Human immunodeficiency virus type 1 (HIV-1) infection alters the human intestinal microbiome; however, behavioral factors driving these changes remain poorly defined. We show that drug use and sex behavior are important factors associated with intestinal dysbiosis during chronic HIV-1 infection among young men who have sex with men.
Keywords: Microbiome, substance use, HIV-1 infection, men who have sex with men
Abstract
Background
Human immunodeficiency virus type 1 (HIV-1) infection alters the human intestinal microbiome; however, behavioral factors driving these changes remain poorly defined. Here we examine the effects of substance use and sex behavior on the microbiome during HIV-1 infection.
Methods
Archival rectal swab specimens, urine drug test results, and responses to substance use and sex behavior questionnaires were obtained from 37 HIV-positive participants at 2 time points, separated by 6 months, in a cohort examining the effects of substance use in men who have sex with men (MSM). Microbiome profiling was performed using 16S ribosomal RNA gene sequencing, and associations with behavioral factors were examined using 0-inflated negative binomial regression. Further analysis of selected variables of interest was performed using propensity scores to account for multiple confounders.
Results
Using permutational multivariate analysis of variance, we found that receptive anal intercourse, methamphetamine use, and marijuana use were among the most important drivers of microbiome variation. Propensity score–adjusted analyses revealed that methamphetamine use and marijuana use displayed unique associations; methamphetamine use was associated with an increased abundance of Porphyromonas and Granulicatella organisms and a decreased abundance of Ruminococcus, Collinsella, and Parabacteroides organisms, whereas marijuana use was associated with an increased abundance of Ruminococcus, Clostridium cluster IV, Solobacterium, and Fusobacterium organisms and a decreased abundance of Acidaminococcus, Prevotella, Dialister, Anaerostipes, and Dorea organisms.
Conclusions
Drug use and sex behavior are important factors associated with intestinal dysbiosis during chronic HIV-1 infection among young MSM.
The human intestinal microbiome comprises trillions of bacteria, fungi, and viruses, and appreciation of their functions in host biology and disease is increasing [1]. In the gastrointestinal tract, the microbiome helps maintain gut immune homeostasis, and perturbations in the microbiome affect mucosal barrier integrity and immune function [2, 3]. Numerous factors affect microbiome composition, including age, diet, and geography [4–6], making studies of the microbiome challenging to control.
Human immunodeficiency virus type 1 (HIV-1) infection is associated with alterations in the intestinal microbiome, although specific changes observed can differ, based on study type, population, and sampling method. Some studies showed that HIV-infected individuals have a greater abundance of Prevotella organisms and fewer Bacteroides organisms when compared to uninfected controls [7, 8]. However, these findings have not been replicated in other studies [9–11], suggesting that confounding factors may be present. Without considering behavioral factors that may be driving microbial composition, the nature and scope of HIV-associated dysbiosis is not fully defined. Noguera-Julian et al began to address this question by examining microbiome composition among HIV-positive and HIV-negative men who have sex with men (MSM) and found a similar, increased abundance of Prevotella organisms and decreased abundance of Bacteroides organisms among MSM as compared to participating non-MSM, regardless of HIV-1 infection [11], suggesting that sex practices may also influence microbiome composition.
Substance use frequently coexists with HIV-1 infection, particularly among young MSM. The effects of substance use on the microbiome is not fully understood. Substance use disorders are associated with changes in intestinal microbiome diversity when compared to healthy controls, specifically increasing the Prevotella and Ruminococcus abundances while decreasing the Bacteroides abundance, among other genera [12]. One study examining the direct effects of methamphetamine in a rat model similarly found increased Ruminococcaceae numbers [13]. Other studies have shown associations between chronic prescription opioid use and cocaine use and dysbiosis [14, 15], but the detailed effects of other substances are not known. Only one of these studies examined these effects in the context of HIV-1 infection, showing that cocaine-associated dysbiosis was exponentiated in HIV-positive participants [15].
To examine the association between selected behavioral factors and HIV-associated dysbiosis, we characterized the microbiome in a cohort of HIV-seropositive MSM and then analyzed the relative effects of self-reported sex practices and substance use on the microbiome. We hypothesize that behavioral factors significantly influence HIV-associated dysbiosis.
METHODS
Study Design and Participants
This is a retrospective study that used archived biospecimens and clinical data collected from August 2014 to March 2016 through a prospective cohort study, the mSTUDY (National Institute of Drug Abuse project U01 DA036267). The mSTUDY was approved by the UCLA Office of the Human Research Protection Program Institutional Review Board, and all subjects provided written informed consent at the time of enrollment. Participants in the mSTUDY attend study visits approximately every 6 months, during which clinical data and laboratories were collected, peripheral blood mononuclear cells, plasma, and rectal swab specimens were collected and stored until analysis, urine drug screening was performed, and computer-assisted self-interview questionnaires about sex behavior and substance use were completed. Inclusion in the current study was based on HIV-seropositive status and availability of data from at least 2 consecutive study visits. In total, 37 of 151 mSTUDY cohort participants at the time of this study met criteria and were included.
Specimen Collection and DNA Preparation
Rectal sponge specimens (Merocel, BVI, Waltham, MA) were collected via anoscopy under direct visualization and mucosal contact for 2 minutes. Sponges were frozen neat at −80°C until processing in bulk. The samples were transferred to Lysing Matrix E tubes (MP Biomedicals, Burlingame, CA) containing RLT lysis buffer (Qiagen, Hilden, Germany) and bead beated on a TissueLyser (Qiagen). DNA was extracted using the AllPrep DNA/RNA/Protein kit (Qiagen) per the manufacturer’s protocol.
16S Ribosomal RNA (rRNA) Sequencing and Data Processing
Microbiome profiling was performed by sequencing the 16S rRNA gene, and Golay-barcoded primers 27F and 338R were used to amplify the V1V2 region in triplicate reactions as previously described [16]. PCR products were then pooled and sequenced on the Illumina MiSeq platform, using 2 × 250-bp output version 2 chemistry.
Sequences were demultiplexed with Golay error correction, using QIIME, version 1.9.1 [17]. Divisive amplicon denoising algorithm, version 2, was used for error correction, exact sequence inference, read merging, and chimera removal [18]. The resultant amplicon sequence variant table comprised 9732757 merged read pairs (mean number per sample, 131524; range, 59938–557398). Rarefaction was performed at a depth of 59938 reads for the corresponding analyses (0-inflated negative binomial/negative binomial regression, α diversity). For all other analyses, counts were transformed to relative abundances. Taxonomic assignment was done using RDP training set 16 (available at: https://doi.org/10.5281/zenodo.810827). All sequence data has been deposited into BioProject with the accession number PRJNA422134.
Microbiome Analysis
Statistical analyses of microbiome data were performed using the phyloseq (version 1.19.1), vegan (version 2.4-2), and pscl (version 1.4.9) R packages [19, 20]. A marginal model for permutational multivariate analysis of variance (PERMANOVA) as implemented in the “adonis2” R function with by = “margin” was used to identify significant contributors to variation in the overall microbiome composition. Factors with a P value of <.15 were considered for further analysis with propensity scores (Supplemental Table 1). To test for associations between clinical covariates and microbial abundances, 0-inflated negative binomial regression was used to fit sample counts of each bacterial genus with a clinical covariate (eg, drug use; Supplemental Table 2). Genera detected in <20% of the samples at a depth of at least 100 reads were excluded prior to analysis. Since the distribution among ordinal responses was limited, we grouped data into dichotomous categories for all variables, whereby any response of “never” was grouped into the “no” category and all other responses (ie, “rarely,” “sometimes,” “often,” “most of the time,” and “always”) were grouped into the “yes” category. To account for repeated measures in our study, we used a fixed-effects approach by generating a subject-specific dummy variable for inclusion as a covariate in the regression models. Log plasma HIV RNA copies (continuous) and binned CD4+ T-cell counts (ie, 0–200 cells/μL, 200–350 cells/μL, and >350 cells/μL) were also included as covariates, to control for possible confounding from variation in HIV disease severity. Missing data (CD4+ T-cell counts were missing for 6 participants) were predicted using multiple imputation methods [21]. If the 0-inflated negative binomial model produced an error or failed to converge, negative binomial regression was used instead. All P values from the 0-inflated negative binomial/negative binomial analyses were adjusted for multiple hypotheses, using the Benjamini-Hochberg false-discovery rate method.
Propensity Score Analysis
For further analysis of variables of interest, we used propensity scores to control for multiple confounders. Propensity scores are used frequently to isolate an exposure from confounding variables to create a randomization effect [34]. Based on the data from our initial PERMANOVA analysis, we focused our further analysis on the effects of methamphetamine use, marijuana use, and recent receptive anal intercourse. Propensity scores were calculated to balance ethnicity, smoking, alcohol use, other substance use, log HIV RNA copy number, binned CD4+ T-cell count, and any oral sex. A separate propensity score was calculated for methamphetamine use, marijuana use, and receptive anal intercourse. The other 2 exposures of interest were included in each propensity score model (eg, the propensity score for methamphetamine use included marijuana and receptive anal intercourse). Propensity scores were estimated using both generalized boosted models and logistic regression, retaining whichever estimates resulted in better balance between exposure groups [22]. Balance was assessed by computing a Kolmogorov-Smirnov statistic for each covariate in an inverse probability of treatment–weighted sample [23]. If the Kolmogorov-Smirnov statistic was >0.2 for an individual variable in a weighted sample (indicating suboptimal balance), then that variable was included as an additional covariate in the analysis of microbial abundance. Propensity score models were fit using R, version 3.3.2 (R Core Team), using the twang package [24].
Statistical Analyses
For demographic and clinical characteristics, P values for changes between visits were computed using the McNemar test for binary variables, the Wilcoxon signed-rank test for continuous variables, mixed ordinal logistic regression with person-level random intercepts, and mixed multinomial logistic regression models with random intercepts for N×N (N > 2) nominal variables. For the last category of models, the P value represents a likelihood ratio test of all coefficients versus an intercepts-only model.
RESULTS
Characteristics of Study Participants
Demographic characteristics of the study population are shown in Table 1. Of 151 potential participants, only 37 met inclusion criteria for this retrospective study that evaluated archived biospecimens and clinical data collected from August 2014 to March 2016. In general, this is a relatively young cohort (median age 36 years). The median time since HIV-1 diagnosis is 4 years, and approximately 81% of participants report recent history of antiretroviral therapy. The plasma HIV RNA concentration and CD4+ T-cell count in specimens collected at each of 2 consecutive visits are shown in Table 1, and only median CD4+ T-cell counts differed between the 2 time points.
Table 1.
Demographic and Clinical Characteristics of 37 Study Participants, by Study Visit
| Visit 1 | Visit 2 | P a | |
|---|---|---|---|
| Age, y | 36 (28–39) | … | |
| Ethnicity | |||
| Black, non-Hispanic | 17 (46) | … | |
| Black, Hispanic | 1 (3) | … | |
| Hispanic, all races | 17 (46) | … | |
| White, non-Hispanic | 0 (0) | … | |
| Refuse to answer | 2 (5) | … | |
| Employment status | |||
| Disabled | 11 (30) | … | |
| Unemployed | 11 (30) | … | |
| Employed full time | 5 (13) | … | |
| Employed part time | 8 (22) | … | |
| Student | 2 (5) | … | |
| Experienced homelessness in past 6 mo | 12 (32) | 7 (19) | .06 |
| Body mass indexb | 25.8 (22.6–28.5) | 25.3 (21.9–29.2) | .87 |
| Time since HIV infection diagnosis, y | 4 (2–12) | … | |
| HIV-1 RNA level, log10 copies/mL | 3.41 (1.28–4.56) | 2.56 (1.30–4.05) | .24 |
| CD4+ T-cell count | |||
| Overall, cells/μL | 427 (319–568) | 532 (302–713) | .01 |
| By binc | .10 | ||
| 0–200 cells/μL | 8 (22) | 4 (11) | |
| 200–350 cells/μL | 6 (16) | 7 (19) | |
| >350 cells/μL | 23 (62) | 26 (70) | |
| Currently receiving ART | 30 (81) | 30 (81) | >.99 |
| ART regimen | .99 | ||
| NNRTI | 10 (27) | 9 (24) | |
| PI | 8 (22) | 9 (24) | |
| INSTI | 10 (27) | 10 (27) | |
| NNRTI + INSTI | 1 (3) | 1 (3) | |
| PI + INSTI | 1 (3) | 1 (3) | |
| STI prevalence | |||
| Syphilisd | 0 | 2 (5) | .16 |
| N. gonorrhoeae infection | |||
| Rectal | 3 (8) | 4 (11) | .66 |
| Urinary | 0 | 0 | |
| Pharyngeal | 2 (5) | 2 (5) | >.99 |
| C. trachomatis infection | |||
| Rectal | 5 (14) | 2 (5) | .08 |
| Urinary | 2 (5) | 0 | .16 |
| Pharyngeal | 1 (3) | 1 (3) | >.99 |
| Frequency of alcohol use in past 6 mo | <.001 | ||
| Never | 5 (13) | 9 (24) | |
| Monthly | 7 (19) | 10 (27) | |
| 2–4 times/mo | 13 (35) | 9 (24) | |
| 2–3 times/wk | 4 (11) | 2 (5) | |
| ≥4 times/wk | 7 (19) | 5 (14) | |
| Refuse to answer | 1 (3) | 2 (5) | |
| Frequency of smoking in past 6 mo | <.001 | ||
| Never | 5 (13) | 9 (24) | |
| Monthly | 7 (19) | 10 (27) | |
| 2–4 times/mo | 13 (35) | 9 (24) | |
| 2–3 times/wk | 4 (11) | 2 (5) | |
| ≥4 times/wk | 7 (19) | 5 (14) | |
| Refuse to answer | 1 (3) | 2 (5) | |
Data are median value (interquartile range) or no. (%) of participants.
Abbreviations: ART, antiretroviral therapy; C. trachomatis, Chlamydia trachomatis; HIV-1, human immunodeficiency virus type 1; INSTI, integrase strand transfer inhibitor; N. gonorrhoeae, Neisseria gonorrhoeae; NNRTI, nonnucleoside reverse transcriptase inhibitor; PI, protease inhibitor; STI, sexually transmitted infection.
aBinary variables were compared using by the McNemar test; continuous variables, using the Wilcoxon signed rank test; ordinal variables, using mixed ordinal logistic regression; and N×N (N > 2) nominal variables, using mixed multinomial logistic regression models.
bCalculated as the weight in kilograms divided by the height in meters squared.
cVisit 1 includes missing data (CD4+ T-cell counts for 6 participants) that were predicted using multiple imputation methods.
dDetected by a rapid plasma reagin screen.
Substance Use and Sex Practices Among Study Participants
Sex practices are shown in Table 2. Nearly all participants reported oral sex (92% at visit 1 and 65% at visit 2), and the majority included oral semen exposure and/or ingestion (partner ejaculation in mouth and/or swallowing). At visits 1 and 2, 41% and 46% of participants, respectively, reported recent (ie, within the past 7 days) receptive anal intercourse. The frequency of receptive anal intercourse was similar between the 2 visits. Self-reported substance use over the last 6 months is shown in Table 2. While most participants reported some substance use at visits 1 and 2 (83% and 65%, respectively), some did not use any drugs. The most commonly used substances were methamphetamine (65% and 57% during visits 1 and 2, respectively) and marijuana (49% and 30%, respectively). Few reported opiate use in this cohort. Urine drug screen results from the time of study visit are also shown (Table 2). Again, methamphetamines and marijuana remained the most commonly used substances. Since urine drug screens have a limited detection window (eg, methamphetamines are detected ≥72 hours after use), self-reported data were used for the microbiome analyses because they would more broadly capture substance use among participants.
Table 2.
Self-Reported Sex Behavior and Substance Use and Findings of Urine Drug Screen, by Study Visit
| Characteristic | Visit 1 | Visit 2 | P a |
|---|---|---|---|
| Sex behavior | |||
| Recent receptive anal intercourse in past 7 d | 15 (41) | 17 (46) | .80 |
| Receptive anal intercourse events in past 30 d, no., mean ± SD | 2.8 ± 5.2 | 3.4 ± 4.9 | .96 |
| Any oral sex in past 90 d | 34 (92) | 24 (65) | .004 |
| Oral sex with partner ejaculation in mouth in past 90 d | .17 | ||
| Never | 9 (27) | 9 (38) | |
| Rarely | 12 (35) | 9 (38) | |
| Sometimes | 4 (12) | 4 (17) | |
| Often | 3 (9) | 0 | |
| Most of the time | 6 (18) | 2 (8) | |
| Oral sex with swallowing partner ejaculate in past 90 d | .62 | ||
| Never | 6 (24) | 3 (20) | |
| Rarely | 6 (24) | 9 (60) | |
| Sometimes | 2 (8) | 0 | |
| Often | 4 (16) | 0 | |
| Most of the time | 5 (20) | 0 | |
| Always | 2 (8) | 3 (20) | |
| Substance useb | |||
| Any use | 31 (83) | 24 (65) | .008 |
| Drug used | |||
| Methamphetamines | 24 (65) | 21 (57) | .08 |
| Ecstasy | 8 (22) | 2 (5) | .01 |
| Cocaine | 9 (24) | 3 (8) | .16 |
| Opiates | 1 (3) | 1 (3) | >.99 |
| Party drugs | 9 (24) | 3 (8) | .03 |
| Marijuana | 18 (49) | 11 (30) | .008 |
| Prescription drugs | 5 (14) | 2 (5) | .08 |
| Erectile dysfunction drugs | 8 (22) | 7 (19) | .74 |
| Poppers | 12 (32) | 10 (27) | .32 |
| Synthetic drugs (e.g. spice, bath salts) | 1 (3) | 3 (8) | .32 |
| Urine drug screen | |||
| Any positive result | 16 (43) | 16 (43) | >.99 |
| Drug detected | |||
| Methamphetamines | 15 (41) | 14 (38) | .74 |
| Opiates | 1 (3) | 0 | .32 |
| Cocaine | 3 (8) | 2 (5) | .56 |
| Ecstasy | 0 | 1 (3) | .32 |
| Marijuana | 3 (8) | 10 (27) | .008 |
| Amphetamines | 6 (16) | 13 (35) | .04 |
Data are no. (%) of participants unless otherwise stated.
aBinary variables were compared using by the McNemar test; continuous variables, using the Wilcoxon signed rank test; ordinal variables, using mixed ordinal logistic regression; and N×N (N > 2) nominal variables, using mixed multinomial logistic regression models.
bIn the past 6 months.
Intestinal Microbiome Composition Is Affected by Substance Use and Receptive Anal Intercourse
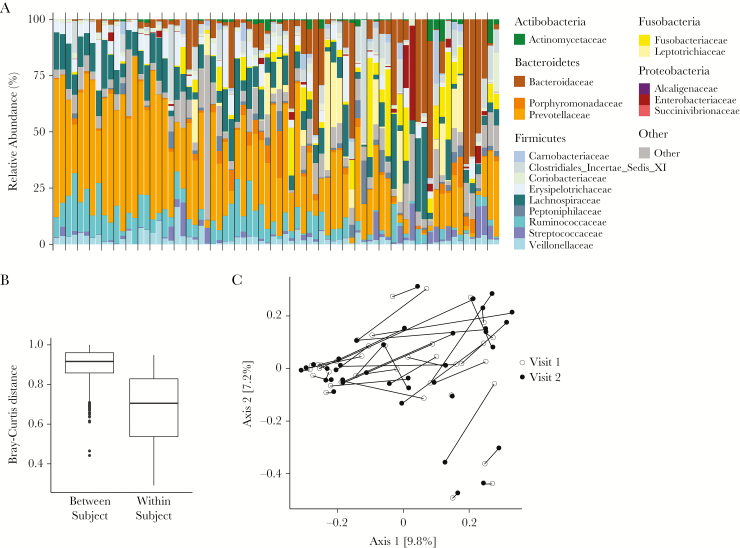
Since we had clinical, substance use, and sex behavior data associated with each visit, we analyzed the intestinal microbiota abundance and distribution at each visit (n = 74) while controlling for a subject-specific identifier. Unsurprisingly, we found that the intestinal microbiome was relatively stable within individuals over the 6-month interval between visits (Figure 1). Using PERMANOVA with Bray-Curtis distances (Supplemental Table 1), we found that, among the most important factors driving microbial variation were receptive anal intercourse (R2 = 0.01, P = .13), methamphetamine use (R2 = 0.01, P = .06), and marijuana use (R2 = 0.01, P = .14). Other contributing factors that were not used for further analysis, owing to limited samples, were gonorrhea (R2 = 0.01, P = .06) and chlamydia (R2 = 0.01, P = .08). We separately examined the influence of each factor on α diversity but found no significant differences (data not shown).
Figure 1.
Taxonomic distribution and similarity between samples across time points. A, Stacked columns show relative abundance as a percentage of total bacterial sequences at the family level. Each vertical bar indicates findings for a sample from 1 time point, and the vertical gray lines delineate different individuals. Families detected in <20% of the samples at a depth of 100 reads are aggregated into the category “Other.” B, Bar plot showing Bray-Curtis distances between participants (left) and between time points within the same participant (right). C, Principal coordinates analysis plot using the Bray-Curtis dissimilarity metric. Lines connect samples from visit 1 (open circle) and visit 2 (closed circle) within the same individual. Numbers in brackets denote the percentage variation explained by each axis.
Oral Sex and Receptive Anal Intercourse Are Associated With Distinct Intestinal Microbiome Bacterial Genera
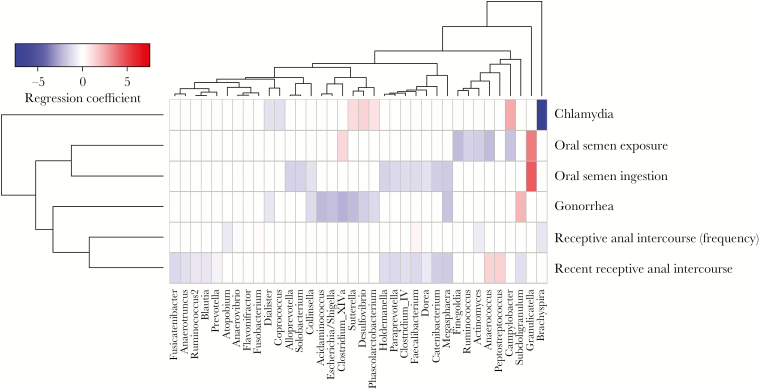
The relationship between self-reported sex behavior and the relative abundance of bacterial genera was examined. To account for variation in HIV disease severity among participants, HIV RNA copy numbers and CD4+ T-cell counts were included as separate covariates in all analyses. Oral sex with partner ejaculation was associated with increased abundance of Granulicatella and Clostridium cluster XIVa organisms and with decreased abundance of Campylobacter, Actinomyces, and Firmicutes organisms, including Finegoldia, Ruminococcus, and Anaerococcus organisms (Figure 2). Receptive anal intercourse was associated with increased Anaerococcus and Peptostreptococcus abundances, as well as a decreased abundance of Firmicutes organisms, including the family Clostridiaceae (ie, Clostridium complex IV, Dorea, Anaerotruncus, Faecalibacterium, and Subdoligranulum organisms; Figure 2). Of note, recency and frequency of receptive anal intercourse were associated with different bacterial genera, suggesting that repeated exposures may have a unique influence on the intestinal microbiota. The presence of a sexually transmitted infection (gonorrhea and/or chlamydia) also displayed unique associations with dysbiosis, though this may be limited by the relatively low prevalence of sexually transmitted infection (<15%) in the sample population.
Figure 2.
Associations between microbiome composition and sex behaviors. Heat map showing associations between self-reported sex behavior (Table 2) and microbiome composition after controlling for participant identifier, CD4+ T-cell counts, and human immunodeficiency virus type 1 RNA copies. Red shading indicates positive associations and blue shading indicates negative associations. Intensity of shading refers to the relative strength of the association determined by regression coefficients from 0-inflated negative binomial regression analysis. Only genera that were associated with a behavioral variable with an adjusted P value of <.05 are shown. “Oral semen exposure” refers to oral sex with partner ejaculation in past 90 days (Table 2). “Oral semen ingestion” refers to oral sex with swallowing of partner ejaculate in past 90 days (Table 2). “Recent receptive anal intercourse” refers to receptive anal intercourse in past 7 days (Table 2).
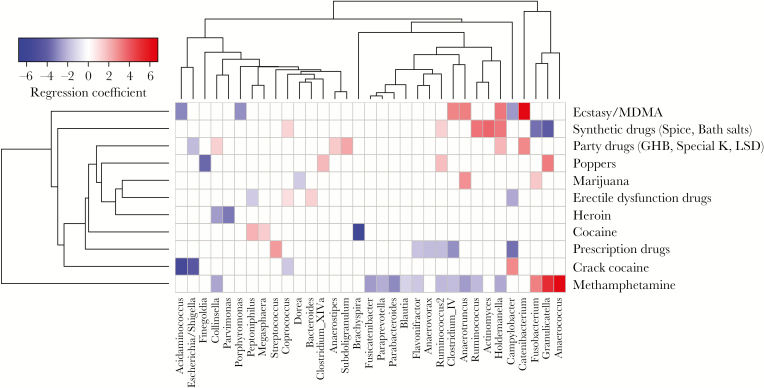
Microbiome Alterations Associated With Substance Use Differ by Drug
The intestinal microbiome signatures associated with substance use are shown in Figure 3. Based on our PERMANOVA results (Supplemental Table 1), we focused additional analyses on 2 substances identified as important drivers of microbiome variation: marijuana and methamphetamine. Marijuana was associated with overabundant Fusobacterium and Anaerotruncus organisms and a decreased abundance of Dorea organisms (Figure 3). Methamphetamine use was associated with increased Fusobacterium, Granulicatella, and Anaerococcus abundances and decreased abundances of Parabacteroides, Collinsella, Paraprevotella, and multiple Clostridiales organisms (ie, species of Fusicatenibacter, Blautia, Ruminococcus, Clostridium complex IV, and Anaerotruncus; Figure 3). These findings are analogous to the microbiome changes associated with receptive anal intercourse (Figure 2). Additionally, some genera with decreased abundances (ie, Parabacteroides, Blautia, and Ruminococcus) are reminiscent of findings associated with HIV-1 infection in other studies [8, 25, 26]. Importantly, CD4+ T-cell count and plasma HIV RNA concentration were explicitly modeled as potential confounding variables in this analysis, suggesting that methamphetamine use and/or receptive anal intercourse may independently exacerbate HIV-associated microbial dysbiosis.
Figure 3.
Associations between microbiome composition and substance use. Heat map showing associations between self-reported substance use and microbiome composition after controlling for participant identifier, CD4+ T-cell counts, and HIV-1 RNA copy numbers. Red shading indicates positive associations, and blue shading indicates negative associations. Intensity of shading refers to the relative strength of the association determined by regression coefficients from 0-inflated negative binomial regression analysis. Only genera that were associated with a substance use variable with an adjusted P value of <.05 are shown.
Propensity Score Analysis Allows Assessment of Microbiome Composition Associated With a Single Behavioral Variable
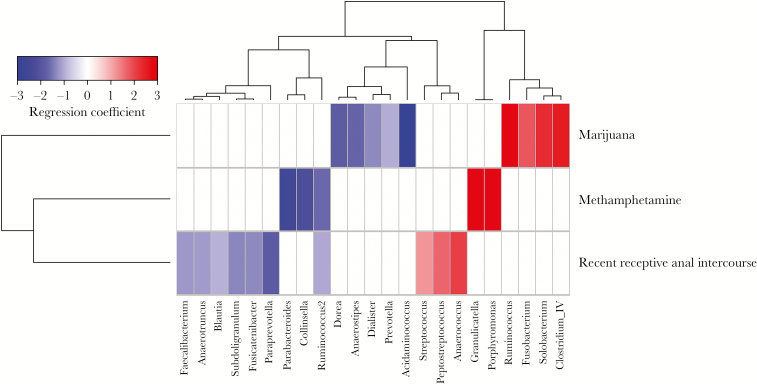
The prior analyses show an overview of the intestinal microbiome changes after controlling for CD4+ T-cell count and HIV RNA copy number but do not take into account the potential role of other factors. Based on the data from our initial analyses (Figures 2 and 3), we focused our further analysis on separately examining the effects of methamphetamine use, marijuana use, and recent receptive anal intercourse. Propensity scores were calculated to balance ethnicity, smoking, alcohol use, other substance use, HIV RNA copy number, CD4+ T-cell count, and any oral sex. Propensity score estimates achieved adequate balance for all 3 variables of interest, with Kolmogorov-Smirnov statistics of <0.2 and no significant differences between any of the covariates (Supplemental Figure 1). The microbiome analysis was then repeated using the propensity score as a covariate.
Associations between bacterial genera and either methamphetamine use, marijuana use, or recent receptive anal intercourse are shown in Figure 4. In general, these findings have some notable differences as compared to those from the initial analysis, likely because propensity score analysis removes the effects of confounding covariates. What remains are the most likely true attributable associations. Marijuana use remained associated with an increased abundance of Fusobacterium organisms and a decreased abundance of Dorea organisms and was now also associated with increased Ruminococcus, Solobacterium, and Clostridium complex IV abundances and decreased Prevotella, Acidaminococcus, Anaerostipes, and Dialister abundances. Methamphetamine use, conversely, remained associated with an increased abundance of Granulicatella organisms and a decreased abundance of Ruminococcus, Parabacteroides, and Collinsella organisms and was now also associated with an increased Porphyromonas abundance. Receptive anal intercourse remained associated with increased abundances of Anaerococcus and Peptostreptococcus organisms and was now also associated with an increased Streptococcus abundance and a decreased abundance of Paraprevotella, Fusicatenibacter, Blautia, and Clostridiaceae organisms (ie, species of Anaerotruncus, Faecalibacterium, and Subdoligranulum). Finally, to examine the potential functional contribution of these microbiome changes, we used PICRUSt for functional inference [27]. Only methamphetamine use displayed significant predicted functional content, with increased Kyoto Encyclopedia of Genes and Genomes pathways involved in the “digestive system” (P = .003; Supplemental Table 3).
Figure 4.
Adjusted associations between microbiome and methamphetamine use, marijuana use, and receptive anal intercourse. Heat map showing adjusted associations between bacterial genera and methamphetamine use, marijuana use, or receptive anal intercourse following balancing of multiple confounders, using propensity scores. Red shading indicates positive associations, and blue shading indicates negative associations. Intensity of shading refers to the relative strength of the association determined by regression coefficients from 0-inflated negative binomial regression analysis. Only genera that were associated with a behavioral/substance use variable with an adjusted P value of <.05 are shown.
DISCUSSION
HIV-associated dysbiosis is an increasingly recognized phenomenon with significant potential clinical implications but unclear drivers. Here we examined the influence of behavioral factors, including substance use and sex practices, on HIV-associated dysbiosis using propensity score models to account for multiple confounding factors.
The presence of rectal microbiome perturbations associated with sex behavior that we identified are in agreement with findings from a larger study [11], although we did not find alterations in richness or diversity in our study. Questions remain to explain the potential mechanisms underlying these observations. Changes associated with receptive anal intercourse could be related to physical trauma, exposure to semen or lubricants, or transfer of microbiota from the insertive partner. This study did not include data on lubricant use with anal intercourse, a known cause of epithelial damage [28–30], but this is certainly a plausible contributor to microbiome alterations associated with receptive anal intercourse that needs additional investigation. Our data also suggest that there may be a difference between recent and frequent receptive anal intercourse (Figure 2), which also warrants additional study. The mechanisms underlying the microbial changes associated with oral sex are less clear. Whether this is related to other sex practices (such as receptive anal intercourse) or is an independent factor needs further study. Using propensity score analysis, we identified 3 genera specifically associated with recent receptive anal intercourse: Anaerococcus, Peptostreptococcus, and Streptococcus (Figure 4). Anaerococcus and Peptostreptococcus organisms are fastidious anaerobes that often require additional nutrients for growth, including bicarbonate [31]. Seminal plasma is a highly nutrient rich environment [32], and therefore its presence via receptive anal intercourse may facilitate abundance of such fastidious organisms. These organisms are also known components of the penile microbiome associated with HIV seroconversion [33]. Anaerococcus organisms have been previously associated with dysbiosis in untreated HIV infection [10]; however, our propensity score–adjusted analysis, which included controlling for HIV RNA copy numbers and CD4+ T-cell counts, may suggest that these associations could be influenced more by sex behavior than HIV-1 infection.
The effects of substance use on the microbiome are largely unexplored, and even less is known in the context of HIV-1 infection. Concordant with findings of a prior study comparing microbiomes in persons with collective substance use disorders to those in controls [12], we found alterations in Ruminococcus, Blautia, Paraprevotella, Parabacteroides, and Fusicatenibacter abundances in our initial analysis (Figure 3). We then performed additional analysis, using propensity score models to account for multiple confounders, including other drugs concurrently used, and examined specific drugs of interest. Our propensity score analysis had some discordant genera as compared to findings by Xu et al [12], which may be due to the influence of uncontrolled confounders accounted for in our analysis, differences in analysis based on grouped substance use versus individual drugs, and the potential influence of HIV-1 status not accounted for by CD4+ T-cell and HIV RNA variables.
We found that methamphetamine use was associated with increased abundance of Porphyromonas organisms (Figure 4), a genus known to have potential inflammatory effects in cardiovascular disease and rheumatoid arthritis [34]. This novel association warrants further investigation because Porphyromonas gingivalis is well known to affect systemic inflammatory cytokines [35, 36] and therefore may contribute to the mechanism underlying increased inflammation in chronic substance use and HIV-1. Differences in bacterial associations with methamphetamine use in our initial unadjusted analysis (Figure 3) and propensity score adjusted model (Figure 4) underscore the importance of this approach. For example, our initial analysis found that methamphetamine use was associated with the Anaerococcus abundance, but after propensity score adjustments this association was only significantly associated with recent receptive anal intercourse, suggesting that the Anaerococcus abundance may be more strongly associated with sex behavior than with drug use, which was accounted for in the propensity score models.
Gastrointestinal functions, including energy metabolism and intestinal motility, are regulated in part by the endocannabinoid system, a collection of endogenous bioactive lipid molecules and cannabinoid receptor proteins 1 and 2, all of which are ubiquitously expressed [37]. The endocannabinoids arachidonoyl-glycerol and arachidonyl-ethanolamide help regulate intestinal permeability and inflammation via cannabinoid receptor 1 [38, 39]. For example, bacterial lipopolysaccharide can induce production of arachidonyl-ethanolamide and arachidonoyl-glycerol [40, 41], and microbiome alterations through dietary modifications in mice affect cannabinoid receptor 1 messenger RNA (mRNA) expression but not cannabinoid receptor 2 mRNA expression [39]. Administration of the cannabinoid receptor 1–agonist Δ9-tetrahydrocannabinol to mice altered ratios of Firmicutes to Bacteroidetes abundances [42]. Our data also found similar alterations associated with marijuana use, most notably increased Ruminococcus, Clostridium cluster IV, and Solobacterium abundances (Figure 4). The inflammatory effects of these changes are not known but could include increased inflammation through microbial translocation via arachidonyl-ethanolamide and cannabinoid receptor 1–agonism, or antiinflammatory effects via arachidonoyl-glycerol production. The role of the endocannabinoid system in HIV-related inflammation and gut permeability is entirely unexplored, but given the associations between marijuana use and the microbiome during HIV-1 infection identified in this study, as well as the prevalence of marijuana use among HIV-infected persons, this is an area of needed study.
With the advancement of the microbiome field comes a greater appreciation for the multitude of factors that influence the microbiome composition. As such, the challenge of designing adequately controlled studies to reduce potential confounders is increasingly recognized. This study faces similar challenges and is subject to limitations. Data regarding diet are not available for these participants, and thus diet remains a potential confounder. Additionally, we were not able to include data on lubricant use during anal intercourse, which is a known cause of epithelial damage and potential contributor to microbiome alterations associated with receptive anal intercourse [28–30]. This study is also limited in size, and while it provides evidence for the effects of sex behavior and substance use on the microbiome in HIV-1 infection, the findings need to be confirmed in larger analyses with greater statistical power to ensure reproducibility. While small studies such as this are valuable to highlight new areas for further investigation, these findings may not be generalizable to broader populations. For example, the lack of an HIV-negative comparison group limits the ability to determine whether these changes in the microbiome are specific to HIV-infected MSM.
Understanding the role of behavioral factors on HIV-associated dysbiosis could lead to novel therapeutic strategies to modulate the immune consequences of chronic HIV-1. This study aimed to increase this understanding by investigating the effects of substance use and sex practices on the microbiome during HIV-1 infection and, in doing so, evaluated a novel analysis strategy that used propensity scores to balance multiple confounders. These findings highlight the importance of considering behavioral factors such as substance use in the design and analysis of future HIV-1 microbiome studies.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank all participants in the mSTUDY, for their generous participation, and Dr Omai Garner, for insightful discussion.
Financial support. This work was supported by the National Institute on Drug Abuse (award U01 DA036267 to S. S.) and the National Institute of Mental Health (award P30 MH058107 to P. M. G. and S. S.); the UCLA Clinical and Translational Science Institute (National Institutes of Health [NIH] award KL2 TR001882 to J. A. F.); the UCLA AIDS Institute and Center for AIDS Research (CFAR) (National Institute of Allergy and Infectious Diseases award P30 AI028697) including the CFAR Microbiome and Mucosal Immunology Core.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: CROI 2018, Boston, Massachusetts, 4–7 March 2018.
References
- 1. Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med 2016; 375:2369–79. [DOI] [PubMed] [Google Scholar]
- 2. Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. Regulation of inflammation by microbiota interactions with the host. Nat Immunol 2017; 18:851–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014; 157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012; 486:222–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 2010; 107:14691–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013; 14:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol 2015; 8:760–72. [DOI] [PubMed] [Google Scholar]
- 9. Yu G, Fadrosh D, Ma B, Ravel J, Goedert JJ. Anal microbiota profiles in HIV-positive and HIV-negative MSM. AIDS 2014; 28:753–60. [DOI] [PubMed] [Google Scholar]
- 10. McHardy IH, Li X, Tong M, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome 2013; 1:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Noguera-Julian M, Rocafort M, Guillén Y, et al. Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016; 5:135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu Y, Xie Z, Wang H, et al. Bacterial diversity of intestinal microbiota in patients with substance use disorders revealed by 16S rRNA gene deep sequencing. Sci Rep 2017; 7:3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ning T, Gong X, Xie L, Ma B. Gut microbiota analysis in rats with methamphetamine-induced conditioned place preference. Front Microbiol 2017; 8:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Banerjee S, Sindberg G, Wang F, et al. Opioid-induced gut microbial disruption and bile dysregulation leads to gut barrier compromise and sustained systemic inflammation. Mucosal Immunol 2016; 9:1418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Volpe GE, Ward H, Mwamburi M, et al. Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. J Stud Alcohol Drugs 2014; 75:347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bender JM, Li F, Martelly S, et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med 2016; 8:349ra100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010; 7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeileis A, Kleiber C, Jackman S. Regression models for count data in R. J Stat Softw 2008; 27:1–25. [Google Scholar]
- 21. Van Buuren S, Brand JPL, Groothuis-Oudshoorn CGM, Rubin DB. Fully conditional specification in multivariate imputation. J Stat Comput Simul 2006; 76:1049–64. [Google Scholar]
- 22. McCaffrey DF, Griffin BA, Almirall D, Slaughter ME, Ramchand R, Burgette LF. A tutorial on propensity score estimation for multiple treatments using generalized boosted models. Stat Med 2013; 32:3388–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morgan SL, Todd JJ. A diagnostic routine for hte detection of consequential heterogeneity of causal effects. Sociol Methodol 2008; 38:231–81. [Google Scholar]
- 24. Ridgeway G, McCaffrey DF, Morral A, Burgette LF, Griffin BA.. Toolkit for weighting and analysis of nonequivalent groups: A tutorial for the Twang package. Santa Monica, CA:RAND Corporation, 2016. [Google Scholar]
- 25. Dillon SM, Lee EJ, Kotter CV, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014; 7:983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mutlu EA, Keshavarzian A, Losurdo J, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 2014; 10:e1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dezzutti CS, Brown ER, Moncla B, et al. Is wetter better? An evaluation of over-the-counter personal lubricants for safety and anti-HIV-1 activity. PLoS One 2012; 7:e48328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fuchs EJ, Lee LA, Torbenson MS, et al. Hyperosmolar sexual lubricant causes epithelial damage in the distal colon: potential implication for HIV transmission. J Infect Dis 2007; 195:703–10. [DOI] [PubMed] [Google Scholar]
- 30. Begay O, Jean-Pierre N, Abraham CJ, et al. Identification of personal lubricants that can cause rectal epithelial cell damage and enhance HIV type 1 replication in vitro. AIDS Res Hum Retroviruses 2011; 27:1019–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marshall R, Yasui VK, Prabhala R, Kaufman AK, Wallace I. Growth of Peptococcus and Peptostreptococcus: effect of variations of culture media on efficiency of recovery. Appl Environ Microbiol 1981; 42:493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bromfield JJ. A role for seminal plasma in modulating pregnancy outcomes in domestic species. Reproduction 2016; 152:R223–32. [DOI] [PubMed] [Google Scholar]
- 33. Liu CM, Prodger JL, Tobian AAR, et al. Penile anaerobic dysbiosis as a risk factor for HIV infection. mBio 2017; 8:e00996–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Slingerland AE, Schwabkey Z, Wiesnoski DH, Jenq RR. Clinical evidence for the microbiome in inflammatory diseases. Front Immunol 2017; 8:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang PL, Ohura K. Porphyromonas gingivalis lipopolysaccharide signaling in gingival fibroblasts-CD14 and Toll-like receptors. Crit Rev Oral Biol Med 2002; 13:132–42. [DOI] [PubMed] [Google Scholar]
- 36. Arimatsu K, Yamada H, Miyazawa H, et al. Oral pathobiont induces systemic inflammation and metabolic changes associated with alteration of gut microbiota. Sci Rep 2014; 4:4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cani PD, Plovier H, Van Hul M, et al. Endocannabinoids–at the crossroads between the gut microbiota and host metabolism. Nat Rev Endocrinol 2016; 12:133–43. [DOI] [PubMed] [Google Scholar]
- 38. Turcotte C, Chouinard F, Lefebvre JS, Flamand N. Regulation of inflammation by cannabinoids, the endocannabinoids 2-arachidonoyl-glycerol and arachidonoyl-ethanolamide, and their metabolites. J Leukoc Biol 2015; 97:1049–70. [DOI] [PubMed] [Google Scholar]
- 39. Muccioli GG, Naslain D, Bäckhed F, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol 2010; 6:392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu J, Bátkai S, Pacher P, et al. Lipopolysaccharide induces anandamide synthesis in macrophages via CD14/MAPK/phosphoinositide 3-kinase/NF-κB independently of platelet-activating factor. J Biol Chem 2003; 278:45034–9. [DOI] [PubMed] [Google Scholar]
- 41. Varga K, Wagner JA, Bridgen DT, Kunos G. Platelet- and macrophage-derived endogenous cannabinoids are involved in endotoxin-induced hypotension. FASEB J 1998; 12:1035–44. [DOI] [PubMed] [Google Scholar]
- 42. Cluny NL, Keenan CM, Reimer RA, Le Foll B, Sharkey KA. Prevention of diet-induced obesity effects on body weight and gut microbiota in mice treated chronically with Δ9-tetrahydrocannabinol. PLoS One 2015; 10:e0144270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.