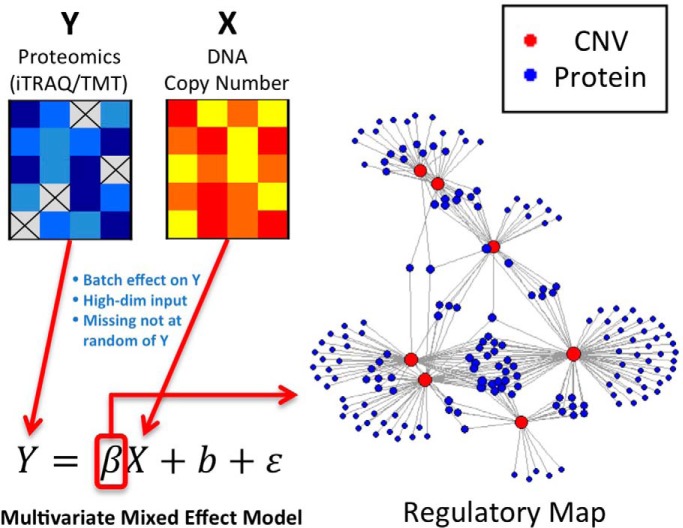
We propose a new method, ProMAP, to characterize regulatory relationships between somatic CNAs and proteins based on proteogenomic data. ProMAP employs a panelized multivariate regression framework to handle high-dimensionality of proteogenomic data and uses proper statistical models to handle multiplex structure and abundance-dependent-missing-data mechanism in proteomics data. Applying ProMAP to breast and ovarian cancer TCGA-CPTAC data sets, we identified novel CNAs associated with many protein/phosphoprotein abundances. The findings help to characterize important CNAs and their functional impact in these tumors.
Keywords: Proteogenomics, phosphoproteome, breast cancer, statistics, mass spectrometry, ovarian cancer, cis-regulation, CNA-protein/phosphosite regulatory map, CNAI, penalized mixed effect model, trans protein/phosphosite hubs
Graphical Abstract
Highlights
ProMAP builds high dimensional regulatory network based on proteogenomic data.
ProMAP properly models non-random missingness in iTRAQ/TMT proteomics data.
Construct CNA-protein/phosphoprotein regulatory map of breast and ovarian cancer.
Detect important breast and ovarian CNA that have ample functional consequences.
Abstract
Recent development in high throughput proteomics and genomics profiling enable one to study regulations of genome alterations on protein activities in a systematic manner. In this article, we propose a new statistical method, ProMAP, to systematically characterize the regulatory relationships between proteins and DNA copy number alterations (CNA) in breast and ovarian tumors based on proteogenomic data from the CPTAC-TCGA studies. Because of the dynamic nature of mass spectrometry instruments, proteomics data from labeled mass spectrometry experiments usually have non-ignorable batch effects. Moreover, mass spectrometry based proteomic data often possesses high percentages of missing values and non-ignorable missing-data patterns. Thus, we use a linear mixed effects model to account for the batch structure and explicitly incorporate the abundance-dependent-missing-data mechanism of proteomic data in ProMAP. In addition, we employ a multivariate regression framework to characterize the multiple-to-multiple regulatory relationships between CNA and proteins. Further, we use proper statistical regularization to facilitate the detection of master genetic regulators, which affect the activities of many proteins and often play important roles in genetic regulatory networks. Improved performance of ProMAP over existing methods were illustrated through extensive simulation studies and real data examples. Applying ProMAP to the CPTAC-TCGA breast and ovarian cancer data sets, we identified many genome regions, including a few novel ones, whose CNA were associated with protein and or phosphoprotein abundances. For example, in breast tumors, a small region in 8p11.21 was recognized as the second biggest hub in the CNA-phosphoprotein regulatory map, and further investigation of the regulatory targets suggests the potential role of 8p11.21 CNA in perturbing oxygen binding and transport activities in tumor cells. This and other findings from our analyses help to characterize the impacts of CNAs on protein activity landscapes and cast light on the genetic regulation mechanisms underlying these tumors.
To improve our ability to diagnose, treat and prevent cancer, it is of great interest to systematically identify proteins perturbed by alterations in cancer genomes. Toward this goal, the Clinical Proteomic Tumor Analysis Consortium (CPTAC)1 of National Cancer Institute (NCI) has undertaken global proteome and phosphoproteome profiling (1, 2) of breast and ovarian cancer samples that have been extensively characterized in the The Cancer Genome Atlas (TCGA) (3, 4). Then, by leveraging genomic analytical outputs from TCGA on these samples (3, 4), we have the unique opportunity to characterize the relationships among protein changes and genomic alterations in a systematic manner. Such integrative proteo-genomic analysis will provide insights into breast cancer etiology and help to identify biomarkers for cancer diagnosis, prevention, and treatment.
Specifically, one question we aim to address in the CPTAC-TCGA study is how the global- and phospho-proteomics profiles were shaped by upstream DNA copy number alterations (CNA) in tumor samples. This question is of interest because these two types of data provide complementary information in characterizing the disease system. It has been shown that CNAs play crucial roles in activating cancer genes and inactivating tumor suppressor genes in breast and ovarian tumors (5, 3, 4). But CNAs observed in tumor samples contains both primary changes driving cancer and passenger changes having no or limited functional consequences. On the other hand, protein profiles characterize proteins' functional activities that directly shape cells' phenotypes. Therefore, integrating CNA data and protein data helps to reveal more subtle yet biologically important genetic regulatory relationships in cancer cells.
To systematically construct a regulatory map between a large number of proteins and CNAs, we face three major challenges: first, this involves assessment of millions of potential regulatory relationships based on data from less than a hundred of tumor samples; second, CNAs profiles have strong spatial correlation; third, protein profiles from mass spectrometry based experiments have unique data characteristics and structures that need to be accounted for in the analysis.
Traditional statistical methods, such as pairwise correlation test or univariate linear regressions, are unlikely to produce satisfactory results for this task, as such methods often lead to high variability and a large number of detected false positives when data is in the high-dimension-low-sample-size regime and with complicated correlation structures (as illustrated through simulation experiments in this article). In the past decade, many penalized multivariate models have been proposed to tackle this problem. Specifically, the regulatory map between a set of CNAs and a set of proteins can be characterized through a penalized multivariate regression model in which protein expressions are responses and CNA levels are predictors. Then, proper statistical penalty can be imposed on the model to handle the high dimensionality through constraining the total number of non-zero parameters in the model (6–10). Specifically, Peng et al. (2010) (11) developed remMap, a penalized multivariate regression model for integrative genomic studies with a focus on identifying master predictors in the regulatory network. In a related work (12), the authors further extended remMap to map expression quantitative trait loci (eQTLs) by incorporating the grouping structure among predictors.
Although the progress in statistical methodology development for integrative genomic analysis is encouraging, these methods are not designed for proteomics data in mind and thus do not account for the unique data structure of proteomics data from mass spectrometry based experiments. In the CPTAC-TCGA breast cancer and ovarian cancer studies, protein profiling was carried out by iTRAQ (isobaric tag for relative and absolute quantitation) LC-MS/MS (liquid chromatography tandem mass spectrometry) experiments. The popularity of labeled proteomics experiment is largely because of its enhancement of the throughput: multiple samples can be processed simultaneously in one iTRAQ or TMT multiplex experiment. However, because of the dynamic nature of MS instruments, the technical variation across different multiplex experiments is generally large. To alleviate this problem, a common practice is to include a common reference sample in each multiplex experiment. For example, the CPTAC breast cancer study used 4-plex iTRAQ experiments in which each multiplex iTRAQ experiment contained 3 breast tumor samples and a common reference sample. The reference sample was created by combining 40 tumor samples with equal representation of the breast cancer subtypes in the CPTAC breast cancer study. Conventional data analyses are usually performed based on the relative abundances of proteins/peptides in the target samples relative to the reference sample (13, 14). However, as we pointed out in another work (15), it is not efficient to purely rely on the relative abundances when analyzing data from labeled MS experiments, because the target samples and the reference sample in one multiplex run usually are subject to slightly different experimental variation because of the complicated process of iTRAQ experiments (14) (e.g. the experimental noises at the quantification of the MS2 step). Thus, in Chen et al. (2017) (15), we proposed to use a mixed effects model to jointly model the absolute protein abundances in both the target and reference samples. Such a strategy enables one to characterize individual-level and experimental-level technical noises separately, and thus to achieve better power in statistical inferences. Moreover, another unique challenge in analyzing proteomics data is the substantial amount of abundance-dependent missing values (16, 17). That is, the lower the abundance of a protein is, the more likely the protein is missing in the output data (supplemental Fig. S1). For iTRAQ-MS experiments, because all the samples in a multiplex experiment are processed together, a given protein is either detected or missing simultaneously in all samples, with the missing probability depending, in large part, on the precursor ion abundances in MS1. Previously, we proposed an exponential function-based probability model to characterize such abundance dependent missing mechanism in labeled MS experiments (17, 15). In this article, we will incorporate these newly developed techniques in the framework of proteogenomic integration analysis.
Specifically, we propose a new method ProMAP—a penalized multivariate linear mixed effects model to directly model the proteins/phosphosites abundances and their variance structures considering multiplex design when performing proteogenomic integrative analysis. In addition, to account for the non-ignorable missing data, we also explicitly model the non-ignorable missing mechanism in the proposed model. Moreover, we use proper penalties to control the sparsity of the multivariate linear mixed effects model and to facilitate the detection of master regulators (11), i.e. CNAs that impact the activities of a large number of proteins phosphoproteins. We demonstrate the advantage of ProMAP over a few competing methods through extensive simulation experiments. In the end, we apply ProMAP on the CPTAC-TCGA breast cancer and ovarian cancer data sets (1, 2) and identify novel regulatory relationships between DNA copy number alterations and the protein expression profiles in both cancer types. We believe the proposed method will greatly enhance the power and accuracy of proteogenomic integrative analysis in a broad range of applications.
EXPERIMENTAL PROCEDURES
Data Description and Preprocessing
In the recent breast and ovarian cancer studies conducted by NCI CPTAC (1, 2), sophisticated quantitative mass spectrometry-based global- and phospho-proteomic analyses were carried out for two groups of breast cancer and ovarian cancer samples that have been genomically annotated by TCGA consortium (3, 4). One key interest in these studies is to characterize how protein activities are regulated by genetic factors in breast and ovarian cancer. Because DNA copy number alterations are key players in tumor initiation and development of both cancer types, we focus on the regulations between CNAs and proteins, and apply ProMAP to the CPTAC global/phospho proteomics data as well as the TCGA CNA data to construct CNA-protein/phosphoprotein regulatory maps.
Proteomics data in both studies were generated using iTRAQ platform, with each multiplex consisting of three tumor samples and one common reference sample. These analyses achieved a substantially greater depth of coverage than previously observed for studies of similar scale. The intensity data of proteins and phosphosites were downloaded from the CPTAC Data portal (https://proteomics.cancer.gov/data-portal) sponsored by the National Cancer Institute. And level-three segmented DNA copy number profiles data of the tumor samples was obtained from the TCGA-GDC web site (https://portal.gdc.cancer.gov/).
Breast Cancer Proteome and Phosphoproteome Data
In the CPTAC breast cancer study (1), 36 iTRAQ LC-MS/MS experiments were employed to quantify protein and phosphosite levels across 108 tumor samples from 105 breast cancer patients. In our analysis, we focus on 77 out of the 108 tumor samples that were suggested to have good quality (e.g. high tumor purity) by CPTAC breast cancer team (1). The intensity data sets downloaded from CPTAC-DCC consist of 15,369 proteins and 62,679 phosphosites. For both the global- and phopsho-proteomics data sets, we first derive protein/phosphosite intensity measurements using both MS1 parent ion peak heights and MS2 spectrums ratios from the output of Spectrum Mill MS Proteomics Workbench (1). We then take log transformation of the intensity measurements and normalize each sample to have the same median and MAD (median absolute deviation).
Although there is no technique difficulty to apply ProMap on the whole proteomic data sets, because of the limited sample sizes in these studies, it is necessary to constrain the dimension of the feature space to avoid the curse of high-dimensionality when deriving regulatory maps (the more features, the lower the power will be). Thus, we chose to focus on the features with (1) small and moderate missing rates, as well as (2) large variation across samples. The reason to use (1) is that information for features with high missing rates is usually very limited, which leads to poor power to test for their associations with CNAs. Because in iTRAQ data missing usually happen at the multiplex level (i.e. one feature is either observed or missing in all samples of one iTRAQ multiplex), we use the missing rate of the reference channels as surrogates of technique missing rates for each feature. At the same time, missing rate of reference channel also indicated the reproducibility of the protein identification, because all reference channels are replicates. The reason to use (2) is that one would not expect features not varying across samples to show strong association with CNAs. Specifically, we set thresholds on IQL (inter-quantile length) such that the numbers of the selected features are about the same across different data sets, so the comparisons of resulting regulatory maps can be more interpretableIn practice, we further filter the proteins/phosphosites that was observed in less than 25 (70%) of the 36 reference runs. We choose to focus on features with large variability across samples and select the top 5009 proteins and top 5015 phosphosites with their inter-quartile lengths across 77 samples greater than 0.5 and 0.98 respectively.
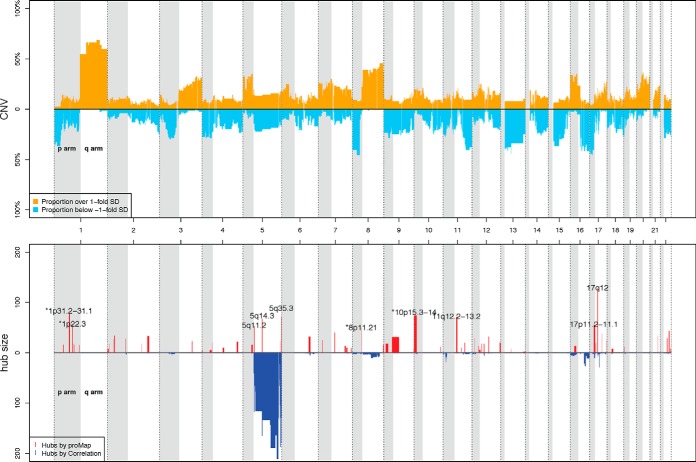
For the preprocessing of the segmented DNA copy number data, We first break the genome using the union of the break-points detected in all tumor samples and filter the small regions with less than 10k base pairs. Then for each region of each sample, we record its copy number based on the inferred DNA copy number of the corresponding segment in the sample, with tail values truncated at ±1.5. Because of the high spatial correlation in DNA copy number profiles, we further condense these regions into 1730 copy number alteration intervals (CNAI) by applying fixed order clustering (FOC) (18), so that DNAs in the same interval tend to have similar CNA patterns in each sample. The copy number of one CNAI in a given sample is then calculated as the mean of the copy number of all regions within the interval in that sample. We exclude CNAI with no variation across the 77 samples, which results in 1184 CNAIs. The amplification/deletion frequencies of these CNAIs among the 77 breast cancer samples is illustrated in Fig. 2.
Fig. 2.
Amplification and deletion frequencies of CNAIs (the upper panel) and CNAI regulatory hubs in CNAI-protein regulatory maps derived from ProMAP and pairwise correlation test (the bottom panel) in breast cancer application. In the upper panel, each CNAI is represented by a pair of yellow and blue bars. The height of the yellow (blue) bar of one CNAI indicates the percentage of samples having CNA values above one standard deviation (below negative one standard deviation) of the whole data set. In the bottom panel, genome location of CNAI regulatory hubs identified by ProMAP and pairwise correlation test are illustrated by red and blue bars respectively. The height of the red (blue) bar of one CNAI represents the number of proteins affected by DNA copy number changes of this CNAI in the corresponding CNAI-protein regulatory maps. Cytoband locations are annotated for the top 10 CNAI hubs regulating the largest number of proteins in ProMAP network.
Ovarian Cancer Proteome Data
In CPTAC-TCGA ovarian cancer study (2), Global proteomics iTRAQ LC-MS/MS (4-plex) experiments were employed to analyze 174 ovarian tumors and 10 quality control samples, with 32 tumor samples having two replicate measurements. The protein abundance data files downloaded from CPTAC-DCC were subjected to the same normalization and preprocessing procedures as described above, resulting in a protein abundance table of 4969 unique proteins.
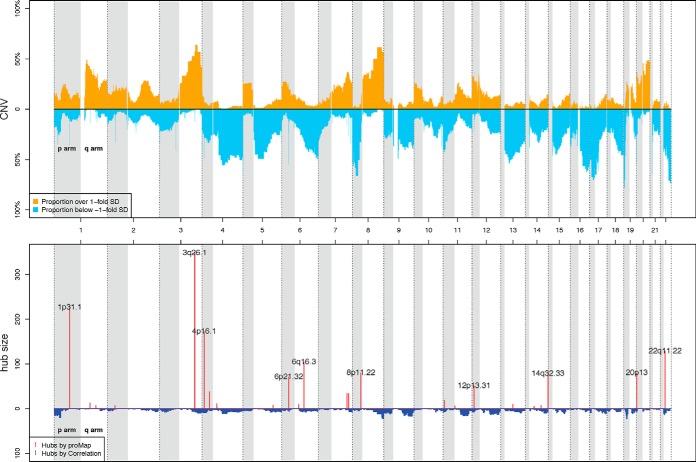
For 169 of the 174 ovarian tumors in the protein data set, level-three segmented DNA copy number profiles were also available from the TCGA website. Following the same FOC analysis as outlined in the breast cancer data analysis, we derived 1351 CNAIs whose amplification/deletion frequencies across the 169 ovarian cancer samples is illustrated in Fig. 7.
Fig. 7.
Amplification and deletion frequencies of CNAIs (the upper panel) and CNAI regulatory hubs in CNAI-protein regulatory maps derived by ProMAP and pairwise correlation test (the bottom panel) in ovarian cancer application. In the upper panel, each CNAI is represented by a pair of yellow and blue bars. The height of the yellow (blue) bar of one CNAI indicates the percentage of samples having CNA values above one standard deviation (below negative one standard deviation) of the whole data set. In the bottom panel, genome location of CNAI regulatory hubs identified by ProMAP and pairwise correlation test are illustrated by red and blue bars respectively. The height of the red (blue) bar of one CNAI represents the number of proteins affected by DNA copy number changes of this CNAI in the corresponding CNAI-protein regulatory maps. Cytoband locations are annotated for the top 10 CNAI hubs regulating the largest number of proteins in ProMAP network.
Note, because phosphoproteomics experiments were only carried out for a subset of tumors in the CPTAC-TCGA ovarian project, we did not include phosphoproteomics data in our analysis and only focused on constructing the CNA-protein regulatory map of ovarian cancer based on data of 169 tumors in this manuscript.
ProMAP
Mixed Effect Model with Abundance Dependent Missing Mechanism
To characterize the dependence of protein activities on CNAs in a systematic manner, a straightforward way is to use a multivariate regression framework, treating protein abundances from iTRAQ experiments as responses and CNA levels as predictors. In addition, because the reference and target samples in one iTRAQ experiment are processed and measured in the same environment, they are exposed to the same experimental noises. This motivates us to further include random effects in the multivariate regression framework to account for the multiplex structure of iTRAQ data.
Suppose we are interested in constructing a regulatory map between L proteins (or phosphosites) and P CNAIs. Let {yli = yl1i, yl2i, L, ylKii}T denote the abundance measurements for the lth (l = 1, …, L) protein of all Ki samples in the ith iTRAQ experiment. Specifically, yl1i represents the protein abundance in the reference sample; and yl2i, L, ylKii represents the protein abundances in the target samples of the ith experiment. Like Chen et al. (2017) (15), we consider the following multivariate mixed effects model
| (1) |
where Wi=(1, 0, L, 0) is an indicator vector for reference samples; Xi = {xpki}p×ki represents the DNA copy number measurements of P genome regions in these samples; βl0, βl1 and αl = {αpl}p×1 are coefficients in the model; bli is a random variable representing the technical noise in the ith iTRAQ experiment; and eli = {elKi}ki×1 is the error term. Here the parameters-of-interest is αl. Specifically, a non-zero αpl suggests that the lth protein is regulated by the copy number changes of the pth CNA. Note, we set xp1i = 0, p = 1, L, P, for the reference samples, so they do not contribute to the estimation of α given all other parameters and random effects in the model.
Because the absolute abundances of the proteins and peptides are modeled in Eq. (1), which is necessary for one to account for the abundance dependent missing mechanisms in iTRAQ/TMT data, βl0 is included to account for different overall mean value of different proteins; βl1 is included to allow proteins to have different mean values in reference samples than in targeted samples (e.g. when reference samples were the pooling of only a subset of tumor samples);bli is a random effect variable accounting for the experimental noise in the ith iTRAQ multiplex for the lth protein. Specifically, bli is introduced to account for the fact that the same peptide precursor in different multiplexing experiments may be isolated at different point of its elution profile and thus was subjected to different technique noises in their quantification measurements across different iTRAQ multiplex. We also provided a comparison of the data density before and after the filtering (supplemental Fig. S2). The distribution of both protein and phosphosites did not change after the filtering.
We assume bli is normally distributed with mean 0 and variance Dl; and eli = {elki }Ki×1 ∼ N(0, Ri) with Ri being a Ki by Ki positive definite diagonal matrix. Specifically, we assume the variances of the error terms are different between reference samples and target samples and let diag(Ri) = (s02, s2, L, s2).
For the common reference samples, s02 was to capture additional technical variations that was not shared across different iTRAQ channels (e.g. those introduced in the MS2 step) and thus was not captured by bli. For targeted samples, σ2 represents not only additional channel specific technical variation but also biological variation that was not explained by CNV across different tumor samples. Thus, we expect s02 to be considerably smaller than σ2.
In iTRAQ LC-MS/MS experiments, whether a peptide can be detected and quantified largely depends on its parent ion abundances in MS1 scans, which reflects the combined abundances of this peptide in all samples of one iTRAQ multiplex. The larger the parent ion abundance in MS1 is, the more likely the peptide is selected to be further examined in MS2 scans. However, if the parent ion fails to be selected for MS2 step, we observe missing values for this peptide in all the samples of the iTRAQ multiplex. In other words, missing events in iTRAQ LC-MS/MS data always happen at the experiment level and the missing probability of one peptide in a multiplex experiment depends on the total abundances of this peptide in all samples of this experiment. To account for this abundance dependent missing mechanism, we follow Chen et al. (2017) (15) and employ a probability model to characterize the missing pattern. Specifically, we denote Mli as the missing event indicator variable, which takes the value of 1 if the abundances of the lth protein feature are missing in the ith experiment; and the value of 0 otherwise. We model the missing probability with an exponential function:
| (2) |
where γ0 and γ are non-negative parameters to describe the missing mechanism. Specifically, γ is a non-negative parameter characterizing the dependence of protein missing probabilities on protein abundances (supplemental Fig. S1). The larger γ is, the bigger the differences between the missing rates of low abundance proteins and that of high abundance proteins. γ = 0 means the probability of missing does not change with protein abundances. γ0 is a nuisance parameter in the exponential probability model and shall take non-negative value when γ = 0.
And 1Ki is a Ki′ 1 vector with all entries being 1. Here, 1KiT yli gives the total abundance of this protein feature in all samples of the ith experiment.
Maximum Penalized Likelihood Estimates
We denote the observed, missing, and complete protein abundance data as Yobs, Ymis, and Y = (Yobs, Ymis) respectively. As mentioned earlier, because missing is not at random in iTRAQ LS-MS/MS data, we need to jointly model the missing patterns M = {Mli} and the observed protein abundance data Yobs in order to perform proper statistical estimation and inference. Denoting Ω = (α, β, D, R), the joint likelihood of M and Yobs can be written as
The conditional probability L(M|Y) is given in (2). In addition, according to model (1), we have
| (3) |
where Σli = Dl′ IKi + Ri and IKi is an identity matrix of dimension Ki. Thus, the likelihood of the complete data L(Y;Ω) can be written out accordingly.
With the joint likelihood being specified, we can estimate the parameters of interest, α, by maximizing the above joint likelihood function. However, because of the high dimensionality of α, maximum likelihood estimate is ill defined. Thus, we propose to add proper penalty terms to the likelihood and calculate MPLE (Maximum penalized likelihood estimate). Specifically, it is reasonable to assume: (1) each protein is regulated by a limited number of CNAs; (2) most CNAs regulate a few (or zero) proteins, whereas some CNAs regulate many proteins. The latter are referred to as master predictors. This motivates us to control the overall sparsity of α, i.e. the total number of non-zero elements in the matrix; while at the same time, encourage the detection of master predictors. In addition, because experimental variations on different proteins are different, the model involves L protein specific standard deviation parameters {Dl}l = 1L for random effects. And because L is generally a big number, it's also important to impose regularization on the high dimension vector {Dl}l = 1L. Thus, we introduce the following penalized likelihood function:
| (4) |
and seek for MPLE
| (5) |
where λ1, λ2, ν1 and ν2 are tuning parameters taking non-negative values; ‖α‖1 is the L1 norm of α; and ‖αp*‖2 is the L2 norm of the pth row of α. In (4), the L1 norm penalty helps to control the sparsity of α. The L2 norm further imposes row sparsity on α, which facilitates the selection of master predictors, i.e. predictors influencing a lot of responses are more likely to be selected in the model than other predictors only influencing a few responses (11). The values of λ1 and λ2 control the level of sparsity of the model.
The remaining penalty terms, n1ål=1L Tr(Dl−1) + n2ål=1L log |Dl|, are amounting to one-dimensional inverse-Wishart priors for {Dl}. They help to bound both {Dl} and {Dl−1} away from 0, and thus increase the stability of these estimates (17). In addition, because the inverse-Wishart distribution is the conjugate prior for the covariance of Gaussian distribution, it is computationally efficient to calculate the MPLE for {Dl}.
We implement an Expectation Conditional-Maximization algorithm to calculate the MPLE. Please see Supplement B for details.
We employ cross validation-based grid search to select optimal tuning parameters. Specifically, when applying ProMAP to the breast cancer TCGA-CPTAC data, we set ν1 = 0.01 and ν2 = 1 based on simulation experiments, and we selected (λ1, λ2) through a 4-fold cross validation. The optimal tuning parameters are (λ1, λ2) = (50, 50) for CNAI-protein analysis, and (λ1, λ2) = (26, 35) for CNAI-phosphosite analysis. After optimal tuning parameters were selected, we applied the majority vote procedure to aggregate the estimation results from different cross validation folds corresponding to the optimal tuning parameters. Please see Supplement C and D for details.
Simulation Experiments
Simulation experiments are conducted to evaluate and compare the performance of the ProMAP to those of two existing methods.
Synthetic Data Sets
The synthetic data sets are generated as follows. Given the dimension of protein features and CNAI (P, L), we first generate a L × P adjacency matrix A by assigning 1 or 0 elements. To mimic the real regulatory network, in all simulation settings we assume only a small subset of CNA regions is associated with the protein features. Among this subset, some CNA regions are associated with many protein features, whereas others may only have a few associations.
Once the adjacency matrix is determined, we simulate the regression coefficients α by setting αpl = 0, if Ap,l; or sampling αp,l from a Uniform distribution, if Ap,l = 1. To be comparable to the real data set, we set the number of samples in one iTRAQ experiment, K, to be 4, and the number of experiments, N, to be 50. For the design matrix X, the first predictor {x1ki}k,i takes value of 1, representing the intercept. The second predictor {x2ki}k,i is an indicator of the reference sample with x21i = 1 and x2ki = 0 for k ≠ 1. CNA predictors are generated from a normal distribution: (x3ki,,xPki ≠ N(0,Σx), where ΣX is the covariance matrix with the (p,p′) element being ρX|p−p′|. Among CNA predictors, elements corresponding to the reference samples are set to 0s. We then generate the protein features based on the linear mixed model (1), with random effects bli ∼ N(0,d) and residuals eli ∼ N(0,R), where R is a diagonal matrix with diagonal elements being (s02, s2, s2, s2). In the end, we generate the missing data following the missing mechanism in (2). We consider three settings with different values of p, l, and ρ.
Simulation 1. Low Dimension In this scenario we set p = 80 and L = 100. When we generate the regulatory map, we simulate 2 large hubs with size 50, 4 median hubs with size 25, 10 small hubs with size 10 and 10 predictor associated with only one of the proteins, hence 310 edges in total. The coefficients at large and median hubs are generated from uniform distributions U(0.3,0.5), and the other coefficients are generated from U(0.5,0.8); We set the parameters in the covariance matrix of design matrix, the variance of the error term and the random effect as: ρx = 0.25, s02 = 0.3, s2 = 1, and d = 0.5. These parameter values result in a signal-to-noise ratio (SNR) of 0.5. In the end, we set γ0 = 0, γ = 0.2, and the overall missing rate is around 24%.
Simulation 2. High Dimension In this scenario, we let p = 1000 and L = 1200. For the regulatory map, we set 4 large hubs with size 50, 4 median hubs with size 25, 20 small hubs with size 10, and 100 predictors associated with only one of the proteins, hence 600 edges in total. The coefficients and covariance parameters are set to be the same as those in the previous simulation setting. The resulting SNR is 0.25. In addition, with the same value of γ0 = 0, γ = 0.2, the overall missing rate under this setting is around 14%.
Simulation 3. High Dimension with High Correlation We consider the third setting where the correlation of the CNA matrix is high, as the CNA profiles usually have strong spatial correlation along the genome. Specifically, the average correlation between the DNA copy numbers of adjacent CNA regions is 0.8 in the TCGA breast cancer data set. Thus, we set ρx = 0.8, while keep the other parameters the same as those in Simulation II.
Comparison with Other Methods
We compare ProMAP with two existing methods: the remMap method (11) and the pairwise correlation test-based method (19, 2, 1). Different from ProMAP which is applied to the abundance measures in the targeted and reference samples, the remMap and pairwise correlation test based method are applied to the relative abundances of proteins/peptides in the target samples relative to the reference samples, as these two methods do not handle experimental variation directly. All three methods are applied to both the complete data sets and the data sets with missingness under each simulation setting. In real data application of breast cancer and ovarian cancer study, ProMAP is applied on the observed MS2 proportional MS1 intensity data set with missingness, and pairwise correlation and remMap are applied on the imputed data with KNN imputation algorithm (20) on the sample to reference ration of the same MS1 intensities.
ProMAP When applying the ProMAP method, we first estimate the missing mechanism parameter Γ following the procedure described in the previous section (we set γ = 0 when analyzing the complete data), and fix the tuning parameter ν1 = 0.01, ν2 = 1. We then employ the 5-fold cross validation to select the tuning parameters (λ1, λ2). At each searching grid point of (λ1, λ2), a summarized adjacency matrix is obtained by the majority voted procedure. Compared with the true coefficients matrix α, we calculate the power and the false discovery rate (FDR) of the estimated adjacency matrix.
remMap We assume the simulated data correspond to the log scale of the raw abundance measurements. So, the relative abundances are calculated as the differences between the abundances in the targeted samples and that of the corresponding reference samples. Because the remMap method do not handle missing values, we perform imputation using a nearest neighbor averaging-based algorithm provided in the R package “pamr” (21, 20). Like ProMAP, we employ a 5-fold CV to select the tuning parameters (λ1,λ2) and use the majority voted procedure to summarize the adjacency matrix.
Pairwise Correlation Test It is common to use marginal pairwise correlation test to determine regulatory relationships in proteogenomic integrative analysis (19, 1). Specifically, based on relative abundances, we calculate the p values for testing non-zero correlations for each protein-CNA pair using either the complete data or the pairwise observed data. We then calculate the FDRs (q-value) using the Benjamini-Hochberg procedure. In the end, we set a cutoff value (0.1) to determine the significance of entries on the adjacency matrix: those entries with q-values less than the cutoff are assigned as 1 and others are assigned as 0.
RESULTS
Simulation Results
We evaluate the performance of three methods, ProMAP, remMap (11) and the pairwise correlation test (19, 1) based on 20 independent data sets under each simulation setting.
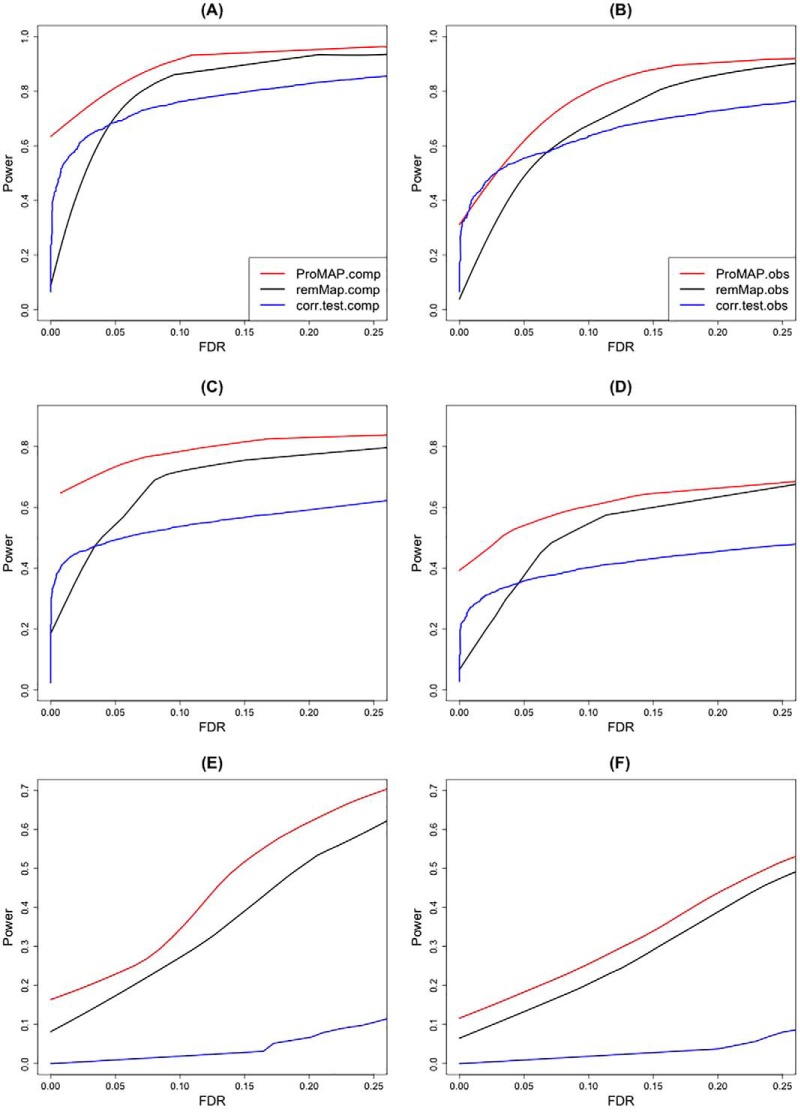
First, for each simulation setting, ROC curves of ProMAP and remMap are derived from the adjacency matrix estimated with the optimal selected λ2, and a variety of λ1. Those of pairwise correlation tests are obtained from calculating power and FDR of the estimated adjacency matrices at different cutoff values. Resulting ROC curves are displayed in Fig. 1. The three plots in the left column are for the results based on the complete data, whereas the ones in the right column are based on the observed data. It is clear from the figure that the performance of all models deteriorates when the number of genes and correlation among CNAs increase, but the ROC curve of ProMAP is always dominantly better than the other two methods. Specifically, for Simulation I ((a) and (b)), ProMAP performs better than remMap and pairwise correlation test, whereas remMap and the correlation test are comparable; for Simulation II ((c) and (d)), remMap becomes slightly better than the pairwise correlation test but not significantly better; whereas for Simulation III ((c) and (d)) with high dimension and high correlation among CNAs, ProMAP and remMap perform much better than pairwise correlation test.
Fig. 1.
Comparison of ROC curve of all models in the three simulation settings: panel (A) and (B) illustrate the results in Simulation I; (C) and (D) illustrate the results in Simulation II; and (E) and (F) illustrate results in Simulation III. ProMAP.comp, remMap.comp, corr.test.comp are the results obtained based on the complete data, and ProMAP.obs, remMap.obs, corr.test.obs are the results obtained based on the observed data.
In addition, performance of all the models with selected tuning parameters/cutoffs in all the three simulation settings is summarized in Table I. For ProMAP and remMap methods, optimal tuning parameters were determined through cross validation. For pairwise correlation test, we set a cutoff on adjusted p value at 0.1. Numbers of false negative, false positive, total false and false discovery rate of the network edge detection are averaged from 20 replicates. Consistent to the trend we observed in the ROC curve, ProMAP always performs better than the other two methods in all the simulation settings in terms of TF (total false count), which is the sum of FN (false negative count) and FP (false positive count). Specifically, in Simulation I and II with low correlation of design matrix, all methods have a good control of FDR, but ProMAP tends to have better power than the other two methods. On the other hand, when CNA profiles have high spatial correlation (Simulation III), pairwise correlation test has extremely high FDR. Although the FDR of ProMAP and remMap are comparable and much lower than that of pairwise correlation test, ProMAP achieves better power than remMap.
Table I. Comparison of Network estimation of the optimal models in all the three simulations.
| Models | FN | FP | TF | FDR |
|---|---|---|---|---|
| Simulation I | ||||
| ProMAP.comp | 43.8 (14.32) | 16.4 (6.72) | 60.2 (12.7) | 0.06 (0.02) |
| remMap.comp | 75.75 (17.76) | 13.55 (5.24) | 89.3 (16.81) | 0.05 (0.02) |
| corr.test.comp | 87.65 (21.81) | 15.75 (6.18) | 103.4 (19.28) | 0.07 (0.02) |
| ProMAP.obs | 65.55 (18.31) | 28.85 (16.5) | 94.4 (17.51) | 0.1 (0.05) |
| remMap.obs | 43.3 (12.57) | 68.15 (14.87) | 111.45 (13.34) | 0.2 (0.03) |
| corr.test.obs | 95.2 (20.37) | 38.35 (10.47) | 133.55 (15.02) | 0.15 (0.03) |
| Simulation II | ||||
| ProMAP.comp | 166.55 (23.03) | 17.3 (5.28) | 183.85 (20.61) | 0.04 (0.01) |
| remMap.comp | 184.3 (26.42) | 42.25 (10.67) | 226.55 (26.47) | 0.09 (0.02) |
| corr.test.comp | 356.4 (13.84) | 2.3 (1.75) | 358.7 (13.55) | 0.01 (0.01) |
| ProMAP.obs | 219.15 (24.57) | 56.6 (9.08) | 275.75 (22.13) | 0.13 (0.02) |
| remMap.obs | 214.5 (24.36) | 80.4 (13.53) | 294.9 (22.08) | 0.17 (0.02) |
| corr.test.obs | 356.25 (12.85) | 29.3 (7.05) | 385.55 (11.66) | 0.11 (0.02) |
| Simulation III | ||||
| ProMAP.comp | 226.3 (21.05) | 83.5 (14.68) | 309.8 (28.37) | 0.18 (0.03) |
| remMap.comp | 265.3 (25.03) | 82.65 (17.32) | 347.95 (31.7) | 0.2 (0.04) |
| corr.test.comp | 303.95 (12.98) | 489.1 (35.96) | 793.05 (31.59) | 0.62 (0.02) |
| ProMAP.obs | 295.25 (21.69) | 84.15 (22.19) | 379.4 (28.36) | 0.21 (0.04) |
| remMap.obs | 313.85 (23.67) | 76.85 (14.85) | 390.7 (30.07) | 0.21 (0.04) |
| corr.test.obs | 304.8 (13.08) | 639.6 (46.69) | 944.4 (40.13) | 0.68 (0.01) |
Network topology: p = 80, L = 100 for Simulation I with 310 edges; p = 1000, L = 1200 for Simulation II & III with 600 edges. FP: false positive; FN: false negative; TF: total false; FDR: false discovery rate. Numbers in parentheses are standard deviations over 20 replicates.
CNAI-Protein/Phosphoprotein Regulatory Maps in Breast Cancer
We applied ProMAP to the TCGA-CPTAC breast cancer data sets (see Section on Data Description and Preprocessing) and constructed two regulatory maps: one for CNAIs and proteins; and the other for CNAIs and phosphosites. We obtain 1554 and 1988 edges corresponding to 61 and 39 CNAIs for the CNAI-protein and CNAI-phosphosite regulatory maps respectively. A selected part of the inferred CNAI-protein/phosphosite regulatory map is illustrated in Fig. 3, in which CNAIs regulating more than 50 genes or cis-regulating at least one gene are shown. Note that, we call a CNAI ∼protein/phosphosite pair to be a cis pair, if the gene corresponding to the protein/phosphosite falls in the ±2Mb neighbor of the CNAI; or otherwise a trans pair. The numbers of regulating targets and the genome locations of the identified CNAI regulatory hubs are shown in Fig. 2 (CNAI-proteome) and supplemental Fig. S2 (CNAI-phosphosite). A few adjacent CNAIs in the 17q12 amplicon are estimated to be the leading hubs, i.e. having the largest number of edges, in both the CNAI-Protein and CNAI-Phosphosite regulatory maps. The second and the third largest hubs in the CNAI-Protein map are CNAIs in 1p31.2–31.1 and 5q36.3 respectively. The second and third largest hubs in the CNAI-Phosphosite maps are CNAIs in 5q13.3 and 8p11.21 respectively (supplemental Fig. S2).
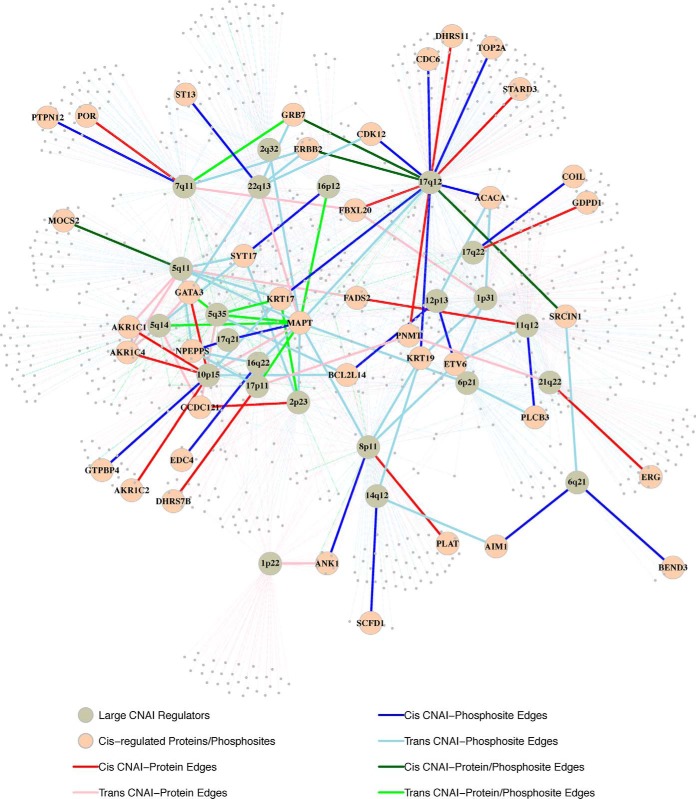
Fig. 3.
CNAI-protein/phosphosite regulatory map. Selected CNAIs, which have cis-edges or at least 50 trans-edges, with their regulated proteins/phosphosites are shown (gray nodes). Proteins/phosphosites that was cis-regulated by one of the selected CNAIs are labeled (pink nodes). The red/pink edges represent cis/trans regulations specific to the CNAI-protein regulatory map; blue/light-blue edges represent cis/trans edges specific to the CAI-phosphosite regulatory map; and the dark-green/green edges represent cis/trans edges shared by the two maps.
Other large CNAI hubs shared by both the CNA-protein and CNA-phosphosite regulatory maps in our analyses include CNAIs in 1p31.2–31.1, 2q32.2–32.3, 5q11.2, 5q14.3, 5q35.3, 7q11.23, 8p11.21, 10p15.3–14, 11q12.2–13.1, and 17p11.2–11.1. More detailed information about inferred regulatory maps and hubs are provided in supplemental Table S1 and S2.
In comparison, we also applied pairwise correlation test and remMap (see Section on Comparison with Other Methods) to construct CNAI-protein regulatory network of breast cancer. 2996 edges corresponding to 225 CNAIs (Fig. 2) and 1505 edges corresponding to 493 CNAIs (supplemental Fig. S2) were identified by pairwise correlation test and remMap respectively. Although the pairwise correlation detected significantly more trans-regulating edges, majority of these edges are for CNAIs in 5q, like the result reported in the CPTAC breast cancer article (1). On the other hand, neither pairwise correlation test nor remMap identified 17q12 amplicon as leading CNAI-hub. Neither methods detected striking CNAI-hub other than 5q, which are clear evidence of lack of power of these two alternative approaches.
CNAI hub in 17q12
As illustrated in Fig. 2 and supplemental Fig. S2, one CNAI in a small region of 17q12 is identified as the largest hub in both the CNAI-protein and CNAI-phosphoprotein regulatory maps by ProMAP. This CNAI is estimated to regulate 128 proteins and 192 phosphoproteins respectively. There are three genes—ERBB2, GRB7, and SRCIN1—whose proteins and phosphoproteins are both cis-regulated by the copy number changes of this CNAI. The detailed genomic and proteomic profiles of these three genes across all the 77 samples are illustrated in Fig. 4. Among the three, ERBB2 and GRB7 are well established oncogenes in the 17q12 amplicon, and the cis-regulations between the copy numbers of ERBB2 and GRB7 and their protein/phosphosite abundances were reported in the original CPTAC breast cancer study (1). The cis-regulations between the copy number of SRCIN1 and its protein/phosphosite abundances, however, is a new observation by ProMAP.
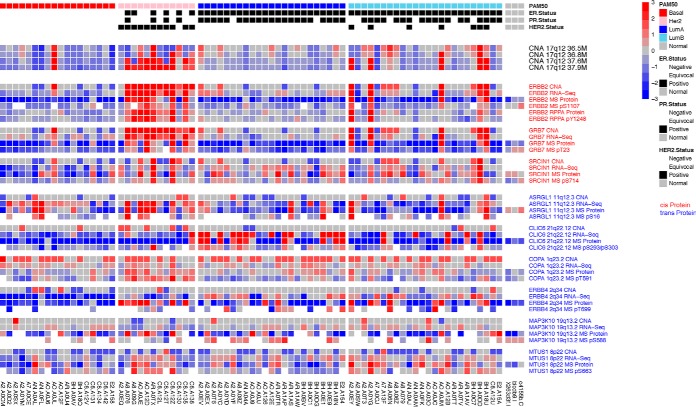
Fig. 4.
Heatmap of selected genes, whose proteins or phosphosites were regulated by the CNAI hub in 17q12. Each column represents a sample, with samples of the same breast cancer subtypes clustered together. RNAs/proteins/phosphosites that were Cis-regulated by this CNAI are labeled in red; whereas the trans-regulated targets are labeled in blue.
In addition to cis-regulations, the 17q12 amplicon is also estimated to trans-regulate a large number of proteins and phosphoproteins. In Fig. 4, detailed omics data of the top three proteins and phosphoproteins who showed the strongest trans-regulation signals with this amplicon in the corresponding regulatory maps are illustrated (see supplemental Table S3 for more details). Interestingly, for ErbB4 and MAP3K10, only their protein abundances were associated with 17q12 amplicon, but their RNA expression levels did not showed similar patterns, suggesting possible trans-regulatory mechanism through post-translational modification.
CNAI in 8p11.21
Besides CNAIs in 17q and 5q, the next largest hub in the CNAI-phosphosite regulatory map is a CNAI in 8p11.21. This CNAI is also a big hub in the CNAI-protein regulatory maps. On the other hand, no or only a limited number of regulatory relationships between CNA of 8p11.21 and proteins/phosphoproteins were detected by pairwise correlation test in the original CPTAC breast cancer study (1).
The short arm of chromosome 8 (8p) is an important region that has been suggested to associate with breast cancer development. In the TCGA-CPTAC data set, 8p11.21 was observed to have copy number losses in more than 40% samples and copy number gains in about 10% samples. ProMAP identifies two genes in 8p11.21—PLAT and ANK1, whose protein or phosphosite abundances were cis-regulated by the copy number changes of this CNAI. The detailed genomic and proteomic data of these two genes across all samples are illustrated in Fig. 5, which clearly demonstrated the concordance between protein/phosphoprotein abundances of these two genes and copy number alterations of 8p11.21.
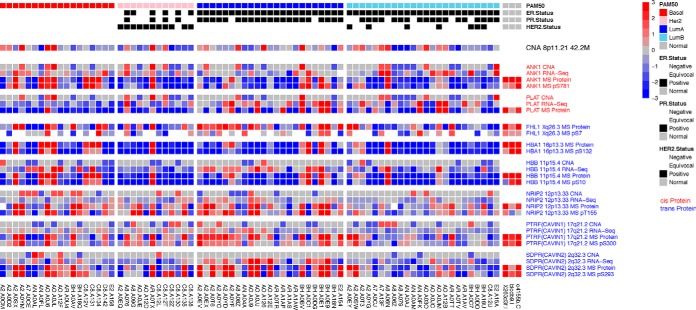
Fig. 5.
Heatmap of selected genes, whose proteins or phosphosites were regulated by the CNAI hub in 8p11. 21. Each column represents a sample, with samples of the same breast cancer subtypes clustered together. RNAs/proteins/phosphosites that were Cis-regulated by this CNAI are labeled in red; whereas the trans-regulated targets are labeled in blue.
Besides cis-regulations, in Fig. 5, we also illustrated the omics data of the top three proteins and phosphoproteins who showed the strongest trans-regulation signals with this CNAI. Specifically, the proteins of HBA1 and HBB showing the strongest trans-association with CNA of 8p11.21. And when we further investigate co-expression patterns between HBA1/HBB and all other proteins, ANK1 appeared to be the one with the highest correlation with both HBA1/HBB. This suggests that the tran-association between CNA of 8p11.21 and HBA1/HBB is likely mediated by protein of ANK1.(see supplemental Table S3 for more details)
Proteins/Phosphosites Trans Hub
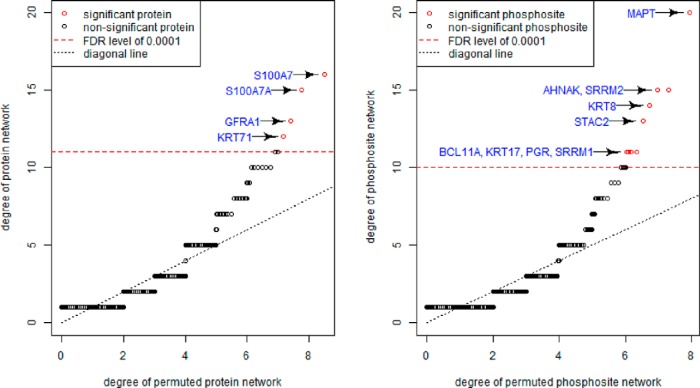
In both regulatory maps, we observe a subset of proteins/phosphosites which are regulated by much more CNAI hubs than the rest. To test whether the large degrees of these hub proteins/phosphosites are because of random chance, we perform statistical inference by comparing the degree of each protein/phosphosite in the inferred regulatory map with the average of 10000 permuted network (each of the permuted network has the fixed CNAI hub sizes as estimated from the real data but edges on each hub were randomly assigned to proteins/phosphosites). Fig. 6 illustrates the degrees of proteins/phosphosites in the inferred breast cancer regulatory map v.s. their empirical null distribution based on permuted networks. At FDR level of 0.001, 4 proteins and 9 phosphosites are detected to have significantly large degrees (see supplemental Table S4)
Fig. 6.
Number of edges (y axis) of protein/phosphosite hubs in each breast cancer regulatory maps versus expected number of ranked degrees in random networks with the same total numbers of nodes and edges (x axis) of the corresponding regulatory map. Proteins/phosphosites whose degrees are significantly larger than the null distribution of ranked degrees in random networks are marked in red (FDR < 0.001).
Among these protein/phosphosite hubs, the most striking one is a phosphosite of MAPT, which was suggested to be simultaneously regulated by more than 20 CNAIs, including the ones in 17q12 and 8p11.21, in the CNAI-phosphosite regulatory map. Genome locations of these CNAIs are illustrated in supplemental Fig. S4. There have been lots of studies investigating the role of MAPT in breast cancer. Under-expression of MAPT was found to be strongly correlated with poor treatment outcomes in breast cancer patients (22), and MAPT has been viewed as one of the most promising prognostic biomarkers in tamoxifen treated patients in a recent study (23). The 20 CNAIs connecting to MAPT phosphosites in our regulatory map harbor 23 kinases, including Limk2 (22q12.2–22q12.3), TTBK1 (6p21.1), and ULK1 (12q24.33), which are known to contribute to the phosphorylation of MAPT in previous studies (24, 25, 26). High-copy-number of these CNAIs will potentially increase these kinase gene expressions, and then impact the activities of MAPT.
CNAI-Protein Regulatory Maps in Ovarian Cancer
We applied ProMAP to the CPTAC-TCGA ovarian cancer data (see Section on Data Description and Preprocessing). The resulting CNAI-protein regulatory map consists of 1706 edges corresponding to 33 CNAIs. Compared with breast cancer CNAI-protein regulatory map, a different set of CNAIs-hubs were detected in the ovarian tumors (Fig. 7). The largest CNAI-hub detected by ProMAP sits in 3q26.1 and is estimated to regulate 348 proteins. The second biggest CNAI-hub is a small region of 40 kb sitting in 1p31.1 (72,539,143–72,574,479 bp), right next to gene NEGR1 (Neuronal Growth Regulator 1), which codes a raft-associated extracellular protein and is a member of the IgLON family. One CNAI from 4p16 was recognized as the third biggest hub and was estimated to regulate 172 proteins. Another large hub identified by ProMAP sits in 22q11 with 128 CNAI-protein edges. Neighbors around selected CNAIs hubs in the inferred CNAI-protein regulatory map are illustrated in supplemental Fig. S5. More detailed information about inferred regulatory maps and hubs of ovarian cancer tumors are provided in supplemental Table S5.
On the other hand, the CNAI-protein map by pairwise correlation test (4267 edges connecting with 928 CNAIs) and that by remMap (1861 edges with 714 CNAIs) on the same data sets failed to reveal any obvious CNAI-hubs (Fig. 7 and supplemental Fig. S3).
DISCUSSION
We developed a new method, ProMAP, to study the regulatory relationship between many CNAs and proteins. ProMAP incorporates mixed effects and missing mechanisms in a penalized multivariate regression model to account for multiplex data structure and abundance dependent missingness in data from labeled proteomics experiments, such as iTRAQ and TMT. We developed an effective ECM algorithm to fit the model and adopt a majority vote procedure to stabilize the variable selection. The resulting ProMAP algorithm achieves favorable computational efficiency. Through extensive synthetic data experiments, we demonstrated that ProMAP has better power to reveal regulatory relationships in proteogenomic integration analysis compared with methods ignoring the multiplex data structure and abundance dependent missingness.
Applying ProMAP to the proteogenomic data from CPTAC-TCGA breast and ovarian cancer study (1, 2), we identified a rich collection of genome regions whose DNA copy number alterations influence the abundances of many protein/phosphoproteins in tumor samples. In comparison, analyses based on existing methods, including pairwise correlation test and remMap, on the same data sets detect much fewer CNA trans-regulatory hub regions which coincides with the contrast in simulation studies.
A big advantage of the ProMAP model is to properly handle the missing values in proteomics data. In the original CPTAC breast cancer and ovarian cancer study articles (1, 2), only proteins/phosphosites with complete measurements were considered.
In the simulation study, we demonstrated more favorable performance of ProMAP over other alternative methods. These simulations, however, were set up assuming linear dependence of functional molecular abundances on DNA alterations. In real biological system, the dependence could be complicated and take non-linear forms. Thus, the performance of the various methods under settings other than linear dependence warrants future investigation.
Breast Cancer CNAI-Protein/Phosphoprotein Regulatory Map
In the original TCGA-CPTAC breast cancer study (1), pairwise correlation test was used to characterize CNAI-protein regulatory patterns and the whole chromosome arm 5q was detected as the the most prominent tran-regulatory hub, as was replicated in our pairwise correlation test result (Fig. 2). One limitation of pairwise correlation test is that it often failed to pinpoint the local genome regions in a chromosome arm that drives trans-regulatory relationships. This is because the high spatial correlation among DNA copy numbers on a chromosome arm causes high FDR in pairwise correlation tests as we demonstrated in simulation III. Instead, ProMAP identified small regions in 5q11.2, 5q14.3, and 5q35.3 as leading CNAI hubs in the two regulatory maps, suggesting this new method could be more effective in pinpointing important local genome regions.
Moreover, different from the network constructed by pairwise correlation test, ProMAP identified the CNAI in 17q12 as the top trans-hub, which regulated 57% more proteins and phosphosites than the second largest CNAI hub in the two breast cancer regulatory maps. Given the known strong biological relevance of 17q12 to breast cancer, it is reasonable to believe that ProMAP revealed more biological important features in the data than pairwise correlation test or remMap did.
ProMAP identified three genes in 17q12: ERBB2, GRB7, and SRCIN1, whose proteins and phosphoites were both cis-regulated by the copy number changes of the corresponding CNAI. Amplification and/or over-expression of ERBB2 is viewed as a trigger event for HER2 subtype of breast cancer (5). Expression of GRB7, which is located adjacent to ERBB2, has been observed to correlate with markers of a more aggressive phenotype, including ER negativity, and p53 positivity in invasive breast cancer (27). Although ERBB2 and GRB7 are well established oncogene in this amplicon, SRCIN1 is a less studied one and the impact of CNA of SRCIN1 was not reported in the literature. The full name of SRCIN1 is SRC Kinase Signaling Inhibitor 1, which codes the protein P140Cap. SRCIN1 has been reported to be expressed in only breast tumors but not in normal breast tissues (28). Recent studies further revealed a strong association between p140Cap and improved survival of ERBB2 patients, and suggested for a key role of p140Cap in curbing the aggressiveness of the ERBB2 tumors, counteracting in vivo tumor growth, epithelial mesenchymal transition and metastatic lesions (29, 30, 31). Results of ProMAP further suggests that the protein activity of SRCIN1 is affected by its copy number gains in the tumor cells.
Besides cis-regulations, ProMAP also reveals interesting targets that were trans-regulated by 17q12 amplicon in breast tumors. One example is ErbB4 (HER4), which sits in 2q34 and is one of the four members in the EGFR subfamily of receptor tyrosine kinases. ErbB4 expression in breast cancers has been reported to correlate with sensitivity to endocrine therapies (32, 33). Study has shown that ErbB2 is necessary for ErbB4 ligands to stimulate oncogenic activities in models of human breast cancer (34), which is consistent with the trans-regulation relationship between the ERBB2 amplicon and ErbB4 detected by ProMAP. Moreover, as shown in Fig. 4, only protein levels of ErbB4 were associated with ERBB2 activities, but neither RNA expression nor DNA copy number of ERBB4 showed similar patterns. This implies that the detected trans-regulation is mostly likely because of the crosstalk between ErbB2 and ErbB4 as a post-translational modification. The fact that ProMAP identified the 17q12 amplicon, which harbors and regulates many known important breast cancer genes, as the leading hub in the inferred regulatory maps suggests that ProMAP captures biologically meaningful signals in the data.
Among new tran-regulatory hubs that were detected by ProMAP but not by the pairwise correlation test or remMap, the CNAI in 8p11.21 is of interest. In a recent research, the loss of heterozygositys at 8p was detected at early stages of breast cancer development and associated with poor patient survival, strongly suggesting a role for 8p LOH in tumor initiation and progression (35). In our study we find two genes: PLAT and ANK1, whose protein or phosphosite abundances were cis-regulated by the copy number changes of 8p11.21. These two genes were firstly studied to be the characterization of the amplification on chromosome region 8p in breast carcinoma, strong physical linkage of them was observed as they appear to be separated by a maximum distance of 700 kb (36). PLAT was suggested to be a candidate biomarker of estrogen-dependent breast cancer (37). In a recent study, PLAT was observed as the highest differentially expressed gene in PRRX2-overexpressing MCF10A cells and is suggested to be a mechanism contributing to TGF-β-induced invasion and EMT in breast cancer (38). The other cis-regulated protein of this CNAI, ANK1, is one of the Ankyrins protein family that link the integral membrane proteins to the underlying spectrin-actin cytoskeleton and play key roles in activities such as cell motility, activation, and the maintenance of specialized membrane domains.
At the same time, ProMAP identified HBA1 and HBB to be the strongest trans-regulating target of CNAs of 8p11.21. Indeed, a very recent study (39), for the first time, reported non-conventional role of hemoglobin beta in breast malignancy. Overexpression of HBB were observed to be associated with lower overall survival; and HBB-overexpressing breast cancer cells appeared to migrate more, invade more, and show HIF-1α up-regulation (39). ProMAP results further suggest that overexpression of HBB was associated with copy number amplification of 8p11.21; and this association was very likely mediated through protein of ANK1. These information casts light on functional consequences of 8p11.21 amplification as well as potential regulatory mechanisms driving oxygen binding and transport activities in breast tumor cells.
Moverover, in the breast cancer application, ProMAP detected a subset of proteins/phosphosites which were regulated by significantly more CNAI hubs than the rest. One possible hypothesis accounting for this interesting phenomenon is, although breast tumors in different patients were triggered by different genome alterations (because of tumor heterogeneity), these different genome alterations interrupted a same set of key biological processes (proteins) in the cells, which then led to tumor. Under such an assumption, the “hub” proteins/phosphatides revealed in the ProMAP network could be essential players in the tumor initiation and/or progression. Indeed, 10 out of the 13 hub proteins/phosphosites have been well documented in the literature to be relevant to breast cancer. The other three genes, SRRM1, SRRM2, and KRT71, which has been barely explored in breast cancer, could also play important roles in these tumors. More investigation about the mechanism of these genes are warranted as future research.
Note, when applying ProMAP on TCGA-CPTAC breast data, we considered samples from all subtypes, as the heterogeneity across different breast cancer subtypes could help to enhance the power to detect regulatory patterns between CNAs and protein/phosphosite abundances. It will also be of great interest to identify subtype specific regulatory patterns, and subtype specific analysis is warranted in future studies.
Ovarian Cancer CNAI-protein Regulatory Map
The application of ProMAP to the TCGA-CPTAC ovarian cancer data, interestingly, revealed a largely different CNAI-protein regulatory map from that of breast cancer, suggesting these two types of tumor were powered by tissue type specific genomics alterations.
The leading CNAI-hub in the ovarian regulatory map sits in 3q26, whose copy number gain was reported to occur in 36–53% ovarian tumors (40). As one of the most frequently amplified regions in ovarian tumors, 3q26 has been suggested to represent oncogenic “drivers” of tumorigenesis by many studies (40). In short, this amplicon harbors a collection of important oncogenes, including SOX2, ECT2, PRKCI and PI3KCA, which influence multiple signaling pathways relating to tumor initiation and progression. Thus, the fact that ProMAP identifies 3q26 amplicon as the leading CNAI-hub in the inferred CNAI-protein regulatory map suggests this analysis captures biological relevant signals in the data. In the contrast, this amplicon was not picked up as a tran-hub in alternative analyses carried out on the same data sets using either pairwise correlation test or remMap (Fig. 7, supplemental Fig. S3), nor in the original CPTAC-TCGA ovarian study (2).
The second CNAI-hub in ovarian regulatory map by ProMAP is a small region in 1p31. Although this region does not harbor any known gene, it has a close neighbor—NEGR1, which is a member of the IgLON family and has been reported to exhibit reduced expression in not only ovarian tumors (41) but also five other tumor types (42). Recent interest in IgLON family has led to the identification of NEGR1 as a potential tumor suppressor which participates in cell recognition and interaction and thus is important in growth control and malignant transformation in ovarian tumors (42). Although NEGR1 is not measured in the proteomics experiments of ovarian cancer and thus does not appear in the inferred regulatory map, it's not unreasonable to hypothesis that copy number deletion of this CNAI in 1p31 contributes to the down-regulation of NEGR1 in ovarian tumors. In addition, the 226 proteins connected to this CNAI in the inferred regulatory map were significantly enriched for genes from Extracellular matrix organization and Innate immune system pathways (p value < 10−9), which could be relevant biological processes influenced by NEGR1 down-regulation in ovarian tumors.
It is worth mentioning that CNAI-hubs from our pairwise correlation test does not overlap much with the result from a similar analysis in the original TCGA-CPTAC ovarian cancer study (2). One key factor that contributes to this discrepancy is the usage of different levels of CNA data: gene-level CNA data was used in the original analysis; whereas segment level (CNAI) data is derived and used in this manuscript. As indicated earlier, using segment level data can help to guard us from being vulnerable to false positive detection because of strong spatial correlation in CNA profiles. More importantly, relying on gene-level CNA data, one ignores local (short region) DNA copy number alterations occurred in gene desert regions. But using segment level CNA data allows one to comprehensively investigate CNA events in both the coding and non-coding regions. For example, the second largest CNAI-hub in ovarian cancer regulatory map by ProMAP corresponds to a local deletion happened in a gene desert region, and thus was missed in the previous study. Further evaluation and validation of these CNAI-hub findings using data from independent cohorts is warranted.
In summary, proteogenomic integrative analysis by ProMAP provides rich and biologically relevant discoveries, which improve our understanding of the genetic regulation mechanisms underlying the disease. We believe this method could enhance the power and accuracy of proteogenomic integrative analysis in a broad range of applications. With the help of this tool, many of the further integrated-omics analysis will be facilitated with our full interests to persue in the future. First, it will be of great value to apply ProMAP to as many tumor types as possible and to carry out a Pan Cancer investigation to compare protein-CNV regulatory maps across different tumor types. Also, it will be of great interest to investigate the mechanism of signaling transduction or interaction network underlying each tran-hub in the regulatory map.
R package ProMAP has been developed and is currently available for public access at github page (https://github.com/WangLab-MSSM/ProMAP/).
Data Availability
PSM files of breast cancer and ovarian cancer data were downloaded from CPTAC public data portal with links (https://cptac-data-portal.georgetown.edu/cptac/s/S039; https://cptac-data-portal.georgetown.edu/cptac/s/S038).
Supplementary Material
Acknowledgments
We acknowledge the computational resources and staff expertise provided by the Department of Scientific Computing at Icahn School of Medicine at Mount Sinai. We thank Drs D. R. Mani and Karl R. Clauser for providing preprocessed breast cancer proteomic data. Data used in this publication were generated by the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC).
Footnotes
* This work was supported by grant U24 CA210993, from the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC). W.M. was also supported by a postdoc fellowship from Roche Inc. P.W. and L.S.C. were partly supported by National Institutes of Health grant R01 GM108711. S.W.H. and H.Z. were partly supported by National Institutes of Health grant R21 GM110450. L.S.C., A.G.P., and P.W. were partly supported by National Institutes of Health grant U01 CA214114.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- CPTAC
- Clinical Proteomics Tumor Analysis Consortium
- TCGA
- The Cancer Genome Atlas
- CNA
- copy number alterations
- iTRAQ
- isobaric tag for relative and absolute quantitation
- FOC
- fixed order clustering
- LC-MS/MS
- Liquid chromatography tandem mass spectrometry
- TMT
- tandem mass tagging
- MS
- mass spectrometry
- eQTL
- expression quantitative trait loci
- CNAI
- copy number alteration intervals
- NCI
- national cancer institute
- MAD
- median absolute deviation
- FDR
- false discovery rate.
REFERENCES
- 1. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., Gatza M. L., Wilkerson M., Perou C. M., Yellapantula V., Huang K. L., Lin C., McLellan M. D., Yan P., Davies S. R., Townsend R. R., Skates S. J., Wang J., Zhang B., Kinsinger C. R., Mesri M., Rodriguez H., Ding L., Paulovich A. G., Fenyö D, Ellis M. J., and Carr S. A.; NCI CPTAC. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang H., Liu T., Zhang Z., Payne S. H., Zhang B., McDermott J. E., Zhou J. Y., Petyuk V. A., Chen L., Ray D., Sun S., Yang F., Chen L., Wang J., Shah P., Cha S. W., Aiyetan P., Woo S., Tian Y., Gritsenko M. A., Clauss T. R., Choi C., Monroe M. E., Thomas S., Nie S., Wu C., Moore R. J., Yu K. H., Tabb D. L., Fenyö D., Bafna V., Wang Y., Rodriguez H., Boja E. S., Hiltke T., Rivers R. C., Sokoll L., Zhu H., Shih I. M., Cope L., Pandey A., Zhang B., Snyder M. P., Levine D. A., Smith R. D., Chan D. W., and Rodland K. D.; CPTAC Investigators (2016) Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell 166, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumors. Nature 490, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bergamaschi A., Kim Y. H., Wang P., Sørlie T., Hernandez-Boussard T., Lonning P. E., Tibshirani R., Børresen-Dale A. L., and Pollack J. R. (2006) Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes, Chromosomes Cancer 45, 1033–1040 [DOI] [PubMed] [Google Scholar]
- 6. Turlach B. A., Venables W. N., and Wright S. J. (2005) Simultaneous variable selection. Technometrics 47, 349–363 [Google Scholar]
- 7. Lutz R. W., and Buhlmann P. (2006) Boosting for high-multivariate responses in high-dimensional linear regression. Statistica Sinica, 16, 471 [Google Scholar]
- 8. Yuan M., Ekici A., Lu Z., and Monteiro R. (2007) Dimension reduction and coefficient estimation in multivariate linear regression. J. Roy. Statistical Soc. 69, 329–346 [Google Scholar]
- 9. Obozinski G., Wainwright M. J., and Jordan M. I. (2008) Union support recovery in high-dimensional multivariate regression. In 2008 46th Annual Allerton Conference on Communication, Control, and Computing (pp. 21–26). IEEE. [Google Scholar]
- 10. Rothman A., Levina E., and Zhu J. (2010) Sparse multivariate regression with covariance estimation. J. Computational Graphical Statistics 19, 947–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng J., Zhu J., Bergamaschi A., Han W., Noh D. Y., Pollack J. R., and Wang P. (2010) Regularized multivariate regression for identifying master predictors with application to integrative genomics study of breast cancer. Ann. Appl. Statistics 4, 53–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang X., Li Q., Zhang H., Zhang Y., Hsu L., and Wang P. (2015) A regularized multivariate regression approach for eQTL analysis. Statistics Biosci. 7, 129–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mertins P., Udeshi N. D., Clauser K. R., Mani D. R., Patel J., Ong S. E., Jaffe J. D., and Carr S. A. (2012) iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Mol. Cell. Proteomics 11, M111.014423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Karp N. A., Huber W., Sadowski P. G., Charles P. D., Hester S. V., and Lilley K. S. (2010) Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell. Proteomics 9, 1885–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen L. S., Wang J., Wang X., and Wang P. (2017) A mixed-effects model for incomplete data from labeling-based quantitative proteomics experiments. Ann. Appl. Statistics 11, 114–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang P., Tang H., Zhang H., Whiteaker J., Paulovich A. G., and Mcintosh M. (2006) Normalization regarding non-random missing values in high-throughput mass spectrometry data. Pacific Sym. Biocomputing 315–326 [PubMed] [Google Scholar]
- 17. Chen L. S., Prentice R. L., and Wang P. (2014) A penalized EM algorithm incorporating missing data mechanism for Gaussian parameter estimation. Biometrics 70, 312–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang P. (2010) Statistical Methods for CGH Array Analysis; VDM Verlag Dr. MüIIer [Google Scholar]
- 19. Zhang B., Wang J., Wang X., Zhu J., Liu Q., Shi Z., Chambers M. C., Zimmerman L. J., Shaddox K. F., Kim S., Davies S. R., Wang S., Wang P., Kinsinger C. R., Rivers R. C., Rodriguez H., Townsend R. R., Ellis M. J., Carr S. A., Tabb D. L., Coffey R. J., Slebos R. J., and Liebler D. C.; NCI CPTAC (2014) Proteogenomic characterization of human colon and rectal cancer. Nature 513, 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., and Altman R. B. (2001) Missing value estimation methods for DNA microarrays. Bioinformatics 17, 520–525 [DOI] [PubMed] [Google Scholar]
- 21. Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D. and Altman R. B. (2001) Missing value estimation methods for DNA microarrays. Bioinformatics 17, 520–525 [DOI] [PubMed] [Google Scholar]
- 22. Wang D.-Y., Youngson B. J., Miller N., Boerner S., Done S. J., and Leong W. L. (2011) Clinical relevance of DNA microarray analyses using archival formalin-fixed paraffin-embedded breast cancer specimens. BMC Cancer 11, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mihály Z., Kormos M., Lánczky A., Dank M., Budczies J., Szász M. A., and Győrffy B. (2013) A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res. Treatment 140, 219–232 [DOI] [PubMed] [Google Scholar]
- 24. Kavallaris M. (2010) Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer 10, 194–204 [DOI] [PubMed] [Google Scholar]
- 25. Sato S., Cerny R. L., Buescher J. L., and Ikezu T. (2006) Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J. Neurochem. 98, 1573–1584 [DOI] [PubMed] [Google Scholar]
- 26. Parker A. L., Kavallaris M., and McCarroll J. A. (2007) Microtubules and their role in cellular stress in cancer. Front. Oncol. 4, 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bivin W. W., Yergiyev O., Bunker M. L., Silverman J. F., and Krishnamurti U. (2016) GRB7 expression and correlation with HER2 amplification in invasive breast carcinoma. Appl. Immunohistochem. Mol. Morphol. 25, 553–558 [DOI] [PubMed] [Google Scholar]
- 28. Kennedy S., Clynes M., Doolan P., Mehta J., Rani S., Crown J., and O'Driscoll L. (2008) SNIP/p140Cap mRNA expression is an unfavourable prognostic factor in breast cancer and is not expressed in normal breast tissue. Br. J. Cancer 98, 1641–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Damiano L., Le Dévédec S. E., Di Stefano P., Repetto D., Lalai R., Truong H., Xiong J. L., Danen E. H., Yan K., Verbeek F. J., De Luca E., Attanasio F., Buccione R., Turco E., van de Water B., and Defilippi P. (2012) p140Cap suppresses the invasive properties of highly metastatic MTLn3-EGFR cells via impaired cortactin phosphorylation. Oncogene 31, 624–633 [DOI] [PubMed] [Google Scholar]
- 30. Sharma N., Repetto D., Aramu S., Grasso S., Russo I., Fiorentino A., Mello-Grand M., Cabodi S., Singh V., Chiorino G., Turco E., Stefano P. D., and Defilippi P. (2013) Identification of two regions in the p140Cap adaptor protein that retain the ability to suppress tumor cell properties. Am. J. Cancer Res. 3, 290–301 [PMC free article] [PubMed] [Google Scholar]
- 31. Grasso S. Chapelle J., Salemme V., Aramu S., Russo I., Vitale N., Verdun di Cantogno L., Dallaglio K., Castellano I., Amici A., Centonze G., Sharma N., Lunardi S., Cabodi S., Cavallo F., Lamolinara A., Stramucci L., Moiso E., Provero P., Albini A., Sapino A., Staaf J., Di Fiore P. P., Bertalot G., Pece S., Tosoni D., Confalonieri S., Iezzi M., Di Stefano P., Turco E., and Defilippi P. (2017) The scaffold protein p140Cap limits ERBB2-mediated breast cancer progression interfering with Rac GTPase-controlled circuitries. Nat. Communications 8, 14797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naresh A., Long W., Vidal G. A., Wimley W. C., Marrero L., Sartor C. I., Tovey S., Cooke T. G., Bartlett J. M., and Jones F. E. (2006) The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 66, 6412–6420 [DOI] [PubMed] [Google Scholar]
- 33. Ghayad S. E., Vendrell J. A., Larbi S. B., Dumontet C., Bieche I., and Cohen P. A. (2010) Endocrine resistance associated with activated ErbB system in breast cancer cells is reversed by inhibiting MAPK or PI3K/Akt signaling pathways. Int. J. Cancer 126, 545–562 [DOI] [PubMed] [Google Scholar]
- 34. Mill C. P., Zordan M. D., Rothenberg S. M., Settleman J., Leary J. F., and Riese D. J. (2011) ErbB2 is necessary for ErbB4 ligands to stimulate oncogenic activities in models of human breast cancer. Genes Cancer 2, 792–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cai Y., Crowther J., Pastor T., Asbagh L. A., Baietti M. F., De Troyer M., Vazquez I., Talebi A., Renzi F., Dehairs J., Swinnen J. V., and Sablina A. A. (2016) Loss of chromosome 8p governs tumor progression and drug response by altering lipid metabolism. Cancer Cell 29, 751–766 [DOI] [PubMed] [Google Scholar]
- 36. Dib A., Adélaïde J., Chaffanet M., Imbert A., Le Paslier D., Jacquemier J., Gaudray P., Theillet C., Birnbaum D., and Pebusque M. J. (1995) Characterization of the region of the short arm of chromosome 8 amplified in breast carcinoma. Oncogene 10, 995–1001 [PubMed] [Google Scholar]
- 37. Thakkar A. D., Raj H., Chakrabarti D., Saravanan N., Muthuvelan B., Balakrishnan A., and Padigaru M. (2010) Identification of gene expression signature in estrogen receptor positive breast carcinoma. Biomarkers Cancer 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Juang Y. L., Jeng Y. M., Chen C. L., and Lien H. C. (2016) PRRX2 as a novel TGF-β-induced factor enhances invasion and migration in mammary epithelial cell and correlates with poor prognosis in breast cancer. Mol. Carcinogenesis 55, 2247–2259 [DOI] [PubMed] [Google Scholar]
- 39. Ponzetti M., Capulli M., Angelucci A., Ventura L., Delle Monache S., Mercurio C., Calgani A., Sanità P., Teti A., and Rucci N. (2017) Non-conventional role of haemoglobin beta in breast malignancy. Br. J. Cancer 117, 994–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fields A. P., Justilien V., and Murray N. R. (2016) The chromosome 3q26 OncCassette: a multigenic driver of human cancer. Adv. Biol. Regulation 60, 47–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ntougkos E., Rush R., Scott D., Frankenberg T., Gabra H., Smyth J. F., and Sellar G. C. (2005) The IgLON family in epithelial ovarian cancer: expression profiles and clinicopathologic correlates. Clin. Cancer Res. 11, 5764–5768 [DOI] [PubMed] [Google Scholar]
- 42. Kim H., Hwang J. S., Lee B., Hong J., and Lee S. (2014) Newly identified cancer-associated role of human neuronal growth regulator 1 (NEGR1). J. Cancer 5, 598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PSM files of breast cancer and ovarian cancer data were downloaded from CPTAC public data portal with links (https://cptac-data-portal.georgetown.edu/cptac/s/S039; https://cptac-data-portal.georgetown.edu/cptac/s/S038).