Abstract
The kidney uses specialized G protein-coupled receptors, including olfactory receptors (ORs), to act as sensors of molecules and metabolites. In the present study, we cloned and studied seven renal ORs, which we previously found to be expressed in the murine renal cortex. As most ORs are orphan receptors, our goal was to identify ligands for these ORs in the hope that this will guide future research into their functional roles. We identified novel ligands for two ORs: Olfr558 and Olfr90. For Olfr558, we confirmed activation by previously reported ligands and identified 16 additional carboxylic acids that activated this OR. The strongest activation of Olfr558 was produced by butyric, cyclobutanecarboxylic, isovaleric, 2-methylvaleric, 3-methylvaleric, 4-methylvaleric, and valeric acids. The primary in vivo source of both butyric and isovaleric acids is gut microbial metabolism. We also identified 14 novel ligands that activated Olfr90, the strongest of which were 2-methyl-4-propyl-1,3-oxathiane, 1-octen-3-ol, 2-octanol, and 3-octanol. Interestingly, 8 of these 14 ligands are of fungal origin. We also investigated the tissue distribution of these receptors and found that they are each found in a subset of “nonsensory” tissues. Finally, we examined the putative human orthologs of Olfr558 and Olfr90 and found that the human ortholog of Olfr558 (OR51E1) has a similar ligand profile, indicating that the role of this OR is likely evolutionarily conserved. In summary, we examined seven novel renal ORs and identified new ligands for Olfr558 and Olfr90, which imply that both of these receptors serve to detect metabolites produced by microorganisms.
Keywords: G protein-coupled receptors, microbiota, Olfr558, Olfr90, OR51E1
INTRODUCTION
Olfactory receptors (ORs) are the largest gene family in the genome, with nearly 1,000 genes in the mouse (21, 84) and ~400 genes in the human (49, 56). However, these receptors have been understudied, largely because few ORs have known ligands (46, 51, 52). Although ORs were originally thought to be found solely in the olfactory epithelium (6), there are now multiple studies that have described functional roles for ORs in tissues outside of the nose (11, 15, 39). For example, both mouse MOR23 (Olfr16) and human OR17-4 have been shown to play a role in sperm chemotaxis (19, 70). Similarly, Olfr544 is expressed in α-cells in the pancreatic islets and signals through Ca2+ mobilization to regulate glucagon secretion (38).
Since the discovery of the olfactory signaling machinery in the kidney (62), we have been interested in the roles these receptors play in renal physiology. We previously identified 10 renal ORs (62, 64) and have determined physiological functions for 2 of these ORs: Olfr78 (61) and Olfr1393 (68). Although most ORs do not have known ligands, we found that Olfr78 and its human ortholog OR51E2 are both activated by short-chain fatty acids (SCFAs), compounds that are produced by the gut microbiota. These data imply that some ORs play a role in not only detecting signals from the host but also in detecting signals sent from commensals.
As ORs are the largest gene family in the genome, we reasoned that not all renal ORs have been identified. In a recent study (25), we published an RNA sequencing (RNA-Seq) screen of the renal cortex from eight mice [data accessible at the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (14) under Accession No. GSE117249]. To work toward identifying the full complement of renal ORs, we analyzed data from this RNA-Seq screen to select a subset of ORs that are consistently expressed in the renal cortex. We found five ORs that we had previously identified, four of which we have previously screened for potential ligands (62, 64). In addition, we identified and confirmed expression of six “novel” renal ORs expressed in all eight animals screened. In the present study, we sought to characterize these novel and previously uncharacterized ORs in terms of their in vitro trafficking, their ligand profiles, and their tissue distribution, to shed light on potential physiological roles for these receptors.
MATERIALS AND METHODS
Organ harvesting and RNA isolation.
Four- to five-month-old C57BL/6J mice were euthanized via CO2 followed by cervical dislocation. Experiments were in compliance with the policies and procedures of the Johns Hopkins Animal Care and Use Committee. Organs were harvested and then immediately flash frozen in liquid nitrogen followed by storage at −80°C. Tissues were homogenized in TRIzol (catalog no. 15596018, Ambion, Life Technologies) using the MP FastPrep 24 homogenizer and appropriate lysing matrix. For all samples except heart samples, lysing Matrix D (catalog no. 116913050, MP Biomedicals) was used for three runs at 6.0 m/s for 40 s at room temperature, resting on ice for 5 min between each run. Lysing Matrix SS (catalog no. 116921050, MP Biomedicals) was used for heart samples (3 runs at 6.0 m/s for 40 s). There was a 5-min rest between each run. RNA was isolated as previously described (25, 64) and used for reverse transcription.
Cloning.
Olfr90 (MOR256-21) has been previously cloned (62). For all other murine ORs, whole kidney RNA (1 μg) was reverse transcribed using the iScript Select cDNA Synthesis Kit (catalog no. 1708897, Bio-Rad) per the manufacturer’s instructions. Cloning primers, designed for the full-length coding sequence of each OR, began 5–10 bases upstream of the start codon and spanned to 5–10 bases downstream of the stop codon for the first round of amplification. This was followed by amplification with primers that included regions homologous to the destination vector flanking the new OR sequence. Cloning was done by overlap extension (5, 64, 69). Overlap extension was successful for four of the six ORs [Olfr56 (MOR276-1), Olfr461 (MOR120-3), Olfr1034 (MOR227-8P/MOR245-14P), and Olfr1396 (MOR276-2)]. For Olfr558 (MOR18-1), using the amplified product with vector overhangs, we instead cloned by restriction digest (EcoRI/XhoI, catalog nos. R3101S and R0146S, New England Biolabs). The resulting clones of the ORs had 100% sequence identity with reported sequences (NCBI Olfr56: NM_010999.3, Olfr461: NM_146382.1, Olfr558: NM_147093.3, Olfr1034: NM_001011872.2, and Olfr1396: NM_146337.1). Full-length cloning primers were unable to amplify Olfr1033 (MOR199-2) from kidney cDNA or even from genomic DNA (as ORs do not contain introns in the coding sequence, genomic DNA is a positive control for the reaction). Thus, Olfr1033 was amplified from kidney cDNA in two pieces using a combination of the full-length cloning primers with an appropriate complement of internal Olfr1033 screening primers. These two pieces had a 268-bp overlap, which was used to anneal them using PCR. The annealed product was then amplified using full-length cloning primers. Finally, overlap extension PCR was performed, and the resulting clone of Olfr1033 had 932/933 bases which matched the reported sequence (NCBI NM_146578.2); the mismatched nucleotide was changed to the reported sequence via site-directed mutagenesis PCR. All ORs were ultimately cloned into pME18S vector with Lucy-Flag-Rho tags (69). Human ORs were cloned from human genomic DNA in a similar manner: OR2H2 and OR2H1 were cloned using the restriction digest strategy described above and OR51E1 was cloned using overlap extension PCR. The clone for OR2H1 (NCBI NM_030883.4) matched 100% to reported sequences, OR2H2 (NCBI NM_007160.3) had one reported single-nucleotide polymorphism (nucleotide C90T resulting in amino acid L30F) (72), and OR51E1 (NCBI NM_152430.3) had a silent single-nucleotide polymorphism that did not affect the resulting amino acid.
RT-PCR.
One microgram of RNA isolated from tissues was reverse transcribed into cDNA using the iScript Select cDNA Synthesis Kit (catalog no. 1708897, Bio-Rad) using oligo(dT) primers. PCR was then performed using HotStarTaq Master Mix Plus (catalog no. 203443, Qiagen) for 35 cycles with an annealing temperature of 57°C unless otherwise specified. The PCR primers used were as follows: Olfr56, forward 5′-TGAAGTGCCGGCTTTACTGA-3′ and reverse 5′-GGACTGGGCAGAGTGCATAC-3′ (163 bp, 54°C, 40 cycles); Olfr90, forward 5′-AAGCTGACAGTCTCATCCCTCCAAC-3′ and reverse 5′-GCGCAGAGTGGATGGCGTCTG-3′ (528 bp, 40 cycles); Olfr461, forward 5′-CATCCGTGGCTGTCCCTTAT-3′ and reverse 5′-CCTGTTGTACTCTGCTGCCT-3′ (596 bp); Olfr558, forward 5′-GACACACAGGCTCTTTCAGGA-3′ and reverse 5′-GCCAATCTTGGCAACACGAG-3′ (512 bp, 54°C, 40 cycles); Olfr1033, forward 5′-CCACAGCTCAGTAGCCCAAT-3′ and reverse 5′-TGCAAACCACCCTGGACATT-3′ (268 bp); Olfr1034, forward 5′-TTCCTTGCAGAAAACATAGACATT-3′ and reverse 5′-AGCACTGTACCAAGCATCCA-3′ (333 bp); Olfr1396, forward 5′-TTTTCAGATGACCCGGTACAGCA-3′ and reverse 5′-TCCACCTCCCTCAAGCTACA-3′ (557 bp); and β-actin, forward 5′-CGGTTCCGATGCCCTGAGGC-3′ and reverse 5′-AGGGTGTAAAACGCAGCTCAGTAAC-3′ (401 bp). PCRs were run on 1% agarose gels, and bands were excised and gel extracted using the GenElute Gel Extraction Kit (catalog no. NA1111-1KT, Sigma).
Surface immunofluorescence.
Human embryonic kidney (HEK)-293T cells were grown on poly-l-lysine-coated coverslips and transfected with various Flag-tagged OR constructs along with receptor-transporting protein 1S (RTP1S) (64, 69). Cell surface labeling for ORs was done using live cells incubated with rabbit polyclonal anti-Flag antibody (catalog no. F7425, MilliporeSigma) at 1:200 in PBS with 0.1% BSA at 4°C for 1 h. Coverslips were washed, fixed (4% paraformaldehyde), permeabilized (0.3% Triton X-100), and blocked (1% BSA and 0.2% milk). Coverslips were then incubated with mouse monoclonal anti-Flag antibody (M2, catalog no. F1802, MilliporeSigma) at 1:100 in PBS at 4°C overnight. Coverslips were washed and incubated with secondary antibodies at 1:1,000 in PBS (donkey anti-rabbit Ig Alexa-594, catalog no. A21207, and donkey anti-mouse Ig Alexa-488, catalog no. A21202, ThermoFisher) along with Hoechst 33342 (catalog no. LSH3570, Invitrogen Molecular Probes) at 1:2,500 in the dark at room temperature for 45 min and then washed and mounted using VectaShield Hard Set mounting medium (H-1400, Vector Laboratories). Images were acquired using a Keyence digital microscope (BZ-X700, Keyence). To assay surface expression, each coverslip was manually scanned at ×40 and scored based on detectable cell surface labeling (rabbit poly-Flag surface signal) as previously described (69). Images were quantified manually using ImageJ, which quantified both surface and internal flag staining, and the total cell number was then quantified using the Keyence Hybrid Cell Count feature of BZ-X Analyzer software.
Luciferase assay.
HEK-293T cells were plated in black-walled 96-well plates coated with poly-l-lysine at 25,000 cells/well. Twenty-four hours later, cells were transfected with the OR construct of interest along with RTP1S and two luciferase constructs (61, 64, 68, 69, 85). Approximately 20 h later, cells were incubated in CD293 minimal media (catalog no. 11913-019, Life Technologies, with l-glutamine and penicillin-streptomycin added) for 30 min. All compounds were ordered from Sigma-Aldrich unless otherwise indicated (see Supplemental Table S1; Supplemental Data are available online at doi.org/10.5281/zenodo.2442234). Working stocks of compounds were made ~24 h before the assay and diluted in DMSO, ethanol, or PBS. Directly before use, these stocks were then diluted in CD293 media. Cells were exposed to compounds for 4 h, and plates were sealed to prevent chemical wafting. The Dual Luciferase Reporter Assay (catalog no. E1960, Promega) was then used, and plates were read using a FLUOstar Omega plate reader (BMG LabTech) (61, 64, 69).
RESULTS
Identifying novel renal ORs.
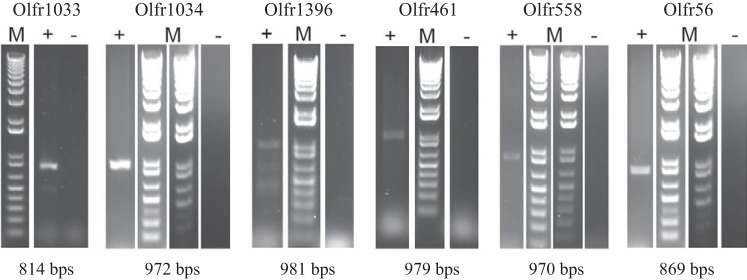
We have previously published an RNA-Seq data set of eight murine cortex samples (25); thus, we referenced this data set to identify ORs expressed in the kidney [data accessible at the NCBI GEO database (14) under Accession No. GSE117249]. Murine ORs are commonly referred to using two different nomenclature systems. The first nomenclature system uses “Olfr” terminology (i.e., Olfr78); the second nomenclature system uses the “MOR#N-#n” system [“mouse OR” in subfamily #N, member #n (i.e, MOR18-2)] (84). In this study, we will primarily use the Olfr terminology. Using our previously generated data set, we aligned the sequences to the mouse mm10 genome using a published set of OR coordinates (29); because we used a separately published set of OR coordinates to map the ORs, ORs are listed as “CUFFOR” in the GEO database file. Using several bioinformatics tools [rsem, as previously described (25), as well as TopHat (v2.0.14) for additional alignments (40, 73) and Cufflinks (v.2.2.1) for additional transcriptome assemblies (74) to both the mm10 build using the previously published coordinates (29) and the mm9 mouse build with established coordinates], we examined ORs that were expressed based on at least one genome build in at least seven of eight samples, and identified a subset of ORs of interest for further study. These ORs (with both CUFFOR and Olfr gene names) are provided in Supplemental Table S2 (available online at doi.org/10.5281/zenodo.2630934). The subset of ORs we focused on included five ORs that we had previously cloned from the kidney (Olfr31, Olfr78, Olfr90, Olfr1392, and Olfr1393) (62, 64), four of five of which we have previously studied [Olfr31 (64), Olfr78 (61), Olfr1392, and Olfr1393 (68)], leaving one OR (Olfr90) for further study. This subset also included 12 new ORs (Olfr56, Olfr212, Olfr267, Olfr461, Olfr558, Olfr613, Olfr920, Olfr1033, Olfr1034, Olfr1396, Olfr1442, and Olfr1443) and 1 OR pseudogene (Olfr1372-ps1). However, it has been previously reported that ORs can form chimeric transcripts in some tissues (15), and our RNA-Seq data did not contain the complete coding sequence (ATG to stop) for all ORs in any of the eight samples. Thus, we sought to confirm the RNA-Seq results using an independent method. We used RT-PCR to amplify the entire coding region of each new OR from renal cortex cDNA. Taking advantage of the fact that ORs do not contain introns within the coding region, we validated all OR primers using murine genomic DNA. Of the 12 previously uncharacterized ORs, we were able to amplify the full coding sequence of 6 ORs from the murine kidney: Olfr56, Olfr461, Olfr558, Olfr1033, Olfr1034, and Olfr1396 (Fig. 1). RT-PCR products were sequenced to confirm their identity. We then cloned each of these six ORs into an expression vector (64, 69) to better characterize these receptors in vitro.
Fig. 1.
RT-PCR of novel olfactory receptors expressed in the renal cortex. RT-PCR confirmed expression of Olfr1033, Olfr1034, Olfr1396, Olfr461, Olfr558, and Olfr56 in the renal cortex. Bands were seen in reverse transcriptase lanes (+) but not in mock controls (−). The band size for each olfactory receptor is indicated below the gel; all bands were sequenced to confirm identity. M, marker.
Determining expression profiles of novel murine renal ORs.
Next, to determine the tissue profile distribution of these previously uncharacterized ORs and thus gain insights into potential functional roles, we used RT-PCR to screen for expression of these ORs in a variety of tissues, as previously described (64). We isolated total RNA from eight different tissues of male and female mice: kidney, heart, skeletal muscle (hindlimb), lung, liver, and stomach as well as testes and uterine horn (Table 1). Kidneys were screened from four male mice and two female mice (n = 6), and two mice of each sex were used to screen the remaining tissues (n = 4). We screened all six new ORs as well as Olfr90 (which had been previously identified in kidney but not yet characterized). Gene-specific primers were used to amplify bands ranging from 100 to 600 bases encompassing unique sequences to each OR screened. Any bands observed in whole kidney samples were gel extracted and sequenced, confirming primer specificity to the OR of interest. All six of the new ORs were positive in all kidneys screened (Table 1). Olfr90 was initially detected in two of six kidneys, and the resulting bands were confirmed via sequencing. The PCR was repeated on the negative samples, and an additional two kidneys were found to be positive for Olfr90 (bands sequenced to confirm). We suspect that the variability in detecting Olfr90 in these samples is due to the low level of this transcript; consistent with this, adding more reverse transcriptase product to the PCR enhanced our ability to detect Olfr90. Each of the six ORs was not only expressed in the kidney but also in several other tissues, each with a unique expression profile. We did not note any sex differences in the distribution of these ORs.
Table 1.
Olfactory receptor PCR-based tissue screen
| Tissues | Olfr56 | Olfr90 | Olfr558 | Olfr461 | Olfr1033 | Olfr1034 | Olfr1396 | n |
|---|---|---|---|---|---|---|---|---|
| Kidney | 6 | 2* | 6 | 6 | 6 | 6 | 6 | 6 (4 male animals and 2 female animals) |
| Liver | 1 | 3 | 3 | 4 (2 male animals and 2 female animals) | ||||
| Lung | 4 | 3 | 2 | 3 | 1 | 4 (2 male animals and 2 female animals) | ||
| Heart | 3 | 1 | 4 | 1 | 4 (2 male animals and 2 female animals) | |||
| Skeletal muscle | 2 | 1 | 4 | 4 (2 male animals and 2 female animals) | ||||
| Stomach | 2 | 1 | 1 | 4 (2 male animals and 2 female animals) | ||||
| Testes | 2 | 2 | 2 | 1 | 1 | 4 (4 male animals and 0 female animals) | ||
| Uterus | 3 | 2 | 4 | 1 | 4 | 4 | 4 | 4 (0 male animals and 4 female animals) |
n, total number of animals screened per tissue. Tissue screening results of all seven olfactory receptors in eight different tissues are shown. Numbers represent the sum of the animals from which a positive result was obtained using PCR screening primers. Blank results indicate that an olfactory receptor was undetected in any samples of that tissue.
Olfr90 was inconsistently observed in an additional two kidneys.
OR surface trafficking in vitro.
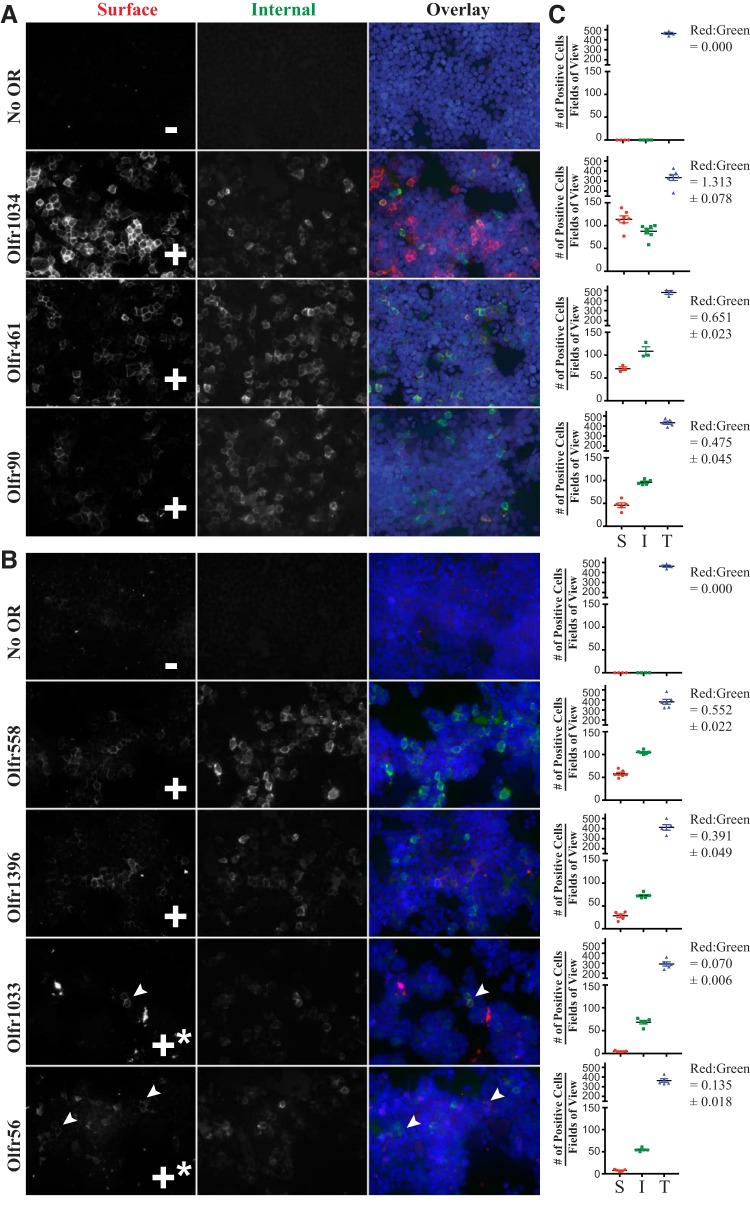
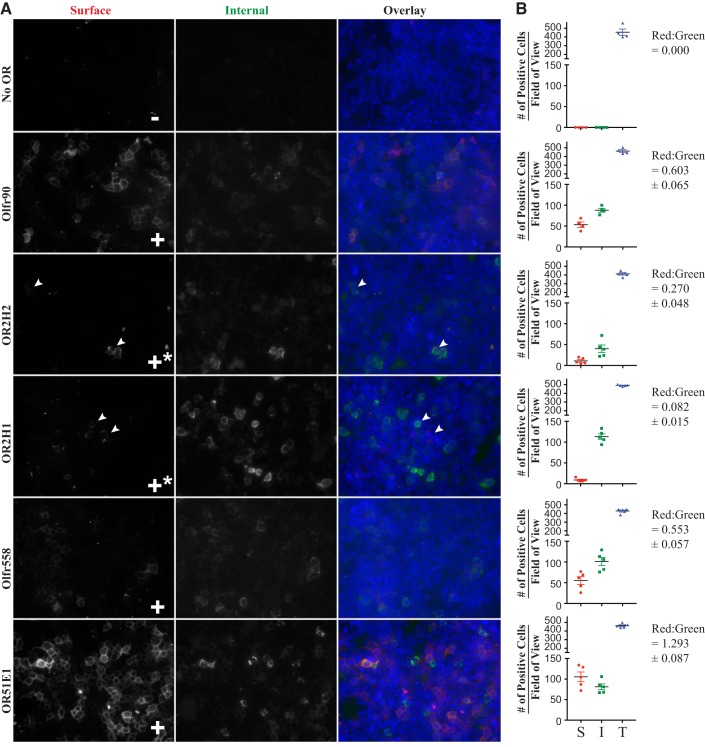
Most of the novel renal ORs are orphan receptors with no known ligand. A prerequisite for ligand screening assays is cell surface expression of the OR; unfortunately, ORs typically fail to traffic to the cell surface when expressed heterologously (35, 42, 46, 50–52, 65, 66, 69, 85, 86). However, we and others have developed strategies [NH2-terminal tags (35, 42, 69) and chaperone protein coexpression (66, 67, 86)] to help bypass this barrier, most notably the chaperone RTP1S (66, 67, 86), the NH2-terminal Rho tag (35, 42), and the NH2-terminal Lucy tag (69). To determine whether the ORs of interest in this study can reach the cell surface when exogenously expressed, we used the NH2-terminal Flag tag on these constructs to assay for both surface and total expression, as previously described (68). Our goal was to determine whether cell surface expression of a given OR was sufficient to warrant ligand screening. After the entire coverslip had been scanned at ×40 by eye, multiple representative fields of view were imaged (Fig. 2, A and B) and then quantified manually using both ImageJ and the Keyence Hybrid Cell Counter feature of BZ-X Analyzer software (Fig. 2C). A field of view was considered positive for surface stain if at least one cell demonstrated surface Flag stain in red with a characteristic “chicken wire-like” pattern typically seen for surface staining in HEK-293T cells (64, 69). This red signal, as well as all the cells demonstrating internal Flag stain in green, were counted using the Multi-Point tool in ImageJ, and the ratio of red cells to green cells was calculated. If this ratio was above 25%, surface expression was considered robust enough to yield detectable signal for subsequent ligand screening. The total number of cells per field of view was quantified using the nuclear stains of each image. The same thresholds and parameters were applied to all images. Concurrently, we noted that for conditions in which we concluded that ligand screening was warranted (red-to-green ratio >25%), the characteristic chicken wire-like pattern of surface staining for the OR was seen in at least 90% of the fields of view on a given coverslip. This aligns well with criteria we have previously used (64, 69), which was based on the percent fields of view demonstrating surface stain. We found that five of seven ORs trafficked to the cell surface in the presence of RTP1S and the Lucy and Rho tags, with red-to-green ratios of >25% (Fig. 2). For the other two receptors, Olfr1033 and Olfr56, surface expression was relatively poor, as indicated by “+*,” and therefore these ORs failed to meet our threshold for ligand screening.
Fig. 2.
Surface trafficking of renal olfactory receptors (ORs) visualized by immunohistochemical staining. Live human embryonic kidney-293T cells, transfected with an OR of interest, were stained with rabbit poly-Flag antibody (surface) and then fixed, permeabilized, and stained with mouse mono-Flag antibody (internal). ORs were tagged with the Lucy and Rho tags and coexpressed with receptor-transporting protein 1S (RTP1S) to help promote surface expression. Representative images are shown for each OR. Images in A were taken at a shorter exposure than those in B. ORs that trafficked to the cell surface in >90% of fields of view are indicated by +, whereas +* indicates surface trafficking visible in <50% of fields of view. C: quantification of OR surface expression, plotting the total number of positive cells per field of view. “S” is surface Flag stain (plotted with red circles), “I” is internal Flag stain (plotted with green squares), and “T” is nuclear stain (plotted in blue triangles). Each data point represents a single field of view. Red-to-green ratios are shown with ±SE. Arrowheads indicate cells with positive staining.
Ligand screening.
For the five ORs that trafficked consistently to the cell surface, ligand screening was performed using a luciferase reporter assay. This assay takes advantage of the fact that ORs are G protein-coupled receptors, which couple to endogenous stimulatory G proteins, and thus OR activation elevates intracellular cAMP. The firefly luciferase construct used in this assay is under the control of a cAMP response element, and thus elevation of intracellular cAMP leads to an increase in firefly luciferase signal. In parallel, cells were transfected with constitutively active Renilla luciferase, and this signal was used to normalize data to control for cell number and viability as well as transfection efficiency. The ratio of firefly-to-Renilla luciferase signal, thus, is an index of OR activation. A prerequisite of this assay is that the OR must be expressed on the cell surface to interact with compounds placed in the assay media, and thus we screened only the five ORs that had met our surface expression criteria shown in Fig. 2 (Olfr90, Olfr461, Olfr558, Olfr1034, and Olfr1396). Of these five ORs, one has previously published ligands: Olfr558 has been reported to respond to butyric acid (1) and nonanoic acid (65). In our hands, robust activation of Olfr558 was observed with butyric acid, whereas inconsistent activation was seen with nonanoic acid. Next, we sought to determine if additional odorant molecules could activate Olfr558 or any of the other novel renal ORs.
A library of 107 compounds was used to screen these ORs (Olfr90, Olfr461, Olfr558, Olfr1034, and Olfr1396). Compounds were selected based on five different categories: 1) odorants from our chemical screening library, many of which broadly activate a large percentage of isolated olfactory epithelium (3, 47, 62); 2) compounds that activate two or more siblings (ORs in the same MOR subfamily) of one of these five ORs (12, 24, 44, 65); 3) ligands for sibling ORs that are found in biofluids by the Human Metabolome Database (HMDB; blood, urine, etc.) (17, 78–80); 4) molecules classified as “odorants” by the HMDB, also present in biofluids (17, 78–80); and 5) small molecules produced by commensal (61, 81) or environmental (41, 53, 63) microorganisms. A complete list of ligands along with the primary reason for their selection (i.e., sibling ligand, etc.) is provided in Supplemental Table S1 (available online at doi.org/10.5281/zenodo.2442234). Compounds were tested at 500 μM or at the highest tolerated dose for molecules toxic at 500 μM (Supplemental Table S1, available online at doi.org/10.5281/zenodo.2442234). None of these 107 compounds activated Olfr461, Olfr1034, or Olfr1396.
Olfr558.
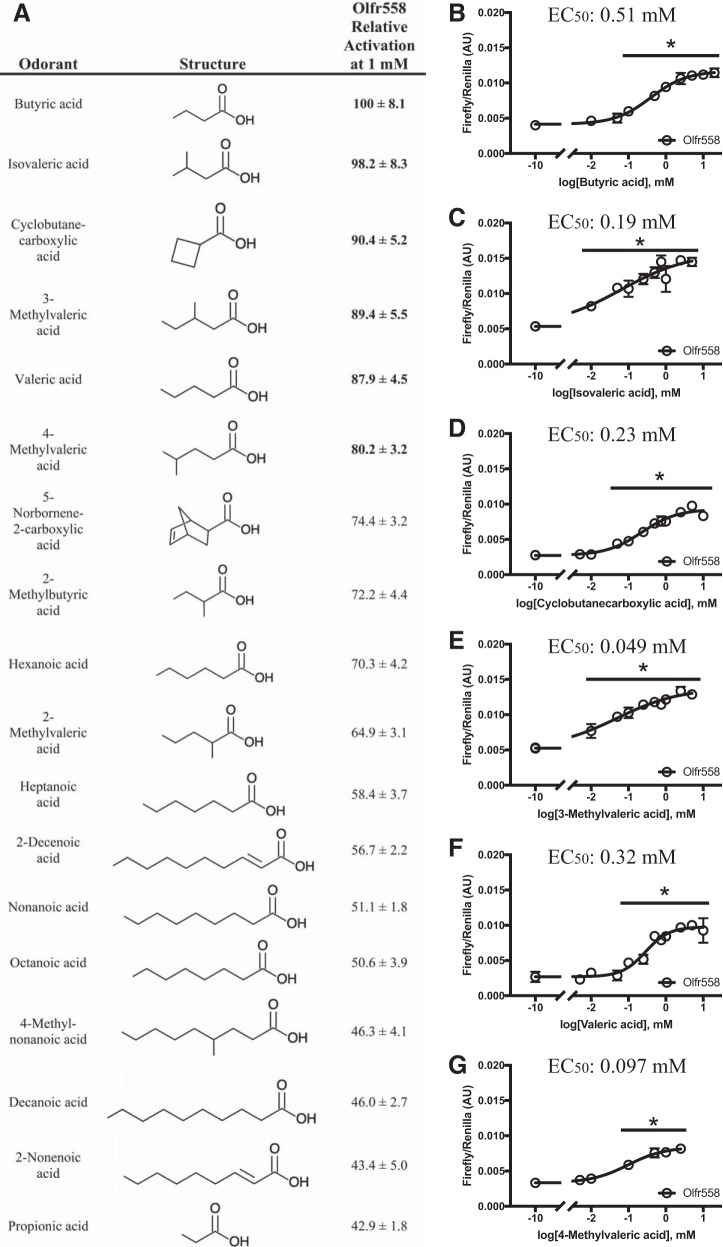
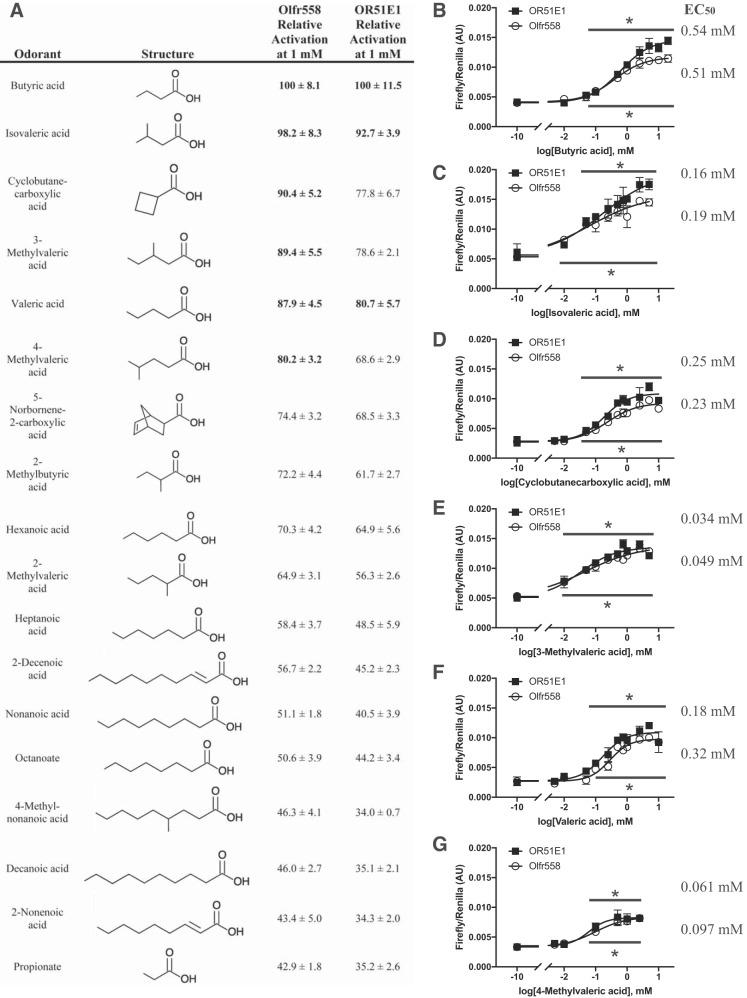
Olfr558 was activated by 18 compounds, all of which contained a carboxylic acid. We compared all Olfr558 ligands at the same dose (1.0 mM; Fig. 3A) and found that the previously reported ligand butyric acid produced the strongest activation (3-fold above baseline). By comparing all ligands at 1.0 mM and setting butyric activation to 100%, we identified six ligands that activated Olfr558 to at least 80% of the butyric acid response. These ligands were butyric acid, cyclobutanecarboxylic acid, valeric acid, isovaleric acid, 3-methylvaleric acid, and 4-methylvaleric acid. For these six ligands, we then determined dose-response curves (Fig. 3, B–G). Olfr558 had an EC50 value of 0.51 mM for butyric acid, an EC50 value of 0.19 mM for isovaleric acid, an EC50 value of 0.23 mM for cyclobutanecarboxylic acid, an EC50 value of 0.049 mM for 3-methylvaleric acid, an EC50 value of 0.32 mM for valeric acid, and an EC50 value of 0.097 mM for 4-methylvaleric acid. We also compared the activation of Olfr558 to butyric acid versus sodium butyrate when both compounds were adjusted to similar pH values and found similar EC50 values as well as similar absolute values of activation, with butyrate eliciting a slightly higher maximum activation. Of note, the two ligands that gave the strongest Olfr558 activation were butyric acid and isovaleric acid. Both of these molecules are found in biofluids (17, 78–80) and arise from gut bacterial fermentation of dietary fiber (butyric acid) or amino acids, particularly leucine (isovaleric acid) (13).
Fig. 3.
Relative activation and dose responses of Olfr558 ligands. A: ligands found to activate Olfr558 are listed in the order of degree of activation relative to butyric acid. All ligands were tested at 1.0 mM; compound structures are shown for reference. Those listed in bold demonstrated activation above 80% of maximal (butyric acid). B−G: dose-response curves are shown for the Olfr558 (○) response to butyric acid (B), isovaleric acid (C), cyclobutanecarboylic acid (D), 3-methylvaleric acid (E), valeric acid (F), and 4-methylvaleric acid (G). Olfr558 was tagged with Rho and Lucy and coexpressed with receptor-transporting protein 1S (RTP1S). Olfr558 demonstrated the highest affinity for 3-methylvaleric acid, with an EC50 value of ~50 μM. AU, arbitrary units. *P < 0.02 vs. no compound using one-way ANOVA.
Olfr90.
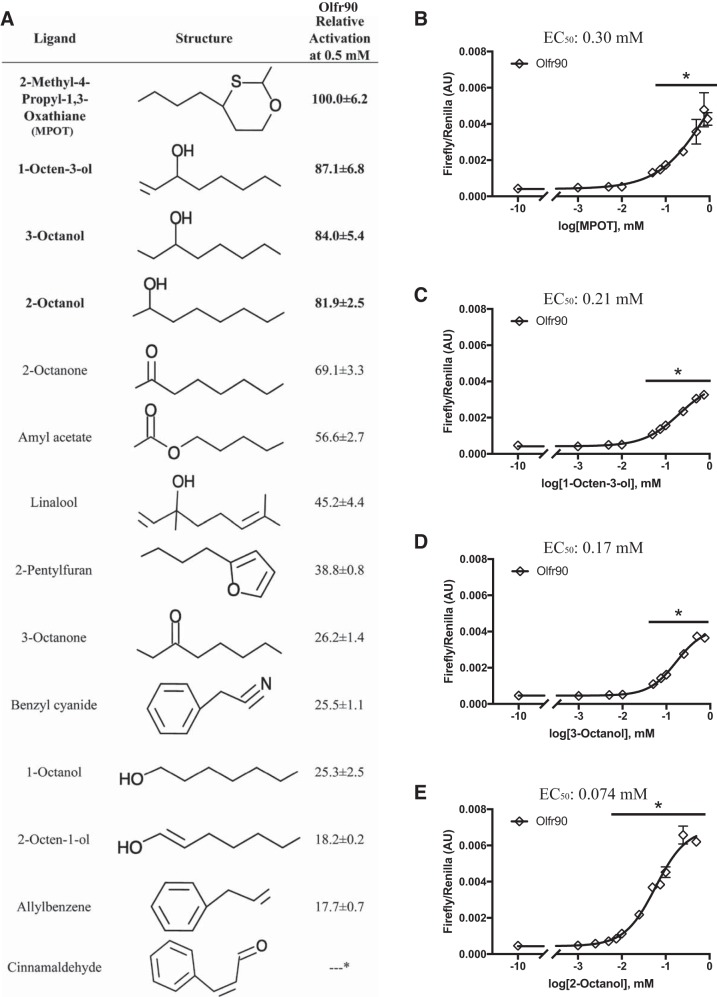
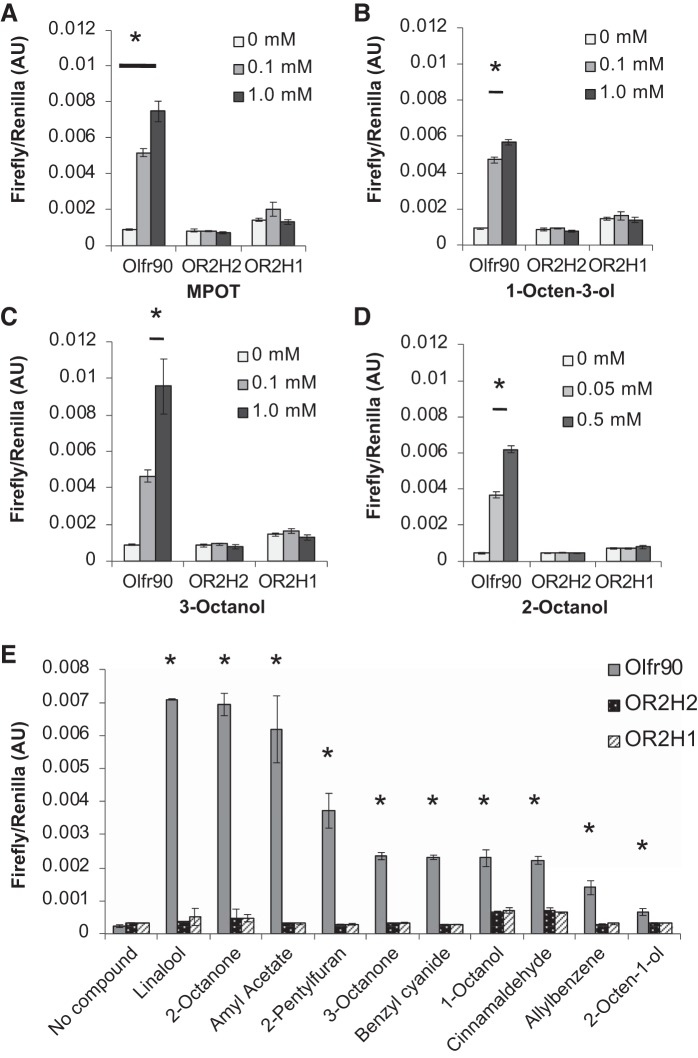
Fourteen compounds in the screen activated Olfr90 in a statistically significant and dose-dependent manner (Fig. 4A), the strongest of which [2-methyl-4-propyl-1,3-oxathiane (MPOT)] activated the receptor 7- to 8.5-fold above baseline. For these 14 ligands, relative activations were determined at 500 μM and are reported relative to MPOT (100%; Fig. 4A). The strongest ligands (at least 80% activation) were MPOT, 1-octen-3-ol, 3-octanol, and 2-octanol. Although MPOT gave the highest activation at the 0.5 mM dose, 2-octanol gave an almost maximal response at a lower dose (0.05 mM), indicating that Olfr90 may have a stronger affinity for 2-octanol. Dose-response curves to determine EC50 values found that Olfr90 had an EC50 value of 74 μM for 2-octanol, an EC50 value of 170 μM for 3-octanol, an EC50 values of 210 μM for 1-octen-3-ol, and an EC50 value of 300 μM for MPOT (Fig. 4, B–E). Concentrations used for dose-response curves were limited by compound toxicity at higher doses; dose-response curves were limited to doses at which toxicity was not observed. Unlike Olfr558, which was activated by ligands that all shared carboxylic acids, ligands for Olfr90 did not have a clear structural commonality. Therefore, we sought to identify any characteristics these compounds do share, including source. Interestingly, 8 of 14 of the Olfr90 ligands are produced by fungus, namely, 1-octen-3-ol (32, 41, 53, 63, 76), 2-pentylfuran (8, 27, 53), benzyl cyanide, linalool, 3-octanol, 2-octanone, 3-octanone, and 2-octen-1-ol (32, 41, 53, 63). Fungi make up a small but significant part of the microbiome (20, 28, 31), and therefore it is possible that Olfr90 may be detecting fungal metabolites in a similar way to the potential detection of bacterial metabolites by Olfr558.
Fig. 4.
Relative activation and dose responses of Olfr90 ligands. A: ligands found to activate Olfr90 are listed in order of the degree of activation relative to 2-methyl-4-propyl-1,3-oxathiane (MPOT). Odorant molecules were all tested at 0.5 mM; compound structures are shown for reference. Those listed in bold demonstrated activation above 80%. B−E: dose-response curves for Olfr90 (◇; tagged with Rho and Lucy) are shown for MPOT (B), 1-octen-3-ol (C), 3-octanol (D), and 2-octanol (E) when coexpressed with RTP1S, receptor-transporting protein 1S (RTP1S). Olfr90 demonstrated the highest affinity for 2-octanol with an EC50 value of ~75 μM. AU, arbitrary units. *P < 0.02 vs. no compound using one-way ANOVA.
Human orthologs of receptors with newly identified ligands.
We next aimed to identify the human orthologs of Olfr558 and Olfr90. Because ORs are the largest gene family in the genome in both species, and all ORs have high levels of homology to one another, primary sequence identity alone is not sufficient to conclusively identify an ortholog. Rather, it is essential to determine functional orthology [i.e., that potential orthologs are activated by similar ligands (64)]. Thus, we cloned the putative human orthologs of Olfr558 and Olfr90 and assayed whether they trafficked to the cell surface (Fig. 5). Human ORs use a different nomenclature: “OR” followed by a number-letter combination denoting the subfamily, with a final number determining the family member (58). We found that two human ORs that share a similar level of sequence identity with murine Olfr90: OR2H2 (84% sequence identity) and OR2H1 (81% sequence identity). Both of these ORs trafficked to the cell surface to a limited extent. Although the surface stain of OR2H2 was stronger than that of OR2H1 with a red-to-green ratio of 27% for OR2H2 and 8.2% for OR2H1, both were noticeably weaker than Olfr90 with a red-to-green ratio of 60%. Of note, OR2H1 did not meet our criteria for ligand screening but was included here for comparison with OR2H2, as this was a directed screen with only 14 ligands; thus, we emphasize that any negative results for OR2H1 must be interpreted with caution. OR2H1 and OR2H2 were cloned and screened against the 14 ligands identified for Olfr90. Of these 14 ligands, none were able to activate either OR2H2 or OR2H1 (Fig. 6). Thus, we conclude that neither OR2H2 nor OR2H1 are likely the functional ortholog of Olfr90, although we cannot rule out that the relatively poor surface expression of OR2H1 has affected our result.
Fig. 5.
Surface trafficking of murine and human olfactory receptor (ORs) visualized by immunohistochemical staining. A: representative images of murine ORs and their putative human orthologs in human embryonic kidney (HEK)-293T cells. Live HEK-293T cells, transfected with the OR of interest, were stained with rabbit poly-Flag antibody (surface) and then fixed, permeabilized, and stained with mouse mono-Flag antibody (internal). ORs were tagged with Lucy and Rho tags and coexpressed with receptor-transporting protein 1S (RTP1S) to help promote surface expression. All ORs trafficked to the surface to some degree; ORs that trafficked the cell surface in >90% of fields of view are indicated by +, whereas +* indicates surface trafficking visible in <50% of fields of view. B: quantification of OR surface expression. The total number of positive cells per field of view are plotted. “S” is surface Flag stain (plotted in red circles), “I” is internal Flag stain (plotted in green squares), and “T” is nuclear stain (plotted in blue triangles). Each data point represents a single field of view. Red-to-green ratios are shown with ±SE. Arrowheads indicate cells with positive staining.
Fig. 6.
OR2H2 and OR2H1 fail to respond to Olfr90 ligands. A−D: the dose-dependent responses seen with Olfr90 were not reproduced by OR2H2 or OR2H1 for any ligand, including 2-methyl-4-propyl-1,3-oxathiane (MPOT; A), 1-octen-3-ol (B), 3-octanol (C), or 2-octanol (D). E: ligands were tested at doses that yield maximal Olfr90 activation via firefly induction without decreasing Renilla. These doses included 5 mM (amyl acetate), 2 mM (allylbenzene, pentylfuran), 1 mM (linalool, 2-octanone, 3-octanone, benzyl cyanide, and 1-octanol), 0.1 mM (2-octen-1-ol), and 0.05 mM (cinnamaldehyde). AU, arbitrary units. *P < 0.05 vs. no compound using Student’s t-test.
Although most ORs do not have a clear ortholog across mammalian species, there are 3 unique orthologous OR gene groups that have highly conserved sequence identity across 13 different classes of placental mammals (55). One of these gene groups includes murine Olfr558, for which the putative human ortholog is OR51E1 (94% sequence identity). Thus, we cloned OR51E1 and screened it against all compounds that we found to activate Olfr558. Olfr558 and OR51E1 demonstrated similar activation profiles for all 18 ligands (Fig. 7A), and butyric acid demonstrated the highest relative activation for OR51E1 at nearly 4-fold above baseline. We next constructed dose-response curves for all compounds that activated OR51E1 above 80% of maximal (butyric acid) activation (Fig. 7, B–G). Of note, only butyric acid (EC50 value of 0.54 mM), isovaleric acid (EC50 value of 0.16 mM), and valeric acid (EC50 value of 0.25 mM) activated OR51E1 above this threshold. Interestingly, the fold activation of OR51E1 by butyric acid was statistically significantly higher than that for Olfr558, although this is likely due to increased surface expression of OR51E1 (Fig. 5). In addition, we performed dose-response curves to calculate EC50 values for OR51E1 for three additional compounds that activated Olfr558 to >80% of maximal but fell short of this criteria (68–78%) for OR51E1. We found that cyclobutanecarboxylic acid (EC50 value of 0.18 mM), 3-methylvaleric acid (EC50 value of 0.034 mM), and 4-methylvaleric acid (EC50 value of 0.061 mM) all had similar affinities for OR51E1 versus Olfr558 (Fig. 7). Thus, we conclude that OR51E1 is the functional ortholog of Olfr558.
Fig. 7.
Relative activation and dose responses of Olfr558 and OR51E1 ligands. A: short- and medium-chain fatty acids that activate Olfr558 and OR51E1 are listed in the order of degree of activation relative to butyric acid. All ligands were tested at 1.0 mM. Compound structures are shown for reference. Data for Olfr558 are repeated from Fig. 3 for reference. Compounds listed in bold demonstrated Olfr558 or OR51E1 activation above 80% of maximal. B−G: dose-response curves for OR51E1 (○) and Olfr558 (■) are shown for butyric acid (B), isovaleric acid (C), cyclobutanecarboylic acid (D), 3-methylvaleric acid (E), valeric acid (F), and 4-methylvaleric acid (G). Both olfactory receptors were tagged with Rho and Lucy and coexpressed with receptor-transporting protein 1S (RTP1S). Olfr558 data from Fig. 3 were replotted here for reference. EC50 values listed are for OR51E1 (top) and Olfr558 (bottom). OR51E1 demonstrated the highest affinity for 3-methylvaleric acid with an EC50 value of ~35 μM. AU, arbitrary units. *P < 0.02 vs. no compound using one-way ANOVA for both OR51E1 (top) and Olfr558 (bottom).
DISCUSSION
Recent studies have underscored the functional roles of ORs in nonsensory tissues (2, 11, 15, 22, 38, 39, 70, 82), including the kidney (61, 64, 68). The goal of the present study was to gain insights into ORs expressed in the renal cortex, with a focus on characterizing the expression pattern and ligands for renal ORs that we identified in a recent RNA-Seq screen (25). We identified novel ligands for two of these ORs, Olfr558 and Olfr90, and unexpectedly found that both of these receptors are activated by metabolites produced by commensals.
Selection of ORs for study.
Our recent RNA-Seq data revealed that five of the ORs we have previously found in the whole murine kidney (62, 64) are expressed in the murine renal cortex. We have already published ligand screens for four of these ORs (Olfr31, Olfr78, Olfr1392, and Olfr1393) (64, 68) but not Olfr90. The RNA-Seq data (25) also identified 12 novel ORs expressed in the renal cortex. We were able to confirm expression of 6 of these 12 ORs by PCR of the full coding region. The failure to detect the full-length transcripts for all ORs determined by RNA-Seq likely reflects either the previously identified “chimera” transcript phenomenon (15) or that some transcripts are expressed below the threshold of detection for PCR, perhaps due to expression being limited to a minority cell type. Closer examination of the alignment maps reveals reads mapping to a large portion of the coding region in at least one genome build for all ORs successfully cloned in at least one genome build, whereas the remaining ORs (those unable to be cloned) had reads mapping to primarily untranslated regions, suggesting the OR may not actually be expressed in the kidney (coordinates and fragments per kilobase of transcript per million mapped reads values are available online at doi.org/10.5281/zenodo.2630934). Thus, we focused on characterizing the six novel renal ORs that we were able to confirm by PCR: Olfr56, Olfr461, Olfr558, Olfr1033, Olfr1034, and Olfr1396. In addition, we included Olfr90, which we had previously identified in the kidney both by PCR (62) and RNA-Seq (25) but had not yet been characterized. We found that five of these seven ORs trafficked to the cell surface, and we were able to identify novel ligands for two of them: Olfr90 and Olfr558.
We found that expression of these ORs was not limited to the kidney, implying that these receptors may be playing physiological roles in multiple tissues, and determining activating ligands could provide hints as to those roles. Ligands have been previously identified for Olfr56 (42) and Olfr558 (1, 65) (discussed further below). The previously published RNA-Seq data set of microdissected tubule segments shows that Olfr1396 is expressed in the thick ascending limb and cortical collecting duct at ~0.6–0.7 fragments per kilobase of transcript per million mapped reads (10).
Olfr90.
Olfr90 (MOR256-21) is a member of the MOR256 subfamily, the largest subfamily of mouse ORs (21, 84). Six members of this subfamily have been deorphanized (12, 24, 44, 54, 65, 68, 83), revealing 129 unique ligands. The chemical library used for our ligand screen included 13 ligands that activate >1 MOR256 receptor (Supplemental Table S1, available online at doi.org/10.5281/zenodo.2442234); two of these (amyl acetate and cinnamaldehyde) activated Olfr90. Three of the Olfr90 ligands (1-octanol, 2-octanol, and 2-pentylfuran) are listed as present in biofluids by the HMDB (17, 78–80). Intriguingly, 8 of the 14 ligands that activate Olfr90 are metabolites produced by fungus. This includes some of the strongest ligands, 1-octen-3-ol and 3-octanol, as well as benzyl cyanide, linalool, 2-octanone, 3-octanone, 2-octen-1-ol, and 2-pentylfuran (8, 27, 32, 41, 53, 63, 76). Although the HMDB indicates that many of these compounds have been detected in human biofluids (presence vs. absence), in most cases levels have not been quantified. Thus, we are unable to speculate whether these compounds circulate at concentrations that could be detected by Olfr90. We have previously shown that Olfr90 is expressed in a macula densa cell line (62), implying that this receptor may be expressed in the macula densa in vivo (expression in this minority cell type may also explain why the OR was not easily detected in all kidney samples by conventional PCR). The canonical downstream olfactory signaling machinery proteins adenylate cyclase 3 and olfactory neuron-specific G protein are also expressed in the macula densa (62), and this signaling cascade could be initiated through activation of Olfr90 by circulating fungal products. Fungi are known to exist in the body as commensals (20, 28, 30) and can also be inhaled or ingested from the environment (28, 31). These fungal products can be toxic (31, 32), and, therefore, as the macula densa is known to play a key role in regulating the glomerular filtration rate, we speculate that Olfr90 may be acting as a sensor to modulate the glomerular filtration rate in response to changes in circulating levels of these compounds. Olfr90 may also be a signaling component within innate immunity, working in conjunction with pattern recognition receptors or other membrane-bound receptors to incite an immune or inflammatory response (77).
We should note that there is currently no known human functional ortholog for Olfr90. Based on protein sequence identity, there were two potential human orthologs for Olfr90: OR2H2 (84% identity) and OR2H1 (81% identity). Although neither OR2H2 nor OR2H1 responded to any of the Olfr90 ligands (Fig. 6), it is worth noting that OR2H1 in particular had poor surface expression, making it difficult to fully rule out OR2H1 as an ortholog. Methional was reported to activate OR2H1 using a calcium assay (16, 45); unfortunately, we were unable to confirm OR2H1 (or Olfr90) activation with methional in our luciferase assay, likely because of compound toxicity. A recent report (36), published while this study was in review, demonstrated using calcium imaging that OR2H2 is activated by aldehyde 13-13. Future studies comparing Olfr90 and OR2H2 responses using aldehyde 13-13 could be used as an additional tool to determine if Olfr90 and OR2H2 are “functional” orthologs.
Olfr558.
Olfr558 (MOR18-1) is a member of the MOR18 subfamily, a relatively small subfamily with three total members (1, 61, 65). Olfr558 has been reported by microarray to be expressed in isolated juxtaglomerular cells (4), and its rat ortholog Olr63 has been reported to be expressed in renal glomeruli (43). Olfr558 is one of two receptors in this family with published ligands and has previously been reported to be activated by butyric acid (1), while some groups (65), but not others (23), have reported activation with nonanoic acid. In the present study, we confirmed butyric acid activation of Olfr558 but did not see consistent activation with nonanoic acid. In addition, we identified 16 new ligands for Olfr558 (Fig. 3). Many of these compounds are reported agonists of human receptor OR51E1 (7, 18, 34), discussed further below. In contrast to our findings, Saito et al. (65) reported that Olfr558 was not activated by propionic acid. In our hands, propionate activation was not detectable until concentrations were in the high micromolar to millimolar range, concentrations above the upper limit in the Saito et al. screen.
The compounds that elicited the strongest activation of Olfr558 were butyric acid and isovaleric acid; both compounds are well-established bacterial metabolites (13, 26, 48, 57, 81). Interestingly, butyric acid is one of the three primary SCFAs produced by commensal gut bacteria (61, 71, 81). These microbial SCFAs are known to affect host physiology, particularly blood pressure (33, 59, 61). The closest sibling to Olfr558 is Olfr78, which responds to the two other SCFAs, propionate and acetate (61). Both Olfr78 and Olfr558 have been shown to be expressed in the juxtaglomerular apparatus (4, 61), which could provide evidence that these two ORs are working together to sense circulating SCFAs and signaling to adjust renal function accordingly. Similarly, both Olfr78 and Olfr558 are expressed in the carotid body (9), demonstrating again that these two receptors may be working in tandem to sense SCFAs produced by the gut microbiota and help regulate host physiology. The reported ortholog for Olfr558 is OR51E1 (55). OR51E1 has been reported to be activated by 22 different carboxylic acids (7, 16, 18, 34, 37, 65), and we were able to confirm OR51E1 activation by 17 of these compounds (Fig. 7A) using similar doses. An initial report (18) demonstrated inconsistent OR51E1 activation by isovaleric acid, yet in our hands isovaleric acid strongly activated both OR51E1 and Olfr558 (Fig. 7C), which others have also reported (37). Maximal activation of Olfr558 was not as strong as that of OR51E1, likely because of more robust in vitro surface expression of OR51E1 (Fig. 5). These similar ligand profiles further demonstrate that Olfr558 and OR51E1 are “functional” orthologs. Additionally, OR51E1 has been shown to be expressed in human kidney cells where it colocalizes with olfactory signaling machinery (37), providing further evidence that the role of this OR in vivo is evolutionarily conserved.
Butyrate concentrations have been reported in mice, with plasma levels of roughly 10–30 μM (60). In humans, the reported circulating levels of butyric acid range from 0.3 to 1.5 μM, whereas isovaleric acid levels range from 0.3 to 5 μM. Although these values are below the EC50 values of Olfr558/OR51E1, as the pro-urine is concentrated, the local concentrations in various regions of the tubule lumen may reach the activation threshold of Olfr558. Finally, as a technical note, in light of the fact that the ligands we identified for Olfr558/OR51E1 were derived from bacteria (and Olfr90 ligands were derived from fungi), it is worth noting that the luciferase assay media contained antibiotics derived from fungi and bacteria; we cannot rule out that these compounds may somehow have interfered with ligand screening.
Ligand screening of other ORs.
We were unable to identify ligands that activated Olfr461 (MOR120-3), Olfr1034 (MOR227-8P/MOR245-14P), or Olfr1396 (MOR276-2), and they remain orphan receptors. Examination of sibling ORs in these subfamilies revealed only two deorphanized receptors among all four subfamilies: Olfr142 (MOR227-2) may weakly respond to acetophenone (75), and Olfr56 (MOR276-1) is activated by limonene (42). We were unable to confirm Olfr56 activation by limonene, likely because of poor receptor surface trafficking (Fig. 2). Although we were able to clone and express Olfr1034 (as evidenced by Flag staining; Fig. 2), Olfr1034 is classified as a pseudogene, and thus we cannot rule out the possibility that Olfr1034 failed to respond to compounds because it is a pseudogene encoding a nonfunctional OR. Of note, in our hands, a few compounds gave a nonspecific signal in the luciferase assay, activating all ORs (and empty vector negative controls) to a similar extent (Supplemental Table S1, available online at doi.org/10.5281/zenodo.2442234). This includes butyl butyryllactate, which has been previously reported to activate OR51E1 (65).
Conclusions.
In summary, we characterized seven ORs expressed in the renal cortex. We identified new ligands for two ORs (Olfr90 and Olfr558) and demonstrated that the ligands for Olfr558 also activate its human ortholog, OR51E1. Intriguingly, both Olfr90 and Olfr558 are activated by ligands that are produced by microbes, indicating that these receptors may help mediate microbial influence on host physiology. In future studies, it will be important to uncover how Olfr90 and Olfr558 activation can modulate renal physiology.
GRANTS
This work was supported in part by National Institute of Diabetes and Digestive and Kidney Diseases Grants R01-DK-107726 and F31-DK-104454.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
V.L.H.K. and J.L.P. conceived and designed research; V.L.H.K., J.S., D.C.S., Z.K., and P.R. performed experiments; V.L.H.K., J.S., D.C.S., Z.K., and P.R. analyzed data; V.L.H.K., J.S., D.C.S., Z.K., P.R., K.A.M., and J.L.P. interpreted results of experiments; V.L.H.K. prepared figures; V.L.H.K. drafted manuscript; V.L.H.K., J.S., D.C.S., Z.K., P.R., K.A.M., and J.L.P. edited and revised manuscript; V.L.H.K., J.S., D.C.S., Z.K., P.R., K.A.M., and J.L.P. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the Johns Hopkins Sidney Kimmel Cancer Center Next Generation Sequencing Core, the Johns Hopkins Center for Computational Biology, Dr. Blythe Shepard (Georgetown/Johns Hopkins University), William Aisenberg, Dr. Kausik Datta, and other members of the Pluznick and Marr laboratories for advice and helpful discussions.
REFERENCES
- 1.Adipietro KA, Mainland JD, Matsunami H. Functional evolution of mammalian odorant receptors. PLoS Genet 8: e1002821, 2012. doi: 10.1371/journal.pgen.1002821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aisenberg WH, Huang J, Zhu W, Rajkumar P, Cruz R, Santhanam L, Natarajan N, Yong HM, De Santiago B, Oh JJ, Yoon AR, Panettieri RA, Homann O, Sullivan JK, Liggett SB, Pluznick JL, An SS. Defining an olfactory receptor function in airway smooth muscle cells. Sci Rep 6: 38231, 2016. doi: 10.1038/srep38231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozza T, Feinstein P, Zheng C, Mombaerts P. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci 22: 3033–3043, 2002. doi: 10.1523/JNEUROSCI.22-08-03033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunskill EW, Sequeira-Lopez ML, Pentz ES, Lin E, Yu J, Aronow BJ, Potter SS, Gomez RA. Genes that confer the identity of the renin cell. J Am Soc Nephrol 22: 2213–2225, 2011. doi: 10.1681/ASN.2011040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryksin AV, Matsumura I. Overlap extension PCR cloning: a simple and reliable way to create recombinant plasmids. Biotechniques 48: 463–465, 2010. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65: 175–187, 1991. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- 7.Bushdid C, de March CA, Fiorucci S, Matsunami H, Golebiowski J. Agonists of G-protein-coupled odorant receptors are predicted from chemical features. J Phys Chem Lett 9: 2235–2240, 2018. doi: 10.1021/acs.jpclett.8b00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers ST, Syhre M, Murdoch DR, McCartin F, Epton MJ. Detection of 2-pentylfuran in the breath of patients with Aspergillus fumigatus. Med Mycol 47: 468–476, 2009. doi: 10.1080/13693780802475212. [DOI] [PubMed] [Google Scholar]
- 9.Chang AJ, Ortega FE, Riegler J, Madison DV, Krasnow MA. Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature 527: 240–244, 2015. doi: 10.1038/nature15721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Lee JW, Chou CL, Nair AV, Battistone MA, Păunescu TG, Merkulova M, Breton S, Verlander JW, Wall SM, Brown D, Burg MB, Knepper MA. Transcriptomes of major renal collecting duct cell types in mouse identified by single-cell RNA-seq. Proc Natl Acad Sci USA 114: E9989–E9998, 2017. doi: 10.1073/pnas.1710964114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Zhao H, Fu N, Chen L. The diversified function and potential therapy of ectopic olfactory receptors in non-olfactory tissues. J Cell Physiol 233: 2104–2115, 2018. doi: 10.1002/jcp.25929. [DOI] [PubMed] [Google Scholar]
- 12.Dahoun T, Grasso L, Vogel H, Pick H. Recombinant expression and functional characterization of mouse olfactory receptor mOR256-17 in mammalian cells. Biochemistry 50: 7228–7235, 2011. doi: 10.1021/bi2008596. [DOI] [PubMed] [Google Scholar]
- 13.Dai ZL, Wu G, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci 16: 1768–1786, 2011. doi: 10.2741/3820. [DOI] [PubMed] [Google Scholar]
- 14.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210, 2002. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flegel C, Manteniotis S, Osthold S, Hatt H, Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS One 8: e55368, 2013. doi: 10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flegel C, Vogel F, Hofreuter A, Schreiner BS, Osthold S, Veitinger S, Becker C, Brockmeyer NH, Muschol M, Wennemuth G, Altmüller J, Hatt H, Gisselmann G. Characterization of the olfactory receptors expressed in human spermatozoa. Front Mol Biosci 2: 73, 2016. doi: 10.3389/fmolb.2015.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsythe IJ, Wishart DS. Exploring human metabolites using the human metabolome database. Curr Protoc Bioinformatics Chapter 14: 8, 2009. doi: 10.1002/0471250953.bi1408s25. [DOI] [PubMed] [Google Scholar]
- 18.Fujita Y, Takahashi T, Suzuki A, Kawashima K, Nara F, Koishi R. Deorphanization of Dresden G protein-coupled receptor for an odorant receptor. J Recept Signal Transduct Res 27: 323–334, 2007. doi: 10.1080/10799890701534180. [DOI] [PubMed] [Google Scholar]
- 19.Fukuda N, Yomogida K, Okabe M, Touhara K. Functional characterization of a mouse testicular olfactory receptor and its role in chemosensing and in regulation of sperm motility. J Cell Sci 117: 5835–5845, 2004. doi: 10.1242/jcs.01507. [DOI] [PubMed] [Google Scholar]
- 20.Ghannoum MA, Jurevic RJ, Mukherjee PK, Cui F, Sikaroodi M, Naqvi A, Gillevet PM. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6: e1000713, 2010. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godfrey PA, Malnic B, Buck LB. The mouse olfactory receptor gene family. Proc Natl Acad Sci USA 101: 2156–2161, 2004. doi: 10.1073/pnas.0308051100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffin CA, Kafadar KA, Pavlath GK. MOR23 promotes muscle regeneration and regulates cell adhesion and migration. Dev Cell 17: 649–661, 2009. doi: 10.1016/j.devcel.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grison A, Zucchelli S, Urzì A, Zamparo I, Lazarevic D, Pascarella G, Roncaglia P, Giorgetti A, Garcia-Esparcia P, Vlachouli C, Simone R, Persichetti F, Forrest AR, Hayashizaki Y, Carloni P, Ferrer I, Lodovichi C, Plessy C, Carninci P, Gustincich S; FANTOM Consortium . Mesencephalic dopaminergic neurons express a repertoire of olfactory receptors and respond to odorant-like molecules. BMC Genomics 15: 729, 2014. doi: 10.1186/1471-2164-15-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, Mombaerts P, Ma M. SR1, a mouse odorant receptor with an unusually broad response profile. J Neurosci 29: 14545–14552, 2009. doi: 10.1523/JNEUROSCI.2752-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halperin Kuhns VL, Pluznick JL. Novel differences in renal gene expression in a diet-induced obesity model. Am J Physiol Renal Physiol 314: F517–F530, 2018. doi: 10.1152/ajprenal.00345.2017. [DOI] [PubMed] [Google Scholar]
- 26.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27: 104–119, 2008. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 27.Heddergott C, Calvo AM, Latgé JP. The volatome of Aspergillus fumigatus. Eukaryot Cell 13: 1014–1025, 2014. doi: 10.1128/EC.00074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huffnagle GB, Noverr MC. The emerging world of the fungal microbiome. Trends Microbiol 21: 334–341, 2013. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibarra-Soria X, Levitin MO, Saraiva LR, Logan DW. The olfactory transcriptomes of mice. PLoS Genet 10: e1004593, 2014. doi: 10.1371/journal.pgen.1004593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, Rotter JI, Wang HL, McGovern DP, Brown GD, Underhill DM. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336: 1314–1317, 2012. doi: 10.1126/science.1221789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iliev ID, Underhill DM. Striking a balance: fungal commensalism versus pathogenesis. Curr Opin Microbiol 16: 366–373, 2013. doi: 10.1016/j.mib.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inamdar AA, Masurekar P, Bennett JW. Neurotoxicity of fungal volatile organic compounds in Drosophila melanogaster. Toxicol Sci 117: 418–426, 2010. doi: 10.1093/toxsci/kfq222. [DOI] [PubMed] [Google Scholar]
- 33.Jose PA, Raj D. Gut microbiota in hypertension. Curr Opin Nephrol Hypertens 24: 403–409, 2015. doi: 10.1097/MNH.0000000000000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jovancevic N, Dendorfer A, Matzkies M, Kovarova M, Heckmann JC, Osterloh M, Boehm M, Weber L, Nguemo F, Semmler J, Hescheler J, Milting H, Schleicher E, Gelis L, Hatt H. Medium-chain fatty acids modulate myocardial function via a cardiac odorant receptor. Basic Res Cardiol 112: 13, 2017. doi: 10.1007/s00395-017-0600-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci 21: 6018–6025, 2001. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalbe B, Osterloh M, Schulz VM, Altmüller J, Becker C, Osterloh S, Hatt H. OR2H2 regulates the differentiation of human myoblast cells by its ligand aldehyde 13-13. Arch Biochem Biophys 645: 72–80, 2018. doi: 10.1016/j.abb.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 37.Kalbe B, Schlimm M, Wojcik S, Philippou S, Maßberg D, Jansen F, Scholz P, Luebbert H, Ubrig B, Osterloh S, Hatt H. Olfactory signaling components and olfactory receptors are expressed in tubule cells of the human kidney. Arch Biochem Biophys 610: 8–15, 2016. doi: 10.1016/j.abb.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 38.Kang N, Bahk YY, Lee N, Jae Y, Cho YH, Ku CR, Byun Y, Lee EJ, Kim MS, Koo J. Olfactory receptor Olfr544 responding to azelaic acid regulates glucagon secretion in α-cells of mouse pancreatic islets. Biochem Biophys Res Commun 460: 616–621, 2015. doi: 10.1016/j.bbrc.2015.03.078. [DOI] [PubMed] [Google Scholar]
- 39.Kang N, Koo J. Olfactory receptors in non-chemosensory tissues. BMB Rep 45: 612–622, 2012. doi: 10.5483/BMBRep.2012.45.11.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36, 2013. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korpi A, Järnberg J, Pasanen AL. Microbial volatile organic compounds. Crit Rev Toxicol 39: 139–193, 2009. doi: 10.1080/10408440802291497. [DOI] [PubMed] [Google Scholar]
- 42.Krautwurst D, Yau KW, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 95: 917–926, 1998. doi: 10.1016/S0092-8674(00)81716-X. [DOI] [PubMed] [Google Scholar]
- 43.Lee JW, Chou CL, Knepper MA. Deep sequencing in microdissected renal tubules identifies nephron segment-specific transcriptomes. J Am Soc Nephrol 26: 2669–2677, 2015. doi: 10.1681/ASN.2014111067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Haddad R, Chen S, Santos V, Luetje CW. A broadly tuned mouse odorant receptor that detects nitrotoluenes. J Neurochem 121: 881–890, 2012. doi: 10.1111/j.1471-4159.2012.07740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López del Castillo-Lozano M, Delile A, Spinnler HE, Bonnarme P, Landaud S. Comparison of volatile sulphur compound production by cheese-ripening yeasts from methionine and methionine-cysteine mixtures. Appl Microbiol Biotechnol 75: 1447–1454, 2007. doi: 10.1007/s00253-007-0971-3. [DOI] [PubMed] [Google Scholar]
- 46.Lu M, Echeverri F, Moyer BD. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic 4: 416–433, 2003. doi: 10.1034/j.1600-0854.2003.00097.x. [DOI] [PubMed] [Google Scholar]
- 47.Ma M, Shepherd GM. Functional mosaic organization of mouse olfactory receptor neurons. Proc Natl Acad Sci USA 97: 12869–12874, 2000. doi: 10.1073/pnas.220301797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macfarlane GT, Gibson GR, Beatty E, Cummings JH. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements. FEMS Microbiol Ecol 10: 81–88, 1992. doi: 10.1111/j.1574-6941.1992.tb00002.x. [DOI] [Google Scholar]
- 49.Malnic B, Godfrey PA, Buck LB. The human olfactory receptor gene family. Proc Natl Acad Sci USA 101: 2584–2589, 2004. [Erratum in Proc Natl Acad Sci USA 101: 7205, 2004.] doi: 10.1073/pnas.0307882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsunami H, Mainland JD, Dey S. Trafficking of mammalian chemosensory receptors by receptor-transporting proteins. Ann N Y Acad Sci 1170: 153–156, 2009. doi: 10.1111/j.1749-6632.2009.03888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McClintock TS, Sammeta N. Trafficking prerogatives of olfactory receptors. Neuroreport 14: 1547–1552, 2003. doi: 10.1097/00001756-200308260-00001. [DOI] [PubMed] [Google Scholar]
- 52.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci 5: 263–278, 2004. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 53.Müller A, Faubert P, Hagen M, Zu Castell W, Polle A, Schnitzler JP, Rosenkranz M. Volatile profiles of fungi—chemotyping of species and ecological functions. Fungal Genet Biol 54: 25–33, 2013. doi: 10.1016/j.fgb.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Nara K, Saraiva LR, Ye X, Buck LB. A large-scale analysis of odor coding in the olfactory epithelium. J Neurosci 31: 9179–9191, 2011. doi: 10.1523/JNEUROSCI.1282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niimura Y, Matsui A, Touhara K. Extreme expansion of the olfactory receptor gene repertoire in African elephants and evolutionary dynamics of orthologous gene groups in 13 placental mammals. Genome Res 24: 1485–1496, 2014. doi: 10.1101/gr.169532.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci USA 100: 12235–12240, 2003. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nishitsuji K, Xiao J, Nagatomo R, Umemoto H, Morimoto Y, Akatsu H, Inoue K, Tsuneyama K. Analysis of the gut microbiome and plasma short-chain fatty acid profiles in a spontaneous mouse model of metabolic syndrome. Sci Rep 7: 15876, 2017. doi: 10.1038/s41598-017-16189-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olender T, Lancet D, Nebert DW. Update on the olfactory receptor (OR) gene superfamily. Hum Genomics 3: 87–97, 2008. doi: 10.1186/1479-7364-3-1-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Padmanabhan S, Joe B. Towards precision medicine for hypertension: a review of genomic, epigenomic, and microbiomic effects on blood pressure in experimental rat models and humans. Physiol Rev 97: 1469–1528, 2017. doi: 10.1152/physrev.00035.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature 534: 213–217, 2016. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J, Brunet I, Wan LX, Rey F, Wang T, Firestein SJ, Yanagisawa M, Gordon JI, Eichmann A, Peti-Peterdi J, Caplan MJ. Olfactory receptor responding to gut microbiota-derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA 110: 4410–4415, 2013. doi: 10.1073/pnas.1215927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pluznick JL, Zou DJ, Zhang X, Yan Q, Rodriguez-Gil DJ, Eisner C, Wells E, Greer CA, Wang T, Firestein S, Schnermann J, Caplan MJ. Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci USA 106: 2059–2064, 2009. doi: 10.1073/pnas.0812859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Polizzi V, Adams A, Malysheva SV, De Saeger S, Van Peteghem C, Moretti A, Picco AM, De Kimpe N. Identification of volatile markers for indoor fungal growth and chemotaxonomic classification of Aspergillus species. Fungal Biol 116: 941–953, 2012. doi: 10.1016/j.funbio.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 64.Rajkumar P, Aisenberg WH, Acres OW, Protzko RJ, Pluznick JL. Identification and characterization of novel renal sensory receptors. PLoS One 9: e111053, 2014. doi: 10.1371/journal.pone.0111053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci Signal 2: ra9, 2009. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. RTP family members induce functional expression of mammalian odorant receptors. Cell 119: 679–691, 2004. doi: 10.1016/j.cell.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 67.Sharma R, Ishimaru Y, Davison I, Ikegami K, Chien MS, You H, Chi Q, Kubota M, Yohda M, Ehlers M, Matsunami H. Olfactory receptor accessory proteins play crucial roles in receptor function and gene choice. eLife 6: e21895, 2017. doi: 10.7554/eLife.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shepard BD, Cheval L, Peterlin Z, Firestein S, Koepsell H, Doucet A, Pluznick JL. A renal olfactory receptor aids in kidney glucose handling. Sci Rep 6: 35215, 2016. doi: 10.1038/srep35215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shepard BD, Natarajan N, Protzko RJ, Acres OW, Pluznick JL. A cleavable N-terminal signal peptide promotes widespread olfactory receptor surface expression in HEK293T cells. PLoS One 8: e68758, 2013. doi: 10.1371/journal.pone.0068758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spehr M, Gisselmann G, Poplawski A, Riffell JA, Wetzel CH, Zimmer RK, Hatt H. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science 299: 2054–2058, 2003. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- 71.Tahara Y, Yamazaki M, Sukigara H, Motohashi H, Sasaki H, Miyakawa H, Haraguchi A, Ikeda Y, Fukuda S, Shibata S. Gut microbiota-derived short chain fatty acids induce circadian clock entrainment in mouse peripheral tissue. Sci Rep 8: 1395, 2018. doi: 10.1038/s41598-018-19836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson EE, Haller G, Pinto JM, Sun Y, Zelano B, Jacob S, McClintock MK, Nicolae DL, Ober C. Sequence variations at the human leukocyte antigen-linked olfactory receptor cluster do not influence female preferences for male odors. Hum Immunol 71: 100–103, 2010. doi: 10.1016/j.humimm.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111, 2009. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578, 2012. [Erratum in Nat Protoc 9: 2513, 2014.] doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.von der Weid B, Rossier D, Lindup M, Tuberosa J, Widmer A, Col JD, Kan C, Carleton A, Rodriguez I. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat Neurosci 18: 1455–1463, 2015. doi: 10.1038/nn.4100. [DOI] [PubMed] [Google Scholar]
- 76.Wålinder R, Ernstgård L, Norbäck D, Wieslander G, Johanson G. Acute effects of 1-octen-3-ol, a microbial volatile organic compound (MVOC)−an experimental study. Toxicol Lett 181: 141–147, 2008. doi: 10.1016/j.toxlet.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 77.Wang YH, Zhang YG. Kidney and innate immunity. Immunol Lett 183: 73–78, 2017. doi: 10.1016/j.imlet.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 78.Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C, Karu N, Sayeeda Z, Lo E, Assempour N, Berjanskii M, Singhal S, Arndt D, Liang Y, Badran H, Grant J, Serra-Cayuela A, Liu Y, Mandal R, Neveu V, Pon A, Knox C, Wilson M, Manach C, Scalbert A. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res 46: D608–D617, 2018. doi: 10.1093/nar/gkx1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. HMDB 3.0−the Human Metabolome Database in 2013. Nucleic Acids Res 41: D801–D807, 2013. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. HMDB: the Human Metabolome Database. Nucleic Acids Res 35: D521–D526, 2007. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40: 235–243, 2006. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 82.Wu C, Hwang SH, Jia Y, Choi J, Kim YJ, Choi D, Pathiraja D, Choi IG, Koo SH, Lee SJ. Olfactory receptor 544 reduces adiposity by steering fuel preference toward fats. J Clin Invest 127: 4118–4123, 2017. doi: 10.1172/JCI89344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu Y, de March CA, Ni MJ, Adipietro KA, Golebiowski J, Matsunami H, Ma M. Responsiveness of G protein-coupled odorant receptors is partially attributed to the activation mechanism. Proc Natl Acad Sci USA 112: 14966–14971, 2015. doi: 10.1073/pnas.1517510112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci 5: 124–133, 2002. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 85.Zhuang H, Matsunami H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nat Protoc 3: 1402–1413, 2008. doi: 10.1038/nprot.2008.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhuang H, Matsunami H. Synergism of accessory factors in functional expression of mammalian odorant receptors. J Biol Chem 282: 15284–15293, 2007. doi: 10.1074/jbc.M700386200. [DOI] [PubMed] [Google Scholar]