 and TGFβ1, which play significant roles in WNT pathway, are also down-regulated. An increase in VHL expression was also found. It further down-regulates the genes involved in inflammation such as CXCL8, MYC, IL2 and IL1A (Figure 7D).
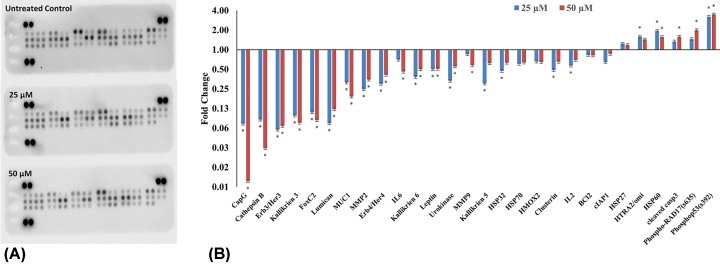
Quercetin modulates expression of pro- and anti-apoptotic proteins
Proteome profiler-based quantitation of the proteins involved in apoptosis, cell cycle regulation showed modulation consistent with mRNA expression. Cleaved caspase3, FAS, HTRA2/omi, phospho53(s392), phospho-RAD17(s635) and endolglin are up-regulated, whereas cIAP1, clusterin, HSP32, HMOX2, HSP70, CapG, cathepsin B, Erb3/Her3, Erb4/Her4, FoxC2, IL2, IL6, kallikrien 3, kallikrien 5, kallikrien 6, leptin, lumican, MMP2, MMP9, MUC1 and urokinase are down-regulated. The fold changes with respect to untreated control are plotted in Figure 8.
Figure 8. Quercetin induces apoptosis in HeLa cells.
Proteome profiler: (A) image of the proteome profiler membrane showing differential protein expression. (B) Proteins involved in apoptosis and regulatory pathways were quantitated after treating HeLa cells with 25, 50 µM of quercetin for 48 h and represented as fold change over control. Quercetin increased pro-apoptotic proteins and decreased anti-apoptotic proteins (*P≤0.05).
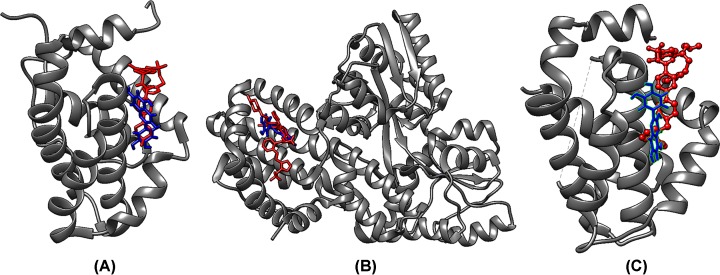
Quercetin modulates anti-apoptotic proteins
Quercetin was docked with BCL2, BCL-xl and MCL1 proteins. The docked poses were selected on the basis of least energy (lowest fullfitness) values. The docked pose of quercetin was compared with the co-crystallized known inhibitor bound to the protein (Figure 9). The interaction between quercetin and protein in each case was similar to that of the established inhibitor in the X-ray crystal structure; with high degree of shared amino acid residues. The region in which the co-crystallized inhibitor and quercetin are bound is responsible for interacting with the pro-apoptotic BCL2 proteins. These anti-apoptotic proteins bind to and sequester the pro-apoptotic proteins thereby limiting their activation [32,33,37]. Quercetin engages with these anti-apoptotic proteins in the same manner as the known inhibitors and could prevent their sequestering of the pro-apoptotic proteins. This will aid in apoptotic response.
Figure 9. Docking analysis of anti-apoptotic proteins with quercetin (blue) co-crystallized inhibitor (red) shows that quercetin occupies the same region as the inhibitor and could inhibit the proteins.
(A) BCL2; (B) BCLxl; (C) MCL1.
Discussion
Worldwide, the prevalence and mortality from cancer has been growing, even though our knowledge and treatment of diseases has progressed tremendously. Therefore, chemoprevention using natural dietary agents presents itself as a rational and appealing strategy. In the present study, we comprehensively analyze the anti-proliferative, pro-apoptotic and anti-migratory effect of quercetin on HeLa cells by modulating various signaling pathways.
Deregulation of the cell cycle with rampant proliferation while evading apoptosis and promoting metastasis are characteristics of cancer cells. An effective drug candidate should be able to limit cell proliferation, induce apoptosis and restrict migration. In the present study, the ability of quercetin to induce cytotoxicity in HeLa cells was established through the MTT assay and its EC50 was found to be 100 µM in 24 h. Whereas, other research groups have reported 50% cell viability in HeLa cells at 110.38 μM of quercetin in 18 h and 80 μM at 24 h [38,39]. Further, anti-proliferative and cytostatic ability of quercetin was demonstrated by a dose- and time-dependent inhibition of colony formation on quercetin treated HeLa cells (Figure 2). In comparison with the control it was found that quercetin limits colony formation after 24 h and almost no colonies were formed after 48 h of treatment. Earlier studies in different cell lines also showed similar findings [40–43]. Morphological analysis of quercetin treated HeLa cells by light microscopy and fluorescent microscopy (after PI staining) (Figures 1C and 3A) showed the characteristic changes associated with apoptosis including shrinkage, nuclear fragmentation, rounding off of dying cells and apoptotic bodies. To further assess the anti-proliferative and apoptosis-inducing effect of quercetin on Hela cells, flow cytometry and DNA fragmentation assay was performed. In our study, quercetin was found to induce G2-M arrest with accumulation of sub-G0 apoptotic population which is in line with previous findings of cell cycle arrest at G2-M in HeLa, breast carcinoma, leukemia and esophageal adenocarcinoma cell lines [39,44–46]. In leukemic cell line, NALM6 while sub-G0 accumulation of apoptotic population was found with increasing doses of quercetin, at lower concentrations, S-phase arrest was observed [47]. This shows that in some cell types, a variable response is observed. Our flow cytometry results are consistent with the observed morphological changes. Apoptotic induction was further supported by the DNA fragmentation assay. A clear DNA laddering pattern with bands was observed in quercetin treated HeLa cells in sharp contrast with the untreated control. Apoptosis can be initiated by the onset of DNA damage; in order to ascertain whether DNA damage could be partly responsible for the observed apoptotic response, Comet assay was performed. Mild DNA damage was mediated by quercetin in a dose- and time-dependent manner with marked increase in comet tail length by 50 µM quercetin in 48 h. Earlier studies also support both these observations [43,48,49].
Apoptosis can be induced by one of two core pathways, the extrinsic and the intrinsic pathways, with both requiring the activation of caspase proteins [50–53]. Quercetin increases the transcription of the extrinsic pathway death receptors and ligands including TNF, DR4, DR5, FAS as well as FASL and TRAIL. TRAIL and TNF receptors are important starting points for extrinsic apoptosis [51]. The initiator caspases consisting of caspase 2, -8, -9 and -10 were up-regulated by quercetin in a dose-dependent manner with caspase 8 reaching an RQ of 3.6 when treated with 50 μM quercetin for 48 h. The function of caspase 8 is an important step in TNF-induced extrinsic apoptotic pathway, which was also found to be up-regulated. The up-regulation of the receptors and caspases indicated that quercetin induces the extrinsic pathway of apoptosis. The intrinsic pathway involves the release of cytochrome c from the mitochondria, activation of caspase 9, eventually activating caspase 3 and other pro-apoptotic molecules [54,55]. Quercetin increases caspase 9 gene expression and cytochrome c protein levels marginally; indicating perhaps a lower emphasis on mitochondrial pathway. The effector caspases (caspase 3, 6 and 7) which produce the apoptotic indicators are up-regulated by quercetin in a dose-dependent manner. The increase in the levels of initiator and executioner caspases highlight the concordance in the apoptotic response. Of all the effector caspases, caspase 3 has a central role and is important for both PARP cleavage and DNA fragmentation [55]. It is significant therefore that quercetin increases both the transcription of caspase 3, and the functional cleaved caspase 3 protein in a dose-responsive manner (Figures 7 and 8). Further, the activity of caspase 3 in quercetin treated HeLa cells showed a steep increase, reiterating the evident activation of the caspase cascade and onset of apoptosis (Figure 6). Caspase 2 is required for DNA damage and is a substrate for both caspase 3 and caspase 8 [55]. The up-regulation of caspase 2 by quercetin is significant in that it further reiterates the functional activation of caspase 3 and caspase 8, as well as substantiates the results of the DNA fragmentation and comet assay.
The caspases are supported by the increased transcription of several genes involved in the apoptotic response (Figure 7). Quercetin was found to up-regulate the pro-apoptotic members, Bax, Hrk, Noxa, Bim, BCL10 and BCL2L14. It also mediates an increase in several pro-apoptotic proteins (Figures 7 and 8) including HTRA2/omi and endolglin. HTRA2/omi functions as an antagonist to inhibitors of apoptosis (IAPs) and aids in apoptosis [56]. It is further significant that quercetin increases s635 phosphorylated Rad17 (s635). The ability of Rad17 to trigger G2-M arrest and DNA damage induced apoptosis is dependent on s635 phosphorylation [57]. This further strengthens the earlier observations. Quercetin also up-regulates transcription of p53, p73 and p5313 and protein levels of phospho53(s392). P53 and its homolog p73 are silenced by HPV-E6 and play an important role in cell cycle and apoptosis [58–60]. Anti-apoptotic proteins such as cIAP1, Clusterin, HSP32, HMOX2, HSP70, CapG, Cathepsin B are down-regulated. Further, molecular docking experiments suggest that anti-apoptotic BCL2 family proteins (BCL2, BCL-xl and MCL1) could be directly inhibited by quercetin (Figure 9 and Table 2). A recent study showed that a quercetin–alanine conjugate directly binds to BCL2 and enhances apoptosis [61]. We believe that a similar interaction may be at play here.
Table 2. Interaction of least energy docked pose of quercetin with anti-apoptotic proteins.
| Protein (PDB ID) | Full fitness value | Interacting residues within 5A of quercetin | Predicted pattern of interaction |
|---|---|---|---|
| BCL2, (PDB ID 2O22) | −1446.2208 | PHE 101, TYR 105, ASP 108, PHE 109, MET 112, LEU 134, ASN 140, GLY 142, ARG 143, ALA 146, PHE 150 | Inhibitory. Similar to co-crystallized inhibitor |
| BCL xl, (PDB ID 1R2D | −869.9743 | PHE 97, TYR 101, ARG 103, PHE 105, ASP 107, LEU 108, GLN 111, GLU 129, LEU 130, PHE 131, ARG 132, ASP 133, GLY 134, ARG 139, ALA 142 | Inhibitory. Similar to co-crystallized inhibitor |
| MCL1, (PDB ID 5LOF) | −2807.5872 | HIS 224, ALA 227, PHE 228, MET 231, MET 250, VAL 253, PHE 254, ARG 263, ILE 264, THR 266, LEU 267, PHE 270 | Inhibitory. Similar to co-crystallized inhibitor |
This lends further support to the apoptotic outcome mediated by quercetin. In the present study, while both intrinsic and extrinsic pathways are activated by quercetin, a comparatively higher folds increase in caspase 8 and other proteins of the extrinsic pathway allow us to conclude that the extrinsic pathway could have the lead role with the mitochondrial pathway playing a supportive role. In leukemia cells (NALM6), quercetin was found to bring about mitochondrial pathway of apoptosis by increasing cytochrome c, caspase 9 and depolarization of mitochondrial membrane potential [47]. In breast cancer cells, quercetin induced caspase-dependent extrinsic apoptosis by up-regulating the levels of cleaved caspase-8 and caspase-3 without altering the mitochondrial membrane potential [62].
TERT, overexpressed in cervical cancer cells, determines telomere length and facilitates cancer cells to evade apoptosis and continue proliferation [63–65]. As further evidence to explain the anti-proliferative and cell cycle arrest mediated by quercetin, it was observed that the tested doses of quercetin promote a significant down-regulation in TERT transcript expression as well as down-regulates CCNB1, CCNB2 and CDK2 which are involved in cell cycle regulation.
Another hallmark of cancer, invasion and metastasis is responsible for most cancer-related mortality and morbidity and is thus an important therapeutic target. Quercetin’s ability to inhibit the migration is evidenced by the results of the scratch wound assay (Figure 5). At higher treatment dosages and durations, a cell-free line was maintained. This outcome is explained by the observed down-regulation of MMP14, MMP9, MTA1 and TWIST1 with simultaneous up-regulation of CDH1, TIMP3 and TIMP4. CDH1 is important for cell adhesion and when usually silenced by methylation can lead to metastasis and tumor progression [66–68]. Several studies have documented that increase in CDH1 can inhibit metastasis and cell growth [69,70]. TWIST1 acts as an inhibitor of CDH1 and is seen to be steeply reduced. MMPs promote tumor invasion and metastasis, while TIMPs oppose this action [71]. Thus, the concordant modulation of these genes by quercetin highlights the thoroughness of its anti-migratory effect.
The dysregulation of several signaling pathways such as the PI3K, WNT, MAPK, JAK/STAT help in cancer progression by promoting proliferation through growth stimulating signals, suppressing growth inhibitors, evading apoptosis and promoting metastasis. Quercetin was found to modulate the expression of several genes involved in these pathways; effectively causing inhibition of proliferation, migration and apoptosis (Figure 7 and Table 1). In the PI3K pathway, quercetin brings about decrease in transcript expression of AKT1, AKT2, MTOR, PI3KCTB and PI3KCD. AKT is an important molecule that can further activate other pathways including mTORC1 [72]. AKT is activated by several growth factors and cytokines through the receptor tyrosine kinases like HER by binding their cognate receptor tyrosine kinase and promotes cell survival by inactivating pro-apoptotic proteins and the forkhead (FoxO1/3a) transcription factors [73]. Interestingly, quercetin down-regulates the protein levels of HER3 and HER4 and a marked increase in the levels of the FOXO1/3 transcription factors. FOXO1/3 are tumor suppressors that induce the transcription of pro-apoptotic genes and death receptors involved in apoptosis, cell cycle regulation and DNA damage repair [74]. FOXO1 expression inhibits cervical cancer development by bringing about cell cycle arrest and apoptosis and is a favorable prognostic factor [75].
Table 1. Table of the genes modulated by quercetin to bring about its anticancer effect.
| Effect | Molecular target | Up-regulation | Down-regulation |
|---|---|---|---|
| Apoptosis | Caspases | CASP9, CASP7, CASP3, CASP6, CASP14, CASP8AP2, CASP10, CASP2, CASP5, CASP8 | |
| Pro-apoptotic BCL2 family | BAK1, HRK, PMAIP1, BCL2L14, BCL2L10, BCL2L11 | ||
| Death receptors and ligands | TNFRSF10B, FADD, FAS, TNFRSF10A, TNFRSF21, TNFRSF1A, DEDD, CRADD, TNF, TNFRSF25, FASLG, TNFSF10, TRADD | ||
| Other pro-apoptotic proteins | BNIP3, BNIP3L, LTA, PYCARD, RIPK2 | ||
| Signaling pathway and TSG | FOXO3, TP53, FOXO1, TIMP4, MLH1, PTPRR, TP53I3, TP73, CDH1, SOCS1 | TERT, CCNB2, PIK3CD, MYC, SMAD3, CDKN2D, CDK2, TWIST1, MAPK3, TGFB1, CXCL8, IL2, WNT1, AKT2, ELK1, CDKN1A, MAPK1, MTA1, MMP1, MMP10, CTNNB1, CCND3, AKT1, CCNB1, MTOR, MAPK14, SMAD4 | |
| Protein expression | Cleaved CASPASE3, FAS, HTRA2/OMI, phospho53 (S392), phospho-RAD17 (S635), Endolglin | CIAP1, CLUSTERIN, HSP32, HMOX2, HSP70, CAPG, CATHEPSIN B, ERB3/HER3, ERB4/HER4, FOXC2, IL2, IL6, KALLIKRIEN 3, KALLIKRIEN 5, KALLIKRIEN 6, LEPTIN, LUMICAN, MMP2, MMP9, MUC1, UROKINASE | |
| Cell cycle regulation and anti-proliferation | Cell cycle regulatory genes | CCNB2, CDKN2D, CDK2, CDKN1A, CCND3 | |
| Anti-proliferation genes | TERT | ||
| Anti-migration | Anti-metastatic genes | CDH1, TIMP4, SOCS1 | MMP1, MMP10 |
| Anti-proliferation, | |||
| anti-metastatic | PI3K pathway | AKT2, AKT1, MTOR | |
| WNT pathway | CTNNB1, TGFB1, WNT1, SMAD4 | ||
| MAPK pathway | PTPRR | MAPK3, MAPK1, MAPK14, ELK1 | |
| Anti-inflammation | Inflammation markers | CXCL8, IL2, MYC |
WNT signaling pathway plays an important role in cervical cancer and regulates tumor progression, particularly migration [76]. WNT2 expression is up-regulated in cervical cancer and is associated with tumor size, cell motility and invasion [77]. Quercetin reduces expression of WNT2 and CTNNB1, which are important moieties in the WNT pathway. TGFβ1, SMAD2, SMAD3 and SMAD4 are also down-regulated by quercetin. TGFβ/SMAD signaling is linked to EMT, migration and invasion [78]. The TWIST gene, part of the WNT pathway inhibits CDH1 [79,80]. As noted earlier, quercetin decreases TWIST1 and increases CDH1.
MAPK pathway is centrally involved in cell proliferation; while several members showed a decreasing trend, significant gene expression reduction was observed with ELK1 and MEKK/MAP3K5, alongside increase in PTPRR, which is an inhibitor of MAP pathway. Cervical cancer cells carry aberrantly high methylation rates of PTPRR [81]. ELK1 is involved in up-regulating the oncogene, c-fos and activating the cell cycle [82]. Therefore, its notable that these are down-regulated by quercetin.
Inflammation and inflammatory responses are negative regulators of cancer therapeutics and it is pertinent that the tested dosages of quercetin down-regulate the expression of tumor markers and proteins involved in inflammation. Quercetin reduces the expression of CXCL8, IL2, IL8 and IL6. CXCL8, a proinflammatory oncogene is highly expressed in cervical cancer tissues [83]. IL-2, IL-8 and IL-6 expression is significantly correlated with poor prognosis [84–86]. Caspase 1 and 4 are involved in inflammatory response and is not central to the apoptotic response [87]. In this regard, it is interesting that quercetin does not change the expression of caspase 1 as well as 4.
Additionally, we found that quercetin is differentially cytotoxic and does not affect the viability of lymphocytes (Figure 1B). The safe profile of quercetin in normal cell lines and animal models has been validated in other studies as well [47,48,88,89]. The specificity of quercetin’s cytotoxic action against tumor cells while not impacting normal cells, makes it an ideal drug candidate.
Conclusion
The findings of the present study show that quercetin systematically alters the PI3K, MAPK and WNT pathways by modulating the expression of several proteins leading to the inhibition of cell proliferation, cell cycle arrest, DNA damage and apoptosis in cervical cancer (HeLa) cells. A promising alternate route to cancer chemoprevention and treatment strategies appears to be the use of dietary polyphenols such as, quercetin. The present study provides emphatic evidence for the potential use of quercetin as a multipronged anticancer therapeutic agent.
Acknowledgments
The authors are grateful to Dr. Kota Reddy, Academic President and Chairperson, School of Life Sciences, Manipal Academy of Higher Education, Dubai, UAE for his constant support and encouragement.
Abbreviations
- EMT
epithelial to mesenchymal transition
- FASL
Fas ligand
- PARP
Poly (ADP ribose) polymerase 1
- PI
propidium iodide
- PI3KCD
phosphotidyl-inositol-4,5-bisphosphate 3-kinase catalytic subunit delta
- PTPRR
protein tyrosine phosphatase receptor type R
- qPCR
quantitative real time polymerase chain reaction
- TERT
telomerase reverse transcriptase
- TIMP
tissue inhibitors of metalloproteinases
- Wnt
Wingless
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was supported by the MAHE Internal Research Grant [grant number R&DP/MUD/RL-06/2017].
Author Contribution
M.K.S. and A.H. designed the study and wrote the manuscript. M.H. and K.B. performed the analysis. R.R., N.A. and S.H. searched the literature, collected the articles and extracted the relevant data. All the authors reviewed and approved the final manuscript.
References
- 1.Di Muzio M., Marinucci A., De Benedictis A. and Tartaglini D. (2017) A comparative study of data collection methods in the process of nursing : detection of chemotherapy side effects using a self-reporting questionnaire. Acta Clin. Croat. 56, 765–772 10.20471/acc.2017.56.04.26 [DOI] [PubMed] [Google Scholar]
- 2.Farvid M.S., Chen W.Y., Michels K.B., Cho E., Willett W.C. and Eliassen A.H. (2016) Fruit and vegetable consumption in adolescence and early adulthood and risk of breast cancer: population based cohort study. BMJ i2343. 10.1136/bmj.i2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amararathna M., Johnston M.R. and Vasantha Rupasinghe H.P. (2016) Plant polyphenols as chemopreventive agents for lung cancer. Int. J. Mol. Sci. 17, 10.3390/ijms17081352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turati F., Rossi M., Pelucchi C., Levi F. and La Vecchia C. (2015) Fruit and vegetables and cancer risk: a review of southern European studies. Br. J. Nutr. 113, S102–S110 10.1017/S0007114515000148 [DOI] [PubMed] [Google Scholar]
- 5.Taylor P., Zanini S., Marzotto M. and Giovinazzo F. (2014) Digestive system effects of dietary components on cancer of the digestive system. Crit. Rev. Food Sci. Nutr. 55, 37–41 [DOI] [PubMed] [Google Scholar]
- 6.Wu Q.-J., Wu L., Zheng L.-Q., Xu X., Ji C. and Gong T.-T. (2016) Consumption of fruit and vegetables reduces risk of pancreatic cancer: evidence from epidemiological studies. Eur. J. Cancer Prev. 25, 196–205 10.1097/CEJ.0000000000000171 [DOI] [PubMed] [Google Scholar]
- 7.Link A., Balaguer F. and Goel A. (2010) Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem. Pharmacol. 80, 1771–1792 10.1016/j.bcp.2010.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundaram M.K., Ansari M.Z., Al Mutery A., Ashraf M., Nasab R., Rai S.. et al. (2017) Genistein induces alterations of epigenetic modulatory signatures in human cervical cancer cells. Anticancer Agents Med. Chem. 17, 1–11 [DOI] [PubMed] [Google Scholar]
- 9.Khan M.A., Sundaram M.K., Hamza A., Quraishi U., Gunasekera D., Ramesh L.. et al. (2015) Sulforaphane reverses the expression of various tumor suppressor genes by targeting DNMT3B and HDAC1 in human cervical cancer cells. Evid. Based Complement. Alternat. Med. 2015, 10.1155/2015/412149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahmoud A.M., Yang W. and Bosland M.C. (2014) Soy isoflavones and prostate cancer: a review of molecular mechanisms. J. Steroid Biochem. Mol. Biol. 140, 116–132 10.1016/j.jsbmb.2013.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Recio M.C, Andujar I. and Rios J.L (2012) Anti-inflammatory agents from plants: progress and potential. Curr. Med. Chem. 19, 2088–2103 10.2174/092986712800229069 [DOI] [PubMed] [Google Scholar]
- 12.Sharma C., Nusri Qel-A., Begum S., Javed E., Rizvi T.A. and Hussain A. (2012) (-)-Epigallocatechin-3-gallate induces apoptosis and inhibits invasion and migration of human cervical cancer cells. Asian Pac. J. Cancer Prev. 13, 4815–4822 10.7314/APJCP.2012.13.9.4815 [DOI] [PubMed] [Google Scholar]
- 13.Hussain A., Brahmbhatt K., Priyani A., Ahmed M., Rizvi T.A. and Sharma C. (2011) Eugenol enhances the chemotherapeutic potential of gemcitabine and induces anticarcinogenic and anti-inflammatory activity in human cervical cancer cells. Cancer Biother. Radiopharm. 26, 519–527 10.1089/cbr.2010.0925 [DOI] [PubMed] [Google Scholar]
- 14.Tortorella S.M., Royce S.G., Licciardi P.V. and Karagiannis T.C. (2015) Dietary sulforaphane in cancer chemoprevention: the role of epigenetic regulation and HDAC inhibition. Antioxid. Redox Signal. 22, 1382–1424 10.1089/ars.2014.6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arivazhagan L. and Pillai S.S. (2014) Tangeretin, a citrus pentamethoxyflavone, exerts cytostatic effect via p53/p21 upregulation and suppresses metastasis in 7,12-dimethylbenz (α) anthracene induced rat mammary carcinoma. J. Nutr. Biochem. 1140–1153 10.1016/j.jnutbio.2014.06.007 [DOI] [PubMed] [Google Scholar]
- 16.Kim M.Y., Trudel L.J. and Wogan G.N. (2009) Apoptosis induced by capsaicin and resveratrol in colon carcinoma cells requires nitric oxide production and caspase activation. Anticancer Res. 3740, 3733–3740http://ar.iiarjournals.org/content/29/10/3733.long [PubMed] [Google Scholar]
- 17.Pratheeshkumar P., Sreekala C., Zhang Z., Budhraja A., Ding S., Son Yoet al. (2012) Cancer prevention with promising natural products: mechanisms of action and molecular targets. Anticancer Agents Med Chem. 12, 1159–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asati V., Mahapatra D.K. and Bharti S.K. (2016) PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: structural and pharmacological perspectives. Eur. J. Med. Chem. 109, 314–341 10.1016/j.ejmech.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 19.Hao W., Yuan X., Yu L., Gao C., Sun X., Wang D.. et al. (2015) Licochalcone A-induced human gastric cancer BGC-823 cells apoptosis by regulating ROS-mediated MAPKs and PI3K/AKT signaling pathways. Sci. Rep. 5, 1–8 10.1038/srep10336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan M., Maryam A., Qazi J.I. and Ma T. (2015) Targeting apoptosis and multiple signaling pathways with icariside II in cancer cells. Int. J. Biol. Sci. 11, 1100–1112 10.7150/ijbs.11595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunnumakkara A.B., Bordoloi D., Harsha C., Banik K., Gupta S.C. and Aggarwal B.B. (2017) Curcumin mediates anticancer effects by modulating multiple cell signaling pathways. Clin. Sci. 131, 1781–1799 10.1042/CS20160935 [DOI] [PubMed] [Google Scholar]
- 22.Szliszka E. and Krol W. (2015) Natural Polyphenols Target the Tumor Necrosis Factor-related Apoptosis-inducing Ligand (TRAIL) Signaling Pathway for Cancer Chemoprevention, Elsevier Inc. [Google Scholar]
- 23.Cossarizza A., Gibellini L., Pinti M., Nasi M., Montagna J.P., De Biasi S.. et al. (2011) Quercetin and cancer chemoprevention. Evid. Based Complement. Altern. Med. 2011, 10.1093/ecam/neq053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chirumbolo S. (2013) Quercetin in cancer prevention and therapy. Integr. Cancer Ther. 12, 97–102 10.1177/1534735412448215 [DOI] [PubMed] [Google Scholar]
- 25.Alrawaiq N.S. and Abdullah A. (2014) A review of flavonoid quercetin : metabolism, bioactivity and antioxidant properties. Int. J. PharmTech Res. 6, 933–941 [Google Scholar]
- 26.D’Andrea G. (2015) Quercetin: a flavonol with multifaceted therapeutic applications? Fitoterapia 106, 256–271 10.1016/j.fitote.2015.09.018 [DOI] [PubMed] [Google Scholar]
- 27.Shah P.M., Vishnu P.V and Gayathri R. (2016) Quercetin-a flavonoid: a systematic review. J. Pharm. Sci. Res. 8, 870–880 [Google Scholar]
- 28.Crowley L.C., Christensen M.E. and Waterhouse N.J. (2016) Measuring survival of adherent cells with the colony-forming assay. Cold Spring Harb. Protoc. 2016, 721–724 [DOI] [PubMed] [Google Scholar]
- 29.Franken N.A.P., Rodermond H.M., Stap J., Haveman J. and van Bree C. (2006) Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319 10.1038/nprot.2006.339 [DOI] [PubMed] [Google Scholar]
- 30.Thiagarajan P.S., Hsu D., Clement M.-V., Gyori B.M. and Venkatachalam G. (2014) OpenComet: an automated tool for comet assay image analysis. Redox Biol. 2, 457–465 10.1016/j.redox.2013.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuura N., Miyamae Y., Yamane K., Nagao Y., Hamada Y., Kawaguchi N.. et al. (2006) Aged garlic extract inhibits angiogenesis and proliferation of colorectal carcinoma cells. J. Nutr. 136, 842S–846S 10.1093/jn/136.3.842S [DOI] [PubMed] [Google Scholar]
- 32.Bruncko M., Oost T.K., Belli B.A., Ding H., Joseph M.K., Kunzer A.. et al. (2007) Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J. Med. Chem. 50, 641–662 10.1021/jm061152t [DOI] [PubMed] [Google Scholar]
- 33.Lee E.F., Czabotar P.E., Smith B.J., Deshayes K., Zobel K., Colman P.M.. et al. (2007) Crystal structure of ABT-737 complexed with Bcl-xL: implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ. 14, 1711–1713 10.1038/sj.cdd.4402178 [DOI] [PubMed] [Google Scholar]
- 34.Caenepeel S., Brown S.P., Belmontes B., Moody G., Keegan K.S., Chui D.. et al. (2018) a selective MCL1 inhibitor, is effective in hematologic cancer models alone and in combination with established therapies. Cancer Discov. 8, 1582–1597 [DOI] [PubMed] [Google Scholar]
- 35.Grosdidier A., Zoete V. and Michielin O. (2011) SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277 10.1093/nar/gkr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C.. et al. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 37.Kotschy A., Szlavik Z., Murray J., Davidson J., Maragno A.L., Le Toumelin-Braizat G.. et al. (2016) The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482 10.1038/nature19830 [DOI] [PubMed] [Google Scholar]
- 38.Bishayee K., Ghosh S., Mukherjee A., Sadhukhan R., Mondal J. and Khuda-Bukhsh A.R. (2013) Quercetin induces cytochrome-c release and ROS accumulation to promote apoptosis and arrest the cell cycle in G2/M, In cervical carcinoma: signal cascade and drug-DNA interaction. Cell Prolif. 46, 153–163 10.1111/cpr.12017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidya Priyadarsini R., Senthil Murugan R., Maitreyi S., Ramalingam K., Karunagaran D. and Nagini S. (2010) The flavonoid quercetin induces cell cycle arrest and mitochondria-mediated apoptosis in human cervical cancer (HeLa) cells through p53 induction and NF-κB inhibition. Eur. J. Pharmacol. 649, 84–91 10.1016/j.ejphar.2010.09.020 [DOI] [PubMed] [Google Scholar]
- 40.Su Q., Peng M., Zhang Y., Xu W., Darko K.O., Tao T.. et al. (2016) Quercetin induces bladder cancer cells apoptosis by activation of AMPK signaling pathway. Am. J. Cancer Res. 6, 498–508 [PMC free article] [PubMed] [Google Scholar]
- 41.Seo H.S., Ku J.M., Choi H.S., Choi Y.K., Woo J.K., Kim M.. et al. (2016) Quercetin induces caspase-dependent extrinsic apoptosis through inhibition of signal transducer and activator of transcription 3 signaling in HER2-overexpressing BT-474 breast cancer cells. Oncol. Rep. 36, 31–42 10.3892/or.2016.4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nair H.K., Rao K.V.K., Aalinkeel R., Mahajan S., Chawda R. and Schwartz S.A. (2004) Inhibition of prostate cancer cell colony formation by the flavonoid quercetin correlates with modulation of specific regulatory genes. Clin. Diagn. Lab. Immunol. 11, 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oršolić N., Karač I., Sirovina D., Kukolj M., Kunštić M., Gajski G.. et al. (2016) Chemotherapeutic potential of quercetin on human bladder cancer cells. J. Environ. Sci. Heal. 51, 776–781 10.1080/10934529.2016.1170465 [DOI] [PubMed] [Google Scholar]
- 44.Zhang Q., Zhao X.-H. and Wang Z.-J. (2008) Flavones and flavonols exert cytotoxic effects on a human oesophageal adenocarcinoma cell line (OE33) by causing G2/M arrest and inducing apoptosis. Food Chem. Toxicol. 46, 2042–2053 10.1016/j.fct.2008.01.049 [DOI] [PubMed] [Google Scholar]
- 45.Lee T.-J., Kim O.H., Kim Y.H., Lim J.H., Kim S., Park J.-W.. et al. (2006) Quercetin arrests G2/M phase and induces caspase-dependent cell death in U937 cells. Cancer Lett. 240, 234–242 10.1016/j.canlet.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 46.Choi J.-A., Kim J.-Y., Lee J.-Y., Kang C.-M., Kwon H.-J., Yoo Y.-D.. et al. (2001) Induction of cell cycle arrest and apoptosis in human breast cancer cells by quercetin. Int. J. Oncol. 19, 837–844 [DOI] [PubMed] [Google Scholar]
- 47.Srivastava S., Somasagara R.R. and Hegde M. (2016) Quercetin, a natural flavonoid interacts with DNA, arrests cell cycle and causes tumor regression by activating mitochondrial pathway of apoptosis. Sci. Rep. 6, 24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee Y.J., Jeong J.-H., An J.Y., Kwon Y.T. and Rhee J.G. (2009) Effects of low dose quercetin: cancer cell-specific inhibition of cell cycle progression. J Cell Biochem 106, 73–82 10.1002/jcb.21977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson M.K. and Loo G. (2000) Effects of epigallocatechin gallate and quercetin on oxidative damage to cellular DNA. Mutat. Res. 459, 211–218 10.1016/S0921-8777(99)00074-9 [DOI] [PubMed] [Google Scholar]
- 50.Shalini S., Dorstyn L., Dawar S. and Kumar S. (2015) Old, new and emerging functions of caspases. Cell Death Differ. 22, 526. 10.1038/cdd.2014.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ouyang L., Shi Z., Zhao S., Wang F., Zhou T., Liu B.. et al. (2012) Programmed cell death pathways in cancer : a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 45, 487–498 10.1111/j.1365-2184.2012.00845.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pop C. and Salvesen G.S. (2009) Human caspases: activation, specificity, and regulation. J. Biol. Chem. 284, 21777–21781 10.1074/jbc.R800084200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu H., Che X., Zheng Q., Wu A., Pan K., Shao A.. et al. (2014) Caspases: a molecular switch node in the crosstalk between autophagy and apoptosis. Int. J. Biol. Sci. 10, 1072–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hassan M., Watari H., Abualmaaty A., Ohba Y. and Sakuragi N. (2014) Apoptosis and molecular targeting therapy in cancer. Biomed Res. Int. 2014, 10.1155/2014/150845 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Kiraz Y., Adan A., Kartal Yandim M. and Baran Y. (2016) Major apoptotic mechanisms and genes involved in apoptosis. Tumour Biol. 37, 8471–8486 10.1007/s13277-016-5035-9 [DOI] [PubMed] [Google Scholar]
- 56.Akondi B.R., Challa S.R. and Akula A. (2011) Protective effects of rutin and naringin in testicular ischemia- reperfusion induced oxidative stress in rats. J. Reprod. Infertil. 12, 209–214 [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X., Zou L., Lu T., Bao S., Hurov K.E., Hittelman W.N.. et al. (2006) Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol. Cell. 23, 331–341 10.1016/j.molcel.2006.06.022 [DOI] [PubMed] [Google Scholar]
- 58.Henken F.E., Wilting S.M., Overmeer R.M., Van Rietschoten J.G.I., Nygren A.O.H., Errami A.. et al. (2007) Sequential gene promoter methylation during HPV-induced cervical carcinogenesis. Br. J. Cancer 97, 1457–1464 10.1038/sj.bjc.6604055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S.S., Chan K.Y.K., Cheung A.N.Y., Liao X.Y., Leung T.W. and Ngan H.Y.S. (2006) Expression of ΔNp73 and TAp73α independently associated with radiosensitivities and prognoses in cervical squamous cell carcinoma. Clin. Cancer Res. 12, 3922–3927 10.1158/1078-0432.CCR-05-2573 [DOI] [PubMed] [Google Scholar]
- 60.Roos W.P., Thomas A.D. and Kaina B. (2015) REVIEWS DNA damage and the balance between survival and death in cancer biology. Nat. Publ. Gr. 16, 20–33 [DOI] [PubMed] [Google Scholar]
- 61.Primikyri A., Sayyad N., Quilici G., Vrettos E.I., Lim K., Chi S.W.. et al. (2018) Probing the interaction of a quercetin bioconjugate with Bcl-2 in living human cancer cells with in-cell NMR spectroscopy. FEBS Lett. 592, 3367–3379 10.1002/1873-3468.13250 [DOI] [PubMed] [Google Scholar]
- 62.Granato M., Rizzello C., Montani M.S.G., Cuomo L., Vitillo M., Santarelli R.. et al. (2017) Quercetin induces apoptosis and autophagy in primary effusion lymphoma cells by inhibiting PI3K/AKT/mTOR and STAT3 signaling pathways. J. Nutr. Biochem. 41, 124–136 10.1016/j.jnutbio.2016.12.011 [DOI] [PubMed] [Google Scholar]
- 63.Jagadeesh S. and Banerjee P.P. (2006) Inositol hexaphosphate represses telomerase activity and translocates TERT from the nucleus in mouse and human prostate cancer cells via the deactivation of Akt and PKCα. Biochem. Biophys. Res. Commun. 349, 1361–1367 10.1016/j.bbrc.2006.09.002 [DOI] [PubMed] [Google Scholar]
- 64.Van Doorslaer K. and Burk R.D. (2012) Association between hTERT activation by HPV E6 proteins and oncogenic risk. Virology 433, 216–219 10.1016/j.virol.2012.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H.Y., Park S., Kim S., Lee D., Kim G., Kim Y.. et al. (2015) Use of hTERT and HPV E6/E7 mRNA RT-qPCR TaqMan assays in combination for diagnosing high-grade cervical lesions and malignant tumors. Am. J. Clin. Pathol. 143, 344–351 10.1309/AJCPF2XGZ2XIQYQX [DOI] [PubMed] [Google Scholar]
- 66.Christofori G. (2003) Changing neighbours, changing behaviour: cell adhesion molecule-mediated signalling during tumour progression. EMBO J. 22, 2318–2323 10.1093/emboj/cdg228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widschwendter A., Ivarsson L., Blassnig A., Müller H.M., Fiegl H., Wiedemair A.. et al. (2004) CDH1 AND CDH13 methylation in serum is an independent prognostic marker in cervical cancer patients. Int. J. Cancer. 109, 163–166 10.1002/ijc.11706 [DOI] [PubMed] [Google Scholar]
- 68.How S., Wong M., Fang C.M., Chuah L. and Leong C.O. (2018) Critical reviews in Oncology/Hematology E-cadherin : its dysregulation in carcinogenesis and clinical implications. Crit. Rev. Oncol. Hematol. 121, 11–22 [DOI] [PubMed] [Google Scholar]
- 69.Fan L.-C., Teng H.-W., Shiau C.-W., Tai W.-T., Hung M.-H., Yang S.-H.. et al. (2016) Regorafenib (Stivarga) pharmacologically targets epithelial-mesenchymal transition in colorectal cancer. Oncotarget 7, 64136–64147 10.18632/oncotarget.11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen D., Dai F., Chen Z., Wang S., Cheng X., Sheng Q.. et al. (2016) Dimethoxy curcumin induces apoptosis by suppressing survivin and inhibits invasion by enhancing E-cadherin in colon cancer cells. Med. Sci. Monit. 22, 3215–3222 10.12659/MSM.900802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sartakhti J.S., Manshaei M.H. and Sadeghi M. (2017) MMP-TIMP interactions in cancer invasion: An evolutionary game-theoretical framework. J. Theor. Biol. 412, 17–26 10.1016/j.jtbi.2016.09.019 [DOI] [PubMed] [Google Scholar]
- 72.De Fátima M., Cardoso S., Henrique C. and Castelletti M. (2017) Putative biomarkers for cervical cancer : SNVs, methylation and expression profiles. Mutat. Res. 773, 161–173 10.1016/j.mrrev.2017.06.002 [DOI] [PubMed] [Google Scholar]
- 73.Mayer I.A. and Arteaga C.L. (2016) The PI3K/AKT pathway as a target for cancer treatment. Annu. Rev. Med. 67, 15.1–15.18 10.1146/annurev-med-062913-051343 [DOI] [PubMed] [Google Scholar]
- 74.Song K.H., Woo S.R., Chung J.Y., Lee H.J., Oh S.J., Hong S.O.. et al. (2017) REP1 inhibits FOXO3-mediated apoptosis to promote cancer cell survival. Cell Death Dis. 8, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang B., Gui L.S., Zhao X.L., Zhu L.L. and Li Q.W. (2015) FOXO1 is a tumor suppressor in cervical cancer. Genet. Mol. Res. 14, 6605–6616 10.4238/2015.June.18.3 [DOI] [PubMed] [Google Scholar]
- 76.MacDonald B.T., Tamai K. and He X. (2009) Wnt/β-Catenin signaling: components, mechanisms, and diseases. Dev. Cell. 17, 9–26 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Y., Huang Y., Cao X., Xu J. and Zhang L. (2016) WNT2 promotes cervical carcinoma metastasis and induction of epithelial-mesenchymal transition. PLoS ONE 11, e0160414. 10.1371/journal.pone.0160414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao J., Wang J. and Liu M. (2015) Expression to suppress the G1/S phase transition of the cell cycle and the epithelial to mesenchymal transition in cervical cancer cells. Int. Union Biochem. Mol. Biol. 20, 380–394 [DOI] [PubMed] [Google Scholar]
- 79.Fan Q., Qiu M.T., Zhu Z., Zhou J.H., Chen L., Zhou Y.. et al. (2015) Twist induces epithelial-mesenchymal transition in cervical carcinogenesis by regulating the TGF-β/Smad3 signaling pathway. Oncol. Rep. 34, 1787–1794 10.3892/or.2015.4143 [DOI] [PubMed] [Google Scholar]
- 80.Liu Q., Li H. and Zhang Y. (2016) Twist and YB-1 gene expression in cervical cancer and cervical intraepithelial neoplasia tissue as well as its correlation with epithelial-mesenchymal transition. J. Hainan Med. Univ. 22, 20–23 [Google Scholar]
- 81.Chang C.C., Huang R.L., Wang H.C., Liao Y.P., Yu M.H. and Lai H.C. (2014) High methylation rate of LMX1A, NKX6-1, PAX1, PTPRR, SOX1, and ZNF582 genes in cervical adenocarcinoma. Int. J. Gynecol. Cancer 24, 201–209 10.1097/IGC.0000000000000054 [DOI] [PubMed] [Google Scholar]
- 82.Sun Y., Liu W.Z., Liu T., Feng X., Yang N. and Zhou H.F. (2015) Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. 35, 600–604 10.3109/10799893.2015.1030412 [DOI] [PubMed] [Google Scholar]
- 83.Yan R., Shuai H., Luo X., Wang X. and Guan B. (2017) The clinical and prognostic value of CXCL8 in cervical carcinoma patients : immunohistochemical analysis. Biosci. Rep. 37, BSR20171021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song Z., Lin Y., Ye X., Feng C., Lu Y., Yang G.. et al. (2016) Expression of IL-1α and IL-6 is associated with progression and prognosis of human cervical cancer. Med. Sci. Monit. 22, 4475–4481 10.12659/MSM.898569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valle-Mendiola A., Gutiérrez-Hoya A., Lagunas-Cruz M.D.C., Weiss-Steider B. and Soto-Cruz I. (2016) Pleiotropic effects of IL-2 on cancer: its role in cervical cancer. Mediators Inflamm. 2016, 10.1155/2016/2849523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jia L., Li F., Shao M., Zhang W., Zhang C., Zhao X.. et al. (2018) IL-8 is upregulated in cervical cancer tissues and is associated with the proliferation and migration of HeLa cervical cancer cells. Oncol. Lett. 15, 1350–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Puren A.J., Fantuzzi G., Gu Y., Su M.S. and Dinarello C.A. (1998) Interleukin-18 (IFNgamma-inducing factor) induces IL-8 and IL-1beta via TNFalpha production from non-CD14+ human blood mononuclear cells. J. Clin. Invest. 101, 711–721 10.1172/JCI1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Son Y.-O., Lee K.-Y., Kook S.-H., Lee J.-C., Kim J.-G., Jeon Y.-M.. et al. (2004) Selective effects of quercetin on the cell growth and antioxidant defense system in normal versus transformed mouse hepatic cell lines. Eur. J. Pharmacol. 502, 195–204 10.1016/j.ejphar.2004.09.012 [DOI] [PubMed] [Google Scholar]
- 89.Du G., Lin H., Wang M., Zhang S., Wu X., Lu L.. et al. (2010) Quercetin greatly improved therapeutic index of doxorubicin against 4T1 breast cancer by its opposing eV acts on HIF-1 in tumor and normal cells. Cancer Chemother. Pharmacol. 65, 277–287 [DOI] [PubMed] [Google Scholar]