Abstract
Genetic factors have been postulated to be involved in the etiology of diabetic peripheral neuropathy (DPN), but their identity remains mostly unknown. The aim of this study was to conduct a systematic search for genetic variants influencing DPN risk using two well-characterized cohorts. A genome-wide association study (GWAS) testing 6.8 million single nucleotide polymorphisms was conducted among participants of the Action to Control Cardiovascular Risk in Diabetes (ACCORD) clinical trial. Included were 4,384 white case patients with type 2 diabetes (T2D) and prevalent or incident DPN (defined as a Michigan Neuropathy Screening Instrument clinical examination score >2.0) and 784 white control subjects with T2D and no evidence of DPN at baseline or during follow-up. Replication of significant loci was sought among white subjects with T2D (791 DPN-positive case subjects and 158 DPN-negative control subjects) from the Bypass Angioplasty Revascularization Investigation in Type 2 Diabetes (BARI 2D) trial. Association between significant variants and gene expression in peripheral nerves was evaluated in the Genotype-Tissue Expression (GTEx) database. A cluster of 28 SNPs on chromosome 2q24 reached GWAS significance (P < 5 × 10−8) in ACCORD. The minor allele of the lead SNP (rs13417783, minor allele frequency = 0.14) decreased DPN odds by 36% (odds ratio [OR] 0.64, 95% CI 0.55–0.74, P = 1.9 × 10−9). This effect was not influenced by ACCORD treatment assignments (P for interaction = 0.6) or mediated by an association with known DPN risk factors. This locus was successfully validated in BARI 2D (OR 0.57, 95% CI 0.42–0.80, P = 9 × 10−4; summary P = 7.9 × 10−12). In GTEx, the minor, protective allele at this locus was associated with higher tibial nerve expression of an adjacent gene (SCN2A) coding for human voltage-gated sodium channel NaV1.2 (P = 9 × 10−4). To conclude, we have identified and successfully validated a previously unknown locus with a powerful protective effect on the development of DPN in T2D. These results may provide novel insights into DPN pathogenesis and point to a potential target for novel interventions.
Introduction
Diabetic peripheral neuropathy (DPN)—the most common cause of neuropathy worldwide—is a frequent complication of diabetes that significantly contributes to the increased morbidity and mortality associated with this disease (1). Up to one-fourth of the annual U.S. expenditure on diabetes is due to DPN, a large proportion of which is secondary to type 2 diabetes (T2D) (2,3). The most common presentation of DPN is a distal symmetric polyneuropathy resulting from small and large fiber dysfunction leading to loss of sensory, proprioception, temperature, and pain discrimination, along with symptoms of numbness, tingling, burning, and shooting pain (1,4). Signs and symptoms begin distally and spread proximally.
Known risk factors for DPN include duration and severity of hyperglycemia, age, dyslipidemia, hypertension, obesity, height, and smoking (1,5–9). Because these exposures fail to fully explain interindividual differences in the risk of developing DPN, genetic factors have been postulated to be involved in the etiology of DPN (10), although formal heritability estimates, such as those available for other diabetic complications, including cardiovascular autonomic neuropathy (narrow-sense heritability up to 39%) (11–13), are not available for DPN. Knowledge of the genetic factors that modulate DPN risk may provide insights into the molecular pathways linking the diabetic milieu to nerve damage, which may in turn suggest new pharmacological targets for preventing or treating DPN. However, although many genetic studies have been performed for other diabetic complications (14), data on the genetic determinants of DPN are scant. A few candidate gene studies have been performed, but these were small and not followed by replication attempts (10). A locus on chromosome 8p21.3 was recently found to be associated with painful DPN in a genome-wide association study (GWAS), but this finding did not reach genome-wide significance and is still to be replicated (15).
Here we report the results of a GWAS for DPN conducted among participants with T2D in the Action to Control Cardiovascular Risk in Diabetes (ACCORD) clinical trial. Our findings, which were replicated in the population of the Bypass Angioplasty Revascularization Investigation 2 Diabetes (BARI 2D) trial, point to a locus on chromosome 2q24 as a powerful determinant of DPN risk that may act by affecting a nearby voltage-gated sodium channel, NaV1.2, expressed in peripheral nerves. This is the first GWAS of DPN yielding a genome-wide significant signal with independent replication and biological plausibility of the leading candidate gene in the associated region.
Research Design and Methods
Study Population
Discovery Set: ACCORD Study
The ACCORD clinical trial investigated whether cardiovascular event rates could be reduced by intensively targeting hyperglycemia to HbA1c <6.0% (42 mmol/mol) compared with a standard target of HbA1c between 7 and 7.9% (53–63 mmol/mol). For this purpose, 10,251 participants with T2D and high cardiovascular risk were randomized in a 1:1 ratio to receive intensive or standard glycemic control therapy at 77 clinical sites across the U.S. and Canada (16). The study also investigated the effect of intensive versus standard blood pressure control and fibrate versus placebo therapy on cardiovascular events through the ACCORD blood pressure and lipid subtrials in a double two-by-two factorial design (16). In addition, ACCORD had a rich follow-up of study participants and collected data on other diabetic complications, including DPN, at baseline and during follow-up. Of the 10,251 ACCORD participants, 8,174 consented to genetic studies. To minimize confounding by race, our GWAS for discovery of genetic predictors of DPN was restricted to 5,360 whites. Among them, DPN data were collected from 5,168 participants at baseline and/or during follow-up by means the Michigan Neuropathy Screening Instrument (MNSI) clinical examination score.
Validation Set: BARI 2D Study
BARI 2D was a two-by-two factorial clinical trial enrolling 2,368 participants with T2D and established coronary artery disease (17). For glycemic control, participants were randomly assigned to insulin sensitization or insulin provision strategy to reach the same HbA1c target of <7.0% (53 mmol/mol). For coronary artery disease, participants were randomly assigned to undergo coronary revascularization procedures (percutaneous coronary intervention or coronary artery bypass graft surgery) or medical therapy. The aim was to investigate the cardiovascular event reduction by both treatment strategies. As in ACCORD, other secondary outcomes were obtained in BARI 2D, including DPN, which was evaluated with the same MNSI clinical examination score at baseline and follow-up (18). A total of 1,030 whites consented to genetic studies in BARI 2D; 949 had genetic data that passed internal quality control (QC) and also had information about the presence of DPN at baseline and follow-up. Because participants were recruited in the two studies during the same time period (January–June 2001 and February 2003–October 2005 for ACCORD, and January 2001–March 2005 for BARI 2D) and both studies upheld participation in another clinical trial as an exclusion criterion (16,19), the chances of a person being included in both studies were deemed to be negligible.
Outcome Definition
The ACCORD and BARI 2D trials both defined neuropathy as an MNSI clinical examination score >2.0 (18,20)—a criterion that has been extensively validated as being highly sensitive and specific for the diagnosis of DPN (21,22). The MNSI clinical examination includes a focused examination of the feet to assess skin and structural abnormalities, along with assessment of distal vibration perception with a 128-Hz tuning fork and ankle reflexes. For the purpose of the genetic association study, DPN case subjects were defined as participants having an MNSI >2.0 at study entry and/or at any time during follow-up (4.9 and 5.3 years on average in ACCORD and BARI 2D, respectively), whereas DPN control subjects were defined as participants having an MNSI ≤2.0 at study entry and for the entire duration of follow-up.
DNA Extraction and Genotyping
Discovery Set: ACCORD Study
Detailed DNA extraction, genotyping, QC methods, and imputation in ACCORD can be found in the supplementary material of the article in which the GWAS was first reported (23). In brief, genotyping was performed in two centers using different platforms and chips: 6,085 DNA samples (ACCORD UVA Set) were genotyped at the University of Virginia (UVA) on Illumina HumanOmniExpressExome-8 (v1.0) chips containing 951,117 SNPs; 8,174 samples (ACCORD UNC Set) were genotyped at the University of North Carolina (UNC), on Affymetrix Axiom Biobank1 chips containing 628,679 probes. The ACCORD UVA set included subjects who had given consent to genetic studies by any investigator; the ACCORD UNC included the subjects in the ACCORD UVA set plus those who had given consent only to studies by ACCORD investigators. After extensive QC of the individual genotyping sets and harmonization of genotypes for those samples and SNPs that were in common between the two genotyping sets, data were organized in two nonoverlapping data sets: ANYSET (n = 5,971), including subjects from the ACCORD UVA set, genotyped at either UVA or UNC at a total of 1,263,585 individual SNPs, and ACCSET (n = 2,113), including subjects from the ACCORD UNC set minus those in the ACCORD UVA set, genotyped only at UNC at a total of 572,192 SNPs. Imputation was conducted independently in the ANYSET and ACCSET data sets by means of IMPUTEv2.3.1 (24) using the full 1000 Genomes Phase 3 (October 2014) panel as the reference. After discarding imputed SNPs with an information content <0.3, the total (genotyped + imputed) SNPs were 25,017,489 in ANYSET and 25,012,865 in ACCSET.
Validation Set: BARI 2D Study
DNA was isolated and purified from whole blood using the Qiagen QIAamp DNA purification kit (Qiagen, Germantown, MD), as previously described (25). Genotyping was performed using the Infinium Multi-Ethnic Global Array (Illumina, San Diego, CA) according to the manufacturer’s instructions. After indels, SNPs with call rates <0.99, and multiallelic sites were filtered out, the samples were prephased with SHAPEIT and imputed toward the Phase 3 Cosmopolitan Reference panel of 1000 Human Genomes project using IMPUTE2 software. Imputed genotypes were then converted into dosages using FCGENE software. Imputation quality ranged from 85 to 100% for the 40 variants available for validation in BARI 2D (variants listed in Table 2 and Supplementary Table 6).
Table 2.
Top loci (P < 1 × 10−5) associated with DPN: effects in ACCORD and validation in BARI 2D
| SNP | Position‡ | Closest gene | MA** | Discovery set (ACCORD) |
Validation set (BARI 2D) |
ACCORD + BARI 2D* |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF¶ 1000G | MAF§ case subjects | MAF§ control subjects | OR (95% CI) | P | MAF case subjects | MAF control subjects | OR (95% CI) | P | OR (95% CI) | P | ||||
| rs13417783 | 2:167629849 | XIRP2 | T | 0.14 | 0.14 | 0.20 | 0.64 (0.55–0.74) | 1.9 × 10−9 | 0.14 | 0.20 | 0.57 (0.41–0.80) | 0.0009 | 0.63 (0.55–0.72) | 7.9 × 10−12 |
| rs12988669 | 2:240275570 | HDAC4 | C | 0.15 | 0.16 | 0.20 | 0.71 (0.62–0.82) | 2.7 × 10−6 | 0.16 | 0.13 | 1.22 (0.83–1.78) | 0.32 | 0.76 (0.66–0.87) | 5.1 × 10−5 |
| rs60770880 | 3:8037416 | LOC101–927394 | A | 0.21 | 0.21 | 0.26 | 0.75 (0.66–0.84) | 5.0 × 10−6 | 0.20 | 0.21 | 0.93 (0.68–1.26) | 0.62 | 0.77 (0.68–0.86) | 1.0 × 10−5 |
| rs11932946 | 4:45140214 | GUF1 | G | 0.12 | 0.12 | 0.16 | 0.69 (0.59–0.82) | 9.6 × 10−6 | 0.13 | 0.11 | 1.01 (0.68–1.49) | 0.98 | 0.73 (0.63–0.85) | 4.5 × 10−5 |
| rs1202660 | 7:70658177 | WBSCR17 | T | 0.22 | 0.20 | 0.25 | 0.74 (0.65–0.84) | 6.2 × 10−6 | 0.23 | 0.26 | 0.81 (0.60–1.09) | 0.16 | 0.75 (0.67–0.85) | 2.5 × 10−6 |
| rs13265430 | 8:4165607 | CSMD1 | A | 0.09 | 0.09 | 0.13 | 0.64 (0.52–0.76) | 1.0 × 10−6 | 0.11 | 0.09 | 1.35 (0.84–2.15) | 0.21 | 0.70 (0.59–0.83) | 4.3 × 10−5 |
| rs2491019 | 10:70776987 | KIAA1279 | A | 0.45 | 0.47 | 0.40 | 1.31 (1.17–1.47) | 4.4 × 10−6 | 0.45 | 0.46 | 1.02 (0.79–1.32) | 0.86 | 1.25 (1.13–1.39) | 2.0 × 10−5 |
| rs77494074 | 11:132794801 | OPCML | T | 0.08 | 0.07 | 0.10 | 0.61 (0.50–0.74) | 1.0 × 10−6 | 0.07 | 0.06 | 1.28 (0.71–2.29) | 0.41 | 0.66 (0.54–0.79) | 1.3 × 10−5 |
| rs201655918 | 14:76791306 | ESRRB | C | 0.28 | 0.26 | 0.32 | 0.75 (0.66–0.85) | 6.9 × 10−6 | 0.30 | 0.24 | 1.41 (1.02–1.96) | 0.04 | 0.81 (0.72–0.91) | 5.3 × 10−4 |
| rs11073752 | 15:88423051 | NTRK3 | C | 0.33 | 0.32 | 0.38 | 0.76 (0.67–0.85) | 2.1 × 10−6 | 0.33 | 0.36 | 0.80 (0.61–1.05) | 0.11 | 0.76 (0.68–0.85) | 6.5 × 10−7 |
| rs9948095 | 18:12018665 | IMPA2 | C | 0.14 | 0.15 | 0.20 | 0.71 (0.61–0.82) | 3.6 × 10−6 | 0.14 | 0.16 | 0.88 (0.63–1.24) | 0.46 | 0.73 (0.64–0.84) | 5.4 × 10−6 |
| rs10555080 (aka rs72397229) | 19:32043170 | THEG5 | A | NA | 0.36 | 0.30 | 1.32 (1.17–1.49) | 6.9 × 10−6 | 0.38 | 0.32 | 1.46 (1.10–1.94) | 0.0098 | 1.34 (1.20–1.50) | 2.6 × 10−7 |
| rs34948558 | 21:42825856 | MX1 | A | 0.28 | 0.27 | 0.32 | 0.76 (0.67–0.85) | 4.7 × 10−6 | 0.30 | 0.31 | 1.01 (0.77–1.33) | 0.95 | 0.79 (0.71–0.88) | 3.0 × 10−5 |
In ACCORD, the primary model was adjusted by assignment to interventions, seven clinical center networks, and top three principal components. In BARI 2D, adjustments included assignment to interventions, country of origin, and top three principal components.
*Meta-analysis of results in the discovery and validation sets. ‡Position is chromosome:bp according to the National Center for Biotechnology Information assembly build GRCh37/hg19. **MA is minor or effect allele. §MAF in the discovery set is the average of minor allele frequencies in the two ACCORD genotyping sets (ANYSET and ACCSET). ¶MAF in 1000 Genomes Project Phase 3 European populations was derived from Ensembl GRCh37 Release 93 (http://grch37.ensembl.org/index.html).
Data Analysis
Descriptive Statistics
Analyses were run in SAS 9.4 software (SAS Institute, Cary, NC). Normally distributed continuous variables are presented as mean ± SD and were analyzed by independent t test for difference in means between groups. Nonnormally distributed continuous variables are presented as median (interquartile range) values and were analyzed by t test after log transformation. Categorical variables are presented as counts (percentages) and were analyzed by χ2 tests to examine differences among groups.
GWAS
A GWAS for DPN was performed in 5,168 white ACCORD participants. GWAS analyses were conducted separately in ACCSET (3,554 case subjects/830 control subjects) and ANYSET (600 case subjects/184 control subjects) using SAS 9.4 software on the Harvard Medical School computing cluster, Orchestra. After filtration for minor allele frequency (MAF) ≥5%, 6,847,206 genotyped or imputed SNPs formed the GWAS panel in each data set. For each common variant, minor allele dosage (ranging from 0 to 2) was inferred from the genotypes or computed from the imputed posterior probabilities. The association between minor allele dosage of each variant and DPN was tested by logistic regression, assuming an additive genetic model and adjusting for assignment to interventions, seven clinical center networks, and top three principal components (PC1–PC3). After using genomic control corrections (λ = 1.014 for ANYSET and λ = 1.032 for ACCSET), the GWAS results from the ACCSET and ANYSET sets were meta-analyzed in METAL (Abecasis Laboratory, University of Michigan, Ann Arbor, MI) (26), through a fixed-effects inverse-variance approach. Genome-wide significance was defined as a P value <5 × 10−8, and suggestive (notable) significance as P < 1 × 10−6. Manhattan and quantile-quantile (QQ) plots were generated to illustrate the GWAS results.
Further analyses of SNPs reaching genome-wide significance were carried out by 1) testing whether the SNP had similar effects on prevalent DPN (MNSI >2.0 at study entry) and incident DPN (MNSI ≤2.0 at baseline and MNSI >2.0 during follow-up); 2) testing for interaction with baseline characteristics or, for incident DPN, with trial treatments (intensive vs. standard glycemic control, intensive vs. standard blood pressure control, fenofibrate + statin vs. placebo + statin for hyperlipidemia control) by adding appropriate interaction terms; 3) evaluating the association between SNP and baseline characteristics, individual MNSI components, and other diabetic complications, including cardiovascular autonomic neuropathy and incident DNP and diabetic retinopathy; 4) evaluating the association between DPN and SNPs in African American participants; and 5) evaluating the association between DPN and genotyped low-frequency variants (MAF 0.01–0.05) placed in the linkage disequilibrium (LD) block as the top common SNP in the GWAS.
Continuous and dichotomous outcomes were tested by linear and logistic regression models, respectively. Incident outcomes were tested by means of Cox regression models.
Validation
All GWAS-significant SNPs (28 SNPs at locus 2q24) and lead variants at loci that reached a P value of <1× 10−5 in the discovery set (ACCORD) were tested for association with DPN in a validation set derived from the BARI 2D study. Logistic regression was used for this purpose with adjustments by the following covariates: country of origin, top three principal components, assignments to cardiovascular treatment strata (early medical therapy vs. early revascularization with percutaneous coronary intervention or coronary artery bypass grafting), and assignments to glycemic control strata (insulin sensitization vs. insulin provision). Given that replication was attempted for 13 variants, the significance threshold was set to P < 0.004 based on a Bonferroni correction (0.05/13). Variants that were successfully validated in BARI 2D were tested for their effects on prevalent DPN (MNSI >2.0 at the study entry) and incident DPN (MNSI turned >2.0 during the follow-up), as described for the discovery set.
Meta-analysis of BARI 2D and ACCORD
Results in BARI 2D were meta-analyzed with those in ACCORD by means of METAL (Abecasis Laboratory, University of Michigan) using a fixed-effects inverse-variance approach.
Power of Genetic Analyses
The discovery set (ACCORD) provided 80% power (α = 5 × 10−8) to detect genetic effects corresponding to a relative risk of 1.20 and 1.24 for effect allele frequencies of 0.50 and 0.15, respectively. The validation set (BARI 2D) provided 80% power (α = 0.0038, adjusting for the 13 loci for which replication was sought) to detect genetic effects corresponding to a relative risk of 1.26 and 1.32 for effect allele frequencies of 0.50 and 0.15, respectively.
Expression Quantitative Trait Loci Analysis
The top variant from the GWAS was tested for association with tibial nerve–specific gene expression through in silico analyses using the Genotype-Tissue Expression (GTEx) v7 database (27). GTEx is an online database containing tissue-specific gene expression data obtained from 620 donors (34% females, 85% whites, 68% >50 years of age) by means of Illumina TrueSeq RNA sequencing and the Affymetrix Human Gene 1.1 ST Expression Array and linked to GWAS data obtained from the same individuals by whole-genome sequencing, whole-exome sequencing, or Illumina SNP arrays (www.gtexportal.org). Expression quantitative trait loci (eQTL) analyses of genes within ±2 mega base pairs (Mbp) from the lead SNP were conducted among 361 genotyped individuals for which tibial nerve tissue expression profiles were available. Normalized effect sizes and their 95% CIs were obtained from linear regression models testing the association of the minor allele of the lead SNP with gene expression adjusted by the top three genotyping principal components, sex, genotyping platform, and a set of other covariates described in the GTEx documentation (27).
Gene Expression Analysis for SCN2A
SCN2A is expressed in the human peripheral nervous system in dorsal root ganglia (28). To quantify the level of gene expression for SCN2A in the tibial nerve, RNA sequencing (RNA-seq) samples from several human tissues were contrasted: tibial nerve, hippocampus, and skeletal muscle samples from the GTEx database (27) and dorsal root ganglia samples from the study by North et al. (29). Because library preparation, mapping, and relative abundance quantification in transcripts per million (TPM) were performed differently for these two studies, the meta-analysis was performed as follows: Coding gene relative abundances for 18,876 quantified coding genes were extracted from the North et al. (29) study, and for each sample with nonzero SCN2A abundance, the percentile of SCN2A expression with respect to all coding genes in that sample were calculated. For the GTEx data sets, the uniformly processed RNA-seq abundances for coding genes were extracted from the GTEx companion website database (GTEx_Analysis_2016-01-15_v7_RNASeQCv1.1.8_gene_tpm.gct.gz) and renormalized by constraining the TPMs of all the 19,260 coding genes quantified in the study to sum to 1 million. For each sample with nonzero SCN2A abundance, the percentile of SCN2A expression with respect to all coding genes in that sample was calculated.
Results
Clinical Characteristics of DPN Case Subjects and Control Subjects
The clinical characteristics of the discovery and validation sets are reported in Table 1. To minimize ethnic confounding, both sets included only self-reported whites. The discovery set consisted of 5,168 ACCORD participants: 4,384 who showed signs of DPN at study entry and/or during follow-up (DPN case subjects) and 784 who did not show signs of DPN at any time during the study (DPN control subjects). Compared with the latter, DPN case subjects were significantly older and taller, had longer duration of diabetes, higher HbA1c, BMI, and urinary albumin-to-creatinine ratio (UACR), and lower HDL cholesterol, were more frequently treated with insulin, and had a higher prevalence of self-reported retinopathy. For many of these variables, differences with control subjects were more pronounced for prevalent than incident case subjects (Supplementary Table 1). The validation set consisted of 949 BARI 2D participants, including 791 DPN case subjects and 158 DPN control subjects, defined as above. Overall, the clinical characteristics of the validation set were similar to those of the discovery set, except for lower HbA1c and cholesterol levels (Table 1). Within the validation set, DPN case subjects had lower total and LDL and HDL cholesterol and were more frequently treated with insulin compared with DPN control subjects.
Table 1.
Characteristics of DPN case and control subjects
| Baseline characteristic | Discovery set (ACCORD) |
Validation set (BARI 2D) |
||||
|---|---|---|---|---|---|---|
| DPN case subjects (n = 4,384) | DPN control subjects (n = 784) | P value | DPN case subjects (n = 791) | DPN control subjects (n = 158) | P value | |
| Female | 1,538 (35.1) | 289 (36.9) | 0.34 | 204 (25.8) | 51 (32.3) | 0.09 |
| Age (years) | 63.2 ± 6.5 | 61.6 ± 6.4 | <0.0001 | 63.33 ± 8.47 | 62.09 ± 9.29 | 0.10 |
| T2D duration (years) | 10.9 ± 7.5 | 9.1 ± 6.9 | <0.0001 | 10.5 ± 8.38 | 9.06 ± 8.71 | 0.05 |
| BMI (kg/m2) | 33.2 ± 5.2 | 31.9 ± 4.8 | <0.0001 | 31.8 ± 5.41 | 32.33 ± 5.97 | 0.27 |
| Waist circumference (cm) | 110.0 ± 12.9 | 105.3 ± 12.0 | <0.0001 | 108.99 ± 13.08 | 108.42 ± 13.97 | 0.62 |
| Height (cm) | 171.7 ± 9.5 | 170.0 ± 9.4 | <0.0001 | NA | NA | NA |
| HbA1c (%) | 8.22 ± 0.93 | 8.11 ± 0.90 | 0.02 | 7.54 ± 1.54 | 7.66 ± 1.63 | 0.40 |
| Fasting glucose (mmol/L) | 9.93 ± 2.82 | 9.75 ± 2.76 | 0.09 | NA | NA | NA |
| SBP (mmHg) | 135.2 ± 16.7 | 134.7 ± 16.4 | 0.42 | 131.65 ± 20.6 | 128.75 ± 16.32 | 0.05 |
| DBP (mmHg) | 74.1 ± 10.4 | 75.0 ± 10.7 | 0.03 | 73.82 ± 11.92 | 73.54 ± 11.22 | 0.78 |
| LDL (mmol/L) | 2.67 ± 0.84 | 2.68 ± 0.83 | 0.52 | 2.43 ± 0.83 | 2.59 ± 0.85 | 0.03 |
| HDL (mmol/L) | 1.04 ± 0.27 | 1.06 ± 0.29 | 0.0047 | 0.95 ± 0.23 | 0.98 ± 0.23 | 0.13 |
| Women | 1.16 ± 0.29 | 1.22 ± 0.29 | 0.0009 | 1.06 ± 0.24 | 1.02 ± 0.22 | 0.19 |
| Men | 0.97 ± 0.23 | 0.98 ± 0.23 | 0.54 | 0.91 ± 0.21 | 0.96 ± 0.24 | 0.02 |
| Total cholesterol (mmol/L) | 4.74 ± 1.05 | 4.76 ± 1.04 | 0.33 | 4.35 ± 1.05 | 4.56 ± 1.16 | 0.03 |
| Triglycerides (mmol/L)† | 1.95 (1.37–2.81) | 1.91 (1.29–2.76) | 0.14 | 1.76 (1.24–2.58) | 1.90 (1.28–2.7) | 0.46 |
| eGFR (mL/min/1.73 m2) | 87.8 ± 21.7 | 90.4 ± 20.4 | 0.0016 | NA | NA | NA |
| UACR (mg/mmol)† | 1.4 (0.7–4.2) | 1.1 (0.6–2.8) | <0.0001 | NA | NA | NA |
| Previous CV event* | 1,556 (35.5) | 264 (33.7) | 0.33 | 318 (40.2) | 69 (43.7) | 0.42 |
| Report of retinopathy | 439 (11.3) | 40 (5.6) | <0.0001 | NA | NA | NA |
| Current smoker | 496 (11.3) | 84 (10.7) | 0.8 | 63 (8) | 19 (12.1) | 0.09 |
| Previous smoker | 2,194 (57.0) | 390 (56.2) | 0.8 | 456 (57.8) | 88 (56.1) | 0.69 |
| Insulin therapy | 1,598 (36.5) | 220 (28.1) | <0.0001 | 212 (26.8) | 27 (17.1) | 0.01 |
| ACCORD Glycemia trial | ||||||
| Standard | 2,227 (50.8) | 371 (47.3) | 0.07 | — | — | — |
| Intensive | 2,157 (49.2) | 413 (52.7) | — | — | — | |
| ACCORD BP trial | 1,862 (42.5) | 361 (46.1) | 0.06 | — | — | — |
| Standard | 926 (21.1) | 182 (23.2) | 0.81 | — | — | — |
| Intensive | 936 (21.4) | 179 (22.8) | — | — | — | |
| ACCORD Lipid trial | 2,522 (57.5) | 423 (54.0) | 0.06 | — | — | — |
| Fibrate | 1,294 (29.5) | 219 (27.9) | 0.86 | — | — | — |
| Placebo | 1,228 (28.0) | 204 (26.0) | — | — | — | |
| BARI 2D CV intervention | ||||||
| Medical therapy | — | — | — | 398 (50.3) | 67 (42.4) | 0.07 |
| Early revascularization | — | — | — | 393 (49.7) | 91 (57.6) | |
| BARI 2D diabetic therapy | ||||||
| Insulin provision | — | — | — | 394 (49.8) | 71 (44.9) | 0.26 |
| Insulin sensitization | — | — | — | 397 (50.2) | 87 (55.1) | |
Except where noted, data are means ± SD for continuous variables and counts (%) for categorical data. BP, blood pressure; CV, cardiovascular; DBP, diastolic blood pressure; NA, not available; SBP, systolic blood pressure.
†Median (interquartile range). *Prior CV event: In ACCORD, this includes secondary prevention status or history of myocardial infarction, stroke, angina, and/or ischemic changes (electrocardiogram) on graded exercise tolerance test or positive imaging, coronary revascularization procedures, or other revascularization procedures at baseline; in BARI 2D, this includes history of myocardial infarction, congestive heart failure, stroke, or transient ischemic attack.
GWAS of DPN
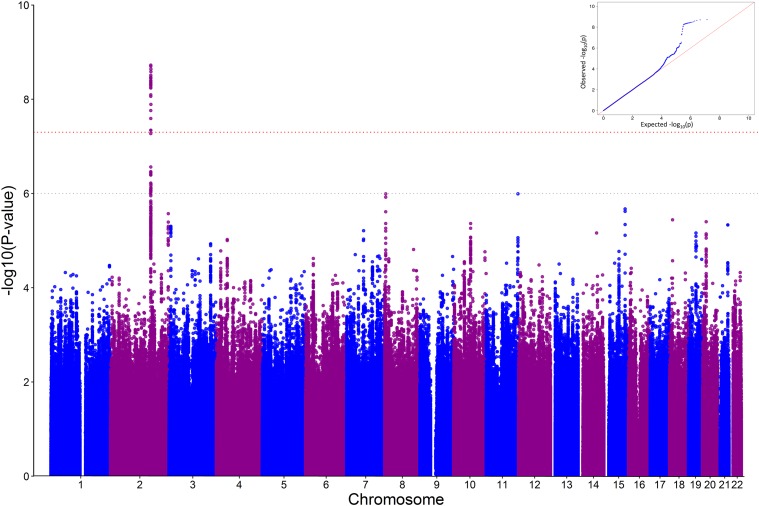
A total of 6,847,206 common variants (MAF >0.05), genotyped or imputed, were tested for association with DPN in the discovery set. Of these, 28 reached genome-wide significance (P < 5 × 10−8), as summarized in the Manhattan and QQ plots shown in Fig. 1. These variants lie in the same region of chromosome 2q24 and are in strong LD with each other in whites (Fig. 2A). When the analysis was conditioned on the lead SNP (rs13417783), the effects of all the other genome-wide significant variants disappeared (Supplementary Fig. 1), indicating that all 28 variants captured the same genetic effect. The lead SNP had a MAF of 0.15 and was associated with a 36% decrease in the odds of DPN per minor allele copy (odds ratio [OR] 0.64, 95% CI 0.55–0.74; P = 1.9 × 10−9) (Table 2). While rs13417783 was an imputed variant (with an excellent quality of imputation), other SNPs in the genome-wide significant cluster were genotyped (Supplementary Table 2). Effects were in the same direction in both genotype sets of ACCORD (Supplementary Table 2). Two other loci, rs13265430 on chromosome 8p23 and rs77494074 on chromosome 11q25, attained P values in the notable range (P = 1 × 10−6 to 5 × 10−8), and 10 more attained P values in the 1 × 10−5 to 1 ×10−6 range (Table 2). All of these associations were only modestly affected by adjustment for differences in age, diabetes duration, and/or BMI (Supplementary Table 3). No evidence of association was observed with the loci on chromosome 8 and 12, where a suggestive association with painful DPN had been detected in a previous GWAS (15) (Supplementary Table 4). Of 14 polymorphisms that were previously reported to be associated with DPN (30) and for which data were available in the ACCORD GWAS, 1 (rs1963645 at the NOS1AP locus) showed a significant association with DPN. However, this was in the opposite direction as that previously reported (Supplementary Table 5).
Figure 1.
GWAS results. A: Manhattan plot. Each dot represents a polymorphism (SNP). The x-axis depicts each chromosome and the y-axis shows the negative log10 P value for association of each SNP with DPN. The red dotted line indicates the genome-wide significance threshold of P = 5 × 10−8; the gray dotted line indicates the notable genome-wide significance threshold of P = 1 × 10−6. B: QQ plot (inset). The black dashed line indicates the null hypothesis. The blue line represents the observed log10 P values corresponding to the expected P values. λ = 0.994.
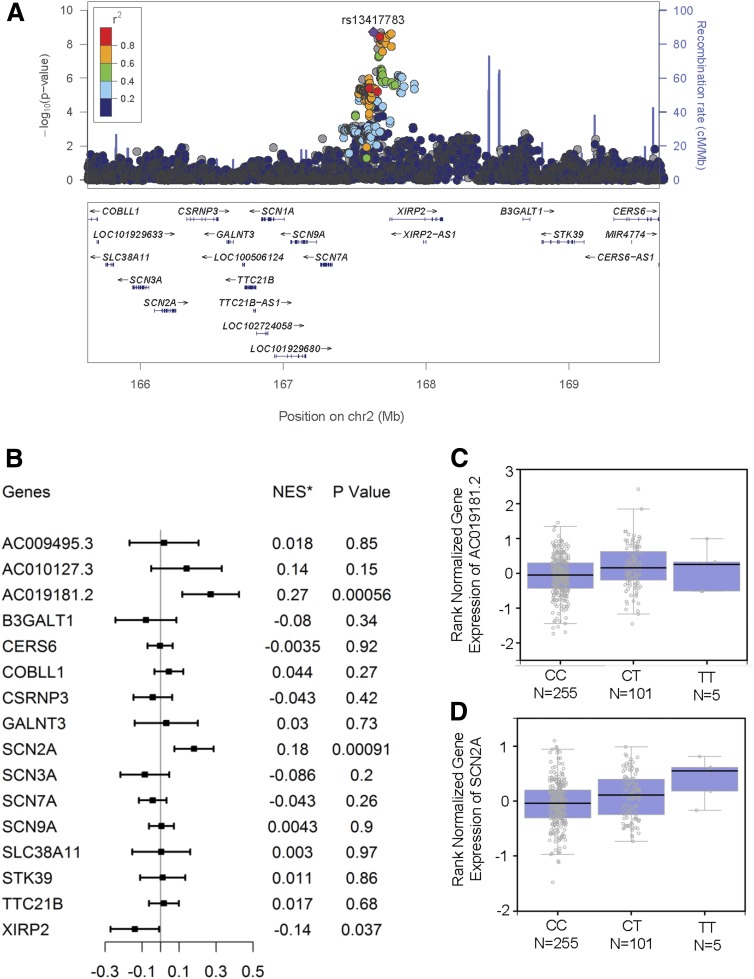
Figure 2.
Characterization of locus 2q24. A: Regional association plot of locus 2q24. Variants are displayed ±2 Mbp upstream and downstream of the lead SNP (rs13417783). The x-axis depicts the chromosomal positions of the SNPs according to National Center for Biotechnology Information Build 37, and the y-axis depicts the −log10 P values for association of these SNPs with DPN in ACCORD. SNPs in strong LD with the lead SNP (purple) are marked in red (r2 > 0.8). The bottom panel depicts genes from the University of California, Santa Cruz (UCSC) Known Genes data set within the region. The locus zoom plot was generated using LocusZoom (Abecasis Laboratory, University of Michigan School of Public Health) through http://locuszoom.sph.umich.edu/genform.php?type=yourdata. The reference database was hg19/1000 Genomes Nov 2014 European. B: eQTL analysis of locus 2q24 showing the association between rs13417783 and tibial nerve–specific expression of neighboring genes. The eQTL analysis was conducted on data from the GTEx database (http://www.gtexportal.org/home/). Genes were those located within ±2 Mbp from the lead SNP rs13417783. *NES is the normalized effect size, i.e., the slope of the linear regression between minor (T) allele dosage and gene expression. This effect size is computed in a normalized space where magnitude has no direct biological interpretation. C: Tibial nerve–specific expression of AC019181.2 stratified by rs13417783 genotypes. D: Tibial nerve–specific expression of SCN2A stratified by rs13417783 genotypes. Box plots were generated in GTEx.
Validation of GWAS Findings
The 13 independent loci that attained P < 1 × 10−5 in the GWAS were investigated for their association with DPN in the validation set. The effect of the top GWAS locus (2q24) was successfully replicated, with an OR of 0.57 per lead SNP (rs13417783) minor allele copy (95% CI 0.41–0.80, P = 9 × 10−4) (Table 2). One of the other 27 SNPs at 2q24 that were GWAS-significant in the discovery set (rs10200297) showed a stronger association than rs13417783 in the validation set (Supplementary Table 6). However, in a meta-analysis of the discovery and validation sets, rs13417783 remained the lead SNP at this locus (OR 0.63, 95% CI 0.55–0.72, P = 7.9 × 10−12). Among the other loci with P < 1 × 10−5 in the discovery set, one (rs10555080, aka rs72397229, on 19q12) showed a nominally significant association with DPN in the validation set (P = 0.0098), resulting in a P value in the notable range (P = 2.6 × 10−7) in the two sets combined. Another locus (rs201655918 on 14q24) was also nominally significant in the validation set, but the effect was in the opposite direction than in the discovery set. Four loci (rs1202660 on 7q11; rs2491019 on 10q22; rs11073752 on 15q25, and rs9948095 on 18p11) showed nonsignificant associations in the validation set that were in the same direction as in the discovery set. One of these SNPs (rs11073752) attained a P value in the notable range (P = 6.5 × 10−7) in the meta-analysis of the discovery and validation sets.
Characteristics of the Association Between 2q24 Locus and DPN
The magnitude of the genetic effect at 2q24 in whites was similar if the analysis was restricted to prevalent or incident cases both in ACCORD and BARI 2D (Table 3). In ACCORD, no significant differences in the strength of the association were observed in relation to clinical characteristics, except for a significantly stronger effect among participants with self-reported diabetic retinopathy than in those without this trait (P = 0.02) (Supplementary Fig. 2). No interaction was found with the intervention tested in ACCORD (i.e., intensive vs. standard glycemic control, intensive vs. standard blood pressure control, and fenofibrate vs. placebo) (Supplementary Fig. 3). When the MNSI score components were analyzed individually, the strongest association was seen with alterations of foot appearance (OR 0.71, P = 2 × 10−7) and ankle jerk (OR 0.76, P = 2 × 10−5), followed by loss of vibration perception (OR 0.88, P = 0.04), whereas no association could be observed with foot ulcer (OR 1.00, P = 0.99). Nominally significant associations were observed between 2q24 locus and triglycerides, estimated glomerular filtration rate (eGFR), and UACR at baseline (Supplementary Table 7). As reported in the Type 2 Diabetes Knowledge Portal (http://www.type2diabetesgenetics.org), an association between triglycerides and the index SNP (rs13417783) as well as the other GWAS-significant SNPs in LD with it had been previously detected in the Genetics of Diabetes Audit and Research in Tayside Scotland (GoDARTS) study (P = 0.00011 for rs13417783). However, adjustment for this variable or for the other two associated with 2q24 (eGFR and UACR) did not attenuate the association of this locus with DPN (Supplementary Table 8). Similarly, adjustment for age and duration of diabetes, which showed significant differences between case subjects and control subjects, had no effect (Supplementary Table 8). In addition, the association between the 2q24 locus and DPN was unaffected (OR 0.64, 95% CI 0.54–0.77) by selecting control subjects who had a longer duration of diabetes (longer than the median of 7 years), and was only modestly reduced (OR 0.73, 95% CI 0.60–0.89) by selecting older control subjects (older than the median age of 61 years). No association was detected between the 2q24 locus and risk of cardiac autonomic neuropathy or risk of micro- and/or macrovascular complications (Supplementary Tables 9 and 10).
Table 3.
Association between top loci and prevalent/incident DPN in ACCORD and BARI 2D
| ACCORD |
BARI 2D |
|||||
|---|---|---|---|---|---|---|
| N case/N control subjects | OR (95% CI) | HR (95% CI) | N case/N control subjects | OR (95% CI) | HR (95% CI) | |
| Case vs. control subjects | ||||||
| Prevalent | 2,547/784 | 0.64 (0.55–0.75) | — | 518/158 | 0.59 (0.42–0.83) | — |
| Incident | 1,837/784 | 0.65 (0.56–0.77) | 0.80 (0.73–0.88) | 271/158 | 0.59 (0.40–0.87) | 0.69 (0.55–0.88) |
2q24 Locus and DPN in African Americans
None of the 28 SNPs at the 2q24 locus showing GWAS-significant association with DPN in whites were associated with DPN in African Americans from ACCORD (n = 1,345), as judged on the basis of a two-sided P < 0.05. However, 15 of these SNPs had two-sided P < 0.20, with effects going in the same direction as in whites (OR 0.75–0.85) (Supplementary Table 6). The strongest association was observed with rs16852735 (OR 0.75, two-sided P = 0.069, one-sided P = 0.034), which in African Americans was not in LD (r2 = 0.02) with the top SNP in whites (rs13417783). The number of African Americans in BARI 2D was too small for a meaningful analysis.
Low-Frequency Variants at 2q24 and DPN
On the basis of the findings with the common variants, five low-frequency variants (MAF 0.01–0.05), which were placed in the same LD block as the lead SNP (rs13417783) and had been genotyped in ACCORD, were tested for association with DPN among white participants. All five variants, which were in high LD with each other (r2 = 0.82–1.00), showed a significant association with DPN (P < 0.01, based on a Bonferroni adjustment for five comparisons) (Table 4). This association was somewhat attenuated but remained significant after adjustment for rs13417783 (Table 4). Conversely, adjustment for any of these low-frequency variants had minimal impact on the magnitude of the association between the common variant (rs13417783) and DPN (Table 4). Two of these variants (rs1509652 and rs6755015) were genotyped in BARI 2D and did not show any association with DPN (OR 0.94 and OR 1.07, respectively, P > 0.05 for both), although power for the analysis of low-frequency variants was limited in this data set due to the smaller sample size.
Table 4.
Association between low-frequency variants at 2q24 and DPN
| Low-frequency variant | Position | Alleles* | MAF | r2‡ | Univariable |
Multivariable† |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low-frequency variant |
rs13417783 |
Low-frequency variant |
rs13417783 |
|||||||||
| OR | P | OR | P | OR | P | OR | P | |||||
| rs16852465 | 2:167524744 | G/A | 0.036 | 0.009 | 1.94 | 3.8E–04 | 0.64 | 1.9E–09 | 1.80 | 1.7E–03 | 0.66 | 2.1E–08 |
| rs1509652 | 2:167589745 | C/T | 0.038 | 0.01 | 1.76 | 1.1E–03 | 0.64 | 1.9E–09 | 1.63 | 5.3E–03 | 0.66 | 2.0E–08 |
| rs11884905 | 2:167611786 | C/T | 0.037 | 0.009 | 1.84 | 6.7E–04 | 0.64 | 1.9E–09 | 1.70 | 3.1E–03 | 0.66 | 2.1E–08 |
| rs6721669 | 2:167636784 | T/C | 0.035 | 0.009 | 1.98 | 2.9E–04 | 0.64 | 1.9E–09 | 1.84 | 1.3E–03 | 0.66 | 2.2E–08 |
| rs6755015 | 2:167666021 | T/C | 0.040 | 0.008 | 1.68 | 2.5E–03 | 0.64 | 1.9E–09 | 1.56 | 1.0E–02 | 0.66 | 1.6E–08 |
*Minor allele/major allele. †Results of regression models including one of the low-frequency variants along with rs13417783 as DPN predictors.
‡r2 with rs13417783.
Association Between 2q24 Locus and Gene Expression
The common variants associated with DPN at 2q24 span 80 kb of noncoding sequence placed between a cluster of genes coding for NaVs (SCN1A, SCN2A, SCN3A, SCN7A, and SCN9A) on the centromeric side and XIRP2 (xin actin binding repeat containing 2) on the telomeric side (Fig. 2A). In an eQTL analysis of GTEx data concerning 16 genes placed in a 2-Mbp radius from the lead SNP, carriers of the rs13417783 minor allele had significantly higher tibial nerve expression of SCN2A, coding for NaVα subunit 2, and AC019181.2 (LOC101929633), coding for an uncharacterized noncoding RNA (P = 9 × 10−4 and P = 6 × 10−4, respectively) (Fig. 2B–D). A nominally significant association (P = 0.03) was also detected with lower expression of the immediately adjacent XIRP2 gene.
To gain further insight into SCN2A gene expression in peripheral nerves, we contrasted SCN2A gene expression in the hippocampus (a central nervous system tissue, serving as a positive control), with dorsal root ganglion (DRG), which contains the cell bodies of the neurons supplying the axons to the sensory portion of the tibial nerve, tibial nerves, and skeletal muscle, an excitable nonnervous system tissue, serving as negative control. Coding gene relative abundances (in TPMs) and their corresponding percentiles are provided for all four tissues in Table 5, which includes samples from both sexes. Abundant RNA signal for SCN2A was found in both the hippocampus and in the DRG. Although at lower levels, SCN2A was also consistently expressed (in all but one sample) in the tibial nerve. By contrast, it was undetectable in 80% of the samples of the skeletal muscle samples and, in the remaining 20% of the samples, was among the lowest in abundance across all tissues.
Table 5.
Relative SCN2A abundance (in TPM) and corresponding percentiles across coding genes
| Tissue | Fraction of samples with detectable (TPM >0) SCN2A expression | Mean SCN2A TPM (± SD) among coding genes in samples with TPM >0 | Mean SCN2A percentile (± SD) among coding genes in samples with TPM >0 |
|---|---|---|---|
| Hippocampus | 123 of 123 | 18.8 ± 15.8 | 71.7 ± 14.9 |
| DRG | 21 of 21 | 18.1 ± 6.0 | 49.0 ± 6.6 |
| Tibial nerve | 413 of 414 | 2.0 ± 1.8 | 30.8 ± 5.7 |
| Skeletal muscle | 112 of 564 | 0.13 ± 0.08 | 22.8 ± 3.3 |
Discussion
Through a GWAS of the ACCORD cohort, we have identified a previously unrecognized locus on chromosome 2q24 having a powerful effect on the risk of DPN in T2D. An association of similar strength was found between this locus and DPN in BARI 2D. In both cohorts, the 2q24 minor allele conferred protection from DPN, being more frequent in DPN-negative control subjects than in DPN case subjects and the general population. In further support of these findings, the same allele was found to be associated with higher tibial nerve expression of a nearby gene (SCN2A) coding for the α subunit of NaV1.2, providing a possible functional basis for the genetic effect observed at the population level. This is the first report of a DPN locus that reaches genome-wide significance in a GWAS and is replicated in an independent study.
The fact that almost identical effects on DPN were found for this locus in the discovery (OR 0.63) and replication (OR 0.57) is remarkable and makes this finding especially robust. Such a high degree of concordance reflects the use of the same DPN definition (clinical examination MNSI >2) in studies and the high sensitivity, specificity, and correlation with abnormal nerve conduction characteristic of this DPN outcome (21,22). Other contributing factors include the similar clinical characteristics of the two cohorts with regard to height, BMI, diabetes duration, blood pressure, and smoking status, and the similar incidence of DPN, despite the administration of different interventions.
There are scant data in the literature to which our findings can be compared because only one other GWAS of DPN has been published to date. That study, based on the GoDARTS data sets, found suggestive evidence (P = 1.8 × 10−7) for a DPN locus on chromosome 8p21 near GFRA2 (15). We could not replicate that finding in our study, and conversely, no significant evidence for the locus 2q24 that we identified in ACCORD and BARI 2D was reported in the GoDARTS study. In addition to the possibility of the 8p21 locus being a false positive, given the lack of genome-wide significance and previous replication, there are important differences between the two GWASs that could explain these discrepancies. First, the outcome considered by the GoDARTS study was painful DPN, defined as a prescription history of at least one of five drugs indicated for the treatment of neuropathic pain along with a positive monofilament test for sensory neuropathy, which is known to have low specificity. By contrast, in our GWAS, DPN was defined by means of a validated instrument (MNSI clinical examination) that does not include pain in its assessment. Thus, the two studies captured different aspects of DPN involving different components of the peripheral nervous system (predominantly small fibers in the case of painful DPN vs. large fibers in the case of MNSI-diagnosed DPN).
Second, both ACCORD and BARI 2D were clinical trials, with close follow-up, standardized outcome acquisition by trained professionals, and strict adjudication, whereas the GoDARTS study was based on medical record data collected as part of routine medical care.
Finally, ACCORD and BARI 2D participants were characterized by a long duration of diabetes (15 years on average by the end of the study), which helped minimize misclassification of control subjects—an essential feature for the identification of protective effects on DPN, such as that associated with the 2q24 locus. Although diabetes duration was not specified in the GoDARTS report, it was likely to be shorter owing to the population-based nature of that cohort.
The DPN locus that we have identified may allow clinicians to focus prevention efforts on patients at higher risk of DPN. However, the real relevance of this finding lies in the great potential for improving our understanding of DPN biology and for identifying novel targets for preventive interventions. It is noteworthy in this regard that the newly identified DPN locus is located in close vicinity to a cluster of genes coding for the α subunits of NaVs, which are responsible for multiple aspects of neuron excitability, including thresholds for depolarizing responses and the amplitude of action potentials (31). From the most proximal to the most distal from the top variant, these genes include SCN7A, SCN9A, SCN1A, SCN2A, and SCN3A, which code for NaVx, NaV1.7, NaV1.1, NaV1.2, and NaV1.3, respectively. In the GTEx database, the protective allele of the 2q24 locus was significantly associated with higher expression of one of these genes (SCN2A) in the tibial nerve, supporting the hypothesis that increased activity of the corresponding NaV1.2, which is expressed in DRG and tibial nerve, may guard against the detrimental effects of the diabetic milieu on peripheral nerves. SCN2A mRNA is expressed by human and murine DRG neurons but not by Schwann cells or other cell types that are found in the tibial nerve (mousebrain.org), which suggests that the SCN2A mRNA in the tibial nerve is contributed by sensory nerve axons.
Rare mutations in the SCN2A gene have been implicated in the etiology of neurodevelopmental diseases such as autism spectrum disorder (loss of function mutations) and infantile seizures (gain of function mutations) (32), but the effect of these variants on susceptibility to DPN has never been investigated due to the their rarity, nor has the effect of NaV1.2 loss or gain of function been studied in animal models of DPN. A relationship with DPN etiology has been instead clearly established for NaV1.3 and NaV1.7 (coded by SCN3A and SCN9A, respectively) (33–36), but no differences in their expression were found among 2q24 genotypes in the GTEx database. Whether this was due to the small size and/or the lack of exposure to diabetes of the samples in the GTEx database, or is related to the fact that NaV1.3 and NaV 1.7 are preferentially expressed in small fibers mediating pain transmission, whereas the MNSI score mainly tests large fibers mediating vibration sense and tendon reflexes, remains to be determined.
Altogether, these findings highlight the need for further studies of the SCN genes in the 2q24 region as functional mediators of the DPN locus and potential targets of novel preventive interventions. These studies would include testing of the association between the 2q24 genotype and the expression of these genes in peripheral nerve samples specifically collected from subjects with diabetes, along with studies assessing the impact of overexpressing or knocking down these genes in peripheral nerves of animal models of DPN (37). The noncoding gene AC019181.2 (LOC101929633), which also showed evidence of association with the 2q24 locus in the GTEx database, should be included in these studies given the potential role of noncoding RNAs as regulators of gene expression (38,39).
The genomic mechanisms linking this locus to differences in gene expression are unclear at this time. The index SNP and the other 27 variants reaching GWAS significance are in a noncoding region with quite subtle epigenetic marks. The index SNP was found by ENCODE to be located in a weak enhancer (as indicated by an H3K4m1 mark) in human embryonic stem cells, but whether this regulatory element is also active in neural cells remains to be determined. Of the other 27 SNPs, the 2 highest ranking in RegulomeDb are rs12993796 and rs16852735. The first SNP is located in a DNA segment that binds BATF (a transcription factor involved in the differentiation of immune cells) in a lymphoblastoid cell line (GM12878). The second SNP is placed in a region showing evidence of binding to REST (RE1 silencing transcription factor) in a neuroectodermic cell line (PFSK-1). REST is a transcription factor that represses neuronal genes, including NaVs (40). Because REST is expressed in peripheral nerves (as shown by GTEx data) and in DRG, one can speculate that the higher expression of SCN2A associated with the 2q24 protective allele in these tissues may be due to decreased binding of this repressor to the DNA segment where rs16852735 is located. The other 25 SNPs showed minimal evidence of being functional in the summary provided by RegulomeDb. Resequencing of the LD block in which the GWAS-significant SNPs are located in large series of case and control subjects, followed by ad hoc functional studies, will be necessary to identify the causal variant(s) and understand its (their) impact on genomic function.
Although the association with DPN was strongest at 2q24, another locus placed on chromosome 15q25 yielded notable GWAS significance in the ACCORD GWAS as well as in the meta-analysis with BARI 2D. Especially interesting is the vicinity of this locus to the NTRK3 gene coding for the receptor of neurotrophin 3, which, by prompting the extension of fibers from proprioceptive DRGs to the muscle spindle and the ventral horn of the spinal cord, is responsible for inducing the synaptic connection between sensory and motor neurons (41). On the basis of this evidence, this locus should be considered as a prime candidate for future studies, although the available GTEx data do not suggest an association between the index SNP at 15q25 and NTRK3 expression.
Strengths of our study include the advantages of clinical trials, such as the rigorous and standardized protocols for data acquisition, the regular follow-up, and access to rich phenotype data, as well as the systematic GWAS approach based on genotyped and imputed data of excellent quality and wide coverage. As discussed above, the use of a validation cohort having very similar clinical characteristics to the discovery cohort was another critical strength.
Nonetheless, some limitations should be acknowledged. Because ACCORD and BARI 2D were both designed to include participants with T2D at high risk of cardiovascular disease, whether our findings could be generalized to individuals with different characteristics or with type 1 diabetes remains to be determined. Similarly, our study was limited to white subjects, and it is not known whether these findings also apply to individuals of other races, although the limited data on African American individuals from ACCORD suggest that this might be the case. Also, our study, while larger than most of the genetic studies of DPN published thus far, was powered to detect only relatively large genetic effects and the fact that control subjects were slightly younger and had slightly shorter diabetes duration than case subjects may have biased results toward the null hypothesis. Therefore, other loci having smaller yet functionally relevant effects may have gone undetected. Finally, because the GWAS was limited to common polymorphisms (MAFs >5%), the existence of additional genetic effects due to less frequent variants cannot be excluded.
In summary, we have identified and successfully validated a locus on chromosome 2q24 having a powerful protective effect on the development of DPN in T2D. Tissue expression analysis suggests that this effect may be mediated by higher expression of the NaV1.2 in the tibial nerve, which is known to increase neuronal excitability. These results may provide novel insights into the pathogenesis of DPN and point to a potential new target for interventions aimed at preventing or treating this complication of diabetes.
Supplementary Material
Article Information
Acknowledgments. The authors thank the investigators, staff, and participants of the ACCORD study and the BARI 2D study for their support and contributions and for providing access to these rich data sets. The authors would like to thank the HMS Research Computing Consultant Group for its consulting services, which facilitated the computational analyses detailed in this paper.
Funding. ACCORD study: The ACCORD genome-wide association analysis was supported by National Institutes of Health (NIH) grants HL-110380 (to J.B.B.) and HL-110400 (to A.D.). The project described was also supported by NIH grant DK-36836 (Advanced Genomics and Genetics Core of the Diabetes Research Center at the Joslin Diabetes Center), NIH grant NS-065926 (to T.J.P.), and the National Center for Advancing Translational Sciences (NCATS), NIH, through grant UL1-TR-001111 (also to J.B.B.). The ACCORD study (ClinicalTrials.gov identifier NCT00000620) was supported by National Heart, Lung, and Blood Institute contracts N01-HC-95178, N01-HC-95179, N01-HC-95180, N01-HC-95181, N01-HC-95182, N01-HC-95183, N01-HC-95184, IAA-Y1-HC-9035, and IAA-Y1-HC-1010. Other components of the NIH, including the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute on Aging, and the National Eye Institute, contributed funding. The Centers for Disease Control and Prevention funded substudies within ACCORD on cost effectiveness and health-related quality of life. General Clinical Research Centers and Clinical and Translational Science Awards provided support at many sites. Portions of this research were conducted on the Orchestra High Performance Computing Cluster, supported by the Research Computing Group, at Harvard Medical School. See http://rc.hms.harvard.edu for more information. BARI 2D study: The efforts of P.A.L. and S.C. were in part supported by NIH (R01-NR-013396 to S.C.). BARI 2D was funded by the National Heart, Lung, and Blood Institute and the National Institute of Diabetes and Digestive and Kidney Diseases, grants U01-HL-061744, U01-HL-061746, U01-HL-061748, and U01-HL-063804. BARI 2D samples were genotyped using the Infinium Multi-Ethnic Global Array (Illumina) at the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine. The Center is partially supported by National Cancer Institute Cancer Center Support Grant P30-CA-91842 to the Siteman Cancer Center and by National Center for Research Resources (NCRR) Institute of Clinical and Translational Sciences/Clinical and Translational Science Award, a component of the NIH, and NIH Roadmap for Medical Research, grant UL1-TR-000448. GTEx: The GTEx Project was supported by the Common Fund of the Office of the Director of the NIH and by the National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. The data used for the analyses described in this article were obtained from the GTEx Portal, https://gtexportal.org/home/ on 27 April 2018.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or other funding entities.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. Y.T., H.S., and A.D. designed the study; acquired, analyzed, and interpreted the data from ACCORD; wrote the initial draft of the manuscript; and revised the manuscript to its final form. P.A.L., H.C., and S.C. acquired, analyzed, and interpreted the data from BARI 2D and revised the manuscript to its final form. R.P.-B., B.A.P., and B.C. contributed to the study design, interpreted the data, and revised the manuscript to its final form. P.R.R. and T.J.P. performed the RNA-seq meta-analysis for SCN2A gene expression, wrote the corresponding sections of the manuscript, and revised the manuscript to its final form. M.J.W., A.A.M.-R., J.B.B., and J.C.M. acquired, analyzed, and interpreted the data and revised the manuscript to its final form. H.S. and A.D. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Data and Resource Availability. The ACCORD database is available upon request from the National Heart, Lung, and Blood Institute Biologic Specimen and Data Repository (https://biolincc.nhlbi.nih.gov/studies/accord/). The ACCORD GWAS data have been deposited in the database of Genotypes and Phenotypes (dbGAP, Study Accession: phs001411.v1.p1).
Prior Presentation. Parts of this study were presented as an oral abstract at the 78th Scientific Sessions of the American Diabetes Association, Orlando, FL, 22–26 June 2018.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db19-0109/-/DC1.
References
- 1.Pop-Busui R, Boulton AJ, Feldman EL, et al. Diabetic neuropathy: a position statement by the American Diabetes Association. Diabetes Care 2017;40:136–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.American Diabetes Association Economic costs of diabetes in the U.S. in 2017. Diabetes Care 2018;41:917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordois A, Scuffham P, Shearer A, Oglesby A, Tobian JA. The health care costs of diabetic peripheral neuropathy in the US. Diabetes Care 2003;26:1790–1795 [DOI] [PubMed] [Google Scholar]
- 4.Callaghan BC, Cheng HT, Stables CL, Smith AL, Feldman EL. Diabetic neuropathy: clinical manifestations and current treatments. Lancet Neurol 2012;11:521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DCCT Research Group Effect of intensive diabetes treatment on nerve conduction in the Diabetes Control and Complications Trial. Ann Neurol 1995;38:869–880 [DOI] [PubMed] [Google Scholar]
- 6.Albers JW, Herman WH, Pop-Busui R, et al.; Diabetes Control and Complications Trial /Epidemiology of Diabetes Interventions and Complications Research Group . Effect of prior intensive insulin treatment during the Diabetes Control and Complications Trial (DCCT) on peripheral neuropathy in type 1 diabetes during the Epidemiology of Diabetes Interventions and Complications (EDIC) Study. Diabetes Care 2010;33:1090–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nathan DM, Genuth S, Lachin J, et al.; Diabetes Control and Complications Trial Research Group . The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 1993;329:977–986 [DOI] [PubMed] [Google Scholar]
- 8.Tesfaye S, Chaturvedi N, Eaton SEM, et al.; EURODIAB Prospective Complications Study Group . Vascular risk factors and diabetic neuropathy. N Engl J Med 2005;352:341–350 [DOI] [PubMed] [Google Scholar]
- 9.Van Acker K, Bouhassira D, De Bacquer D, et al. Prevalence and impact on quality of life of peripheral neuropathy with or without neuropathic pain in type 1 and type 2 diabetic patients attending hospital outpatients clinics. Diabetes Metab 2009;35:206–213 [DOI] [PubMed] [Google Scholar]
- 10.Sandholm N, Groop P-H, Doria A. Genetics of diabetic micro-and macrovascular complications. In The Genetics of Type 2 Diabetes and Related Traits: Biology, Physiology and Translation. Florez JC, Ed. Basel, Switzerland, Springer, 2016, p. 153–180 [Google Scholar]
- 11.Hietala K, Forsblom C, Summanen P, Groop PH; FinnDiane Study Group . Heritability of proliferative diabetic retinopathy. Diabetes 2008;57:2176–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandholm N, Van Zuydam N, Ahlqvist E, et al.; The FinnDiane Study Group; The DCCT/EDIC Study Group; GENIE Consortium; SUMMIT Consortium . The genetic landscape of renal complications in type 1 diabetes. J Am Soc Nephrol 2017;28:557–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osztovits J, Horváth T, Littvay L, et al. Effects of genetic vs. environmental factors on cardiovascular autonomic function: a twin study. Diabet Med 2011;28:1241–1248 [DOI] [PubMed] [Google Scholar]
- 14.Davoudi S, Sobrin L. Novel genetic actors of diabetes-associated microvascular complications: retinopathy, kidney disease and neuropathy. Rev Diabet Stud 2015;12:243–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng W, Deshmukh HA, van Zuydam NR, et al.; Wellcome Trust Case Control Consortium 2 (WTCCC2); Surrogate Markers for Micro- and Macro-Vascular Hard Endpoints for Innovative Diabetes Tools (SUMMIT) Study Group . A genome-wide association study suggests an association of Chr8p21.3 (GFRA2) with diabetic neuropathic pain. Eur J Pain 2015;19:392–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Accord Study Group, Buse JB, Bigger JT, Byington RP, et al. Action to Control Cardiovascular Risk in Diabetes (ACCORD) trial: design and methods. Am J Cardiol 2007;99:21i–33i [DOI] [PubMed] [Google Scholar]
- 17.Frye RL, August P, Brooks MM, et al.; BARI 2D Study Group . A randomized trial of therapies for type 2 diabetes and coronary artery disease. N Engl J Med 2009;360:2503–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pop-Busui R, Lu J, Brooks MM, et al.; BARI 2D Study Group . Impact of glycemic control strategies on the progression of diabetic peripheral neuropathy in the Bypass Angioplasty Revascularization Investigation 2 Diabetes (BARI 2D) Cohort. Diabetes Care 2013;36:3208–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brooks MM, Frye RL, Genuth S, et al.; Bypass Angioplasty Revascularization Investigation 2 Diabetes (BARI 2D) Trial Investigators . Hypotheses, design, and methods for the Bypass Angioplasty Revascularization Investigation 2 Diabetes (BARI 2D) Trial. Am J Cardiol 2006;97(12A):9G–19G [DOI] [PubMed] [Google Scholar]
- 20.Ismail-Beigi F, Craven T, Banerji MA, et al.; ACCORD trial group . Effect of intensive treatment of hyperglycaemia on microvascular outcomes in type 2 diabetes: an analysis of the ACCORD randomised trial. Lancet 2010;376:419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldman EL, Stevens MJ, Thomas PK, Brown MB, Canal N, Greene DA. A practical two-step quantitative clinical and electrophysiological assessment for the diagnosis and staging of diabetic neuropathy. Diabetes Care 1994;17:1281–1289 [DOI] [PubMed] [Google Scholar]
- 22.Herman WH, Pop-Busui R, Braffett BH, et al.; DCCT/EDIC Research Group . Use of the Michigan Neuropathy Screening Instrument as a measure of distal symmetrical peripheral neuropathy in Type 1 diabetes: results from the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications. Diabet Med 2012;29:937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah HS, Gao H, Morieri ML, et al. Genetic predictors of cardiovascular mortality during intensive glycemic control in type 2 diabetes: findings from the ACCORD clinical trial. Diabetes Care 2016;39:1915–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cresci S, Wu J, Province MA, et al.; BARI 2D Study Group . Peroxisome proliferator-activated receptor pathway gene polymorphism associated with extent of coronary artery disease in patients with type 2 diabetes in the bypass angioplasty revascularization investigation 2 diabetes trial. Circulation 2011;124:1426–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Battle A, Brown CD, Engelhardt BE, Montgomery SB; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group . Genetic effects on gene expression across human tissues. Nature 2017;550:204–21329022597 [Google Scholar]
- 28.Ray P, Torck A, Quigley L, et al. Comparative transcriptome profiling of the human and mouse dorsal root ganglia: an RNA-seq-based resource for pain and sensory neuroscience research. Pain 2018;159:1325–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.North RY, Li Y, Ray P, et al. Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons. Brain 2019;142:1215–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Witzel II, Jelinek HF, Khalaf K, Lee S, Khandoker AH, Alsafar H. Identifying common genetic risk factors of diabetic neuropathies. Front Endocrinol (Lausanne) 2015;6:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tibbs GR, Posson DJ, Goldstein PA. Voltage-gated ion channels in the PNS: novel therapies for neuropathic pain? Trends Pharmacol Sci 2016;37:522–542 [DOI] [PubMed] [Google Scholar]
- 32.Sanders SJ, Campbell AJ, Cottrell JR, et al. Progress in understanding and treating SCN2a-mediated disorders. Trends Neurosci 2018;41:442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Li Q, Liu X, Liu S. Roles of voltage-gated tetrodotoxin-sensitive sodium channels Na(V)1.3 and Na(V)1.7 in diabetes and painful diabetic neuropathy. Int J Mol Sci 2016;17:1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev 2005;57:397–409 [DOI] [PubMed] [Google Scholar]
- 35.Cheng KI, Wang HC, Chuang YT, et al. Persistent mechanical allodynia positively correlates with an increase in activated microglia and increased P-p38 mitogen-activated protein kinase activation in streptozotocin-induced diabetic rats. Eur J Pain 2014;18:162–173 [DOI] [PubMed] [Google Scholar]
- 36.Persson A-K, Hoeijmakers JGJ, Estacion M, Black JA, Waxman SG. Sodium channels, mitochondria, and axonal degeneration in peripheral neuropathy. Trends Mol Med 2016;22:377–390 [DOI] [PubMed] [Google Scholar]
- 37.O’Brien PD, Sakowski SA, Feldman EL. Mouse models of diabetic neuropathy. ILAR J 2014;54:259–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beltrami C, Angelini TG, Emanueli C. Noncoding RNAs in diabetes vascular complications. J Mol Cell Cardiol 2015;89:42–50 [DOI] [PubMed] [Google Scholar]
- 39.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong JA, Tapia-Ramírez J, Kim S, et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 1995;80:949–957 [DOI] [PubMed] [Google Scholar]
- 41.Chen HH, Tourtellotte WG, Frank E. Muscle spindle-derived neurotrophin 3 regulates synaptic connectivity between muscle sensory and motor neurons. J Neurosci 2002;22:3512–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.