Abstract
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer and the second leading cause of cancer deaths in the word. Hepatitis B virus (HBV) infection plays an important role in the development of HCC. However, the mechanisms by which HBV integration affects host cells remain poorly understood. HepG2.2.15 cell line is derived from HCC cell line HepG2 with stable transfection HBV expression. In this study, HepG2.2.15 cells showed decreased proliferation, G1 cell cycle arrest and increased apoptosis, when compared to HepG2 cells. HBV capture sequencing was conducted in both genome and transcriptome level, followed by RNA expression sequencing in HepG2.2.15. Here, CAMSAP2/CCDC12/DPP7/OR4F3 were found to be targets for HBV integration in both genome and transcriptome level, accompanied by alteration in their expression when compared to HepG2. Among these genes, DPP7 was the only one gene with HBV integration into its exon, meanwhile DPP7 expression level was also downregulated in HepG2.2.15 as compared to HepG2. Furthermore, DPP7 knockdown resulted in increased apoptosis through upregulation of the Bax/Bcl2 ratio in HepG2 cells. Our results suggest that HBV integration of DPP7 was involved in cell apoptosis.
Keywords: HepG2.2.15, viral integration, Hepatitis B virus, proliferation, cell cycle, apoptosis
Introduction
The most prevalent primary liver cancer is known to be Hepatocellular carcinoma (HCC). It is considered to be the second highest cause of deaths associated with cancer globally 1, 2. Chronic infections with the hepatitis B virus (HBV) or hepatitis C virus (HCV) are the primary risk factors for HCC 3. In China, HBV plays a more prominent role because it may lead to liver fibrosis and cirrhosis, which eventually results in HCC 3-6. Around 50% of HCC patients are associated with HBV infections 7. Carriers of the hepatitis B surface antigen (HBsAg) are at 25-37-fold higher risk of developing HCC as compared to those not expressing the antigen 8, 9.
HBV contains eight known genotypes (A to H) 10. Its genome contains four genes, C, P, S and X, that encode the core protein (HBcAg), DNA polymerase, surface antigen (HBsAg) and protein X (HBx), respectively. Numerous studies have been done to establish the mechanisms of HBV-associated hepatocarcinogenesis 11-14. HBV genome integration, host gene deregulation and many viral factors have already been implied to contribute to HBV-associated HCC. HBx, for example, contributes to intracellular signal transduction, cell cycle and apoptosis regulation, and has been suggested to be an effector of hepatocarcinogenesis 11. Almost 90% of HBx transgenic mice develop HCC 15. HBV Integration leads to the dysfunction of host genes and affects cell proliferation, cell cycle progression, apoptosis, and even chromosomal stability 12, 16-20. Recurrent integrations in TERT and MLL4, for example, are frequently observed in HCC 21-29. HBx-LINE integration, which functions as a long non-coding RNA, has been shown to promote tumorigenesis of HCC 12. With the help of next generation sequencing, it has been found that genome-wide HBV integration is commonly reported in HCC 25, 30.
HepG2.2.15 cells are derived from HepG2 with HBV expression stablely31. Little work has been done in the context of HepG2.2.15 cells with regards to the effects of HBV on cellular function, and HBV integration detection of HepG2.2.15 cells has not been carried out. In this study, we conducted HBV capture sequencing in HepG2.2.15 cells at both the whole genome and transcriptome levels, followed by RNA sequencing. Our results reveal that several genes are targets for HBV integration, leading to altered expression patterns of these genes. Integration in the dipeptidyl peptidase 7 (DPP7) gene causes decreased expression of DPP7 in HepG2.2.15 cells. Furthermore, DPP7 knockdown results in increased apoptosis with upregulation of the Bax/Bcl2 ratio. Our results suggest that HBV integration of DPP7 was involved in cell apoptosis.
Materials and Methods
Cell culture and transfection
HepG2 was cultured with DMEM (HyClone, SH30022.01B) with 10 % FBS (Biological Industries) and incubated at 37℃ with 5% CO2. HepG2.2.15 was cultured with DMEM (HyClone) with 10 % FBS and 200ug/ml G418 (SIGMA). SiRNA transfection was done with Lipofection 3000 reagent according to the manufacturer's protocol. DPP7 siRNA sequence 5'-3': siRNA#1 (CCGAGCACUGCUACGACAUTT), siRNA#2 (GCAACAAUGUGACCGAUAUTT), siRNA#3 (CCUGAGUGCCUCAGUCAUCTT).
Antibody reagents
Protein expression was analyzed by western blot. Rb, pRb780, pRb795, pRb807/811, cyclinD, cyclinE, CDK2, p21, p27, p53, Bcl2, Bax antibody were obtained from Cell Signaling Technology (CST). DPP7 antibody was bought from Santa Cruz Biotechnology. GAPDH antibody was bought from Beyotime.
DNA and RNA isolation
With the use of Genomic DNA Mini Kit (Invitrogen, Life Technologies) and Trizol reagent (Takara), extraction of Genomic DNA and Total RNA from cells were carried out, respectively. Then, quantification of DNA and RNA was carried out with the use of NanoDrop 2000.
HBV capture experiment at genomic DNA level
Firstly, HBV capture probe were designed based on full-length HBV genome of eight types (A to H). The sequencing library construction was conducted as per Illumina protocol. Genomic DNA was sheared to 150-200 bp DNA fragments by Covaris E-210. They were purified, end blunted, “A” tailed, and finally ligated to adaptors. 12 cycle PCR was carried out to amplify the ligation products to build the genomic DNA library. Library quantification was analyzed by Qubit dsDNA HS Assay Kit (Invitrogen). Next, libraries hybridization with HBV probe was carried out at 65℃ for 24 hours. Uncaptured fragments were then removed by washing., The eluted fragments underwent amplification by 16 cycles of PCR to generate sequencing library followed by paired-end index sequencing in the illumina HiSeq 2000 sequencer.
HBV capture experiment at RNA level
Firstly, HBV capture probe were designed based on full-length HBV genome of eight types (A to H). As per Illumina paired-end sequencing protocols, 1 μg of total RNA was used to construct library. With ProtoScript® II Reverse Transcriptase (NEB), First-strand cDNA was synthesized. This was followed by second-strand cDNA synthesis with the help of Second Strand Synthesis Enzyme Mix (NEB) and purified using DNA Clean & Concentrator kit (Zymo Research). The double-strand cDNA products were sheared to 150-200 bp DNA fragments by Covaris E-210. These sheared fragments underwent purification, end blunting, “A” tailing, and finally ligation to adaptors. 12 cycles of PCR amplification were conducted for library construction. Libraries quantification was analyzed by Qubit dsDNA HS Assay Kit (Invitrogen). Then, library hybridization with HBV probe were conducted at 65℃ for 24 hours. Uncaptured fragments were removed by washing. Sequencing libraries were obtained by amplification of eluted fragments with 16 cycles of PCR, and were then subjected to paired-end index sequencing in the illumina HiSeq 2000 sequencer.
Viral-Human Fusion identification by VirusSeq
VirusSeq 32 was used to detect viral-human fusion. Briefly, sequencing raw data was firstly filtered, so that clean reads could be obtained for subsequent analysis. The clean PE reads were then mapped onto both the human genome (NCBI build 37, HG19) and HBV genome (HE815465.1) by Burrows-Wheeler Aligner (BWA). hg19Virus is a novel hybrid reference genome made by combining human genome (H19) and virus genome (Virus). All PE reads are mapped to hg19Virus. Only the PE reads that mapped to both human chromosome and Virus, is considered as a discordant read pair. After annotation with human and viral genes, fusion candidates were reported for further analysis as previously reported 32. The coverage, depth and coverage rate onto HBV genome were measured according to reported reads.
PCR and Sanger sequencing validation
For verification of the HBV integration breakpoints from VirusSeq analysis, Sanger sequencing was conducted. Using the paired-end assembled fragment as a basis, the PCR primers (Table S1) were designed, that one of the primers was located in HBV genome and the other in human genome. PCR was then carried out followed by sequencing.
Detection of HBV gene copy number
HBV gene copy number was detected using Diagnostic Kit for Quantification of Hepatitis B Virus DNA based on manufacturer's protocol. Finally, PCR reactions of the prepared samples were performed with Stepone Real-Time PCR System.
Cell proliferation analysis
We used Cell Counting Kit-8 (CCK-8, Biotool) for analysis of cell proliferation based on manufacturer's instruction. Cells were seeded in 96-well plates at a density of 3.5×103 cells per well and cultured in DMEM with 10% serum. 24h/48h/72h later, the old medium was removed, and then 1/10 volume of CCK-8 was then added into 100ul fresh medium. After incubation at 37℃ for 4h, absorbance was measured at 450 nm.
Cell cycle analysis
We used Cell Cycle and Apoptosis Analysis Kit (Beyotime) to analyze cell cycle based on manufacturer's protocol. Cells (1×106) with different treatments were harvested followed by washing with cold 1 × PBS. Cells were then fixed in 1 ml 70 % cold ethanol at 4 ℃ for one night. Cells were washed three times with cold 1 × PBS, and then resuspended in 500 ul staining buffer (200 mg/ml RNase A and 50 μg/ml PI) and stained in the dark at 37 ℃ for 30min. Finally, Flow Cytometer (ACEA NovoCyteTM) was used for cell cycle detection.
Cell apoptosis analysis
We used Annexin V-FITC/PI apoptosis kit (Multi Science) to analyze cell apoptosis based on manufacturer's protocol. Cells (5 × 105) with different treatments were harvested followed by washing with cold 1 × PBS. The cell were resuspended in 500ul 1 × Binding Buffer, and 5 μl Annexin V-FITC and 10 μl PI were then added to the cell mixture followed by incubation in the dark at 37 ℃ for 5 minutes. Finally, apoptosis was detected by Flow Cytometer (ACEA NovoCyteTM).
RNA extraction, reverse transcription and qPCR
We used Trizol (Takara) to extract RNA from cells. cDNA synthesis was conducted by PrimeScriptTM RT Master Mix kit (Takara). PCR reaction was then done with SYBR Premix Ex TaqTM, (Tli RNaseH Plus) kit (Takara). The primers used in this paper were shown in Table S2. GAPDH was used as a control gene.
Statistics
Data were presented as mean ± SEM based on three independent experiments. Unpaired student's t test analysis was performed in quantification of RT-PCR results. p values that were less than 0.05 were considered to be statistically significant.
Results
Detection of HBV in HepG2.2.15 cells
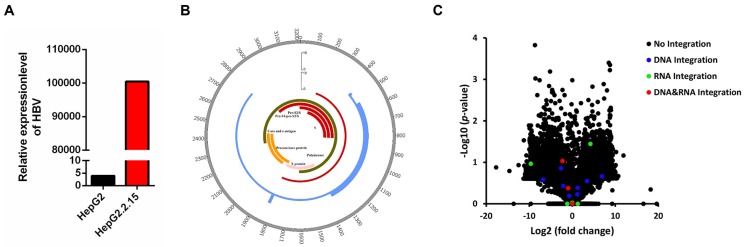
HBV DNA was detected in HepG2.2.15 cells by a fluorescent labeled PCR probe. HepG2 cell was served as a negative control. As shown in Figure 1A, the HBV DNA copy number in HepG2.2.15 cells showed a 25,763-fold increase in comparison with HepG2, confirming that HepG2.2.15 cells are positive for HBV DNA.
Figure 1.
HBV integration breakpoints distribution in HBV genome and gene expression change in HepG2.2.15. (A) Detection of HBV gene copy number by PCR. (B) Distribution of integration breakpoints in the HBV genome in HepG2.2.15 (genome integration breakpoints of HepG2.2.15: blue, transcriptome integration breakpoints of HepG2.2.15: red) are shown. Histograms were constructed for 100bp intervals. HBV genes with different functions are shown. (C) Volcano plot of differently expressed genes between HepG2.2.15 and HepG2. The logarithms of the fold changes of genes (x-axis) are plotted against the negative logarithm of their p-value(y-axis). Black dots represent genes without HBV integration (No integration), blue dots represent genes with HBV integration in genome (DNA integration), green dots represent genes with HBV integration in transcriptome (RNA integration), red dots represent genes with HBV integration in both genome and transcriptome (DNA&RNA integration).
Identification of genome HBV integration sites
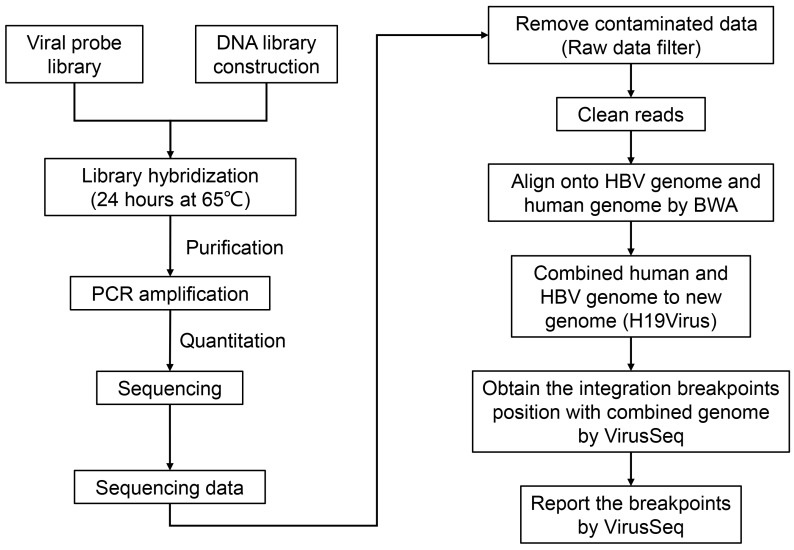
To explore the integration of the HBV genome in HCC, we conducted HBV capture sequencing in HepG2.2.15 cells using the Illumina HiSeq 2000 sequencing system. Identification of the integrated HBV genome was subsequently performed using VirusSeq 32 (Figure 2).
Figure 2.
The pipeline of workflow. Experimental workflow and bioinformatic analysis is performed in this study. BWA: Burrows-Wheeler Aligner.
We obtained 2,374,482 raw reads that had with a 141 bp average read length from HepG2.2.15 genomic DNA samples, of which 192,901 reads were matched to the HBV genome (HBV-D-3) (Table 1). An average depth of 6417.52 and 100% coverage was achieved with 20-fold read mapped to the HBV genome (Table 1). 14 HBV integration breakpoints were identified (Table 1), and the genome HBV integration breakpoints in HepG2.2.15 were distributed in HBV genome (Figure 1B).
Table 1.
HBV capture sequencing data
| Sample | Reads number | Raw bases (Mb) | Clean bases (Mb) | Average read length | No. of the reads mapped to HBV | Target 20X rate (%) | Target mean depth | Coverage rate (%) | No. of integration breakpoints |
|---|---|---|---|---|---|---|---|---|---|
| Genomic DNA | 2374482 | 358.55 | 316.91 | 141 | 192901 | 100 | 8157.27 | 100 | 14 |
| Total RNA | 94192016 | 14222.99 | 12390.66 | 138 | 561510 | 100 | 22604.45 | 100 | 9 |
As shown in Table 2, 14 HBV integration breakpoints are mainly located within the region where the X protein, S protein and core protein are located 33. The HBx protein is the most frequently integrated protein. HBV is preferentially integrated into chromosome 1 (4/14), chromosome 2 (3/14), chromosome 3 (2/14), chromosome 7 (1/14), chromosome 9 (1/14), chromosome 20 (1/14), chromosome 21 (1/14) and chromosome X (1/14) (Table 2). The majority of HBV integrations are intronic and intergenic, while the DPP7 integration sites are exonic (Table 2).
Table 2.
HBV genomic integration breakpoints and gene expression fold change
| Human location | Nearest genes | Location type | HBV location | HBV transcript | Supporting reads number | Nearest gene fold change (log2) |
|---|---|---|---|---|---|---|
| chr1:155104681 | EFNA1 | intronic | 460 | S protein | 14 | -0.693 |
| chr1:200809469 | CAMSAP2 | intronic | 1746 | X protein | 8 | -0.923 |
| chr3:47015291 | CCDC12 | intronic | 1820 | X protein | 71 | 0.023 |
| chr9:140008668 | DPP7 | extronic | 1716 | X protein | 114 | -2.261 |
| chr2:27241835 | MAPRE3 | intronic | 1829 | X protein | 11 | -2.092 |
| chr1:200809321 | CAMSAP2 | intronic | 1746 | precore/core protein | 10 | -0.923 |
| chr1:569999 | OR4F3 | intergenic | 1801 | precore/core protein | 13 | - |
| chr7:25573057 | NPVF | intergenic | 1594 | X protein | 103 | 0 |
| chr3:42696821 | ZBTB47 | intronic | 1829 | X protein | 112 | -2.59583 |
| chrX:9148656 | FAM9B | intergenic | 2082 | precore/core protein | 55 | -6.74895 |
| chr20:62935214 | LINC00266-1 | intergenic | 2639 | polymerase protein | 38 | 3.4362 |
| chr2:27775260 | GCKR | intergenic | 2775 | polymerase protein | 22 | 6.90382 |
| chr21:10007085 | TEKT4P2 | intergenic | 607 | S protein | 45 | 1.28415 |
| chr2:27915039 | SLC4A1AP | intronic | 2567 | polymerase protein | 22 | 1.17871 |
Chr: chromosome; -: no signal test.
Identification of transcriptome HBV integration sites
To further explore HBV integration at the level of the transcriptome, total RNA of HepG2.2.15 cells was obtained, reversed transcribed to cDNA and subjected to HBV capture sequencing (Figure 2).
We obtained 94,192,016 raw reads with an average read length of 138 base pairs. A total of 561,510 reads were matched to the HBV genome (HBV-D-3) (Table 1). We obtained 100% coverage of the targeted region with 20-fold read mapped to the HBV genome (Table 1). Nine HBV integration breakpoints were identified (Table 1), and the transcriptome HBV integration breakpoints in HepG2.2.15 were distributed in HBV genome (Figure 1B).
As shown in Table 3, all nine HBV integration breakpoints are distributed in the region where the X protein and S protein are located within the HBV genome 33 (Table 3). Three integration breakpoints are within the S protein and six are within the X protein. The HBV integration sites are located within chr1, chr3, chr5, chr7, chr9, chr11, chr12, chr17 (Table 3). Most of the integrations are intronic and intergenic, while the HBV integration of DPP7 is exonic (Table 3).
Table 3.
HBV RNA integration breakpoints and gene expression fold change
| Human location | Nearest gene | Location type | HBV location | HBV transcript | Supporting reads number | Nearest gene log2(fold change) |
|---|---|---|---|---|---|---|
| chr11:85195011 | DLG2 | intronic | 407834 | S protein | 9 | 1.283 |
| chr5:71146794 | MAP1B | intergenic | 408128 | S protein | 8 | 4.212 |
| chr17:33478111 | UNC45B | intronic | 407658 | S protein | 6 | -1.162 |
| chr1:200809476 | CAMSAP2 | intronic | 409112 | X protein | 12 | -0.923 |
| chr3:47015261 | CCDC12 | intronic | 409144 | X protein | 41 | 0.023 |
| chr9:140008690 | DPP7 | exonic | 409079 | X protein | 359 | -2.261 |
| chr7:25572926 | MIR148A | intergenic | 408983 | X protein | 16 | 0.003 |
| chr1:570051 | OR4F3 | intergenic | 409270 | X protein | 8 | - |
| chr12:20704358 | PDE3A | intronic | 408967 | X protein | 7 | -9.627 |
Chr: chromosome; -: no signal test.
Expression of target genes with HBV integration
To explore whether HBV integration affects the expression of host genes, we conducted RNA-Seq in HepG2 and HepG2.2.15 cells. Statistical analysis of RNA-Seq data is shown in Table 4. Expression change of genes were shown in Figure 1C, including the genes without or with HBV integration in genome level, transcriptome level and both genome and transcriptome level. Combining the RNA-Seq and HBV integration results, four genes (CAMSAP2/CCDC12/DPP7/OR4F3) are identified with HBV integration in both the genome and transcriptome of HepG2.2.15 cells (Table 5). Expression of DPP7, which shows integration of the X protein transcript, decreases 4.8-fold as compared to expression in HepG2 cells. Changes in expression levels of CAMSAP2/CCDC12/OR4F3, which show intronic or intergenic HBV integration, are lower (Table 5). To confirm HBV integration into the genes, PCR amplification followed by Sanger sequencing was carried out. Integration of HBV into the four genes (CAMSAP2/CCDC12/DPP7/OR4F3) was confirmed by Sanger sequencing (Figure S1), validating the results from HBV capture sequencing.
Table 4.
Statistics information of RNA-seq data
| Sample | Reads number | Raw bases (Mb) | Clean bases (Mb) | Average read length | Average insert size | Mapped reads | Coverage rate(%) | Target mean depth |
|---|---|---|---|---|---|---|---|---|
| HepG2 | 193531404 | 29223.24 | 25439.31 | 130 | 948.7 | 96922102 | 62.76 | 59.55 |
| HepG2.2.15 | 145773306 | 22011.77 | 19359.6 | 126 | 989.9 | 98707630 | 61.12 | 65.79 |
Table 5.
HBV integration genes in both genome and transcriptome
| Gene | Chr | HBV transcript | Integration type | fold change (log2) |
|---|---|---|---|---|
| CAMSAP2 | chr1 | X protein/core protein | intronic | -0.923 |
| CCDC12 | chr3 | X protein | intronic | 0.023 |
| DPP7 | chr9 | X protein | exonic | -2.261 |
| OR4F3 | chr1 | X protein/core protein | intergenic | - |
Chr: chromosome; -: no signal test.
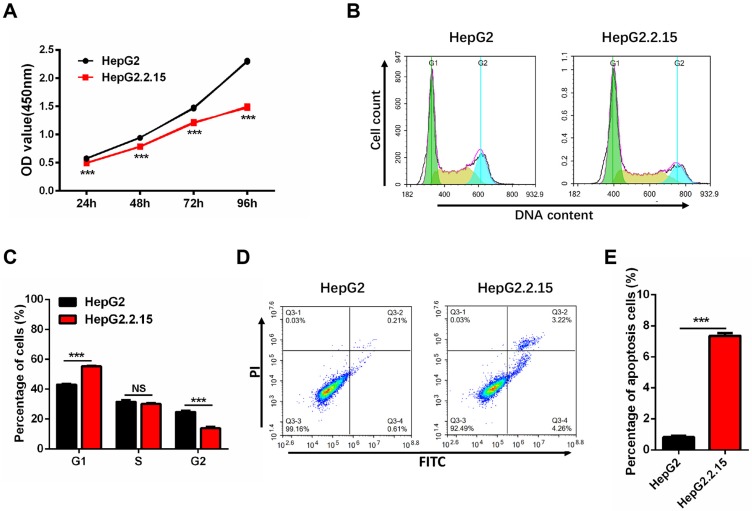
HBV integration decreases cell proliferation, and induces G1 arrest and apoptosis
Since HepG2.2.15 cells originate from HepG2 with HBV expression stablely, we first examined whether the cell phenotype is stable. The cell proliferation assay shows a significant reduction of HepG2.2.15 cells growth rate as compared to HepG2 cells (Figure 3A). Cell cycle analysis was conducted to determine whether cell cycle distribution was changed. As compared to HepG2 cells, there was an obvious increase in the proportion of G1 phase and a decrease in the S and G2 phases in HepG2.2.15 cells (Figure 3B, C). The decreased cell proliferation and G1 cell cycle arrest are consistent with previous reports 34. Cell apoptosis detection was also carried out by flow cytometry, and the rate of apoptosis in HepG2.2.15 cells is increased in comparison with HepG2 (Figure 3D, E).
Figure 3.
HepG2.2.15 cells show decreased proliferation, G1 cell cycle arrest and increased apoptosis. (A) Proliferation of HepG2 and HepG2.2.15 cells. OD450 values were measured at 24 h, 48 h, 72 h and 96h. Data shown represents the mean ± SEM values from three independent experiments (***p < 0.001). (B) Cell cycle analysis of HepG2 and HepG2.2.15 cells. (C) Quantitative cell cycle distribution of HepG2 and HepG2.2.15 cells. HepG2.2.15 cells show G1 cell cycle arrest when compared to HepG2 cells. Data shown represents the mean ± SEM values from three independent experiments (***p < 0.001, NS: none significant). (D) Analysis of cell apoptosis by flow cytometry. Increased apoptosis is observed in HepG2.2.15 cells. (E) Quantitative results of cells undergoing apoptosis in HepG2 and HepG2.2.15 populations. Data shown represents the mean ± SEM values from three independent experiments (***p < 0.001).
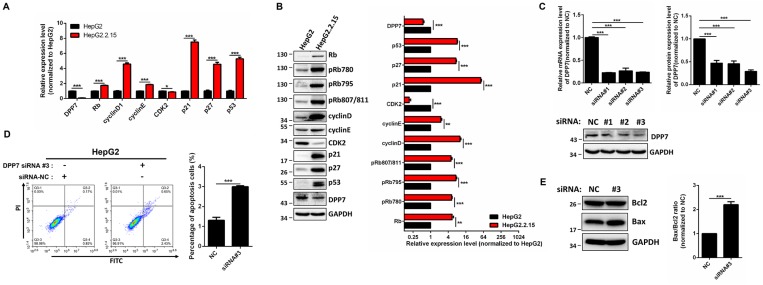
To identify the mechanism of G1 cell cycle arrest, expression of cell cycle-regulated genes were detected. The results obtained from RT-PCR showed that mRNA expression of Rb, cyclin D1, cyclin E1, p21, p27, and p53 is increased in HepG2.2.15 cells, while expression of CDK2 is decreased (Figure 4A) in comparison with HepG2 cells. In addition, the protein expression of Rb, cyclin D1, cyclin E1, p21, p27, p53 is upregulated, while there was a downregulation of CDK2 expression in HepG2.2.15 cells (Figure 4B).
Figure 4.
DPP7 knockdown induces cell apoptosis in HepG2 cells (A) RT-PCR analysis of cell cycle regulated genes in HepG2 and HepG2.2.15 cells. GAPDH was used as a reference gene. Data shown represents the mean ± SEM values from three independent experiments (*p < 0.05, ***p < 0.001). (B) Western blot analysis of cell cycle regulated genes. Quantification results were shown in the right. GAPDH was used as a reference gene. Data shown represents the mean ± SEM values from three independent experiments (**p<0.01, ***p < 0.001). (C) Verification of DPP7 siRNA knockdown efficiency by RT-PCR and western blot. Quantification results were shown in the upper. Data shown represents the mean ± SEM values from three independent experiments (***p < 0.001). (D) Knockdown of DPP7 induces increased cell apoptosis in HepG2 cells. Data shown represents the mean ± SEM values from three independent experiments (***p < 0.001). (E) Knockdown of DPP7 in HepG2 cells leads to increased Bax/Bcl2 ratio as measured by western blot. Quantification results were shown in the right. GAPDH served as a loading control. Data shown represents the mean ± SEM values from three independent experiments (***p < 0.001).
DPP7 knockdown results in cell apoptosis
In order to determine whether the phenotype of HepG2.2.15 cells is linked with HBV integration, we studied DPP7, since it is the only gene with HBV integration at both the genome and transcriptome levels, and decreased expression levels. The mRNA expression of DPP7 is 10-fold decrease in HepG2.2.15 in comparison with HepG2 (Figure 4A).
DPP7, also referred to as quiescent cell proline dipeptidase (QPP, DPP2, DPPII), is a post-proline cleaving aminopeptidase 35, 36. It has been previously reported that DPP7 inhibition in quiescent lymphocytes results in cell apoptosis 37. We reasoned that the HBV integration-mediated decreases in DPP7 expression may be associated with cell apoptosis in HepG2.2.15 cells. Therefore, we used DPP7 siRNAs for knockdown of DPP7 (Figure 4C). In HepG2 cells, DPP7 knockdown induced more cell apoptosis than the siRNA control (Figure 4D). Apoptosis-related proteins such as Bcl2 and Bax play an vital role in cell apoptosis. We then analyzed the expression of apoptosis-related proteins to explore the cellular mechanism of the higher apoptosis of HepG2.2.15 cells. Western blot results show that DPP7 knockdown results in an upregulation of Bax, without affecting Bcl2 (Figure 4E). This leads to an increase in Bax/Bcl2 ratio, which may contribute to the increased levels of apoptosis.
Discussion
Currently, HCC is the second leading cause of cancer-associated deaths globally. In China, HBV has a vital contribution to HCC development. The HepG2.2.15 originates from HepG2 by stable transfection with HBV genome 31. In comparison with HepG2, HepG2.2.15 shows decreased proliferation, G1 cell cycle arrest and increased rate of apoptosis. In our study, HBV capture sequencing was conducted at both the genome and transcriptome levels to discover HBV integration sites in the HepG2.2.15 genome. RNA-Seq was subsequently performed to analyze gene expression. Based on the HBV capture sequencing and RNA-Seq results, a number of genes show HBV integration accompanied by altered expression. In particular, integration of HBV into DPP7 is observed both at the genome and transcriptome levels, and expression of this gene is reduced in HepG2.2.15 cells when compared to HepG2 cells. Furthermore, DPP7 knockdown leads to apoptosis with upregulation of the Bax/Bcl2 ratio in HepG2 cells, which suggest that HBV integration-induced decreases in DPP7 expression can increase apoptosis in HepG2.2.15 cells, indicating that HBV can regulate cell function through integrating into host genes.
Previous studies have revealed a large number of genes with HBV integration in human HCC tissue 30. Although a large amount of data has been obtained in studies of clinical samples, few studies have performed functional analysis. The mechanism by which HBV integration induces HCC remains to be elucidated. One of the challenges in identifying these mechanisms is that HBV was randomly integrated into the genome, which makes choosing a cell line model with specific HBV integration difficult. HBx-LINE1, an HBV-human chimeria contribute to HCC through acting as a LncRNA 12. In the aforementioned study, six HCC cell lines were obtained from patients who were infected with HBV, which paves the way for further functional studies 12. Here, we chose HepG2.2.15 cells as our HBV integration cell model, to explore the relationship between HBV integration and cell function and phenotype.
In a previous study, whole-genome gene expression were profiled in both HepG2 and HepG2.2.15 cells. It showed that 2978 genes are up- or down-regulated, including genes related to cell cycle, transport, signal transduction, cell adhesion and cellular metabolism 38. Two-dimensional gel electrophoresis-mass spectrometry indicated that HBV infection can affect protein expression 39, 40. HepG2.2.15 cells also show reduction in proliferation and G1 cell cycle arrest with alterations in cell cycle-regulated genes 34, 41, 42. However, the association between HBV integration and cell functional and phenotypic changes has not been elucidated. In our study, HepG2.2.15 cells showed reduction in proliferation, G1 cell cycle arrest and increased apoptosis when compared to HepG2 cells. HBV capture sequencing and RNA-Seq revealed that HBV integration occurs in DPP7 accompanied by decreased expression levels of the gene. DPP7, a post-proline cleaving aminopeptidase, has been shown to play role in apoptosis of lymphocytes 37. Consistent with this finding, we show that DPP7 knockdown leads to increased apoptosis with upregulation of the Bax/Bcl2 ratio in HepG2 cells. Together, our data suggests that HBV integration affects the expression of host genes such as DPP7, leading to increased apoptosis in HepG2.2.15 cells.
Although our results reveal alterations of several important genes, it has some limitations. For example, many genes have been identified with HBV integration in HCC tissue, whereas fewer genes were detected here, which may be attributed to the cell model we choose. Since HepG2.2.15 cells originate from HepG2 with HBV expression stablely, they do not exactly mimic the HBV infection process under physiological conditions. In this regard, HepG2.2.15 may not be the best cell line model for HBV integration analysis. Nevertheless, this cell line provides a tool to study HBV integrations and the effects on expression of host genes. Our results suggest that HBV infection decreases host cell activities, such as decreased cell proliferation, and increased cell apoptosis, which are contrary to the predominant hypothesis that HBV infection contributes to hepatocarcinogenesis 4-6. HBV contains eight genotypes (A-H) with different geographic distribution. Increasing evidence has suggested that not all HBV genotypes are linked with HCC 43, 44. The HBV genotype of HepG2.2.15 cells is genotype D, 31 and the clinical impact of genotype D is still controversial. For example, some clinical and epidemiological data suggests that genotype D does not affect the severity and chronicity of the disease 45, while other studies report that genotype D is related to chronic and occult HBV infections 46. In these previous studies, a large amount of data was obtained from clinical specimens and, therefore, more functional studies are needed to clarify these issues. It is possible that the decreased cell proliferation and cell activity induced by HBV integration in HepG2.2.15 is specific for HBV genotype D, as previously reported 34.
Supplementary Material
Supplementary figure and tables.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (no.81772575; 81502463), the Science Technology Department of Zhejiang Province (no. 2016C33116), the key project of Health Bureau of Zhejiang Province (no. 2018274734), the CSCO Merck Serono Oncology Research Fund, SCORE (no. Y-MX2015-038), the Key Research Project of Science Technology Department of Zhejiang Province (no. 2015C03030) and the National High Technology Research and Development Program of China (863 Program, no. 2012AA02A205).
References
- 1.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–55. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16:1. doi: 10.4103/jcar.JCar_9_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iloeje UH, Yang HI, Su J, Jen CL, You SL, Chen CJ. et al. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology. 2006;130:678–86. doi: 10.1053/j.gastro.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 5.Montalto G, Cervello M, Giannitrapani L, Dantona F, Terranova A, Castagnetta LA. Epidemiology, risk factors, and natural history of hepatocellular carcinoma. Ann N Y Acad Sci. 2002;963:13–20. doi: 10.1111/j.1749-6632.2002.tb04090.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen CJ, Yang HI, Su J, Jen CL, You SL, Lu SN. et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295:65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 7.Sherman M. Hepatocellular carcinoma: epidemiology, surveillance, and diagnosis. Semin Liver Dis. 2010;30:3–16. doi: 10.1055/s-0030-1247128. [DOI] [PubMed] [Google Scholar]
- 8.Hassan MM, Hwang LY, Hatten CJ, Swaim M, Li D, Abbruzzese JL. et al. Risk factors for hepatocellular carcinoma: synergism of alcohol with viral hepatitis and diabetes mellitus. Hepatology. 2002;36:1206–13. doi: 10.1053/jhep.2002.36780. [DOI] [PubMed] [Google Scholar]
- 9.Sun CA, Wu DM, Lin CC, Lu SN, You SL, Wang LY. et al. Incidence and cofactors of hepatitis C virus-related hepatocellular carcinoma: a prospective study of 12,008 men in Taiwan. Am J Epidemiol. 2003;157:674–82. doi: 10.1093/aje/kwg041. [DOI] [PubMed] [Google Scholar]
- 10.Kramvis A, Kew M, Francois G. Hepatitis B virus genotypes. Vaccine. 2005;23:2409–23. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 11.Koike K, Moriya K, Iino S, Yotsuyanagi H, Endo Y, Miyamura T. et al. High-level expression of hepatitis B virus HBx gene and hepatocarcinogenesis in transgenic mice. Hepatology. 1994;19:810–9. [PubMed] [Google Scholar]
- 12.Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM. et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335–49. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 13.Neuveut C, Wei Y, Buendia MA. Mechanisms of HBV-related hepatocarcinogenesis. J Hepatol. 2010;52:594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 14.Chaturvedi VK, Singh A, Dubey SK, Hetta HF, John J, Singh MP. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb Pathog. 2019;128:184–94. doi: 10.1016/j.micpath.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–20. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 16.Chami M, Gozuacik D, Saigo K, Capiod T, Falson P, Lecoeur H. et al. Hepatitis B virus-related insertional mutagenesis implicates SERCA1 gene in the control of apoptosis. Oncogene. 2000;19:2877–86. doi: 10.1038/sj.onc.1203605. [DOI] [PubMed] [Google Scholar]
- 17.Lee YH, Oh BK, Yoo JE, Yoon SM, Choi J, Kim KS. et al. Chromosomal instability, telomere shortening, and inactivation of p21(WAF1/CIP1) in dysplastic nodules of hepatitis B virus-associated multistep hepatocarcinogenesis. Mod Pathol. 2009;22:1121–31. doi: 10.1038/modpathol.2009.76. [DOI] [PubMed] [Google Scholar]
- 18.Peng Z, Zhang Y, Gu W, Wang Z, Li D, Zhang F. et al. Integration of the hepatitis B virus X fragment in hepatocellular carcinoma and its effects on the expression of multiple molecules: a key to the cell cycle and apoptosis. Int J Oncol. 2005;26:467–73. [PubMed] [Google Scholar]
- 19.Liang HW, Wang N, Wang Y, Wang F, Fu Z, Yan X. et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol. 2016;64:278–91. doi: 10.1016/j.jhep.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Song X, Tan S, Wu Z, Xu L, Wang Z, Xu Y. et al. HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int J Cancer. 2018;143:3120–30. doi: 10.1002/ijc.31595. [DOI] [PubMed] [Google Scholar]
- 21.Saigo K, Yoshida K, Ikeda R, Sakamoto Y, Murakami Y, Urashima T. et al. Integration of hepatitis B virus DNA into the myeloid/lymphoid or mixed-lineage leukemia ( MLL4 ) gene and rearrangements of MLL4 in human hepatocellular carcinoma †. Human Mutation. 2008;29:703–8. doi: 10.1002/humu.20701. [DOI] [PubMed] [Google Scholar]
- 22.Dong H, Zhang L, Qian Z, Zhu X, Zhu G, Chen Y. et al. Identification of HBV-MLL4 Integration and Its Molecular Basis in Chinese Hepatocellular Carcinoma. PLoS One. 2015;10:e0123175. doi: 10.1371/journal.pone.0123175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N. et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–21. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 24.Toh ST, Jin Y, Liu L, Wang J, Babrzadeh F, Gharizadeh B. et al. Deep sequencing of the hepatitis B virus in hepatocellular carcinoma patients reveals enriched integration events, structural alterations and sequence variations. Carcinogenesis. 2013;34:787–98. doi: 10.1093/carcin/bgs406. [DOI] [PubMed] [Google Scholar]
- 25.Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y. et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–9. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 26.Paterlini-Brechot P, Saigo K, Murakami Y, Chami M, Gozuacik D, Mugnier C. et al. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene. 2003;22:3911–6. doi: 10.1038/sj.onc.1206492. [DOI] [PubMed] [Google Scholar]
- 27.Shiraishi Y, Fujimoto A, Furuta M, Tanaka H, Chiba K, Boroevich KA. et al. Integrated analysis of whole genome and transcriptome sequencing reveals diverse transcriptomic aberrations driven by somatic genomic changes in liver cancers. PLoS One. 2014;9:e114263. doi: 10.1371/journal.pone.0114263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH. et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–4. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 29.Saigo K, Yoshida K, Ikeda R, Sakamoto Y, Murakami Y, Urashima T. et al. Integration of hepatitis B virus DNA into the myeloid/lymphoid or mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in human hepatocellular carcinoma. Hum Mutat. 2008;29:703–8. doi: 10.1002/humu.20701. [DOI] [PubMed] [Google Scholar]
- 30.Zhao LH, Liu X, Yan HX, Li WY, Zeng X, Yang Y. et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. 2016;7:12992. doi: 10.1038/ncomms12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A. 1987;84:1005–9. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Yao H, Thompson EJ, Tannir NM, Weinstein JN, Su X. VirusSeq: software to identify viruses and their integration sites using next-generation sequencing of human cancer tissue. Bioinformatics. 2013;29:266–7. doi: 10.1093/bioinformatics/bts665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Zhang J, Yang Z, Kang J, Jiang S, Zhang T. et al. The function of targeted host genes determines the oncogenicity of HBV integration in hepatocellular carcinoma. Journal of Hepatology. 2014;60:975–84. doi: 10.1016/j.jhep.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Wang T, Zhao R, Wu Y, Kong D, Zhang L, Wu D. et al. Hepatitis B virus induces G1 phase arrest by regulating cell cycle genes in HepG2.2.15 cells. Virol J. 2011;8:231. doi: 10.1186/1743-422X-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leiting B, Pryor KD, Wu JK, Marsilio F, Patel RA, Craik CS. et al. Catalytic properties and inhibition of proline-specific dipeptidyl peptidases II, IV and VII. Biochem J. 2003;371:525–32. doi: 10.1042/BJ20021643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Underwood R, Chiravuri M, Lee H, Schmitz T, Kabcenell AK, Yardley K. et al. Sequence, purification, and cloning of an intracellular serine protease, quiescent cell proline dipeptidase. J Biol Chem. 1999;274:34053–8. doi: 10.1074/jbc.274.48.34053. [DOI] [PubMed] [Google Scholar]
- 37.Chiravuri M, Schmitz T, Yardley K, Underwood R, Dayal Y, Huber BT. A novel apoptotic pathway in quiescent lymphocytes identified by inhibition of a post-proline cleaving aminodipeptidase: a candidate target protease, quiescent cell proline dipeptidase. J Immunol. 1999;163:3092–9. [PubMed] [Google Scholar]
- 38.Ding XR, Yang J, Sun DC, Lou SK, Wang SQ. Whole genome expression profiling of hepatitis B virus-transfected cell line reveals the potential targets of anti-HBV drugs. Pharmacogenomics J. 2008;8:61–70. doi: 10.1038/sj.tpj.6500459. [DOI] [PubMed] [Google Scholar]
- 39.Tong A, Wu L, Lin Q, Lau QC, Zhao X, Li J. et al. Proteomic analysis of cellular protein alterations using a hepatitis B virus-producing cellular model. Proteomics. 2008;8:2012–23. doi: 10.1002/pmic.200700849. [DOI] [PubMed] [Google Scholar]
- 40.Wang J, Jiang D, Zhang H, Lv S, Rao H, Fei R. et al. Proteome responses to stable hepatitis B virus transfection and following interferon alpha treatment in human liver cell line HepG2. Proteomics. 2009;9:1672–82. doi: 10.1002/pmic.200800621. [DOI] [PubMed] [Google Scholar]
- 41.Liu X, Liang J, Li G. Lipopolysaccharide promotes adhesion and invasion of hepatoma cell lines HepG2 and HepG2.2.15. Mol Biol Rep. 2010;37:2235–9. doi: 10.1007/s11033-009-9710-4. [DOI] [PubMed] [Google Scholar]
- 42.Livezey KW, Negorev D, Simon D. Hepatitis B virus-transfected Hep G2 cells demonstrate genetic alterations and de novo viral integration in cells replicating HBV. Mutat Res. 2000;452:163–78. doi: 10.1016/s0027-5107(00)00072-5. [DOI] [PubMed] [Google Scholar]
- 43.Zumbika E, Ruan B, Xu CH, Ni Q, Hou W, Chen Z. et al. HBV genotype characterization and distribution in patients with HBV-related liver diseases in Zhejiang Province, P.R. China: possible association of co-infection with disease prevalence and severity. Hepatobiliary Pancreat Dis Int. 2005;4:535–43. [PubMed] [Google Scholar]
- 44.Tonetto PA, Goncales NS, Fais VC, Vigani AG, Goncales ES, Feltrin A. et al. Hepatitis B virus: molecular genotypes and HBeAg serological status among HBV-infected patients in the southeast of Brazil. BMC Infect Dis. 2009;9:149. doi: 10.1186/1471-2334-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madan K, Batra Y, Sreenivas V, Mizokami M, Tanaka Y, Chalamalasetty SB. et al. HBV genotypes in India: do they influence disease severity? Hepatol Res. 2009;39:157–63. doi: 10.1111/j.1872-034X.2008.00417.x. [DOI] [PubMed] [Google Scholar]
- 46.Chandra PK, Biswas A, Datta S, Banerjee A, Panigrahi R, Chakrabarti S. et al. Subgenotypes of hepatitis B virus genotype D (D1, D2, D3 and D5) in India: differential pattern of mutations, liver injury and occult HBV infection. J Viral Hepat. 2009;16:749–56. doi: 10.1111/j.1365-2893.2009.01129.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figure and tables.