Key Points
Differences in Steap3 expression are a mechanism for genetic variation in RBC storage and oxidative damage in distinct strains of mice.
Steap3 is identified as a novel pathway for increased oxidative stress in mature RBCs leading to metabolic changes and lipid oxidation.
Abstract
Red blood cells (RBCs) are the most numerous cell type in the body and serve a vital purpose of delivering oxygen to essentially all tissues. In addition to the central role of RBCs in health and disease, RBC storage is a requirement for the >90 million units of RBC transfusions given to millions of recipients each year, worldwide. It is well known that there is genetic donor-to-donor variability in how human RBCs store, rendering blood a nonstandardized therapeutic with a wide range of biological properties from unit to unit, by the time it is transfused. As with humans, genetic variation exists in how murine RBCs, from different strains of mice, store and perform after transfusion. The genetic mechanisms for variation, in humans and mice, both remain obscure. Combining advanced metabolomics, genetics, and molecular and cellular biology approaches, we identify genetic variation in six-transmembrane epithelial antigen of prostate 3 (Steap3) expression as a critical and previously unrecognized mechanism of oxidative damage of RBCs during storage. Increased levels of Steap3 result in degradation of cellular membrane through lipid peroxidation, leading to failure of RBC homeostasis and hemolysis/clearance of RBCs. This article is the first report of a role of Steap3 in mature RBCs; it defines a new mechanism of redox biology in RBCs with a substantial effect upon RBC function and provides a novel mechanistic determinant of genetic variation of RBC storage.
Visual Abstract
Introduction
Transfusion of red blood cells (RBCs) is the single most common inpatient therapy, with ∼90 million units of RBCs transfused annually, worldwide. There is substantial donor-to-donor variability in the efficacy of RBC storage ex vivo, as measured by either spontaneous hemolysis in the bag1 or clearance of RBCs in the first 24 hours after transfusion (range, 0%-70%).2 Indeed, transfusion is a higher order of personalized/precision medicine as both the drug and the patient have genetic variability, resulting not only in a therapy with unstandardized efficacy but also in potential toxicities to individuals who receive units that store poorly.3
A substantial amount of variability of RBC storage has been shown to be heritable both in humans4-6 and in mice.7-9 Detailed metabolomics analysis of RBCs from different mouse strains has identified oxidative stress in general, and oxidation/peroxidation of lipid species in particular, as strongly associated with poor RBC storage7; the same general class of compounds (including several specific oxylipins) have been shown to likewise predict posttransfusion RBC survival of stored human RBCs, better than even storage duration.10
The current study combined mouse genetic mapping and metabolomic approaches to identify differential expression of six-transmembrane epithelial antigen of prostate 3 (Steap3) in mature RBCs as causal in biological differences of RBC storage (both metabolic differences and the correlated posttransfusion circulation). To the best of our knowledge, this study represents the first identification of a nonpathologic genetic variant as a mechanism of differential RBC storage biology. Moreover, these findings constitute the first report of a role of Steap3 in mature RBCs and define a novel mechanism of oxidative damage in RBCs.
Materials and methods
Mice
C57BL/6J, CByJ.B6-Tg(UBC-GFP)30Scha/J, and FVB/J mice were purchased from The Jackson Laboratory. FVB mice were bred with CByJ.B6-Tg(UBC-GFP)30Scha/J mice to generate a green fluorescent protein (GFP)–transgenic animal with a B6xFVB F1 genetic background (labeled GFP-F1). HOD mice were generated as previously described.11 All other breeding was conducted as indicated in the article. All mice were housed in the Bloodworks NW Research Institute vivarium, and all procedures were performed under an Institutional Animal Care and Use Committee–approved protocol.
Generation of Steap3 transgenic mice
The whole open reading frame for the FVB variant of Steap3 was synthesized de novo with a Kozak consensus sequence upstream of the start codon and cloned into the BglII site of (µ’LCR β pr δ Nco Bgl; referred to herein as LCB12) (Genewiz). The final construct was linearized, and bacterial elements were removed through digestion with ClaI and SacII and used for pronuclear injection (Cyagen Biosciences Inc.). Founder pups were identified with polymerase chain reaction (PCR) by using transgene-specific primers.
Generation of B6.FVB-A350V mice
Amino acid 350 of Steap3 is an alanine (A) in B6 mice and a valine (V) in FVB mice (single nucleotide polymorphism [SNP] rs30771292). An A350V mutation was introduced into the B6 genome via CRISPR/Cas9 genome engineering of Steap3 exon 5 in partnership with a commercial vendor with expertise in this approach (Cyagen Biosciences Inc.). Founder pups were genotyped by using PCR, followed by subsequent sequence analysis of Steap3 exon 5 to verify that only the intended mutation was introduced (data not shown).
Blood storage and transfusion
All blood storage, transfusion, and determination of posttransfusion recovery were performed as previously described7,9 and shown in the Results. For testing storage of RBCs to provide phenotype-guided congenic backcrossing, the storage system was modified to allow collection and storage of small volumes of blood (50-75 µL) in an attempt to allow living donor RBC storage.
Metabolomics analysis
RBCs were extracted and analyzed by using previously published methods.13 Details are given in the supplemental Materials and methods.
SNP analysis
Genomic DNA was allelotyped at the DartMouse shared resource facility by using the Illumina BeadArray Reader, interrogating 1445 SNPs across the mouse genome; of these, 813 were informative between the B6 and FVB backgrounds. Raw SNP data were analyzed by using DartMouse SNaP-Map and Map-Synth software.
Quantitative trait loci analysis
Using the aforementioned SNPs and the 156 F2 mice, an association test was made at each SNP, using a simple linear model and extracting the P value associated with the F test. This was obtained by using the “lm” function in R (R Foundation for Statistical Computing). These were plotted (on a –log10P scale) over the genome. To account for multiple testing, the false discovery rates were evaluated by using q value methods, using the “qvalue” function in R. A false discovery rate threshold of q = 0.05 was adopted; that is, of the associations deemed to be statistically significant, 5% of these are likely to be false associations.
Steap3 activity assay
The ability of Steap3 to function as a ferrireductase was quantified by use of the chemical 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine-p,p′-disulfonic acid (common name of “ferrozine”) and measuring changes in absorbance at 562 nm, per published methods.14
Western blot
RBC ghosts (membranes) were prepared and subjected to western blot as previously described15 using either rabbit anti-Steap3 polyclonal (Proteintech; catalog #17186-1-AP) or rabbit anti-actin (Cell Signaling Technology; catalog #4970).
RNA isolation and real-time reverse transcriptase–PCR of gene candidates
Erythroid precursors (EPs) were sorted from isolated bone marrow by using fluorescence-activated cell sorting. Real-time quantitative PCR was performed by using TaqMan primers and probes each open reading frame analyzed (supplemental Table 2). Data are presented as fold-change in expression compared with expression in B6 and normalized to housekeeping genes. Details of the assays are provided in the supplemental Materials and methods.
Cloning, mutation, and expression of Steap3
The open reading frames for the B6 variant of Steap3, B6.A350V, B6.N455S, and the FVB variant were each synthesized de novo, cloned into pcDNA3.1/Zeo(+) (Genewiz), and transfected into suspension CHO cells. Details are provided in the supplemental Materials and methods.
Hemolysis determination by Drabkin’s assay
Hemolysis was determined by measuring supernatant hemoglobin with Drabkin’s reagent (RICCA Chemical Co.; catalog #2660-32) by using standard methods.
RBC life span determination by biotinylation was performed as previously described.8
Statistical analyses
Metabolomics graphs and statistical analyses, including partial least squares discriminant analysis and two factor (fresh/stored + strain), were performed with GraphPad Prism 5.0 (GraphPad Software, Inc.) and MetaboAnalyst 3.0. All other data were analyzed by using Excel (Microsoft Corporation) and GraphPad Prism 7.0. Twenty-four–hour recoveries, gene expression, and ferrozine activity differences between mouse strains was analyzed by using either 1-way or 2-way analysis of variance (ANOVA) followed by Bonferroni post hoc analysis. All reported P values have been corrected for multiple comparisons.
Results
Quantitative trait loci mapping of RBC storage phenotypes between B6 and FVB strains
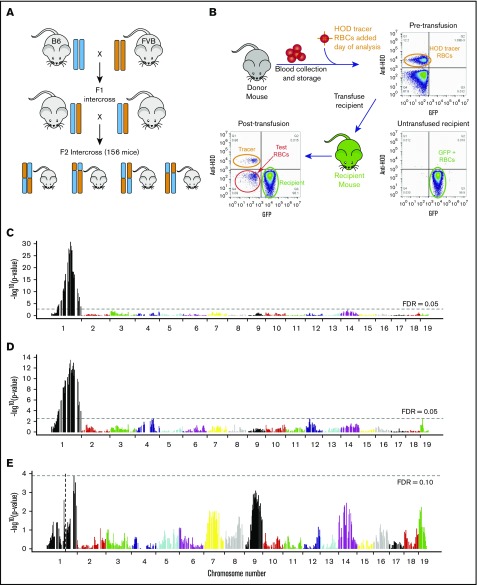
We have previously shown that RBCs from B6 mice store very well, whereas FVB RBCs store poorly.7,8 B6 and FVB mice were bred to generate 156 F2 animals (Figure 1A), and our previously characterized murine model of RBC storage was modified to allow single donor experiments (Figure 1B). Storage of 7 days was observed to be the optimal time to distinguish differences between F2 mice (data not shown), which is consistent with published reports establishing the appropriate storage time to model human RBC storage.7-9 Stored RBCs from individual F2 mice were transfused into GFP-transgenic recipients (UbiC-GFP crossed with wild-type FVB [GFP-F1 mice]). GFP-F1 mice are on a B6xFVB background, and as such, would not be expected to recognize any F2 antigens as foreign. Twenty-four-hour recoveries were calculated by normalizing circulating stored F2 RBCs to fresh HOD tracer RBCs that had been cotransfected. HOD RBCs express a recombinant antigen by which they can be distinguished by staining with anti-HOD antibodies.11 Spontaneous hemolysis was determined in each stored RBC sample by assaying free hemoglobin in the cell-free supernatant.
Figure 1.
QTL mapping of Rbcstor1 and Rbcstor2. (A) B6 mice were crossed with FVB mice to generate an F1 generation. F1 mice were then interbred to generate an F2 generation. A total of 156 F2 mice were analyzed for their genetic backgrounds (SNP-based genotyping) and tested for RBC storage biology both by determining posttransfusion recoveries (depicted in panel B) and by high-resolution metabolomics (detailed in Table 1). (B) Twenty-four–hour recovery of stored RBCs was determined for each individual F2 mouse as depicted. Donor RBCs were stored for 7 days, spiked with a fresh HOD RBC tracer population, and then transfused into GFP mice with a B6xFVB F1 genetic background. Representative flow cytometry plots are shown. (C) QTL analysis using 24-hour recovery as a quantitative phenotype identified a single region of high significance over a broad range of chromosome 1, termed Rbcstor1. (D) QTL analysis using spontaneous hemolysis in the storage tube as a quantitative phenotype identified a single region of high significance over a broad range of chromosome 1, termed Rbcstor2. (E) QTL using 24-hour recovery as a phenotype on the subset of F2 mice containing the same allelotypes at Rbcstor1 did not identify any other contributing QTL. Likewise, taking a similar approach with spontaneous hemolysis and Rbcstor2, no additional contributing QTL was identified (data not shown). FDR, false discovery rate.
RBCs from F2 mice had a wide distribution of posttransfusion 24-hour recoveries and spontaneous hemolysis, with an inverse correlation between the two (supplemental Figure 1). F2 mice were surveyed for 813 SNPs. Quantitative trait loci (QTL) analysis of 24-hour recovery identified a significant QTL (maximum peak P = 2.09 × 10−31), termed Rbcstor1, spanning a ∼149 Mb interval on chromosome 1 (rs13475827 to rs13476300) (Figure 1C). Similarly, QTL analysis of spontaneous hemolysis identified the same region of chromosome 1 (rs13475834 to rs13476300) (Figure 1D). Using a false discovery rate threshold of 0.10, no additional loci were mapped that achieved statistical significance after subtracting RBCstor1 or RBCstor2 (Figure 1E).
Untargeted metabolomics of RBCs stored from F2 donor mice
At the time of transfusion (after 7 days’ storage), a detailed metabolome was generated for stored RBCs from each F2 mouse. Metabolites that were correlated with either 24-hour recoveries or with spontaneous hemolysis were identified by using a cutoff of greater than a correlation of r = 0.50. All correlations had extreme statistical significance, with both P values and q values <1 × 10−15. A list of rank order of metabolites, both positive and negative correlations, is provided in Table 1.
Table 1.
Untargeted metabolomics on RBCs from 156 F2 mice correlating analyte levels to RBC 24-hour recovery and spontaneous hemolysis
| Metabolite | Phenotype 1 | Phenotype 2 | |||||
|---|---|---|---|---|---|---|---|
| P | q value | Correlation | P | q value | Correlation | Subpathway | |
| X-17010 | 0.00E+00 | 0.00E+00 | −0.874753938 | 0.00E+00 | 0.00E+00 | 0.688526854 | |
| 5-Hydroxyhexanoate | 0.00E+00 | 0.00E+00 | −0.864576202 | 0.00E+00 | 0.00E+00 | 0.653912676 | Fatty acid (monohydroxy) |
| 2-Aminoheptanoate | 0.00E+00 | 0.00E+00 | −0.864281519 | 0.00E+00 | 0.00E+00 | 0.636132732 | Fatty acid (amino) |
| Azelate (nonanedioate) | 0.00E+00 | 0.00E+00 | −0.860232367 | 0.00E+00 | 0.00E+00 | 0.626071094 | Fatty acid (dicarboxylate) |
| 2-Hydroxyoctanoate | 0.00E+00 | 0.00E+00 | −0.856764186 | 0.00E+00 | 0.00E+00 | 0.665959185 | Fatty acid (monohydroxy) |
| Undecanedioate | 0.00E+00 | 0.00E+00 | −0.853810808 | 0.00E+00 | 0.00E+00 | 0.627771993 | Fatty acid (dicarboxylate) |
| Glutarate (pentanedioate) | 0.00E+00 | 0.00E+00 | −0.85206903 | 1.00E-15 | 7.00E-15 | 0.602210724 | Lysine metabolism |
| Sebacate (decanedioate) | 0.00E+00 | 0.00E+00 | −0.848597509 | 0.00E+00 | 0.00E+00 | 0.636112085 | Fatty acid (dicarboxylate) |
| X-17438 | 0.00E+00 | 0.00E+00 | −0.8466758 | 0.00E+00 | 0.00E+00 | 0.614796282 | |
| Indole-3-carboxylic acid | 0.00E+00 | 0.00E+00 | −0.846517722 | 3.60E-14 | 2.48E-13 | 0.570818666 | Tryptophan metabolism |
| X-18913 | 0.00E+00 | 0.00E+00 | −0.845652997 | 0.00E+00 | 0.00E+00 | 0.657571074 | |
| 2-Hydroxypalmitate | 0.00E+00 | 0.00E+00 | −0.840877848 | 0.00E+00 | 0.00E+00 | 0.730023848 | Fatty acid (monohydroxy) |
| X-17269 | 0.00E+00 | 0.00E+00 | −0.839764089 | 0.00E+00 | 0.00E+00 | 0.675112373 | |
| 2-Hydroxydecanoate | 0.00E+00 | 0.00E+00 | −0.839426649 | 0.00E+00 | 0.00E+00 | 0.631432459 | Fatty acid (monohydroxy) |
| Suberate (octanedioate) | 0.00E+00 | 0.00E+00 | −0.836743716 | 1.00E-15 | 9.00E-15 | 0.599513973 | Fatty acid (dicarboxylate) |
| X-13879 | 0.00E+00 | 0.00E+00 | −0.834674732 | 2.00E-15 | 1.60E-14 | 0.593612667 | |
| 1-Stearoylglycerophosphoinositol | 0.00E+00 | 0.00E+00 | −0.833827545 | 0.00E+00 | 0.00E+00 | 0.642166753 | Lysolipid |
| 8-Hydroxyoctanoate | 0.00E+00 | 0.00E+00 | −0.832551829 | 0.00E+00 | 3.00E-15 | 0.611349072 | Fatty acid (monohydroxy) |
| 16-Hydroxypalmitate | 0.00E+00 | 0.00E+00 | −0.831001746 | 0.00E+00 | 0.00E+00 | 0.615308123 | Fatty acid (monohydroxy) |
| X-18059 | 0.00E+00 | 0.00E+00 | −0.830245298 | 0.00E+00 | 5.00E-15 | 0.603477289 | |
| Pimelate (heptanedioate) | 0.00E+00 | 0.00E+00 | −0.829496799 | 5.20E-14 | 3.33E-13 | 0.567858806 | Fatty acid (dicarboxylate) |
| 2-Stearoylglycerophosphoinositol | 0.00E+00 | 0.00E+00 | −0.820683766 | 7.00E-15 | 5.20E-14 | 0.583844604 | Lysolipid |
| 3-Hydroxysebacate | 0.00E+00 | 0.00E+00 | −0.816297428 | 2.01E-13 | 1E-12 | 0.556790518 | Fatty acid (monohydroxy) |
| Prostaglandin E2 | 0.00E+00 | 0.00E+00 | −0.808607902 | 4.00E-15 | 3.00E-14 | 0.588704241 | Eicosanoid |
| X-13007 | 0.00E+00 | 0.00E+00 | −0.807024697 | 4.00E-14 | 2.68E-13 | 0.569976362 | |
| 1-Palmitoylglycerophosphoinositol | 0.00E+00 | 0.00E+00 | −0.805798034 | 0.00E+00 | 0.00E+00 | 0.620623355 | Lysolipid |
| X-21892 | 0.00E+00 | 0.00E+00 | −0.804483968 | 0.00E+00 | 0.00E+00 | 0.681145209 | |
| Dodecanedioate | 0.00E+00 | 0.00E+00 | −0.803507011 | 6.65E-13 | 3E-12 | 0.546598982 | Fatty acid (dicarboxylate) |
| α-Hydroxycaproate | 0.00E+00 | 0.00E+00 | −0.801592558 | 1.00E-15 | 1.10E-14 | 0.598051321 | Fatty acid (monohydroxy) |
| 2-Hydroxystearate | 0.00E+00 | 0.00E+00 | −0.796012461 | 0.00E+00 | 0.00E+00 | 0.680287515 | Fatty acid (monohydroxy) |
| X-22810 | 0.00E+00 | 0.00E+00 | −0.787137388 | 0.00E+00 | 0.00E+00 | 0.686363562 | |
| X-21282 | 0.00E+00 | 0.00E+00 | −0.785849133 | 1.13E-13 | 7.02E-13 | 0.56154387 | |
| 7-Hydroxycholesterol (α or β) | 0.00E+00 | 0.00E+00 | −0.778681562 | 2.60E-14 | 1.83E-13 | 0.573517242 | Sterol |
| 9,10-DiHOME | 0.00E+00 | 0.00E+00 | −0.771322126 | 8.00E-15 | 6.00E-14 | 0.582350688 | Fatty acid (dihydroxy) |
| X-14904 | 0.00E+00 | 0.00E+00 | −0.760975525 | 7E-12 | 3.1E-11 | 0.525565498 | |
| 2-Ethylhexanoate | 0.00E+00 | 0.00E+00 | −0.760812755 | 0.00E+00 | 3.00E-15 | 0.606630308 | Xenobiotic |
| Methylmalonate | 0.00E+00 | 0.00E+00 | −0.741753883 | 2.10E-13 | 1E-12 | 0.556392926 | Fatty acid metabolism |
| Caproate (6:0) | 0.00E+00 | 0.00E+00 | −0.735638953 | 0.00E+00 | 0.00E+00 | 0.644196546 | Fatty acid (medium chain) |
| 13-HODE + 9-HODE | 0.00E+00 | 0.00E+00 | −0.733094422 | 2.95E-13 | 2E-12 | 0.553548051 | Fatty acid (monohydroxy) |
| Valerylcarnitine | 0.00E+00 | 0.00E+00 | −0.724840214 | 4.40E-14 | 2.85E-13 | 0.56930096 | Acylcarnitine |
| Pelargonate (9:0) | 0.00E+00 | 0.00E+00 | −0.712763573 | 4.00E-15 | 3.10E-14 | 0.588365111 | Fatty acid (medium chain) |
| Leukotriene B4 | 0.00E+00 | 0.00E+00 | −0.712723538 | 0.00E+00 | 0.00E+00 | 0.631074706 | Eicosanoid |
| X-11538 | 0.00E+00 | 0.00E+00 | −0.70896232 | 4.17E-13 | 2E-12 | 0.550614728 | |
| X-21893 | 0.00E+00 | 0.00E+00 | −0.704328166 | 5.00E-15 | 3.90E-14 | 0.586235524 | |
| X-21343 | 0.00E+00 | 0.00E+00 | −0.698926193 | 3.58E-13 | 2E-12 | 0.551900032 | |
| 3-Hydroxyoctanoate | 0.00E+00 | 0.00E+00 | −0.692620423 | 1.87E-13 | 1E-12 | 0.557379365 | Fatty acid (monohydroxy) |
| X-15220 | 0.00E+00 | 0.00E+00 | −0.684172319 | 2.2E-11 | 9.6E-11 | 0.514701235 | |
| Caprate (10:0) | 0.00E+00 | 0.00E+00 | −0.672583005 | 4.00E-15 | 3.00E-14 | 0.588885093 | Fatty acid (medium chain) |
| Asparagylleucine | 0.00E+00 | 0.00E+00 | −0.670461233 | Dipeptide | |||
| X-17653 | 0.00E+00 | 0.00E+00 | −0.667177701 | ||||
| Hexanoylcarnitine | 0.00E+00 | 0.00E+00 | −0.656722745 | 6E-12 | 2.7E-11 | 0.527206599 | Acylcarnitine |
| 1-Oleoylglycerophosphoinositol | 0.00E+00 | 0.00E+00 | −0.656107572 | 7.34E-13 | 4E-12 | 0.545735856 | Lysolipid |
| 7-Ketocholesterol | 0.00E+00 | 0.00E+00 | −0.617140035 | 2E-12 | 8E-12 | 0.538123834 | Sterol |
| Glutarylcarnitine (C5) | 0.00E+00 | 0.00E+00 | −0.61190476 | Lysine metabolism | |||
| 12,13-DiHOME | 0.00E+00 | 0.00E+00 | −0.610218727 | Fatty acid (dihydroxy) | |||
| Caprylate (8:0) | 0.00E+00 | 1.00E-15 | −0.597848987 | 1.5E-11 | 6.8E-11 | 0.518128627 | Fatty acid (medium chain) |
| Glycerophosphoinositol | 0.00E+00 | 1.00E-15 | −0.594410042 | 7.25E-13 | 4E-12 | 0.545841654 | Phospholipid metabolism |
| X-12860 | 2.00E-15 | 7.00E-15 | −0.581605324 | ||||
| 5-HETE | 2.10E-14 | 8.40E-14 | −0.562842239 | Eicosanoid | |||
| 3-Hydroxypropanoate | 3.00E-14 | 1.21E-13 | −0.559886292 | Fatty acid (monohydroxy) | |||
| X-17654 | 6.00E-14 | 2.38E-13 | −0.554418366 | ||||
| X-22776 | 7.10E-14 | 2.68E-13 | −0.553009309 | 5.8E-11 | 2.39E-10 | 0.505273224 | |
| Valerate | 1.75E-13 | 6.52E-13 | −0.545665626 | Fatty acid (short chain) | |||
| Maleate (cis-Butenedioate) | 5.99E-13 | 2E-12 | −0.535372888 | Fatty acid (dicarboxylate) | |||
| Kynurenine | 1.00E-12 | 4E-12 | −0.529027326 | 1.91E-10 | 7.67E-10 | 0.493180348 | Tryptophan metabolism |
| X-22514 | 5.00E-12 | 1.6E-11 | −0.517642769 | ||||
| 1-Arachidonoylglycerophosphoinositol | 3E-11 | 9.8E-11 | 0.500015447 | Lysolipid | |||
| Docosapentaenoate (n3 DPA; 22:5n3) | 2.1E-11 | 6.9E-11 | 0.503616186 | 1.31E-13 | 7.92E-13 | −0.560357689 | Polyunsaturated fatty acid |
| 2-Linolenoylglycerophosphocholine (18:3n3) | 1.9E-11 | 6.4E-11 | 0.504430281 | Polyunsaturated fatty acid | |||
| 2-Arachidonoyl glycerol | 9E-12 | 3E-11 | 0.511668858 | Monoacylglycerol | |||
| 1-Docosahexaenoylglycerophosphocholine (22:6n3) | 2E-12 | 8E-12 | 0.524271142 | Polyunsaturated fatty acid | |||
| Docosatrienoate (22:3n3) | 4.28E-13 | 2E-12 | 0.538232461 | 1.32E-10 | 5.37E-10 | −0.496995801 | Polyunsaturated fatty acid |
| Adrenate (22:4n6) | 6.90E-14 | 2.65E-13 | 0.553229362 | 3E-12 | 1.6E-11 | −0.532126537 | Polyunsaturated fatty acid |
| 2-Linoleoylglycerophosphocholine | 6.50E-14 | 2.51E-13 | 0.553779147 | Lysolipid | |||
| 2-Linoleoylglycerol (2-monolinolein) | 6.20E-14 | 2.42E-13 | 0.554164088 | 7.00E-15 | 5.20E-14 | −0.583752542 | Monoacylglycerol |
| 2-Arachidonoylglycerophosphocholine | 1.30E-14 | 5.30E-14 | 0.566637034 | Lysolipid | |||
| 12-HETE | 4.00E-15 | 1.70E-14 | 0.575336274 | Eicosanoid | |||
| 2-eicosatrienoylglycerophosphocholine | 2.00E-15 | 1.00E-14 | 0.579615036 | Lysolipid | |||
| Docosahexaenoate (22:6n3) | 1.00E-15 | 4.00E-15 | 0.587074071 | 2.9E-11 | 1.26E-10 | −0.511865462 | Polyunsaturated fatty acid |
| cis-4-decenoyl carnitine | 0.00E+00 | 0.00E+00 | 0.603624362 | 3.9E-11 | 1.65E-10 | −0.509085464 | Acylcarnitine |
| 2-docosahexaenoylglycerophosphocholine | 0.00E+00 | 0.00E+00 | 0.761335073 | 6.57E-13 | 3E-12 | −0.546693703 | Lysolipid |
All analytes that had significant correlation of >0.5 or <−0.5 are reported for 24-hour recovery (phenotype 1) or spontaneous hemolysis (phenotype 2). Analytes with mass spectra that have not yet been elucidated are indicated by a nomenclature of (X-#). For any phenotype, positive correlations are shown in bold type and negative correlations in standard type. Methods of P and q value determination are given in the main text. q values of <1.0 × 10−15 are indicated as 0.00.
For 24-hour recovery, 15 named compounds were identified that met the positive correlation criteria, including polyunsaturated fatty acids, glycolipids, acylcarnitine, 12-HETE, and 2 monoacylglycerols. Forty-six named compounds were negatively correlated with 24-hour recoveries (as well as 20 additional unnamed compounds, the structure of which are unknown). Of the 46 named compounds, 40 fall into the general category of lipid metabolites, including products of lipid oxidation and peroxidation (eg, monohydroxy fatty acids, dihydroxy fatty acids, dicarboxylic acids, hydroxyl and keto cholesterol). Products typically associated with enzymatic oxidation of arachidonic acid (eg, eicosanoids) were also identified. General products of lipid metabolism and breakdown were likewise observed (lysolipids, acylcarnitines, and short- and medium-chain fatty acids). Products of lysine metabolism, tryptophan metabolism, a dipeptide, and a xenobiotic were also identified.
Phenotype-driven backcrossing to further isolate genetic elements responsible for poor RBC storage
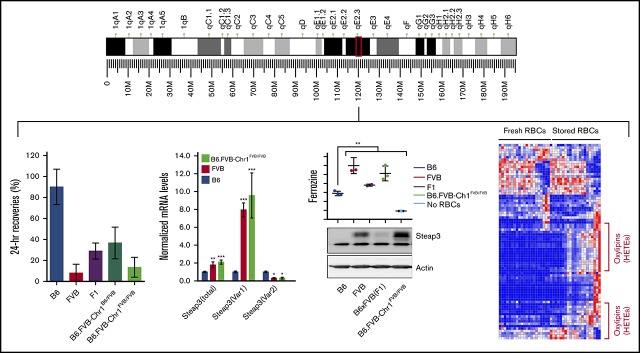
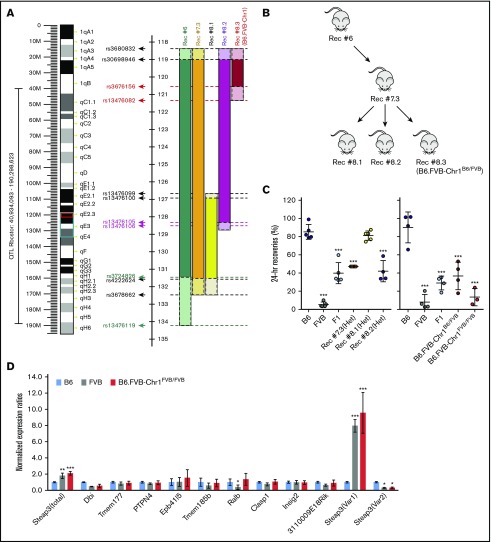
To further narrow down Rbcstor1, F2 mice with the FVB RBC storage phenotype were serially back-crossed onto a B6 background. Recombination within the interval on chromosome 1 was monitored by SNP genotyping and allele-specific PCR flanking the Rbcstor1 interval (Figure 2A; supplemental Table 1). Analysis of progeny from the sixth-generation onward (Rec #7.3, #8.2, and #8.3) narrowed the genetic region to ∼3 Mb (rs3680832 to rs13476082) (Figure 2B). Hereafter, we refer to recombinant Rec#8.3 as “B6.FVB-Chr1.” As predicted, RBCs from fully homozygous B6.FVB-Chr1 mice (B6.FVB-Chr1FVB/FVB) stored poorly, similar to parental FVB RBCs (Figure 2C).
Figure 2.
Phenotype and genotype-guided backcrossing based mapping and generation of congenic mice. (A) Left: map of murine chromosome 1. The black bracket indicates the boundaries of Rbcstor1, the turquoise box indicates the boundaries of the sixth-generation mouse, and the red box indicates the minimal region containing genetic elements sufficient to confer the poor RBC storage phenotype on a B6 background. Right: pictoral representation of the recombinants obtained in generations 6 to 8 of backcrossing, including the SNPs that define these boundaries (supplemental Table 1). (B) Pedigrees of congenic generations 6 to 8 are shown. (C) Twenty-four–hour recoveries of RBCs stored for 7 days. Left and right panels reflect separate experiments, respectively, each with its own B6, FVB, and F1 controls. These data represent means ± standard deviation (SD) from a representative experiment (n = 3-5 mice per group). These data have been replicated in at least 3 experiments. Statistics calculated using 1-way ANOVA with a Bonferroni post hoc analysis. A significant difference from B6 is indicated by ***P < .0001. (D) Quantitative real-time PCR of each of the gene products contained within the mapped genetic region on chromosome 1. Quantitative real-time PCR was performed on EPs isolated by fluorescence-activated cell sorting from each mouse strain using prevalidated TaqMan assays for each of the 20 genes. Amplification was observed for 10 of the 20 genes. Data shown represent the combined means and SD of expression levels (normalized to expression in B6) from 3 independent experiments. Statistics were calculated by using 2-way ANOVA with a Bonferroni post hoc analysis. A significant difference from B6 is indicated by the following: *P < .05, **P < .005, ***P < .0001.
The interval defined in B6.FVB-Chr1 contains 20 open reading frames as well as multiple long noncoding RNAs and microRNAs (supplemental Table 2). Quantitative reverse transcriptase (RT)–PCR on RNA from EPs isolated by fluorescence-activated cell sorting (data not shown; see Materials and methods) was conducted for each of the 20 messenger RNA (mRNA) species. From these 20, 10 yielded detectable amplicons. Of these 10, only 2 showed a significant difference between B6 EP and FVB EP (Ralb and Steap3) (Figure 2D). However, only Steap3 mRNA levels were similar between B6.FVB-Chr1FVB/FVB EP and FVB EP. Two distinct Steap3 mRNA variants have been described in mice, which vary only in untranslated regions.16 Variant-specific quantitative RT-PCR showed that Var1 expression was eightfold to 10-fold higher in B6.FVB-Chr1FVB/FVB EP and FVB EP compared with B6 EP, whereas Var2 expression was threefold to fourfold lower in B6.FVB-Chr1FVB/FVB EP and FVB EP compared with B6 EP.
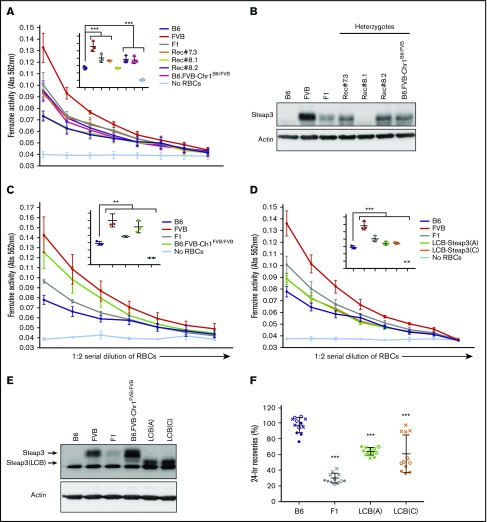
Steap3 activity and protein levels are significantly increased in FVB mice
Consistent with the mRNA differences noted here, ferrireductase activity was high in FVB RBCs and low in B6 RBCs, respectively (Figure 3A). Likewise, Steap3 protein was easily observed in FVB but was barely detectable in B6 RBCs according to western blot analysis (Figure 3B). RBCs from F1 mice had intermediate protein and activity. In all cases, recombinant congenic mice that inherited a single copy of the FVB Steap3 gene had activity and protein levels of Steap3 similar to wild-type mice with a single copy of the FVB Steap3 gene (B6xFVB F1). Rec#8.1 RBCs had a crossover that lost the FVB Steap3 and exhibited ferrireductase activity and Steap3 protein levels similar to B6 RBCs. RBCs from homozygous B6.FVB-Chr1FVB/FVB mice exhibited ferrireductase activity (Figure 3C) and Steap3 protein levels (Figure 3E) similar to FVB RBCs.
Figure 3.
Levels and effects of Steap3 expression on RBCs. (A,C-D). Ferrireductase activity in RBCs as measured by using ferrozine as an indicator of conversion of Fe3+ to Fe2+. Data shown represent combined means from 3 independent experiments (n = 2-4 mice per group, per experiment). Statistics were calculated by using 2-way ANOVA with a Bonferroni post hoc analysis. At the highest concentration of RBCs tested (inset panel in each graph), ferrozine activity of B6 was significantly lower than all other samples (P < .0001) except Rec#8.1 for panel A; ferrozine activity of B6 was significantly lower than all other samples (**P < .005 for F1 and P < .0001 for FVB, B6.FVB-Chr1FVB/FVB) for panel C; and ferrozine activity of B6 was significantly lower than all other samples (P < .0001) for panel D. (B,E) Western blot of RBC ghosts using an anti-Steap3 antibody. After development, membranes were stripped and re-probed with anti-Actin as a control for loading and transfer. (F) Twenty-four–hour recoveries of RBCs stored for 7 days. These data represent combined means and SD from 3 independent experiments (experiment 1 data are shown as a solid circle, experiment 2 data are shown as a hollow circle, and experiment 3 data are shown as an X). Statistics were calculated by using 1-way ANOVA with a Bonferroni post hoc analysis. A significant difference from B6 is indicated by ***P < .0001.
To specifically test if the increased Steap3 expression has a causal role in poor RBC storage, increased Steap3 activity was isolated as a single variable through the generation of transgenic mice (B6 background) with Steap3 expression driven by an RBC-specific regulatory element. As these transgenic mice used an ectopic promoter and had random integration into the genome, their level of expression is expected to vary. RBCs from 2 independent transgenic founder strains (LCB-Steap3A and LCB-Steap3C) exhibited ferrireductase activity and protein levels higher than that of wild-type B6 RBCs but lower than either FVB or F1 mice (Figure 3D-E). Interestingly, for unclear reasons, the transgenic Steap3 protein had a faster mobility than the natural Steap3 in FVB mice; however, the increased activity in RBCs from these strains revealed it to be functional.
Twenty-four-hour recoveries of RBCs from both Steap3 transgenic lines were significantly lower than for wild-type B6 RBCs (Figure 3F). Lower 24-hour recovery was not due to nonspecific effects of the transgenic expression cassette: no such decrease was observed for RBCs from Kel-2N mice that express an unrelated transgene from the same LCB expression construct17 (data not shown). Just as Steap3 activity in the transgenic mice was intermediate between wild-type B6 and F1 mice, so was the 24-recovery of stored RBCs intermediate between B6 and F1 mice. Based on these data, we concluded that increased Steap3 expression is alone sufficient, in a dose-dependent manner, to cause decreased 24-hour recovery of stored RBCs.
Effect of nonsynonymous Steap3 polymorphisms
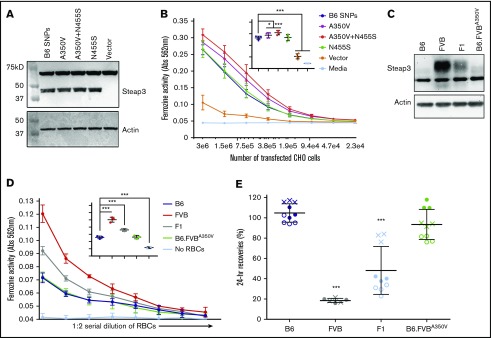
The quantitative RT-PCR and western blot analysis indicated that differential expression of Steap3 protein explains differential ferrireductase activity. However, the predicted B6 and FVB Steap3 open reading frames also differ at 2 amino acids. Expression of each variant in CHO cells showed a modest increased activity of the FVB variant compared with the B6 variant (Figure 4B). Mutation of the alanine (B6) to valine (FVB) at position 350 (SNP1) conferred increased ferrireductase activity, whereas mutation of the asparagine (B6) to serine (FVB) at position 455 (SNP2) conferred no increased activity. Equivalent levels of protein expression were observed for all Steap3 variants; no Steap3 was detected when a control empty vector was used (Figure 4A). Thus, the increased activity of the FVB variant of Steap3 may be due, at least in part, to a more active enzyme resulting from a valine substitution at position 350.
Figure 4.
Effects of Steap3 nonsynonymous polymorphisms between B6 and FVB strains. Western blot of CHO cell lysates from cells transiently expressing recombinant Steap3 variants (A) or RBC ghosts (C). Blots were probed by using an anti-Steap3 antibody; after development, membranes were stripped and re-probed with anti-actin as a control for loading and transfer. (B) Ferrireductase activity in transfected CHO cells as measured by using ferrozine as an indicator of conversion of Fe3+ to Fe2+. Data shown represent combined means from 5 independent experiments. Statistics were calculated by using 2-way ANOVA with a Bonferroni post hoc analysis. At the highest concentration of CHO cells tested (inset panel), ferrozine activity of the transfected B6 Steap3 variant was significantly lower than A350V (*P < .01) and A350V+N455S (***P < .0001) but not different from N455S by itself. (D) Ferrireductase activity in RBCs as measured by using ferrozine as an indicator of conversion of Fe3+ to Fe2+. Data shown represent combined means from 3 independent experiments (n = 2-4 mice per group, per experiment). Statistics were calculated by using 2-way ANOVA with a Bonferroni post hoc analysis. At the highest concentration of RBCs tested (inset panel), ferrozine activity of B6 was not different from B6.FVB-A350V. (E) Twenty-four–hour recoveries of RBCs stored for 7 days. These data represent combined means and SD from 3 independent experiments (experiment 1 data are shown as a solid circle, experiment 2 data are shown as a hollow circle, and experiment 3 data are shown as an X). Statistics were calculated by using 1-way ANOVA with a Bonferroni post hoc analysis. A significant difference from B6 is indicated by ***P < .0001.
CRISPR/Cas9 technology was used to introduce the A350V substitution into the Steap3 gene of B6 mice. The introduction of the A350V substitution and the absence of off-target substitutions at other Steap3 coding sequences were confirmed by using RT-PCR sequencing (data not shown). RBCs from B6-A350V mice did not exhibit a significant increase in Steap3 protein levels (Figure 4C) or in ferrireductase activity (Figure 4D) compared with wild-type B6 RBCs. Likewise, there was no reduction in 24-hour recovery of stored RBCs (Figure 4E). Thus, although transfection of CHO cells shows the potential for differential enzymatic activity due to structural differences between Steap3 variants, such differences are not alone sufficient to confer either increased ferrireductase activity or a poor storage phenotype on RBCs on a B6 background.
Metabolomics analyses of B6.FVB-Chr1 mice and Steap3 transgenic strains
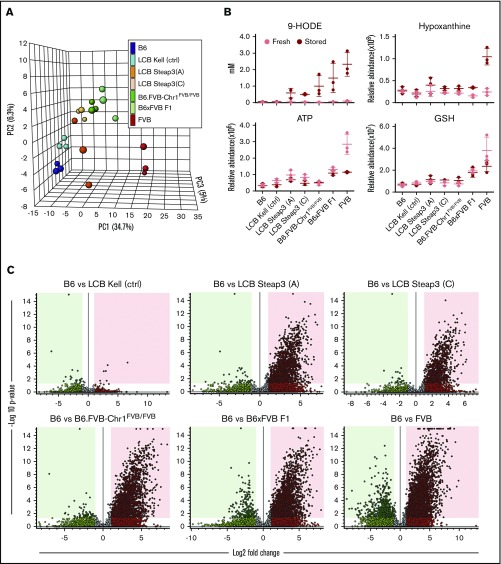
A detailed metabolomics analysis of RBCs (freshly isolated vs stored) was conducted for both Steap3 transgenic strains and compared with RBCs from B6, F1, B6.FVB-Chr1FVB/FVB, and FVB mice. Kel-2N mice were used as a negative control for LCB cassette effects. Over the several years needed to develop congenic mice via backcrossing, metabolomics technology has advanced, allowing use of an untargeted metabolomics platform with relative quantitation of 59 867 different analytes. Principal component analysis showed that the presence of either the Chr1 region from FVB (containing Steap3) or isolated increased expression of Steap3 in transgenic mice on a B6 background resulted in a metabolic trend toward FVB mice (Figure 5A). This effect in transgenic mice was not an artifact of the transgenic process, as the LCB cassette expressing a gene other than Steap3 (Kel-2N) was similar to B6 samples. PC1 accounted for ∼35% of variability across samples.
Figure 5.
Statistical analysis of metabolic clustering and carry forward with congenic and Steap3 transgenic mice. (A) Principle component analysis of 3 separate experiments for each of the indicated mouse strains. (B) Examples of analytes that carry forward (eg, 9-HODE) and those that do not (eg, hypoxanthine, adenosine triphosphate (ATP), and glutathione (GSH). (C) Volcano plots showing the metabolomics of stored RBCs from experimental strains compared with B6, depicting the general extent of clustering of statistically significant analytes that carry forward into congenic and Steap3 transgenic strains.
By focusing on mice expressing increased Steap3 as an isolated variable on a B6 background (LCB-Steap3A and LCB-Steap3C), we were able to identify which metabolic changes were caused due to increased Steap3. There was a progressive decrease in the number of compounds differing between B6 and transgenic RBCs, with a decrease in the number of carried-over compounds from FVB RBCs at each step (volcano plots in Figure 5C). Partial least squares discriminant analysis confirmed the diversity of FVB and B6 RBC samples and revealed a progressive loss of FVB background for the intermediate transgenic mice, whereas Kel-2N mice clustered close to B6 controls (data not shown).
Variable importance in projection analysis identified a series of key analytes that best fit the pattern of carryover; many of these are products of lipid oxidation (supplemental Figure 2). As an example, 9-HODE exhibited strong differences between various mouse strains and emerged as strong correlates in early analyses (Figure 5B).7,18 Indeed, 9-HODE had a carry-forward pattern that correlated with RBC storage and Steap3 activity. In contrast, hypoxanthine, adenosine triphosphate, and glutathione failed to carry forward into B6.FVB-Chr1 mice and were not observed in Steap3 transgenic mice, indicating that although correlative in parental strains, they are not required for poor RBC storage. A detailed heat map across strains tested (with clustering based upon analytes that carried forward vs those that did not) and with assignments to general metabolic pathways is provided (supplemental Figure 3).
Distribution of Steap3 in other inbred strains of mice; effects on in vivo RBC life span
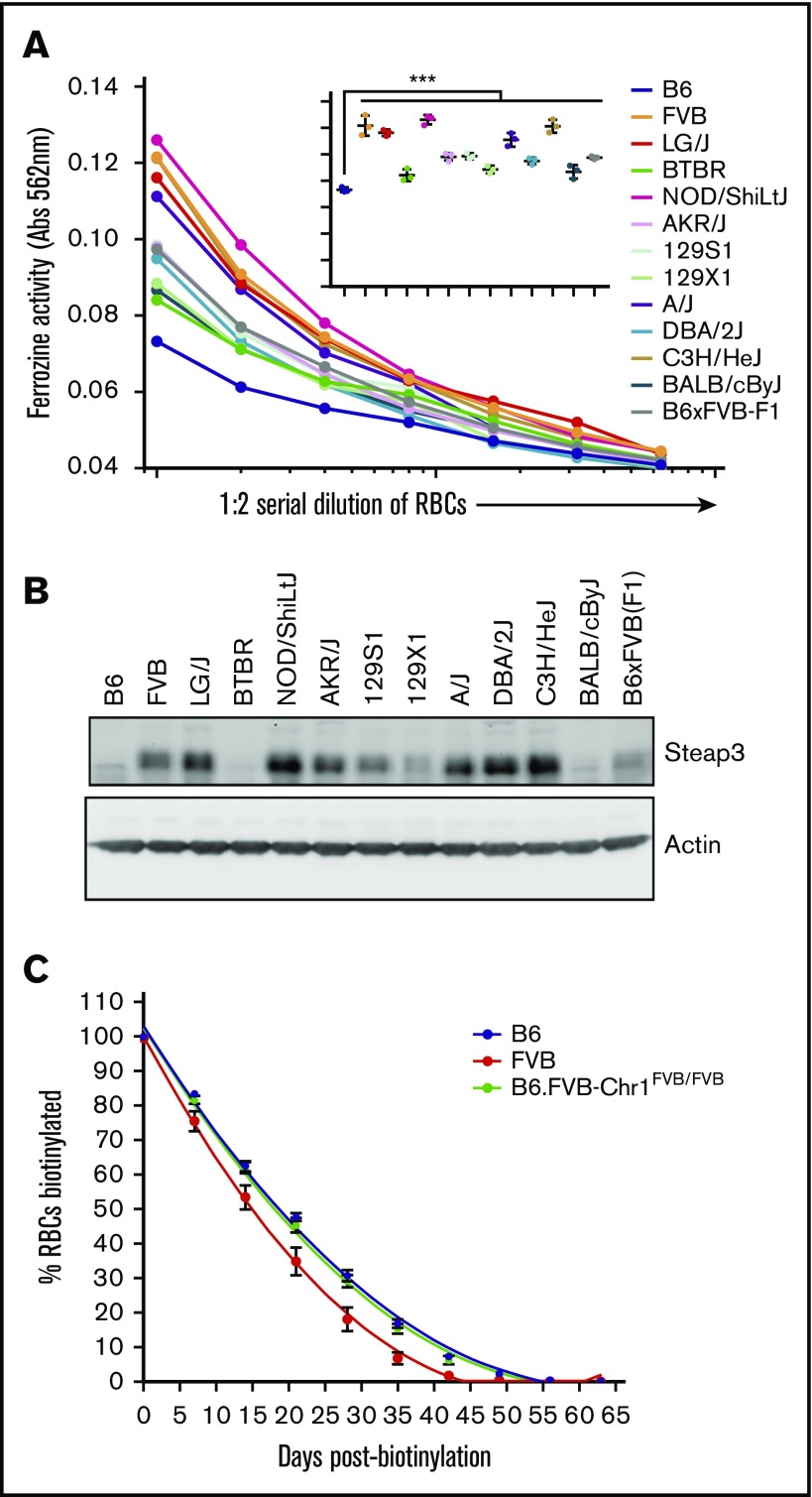
We have previously reported a rank order of RBC storage across 13 strains of mice.7 Analysis of the known genomic sequence of these strains indicates that the 3 best-storing strains (B6, BALB/cByJ, and BTBR) harbored the B6 Steap3 variant, whereas the other 10 strains harbored the FVB Steap3 variant. As predicted, strains B6, BALB/cByJ, and BTBR had generally lower ferrireductase activity (Figure 6A) than other strains and only faintly detectable Steap3 bands according to western blot (Figure 6B). Among the other strains tested, those previously shown to store the most poorly (eg, LG/J, A/J, FVB) had the highest ferrireductase activity and the most intense western blot bands.
Figure 6.
General correlation of Steap3 to RBC storage across strains and in vivo RBC life span. (A) Ferrireductase activity in RBCs as measured by using ferrozine as an indicator of Fe2+. Data shown represent combined means from 3 independent experiments (n = 2-4 mice per group, per experiment). Statistics were calculated by using 2-way ANOVA with a Bonferroni post hoc analysis. At the highest concentration of RBCs tested (inset graph), ferrozine activity of B6 is significantly lower than all other strains tested (***P < .0001). (B) Western blot of RBC ghosts using an anti-Steap3 antibody. After development, membranes were stripped and re-probed with anti-actin as a control for loading and transfer. (C) RBC life span determination by in vivo biotinylation. These data represent means ± SD from a representative experiment (n = 5 mice per group). These data have been replicated in at least 3 experiments.
We have reported that the in vivo life span of RBCs in FVB mice is shorter than that of RBCs in B6 mice, leading us to previously speculate that the genetic determinants of poor RBC storage may also regulate in vivo RBC life span.8 This hypothesis predicts that B6.FVB-Chr1 mice should have a shortened RBC life span compared with B6 mice. To test this theory, we performed an in vivo pulse-chase experiment by biotinylation of the whole RBC mass of B6, FVB, or B6.FVB-Chr1 mice. Mice were bled weekly, and the clearance of labeled RBCs was used to track RBC life span. As previously observed, the life span of FVB RBCs was shorter than that of B6 RBCs. However, RBCs from B6.FVB-Chr1 mice had the same life span as those from B6 mice and not FVB mice (Figure 6C). Thus, the FVB genetic elements in B6.FVB-Chr1 that cause poor in vitro RBC storage are not sufficient to cause decreased in vivo RBC life span.
Discussion
We report that differences in expression of Steap3 cause the altered RBC storage biology between B6 and FVB strains. Multiple lines of evidence, presented in the current article, support this conclusion. The same genetic locus was identified through 2 separate genetic mapping approaches (QTL and congenic mapping). Among potential genes in the isolated 3 Mb region on chromosome 1, Steap3 was the only gene with substantial strain-dependent differences in mRNA expression, also reflected in both protein levels and Steap3 activity on mature RBCs. To test the causal role of Steap3, transgenic mice were made on a B6 background such that increased expression of Steap3 was the only difference compared with wild-type mice. These mice had both decreased posttransfusion recoveries and also acquired the same metabolic characteristics (eg, oxylipin generation) as RBCs from FVB mice. Taken together, these data establish a causal role of Steap3 expression in RBC storage and establish a hitherto unrecognized mechanistic pathway of oxidative damage to RBCs.
Steap3 was first identified in 2005 through the analysis of a mutant mouse strain (nm1054) with hypochromic microcytic anemia.14,19 Steap3 plays a vital role in erythropoiesis by reducing Fe3+ to Fe2+, allowing Fe2+ to be loaded into the heme pocket.20 The defect is both quantitative and qualitative because RBCs from the Steap3 knockout mice had altered rigidity as measured by using ektacytometry, but there was no difference in RBC life span.
As with all animal studies, it is possible that the observed effects do not translate into human biology. Regardless of whether they do, the current report at the very least identifies an important new pathway relevant to mammalian biology. Nevertheless, determining the extent to which these findings translate to humans will be critical. Murine and human Steap3 have 85% identity at the amino acid level, and a human family with a genetic Steap3 deficiency has anemia that resembles what is observed in Steap3 knockout mice,21 suggesting a similar effect of Steap3 on RBCs in mice and humans. A small (n = 10) sampling in a published article shows that Steap3 mRNA expression varies up to threefold among healthy human control subjects, and as such, there is likely variation among blood donors.21 We therefore predict that RBCs from humans with moderately decreased Steap3 function will manifest superior storage compared with RBCs from donors with full Steap3 activity. It is important to note that both B6 and FVB mice have normal RBC hematology in the healthy state and that the phenotype does not emerge until RBCs are stored, consistent with the report that humans with a heterozygous loss of function variant of Steap3 have normal baseline hematologic parameters.20 As such, current routine RBC donor screening would not detect any Steap3 differences.
The current studies identify levels of Steap3 expression as a mechanistic determinant in RBC storage and oxidative damage. This identification leads to the question of why increased levels of a ferrireductase would lead to increased oxidant damage. Ongoing study will be required to elucidate this deeper mechanistic question, but we present the following hypothesis, which is consistent with known Fe biology. Most RBC Fe is contained in hemoglobin, for which there is a dedicated methemoglobin reductase that converts hemoglobin-bound Fe3+ to Fe2+. However, some iron is non-hemoglobin bound, likely complexed with sulfhydryl groups on proteins. H2O2 (generated as part of normal metabolism) combines with Fe2+ to generate reactive oxygen species via the Fenton reaction. This action would be self-limiting if Fe3+ could not be converted back to Fe2+. By converting Fe3+ back to Fe2+, Steap3 catalyzes redox cycling and the generation of reactive oxygen species. Fe3+ can also be converted back to Fe2+ by using the Haber-Weiss reaction (combining with O2-•). Thus, the extent of the Haber-Weiss reaction may affect susceptibility of RBCs to Steap3-facilitated damage. The Fenton reaction allows redox cycling in response to increased Steap3 activity, because such cycling does not occur with other “reductases,” then Steap3 has the seemingly ironic role of increasing oxidative damage with increased reductase activity. It is essential to note that Fe complexed with hemoglobin does not participate in the Fenton reaction, and as such, differences in methemoglobin reductase would not be expected to have the same effect. Moreover, as hemoglobin breaks down, more Fe is released that can be acted on by Steap3, which may explain why the phenotype emerges during RBC storage.
It is interesting that increased Steap3 activity does not decrease RBC life span in vivo in otherwise unchallenged mice. However, it remains to be seen if physiological oxidative stress alters in vivo RBC life span. Other mechanistic hypotheses may also explain the effects of Steap3 on RBC storage. Increased Fe absorption during erythropoiesis due to increased Steap3 activity may alter RBC phenotypes in a way that makes them more susceptible to storage-based damage. Alternatively, in addition to reducing Fe3+, Steap3 also acts on copper, which is known to be capable of affecting RBC redox biology.
In addition to its high direct medical relevance in transfusion, RBC storage can also be viewed as a form of experimental oxidative stress which models other sources of stress that RBCs may encounter in health and disease. Thus, the identification of Steap3 expression as a novel mechanism of oxidant stress in RBCs may have relevance to numerous disease states in which RBCs encounter oxidative stress.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors recognize and acknowledge the excellent technical assistance from Nicole Smith and also thank Jill Johnsen for many useful discussions and guidance.
These studies were funded in part by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (R01 HL095470 and P01 HL086773).
Authorship
Contribution: H.L.H., A.M.H., K.d.W., H.W., J.L., X.F., R.C.-H., A.D., M.S.R., and J.C.Z. each conducted experiments and acquired data; J.D.G. and P.C.T. analyzed and interpreted data; and all authors contributed to the writing of the manuscript.
Conflict-of-interest disclosure: Bloodworks NW has filed a patent on using Steap3 as a predictor of RBC storage. J.C.Z. and K.d.W. are inventors on this application (US20170276665A1). The remaining authors declare no competing financial interests.
Correspondence: James C. Zimring, Department of Pathology, Carter Immunology Center, University of Virginia School of Medicine, PO Box 801386, Charlottesville, VA 22903; e-mail: jcz2k@virginia.edu.
References
- 1.Kanias T, Lanteri MC, Page GP, et al. Ethnicity, sex, and age are determinants of red blood cell storage and stress hemolysis: results of the REDS-III RBC-Omics study. Blood Adv. 2017;1(15):1132-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dumont LJ, AuBuchon JP. Evaluation of proposed FDA criteria for the evaluation of radiolabeled red cell recovery trials. Transfusion. 2008;48(6):1053-1060. [DOI] [PubMed] [Google Scholar]
- 3.Hod EA, Francis RO, Spitalnik SL. Red blood cell storage lesion-induced adverse effects: more smoke; is there fire? Anesth Analg. 2017;124(6):1752-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van ’t Erve TJ, Doskey CM, Wagner BA, et al. Heritability of glutathione and related metabolites in stored red blood cells. Free Radic Biol Med. 2014;76:107-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van ’t Erve TJ, Wagner BA, Martin SM, et al. The heritability of metabolite concentrations in stored human red blood cells. Transfusion. 2014;54(8):2055-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van ’t Erve TJ, Wagner BA, Martin SM, et al. The heritability of hemolysis in stored human red blood cells. Transfusion. 2015;55(6):1178-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Wolski K, Fu X, Dumont LJ, et al. Metabolic pathways that correlate with post-transfusion circulation of stored murine red blood cells. Haematologica. 2016;101(5):578-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimring JC, Smith N, Stowell SR, et al. Strain-specific red blood cell storage, metabolism, and eicosanoid generation in a mouse model. Transfusion. 2014;54(1):137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilson CR, Kraus TS, Hod EA, et al. A novel mouse model of red blood cell storage and posttransfusion in vivo survival. Transfusion. 2009;49(8):1546-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Alessandro AD, Rapido F, Thomas T, et al. Metabolic pathways that correlate with in vivo hemolysis following storage of human RBCs [abstract]. Transfusion. 2017;57(suppl 3):11A. Abstract B16 AO-13I.
- 11.Desmarets M, Cadwell CM, Peterson KR, Neades R, Zimring JC. Minor histocompatibility antigens on transfused leukoreduced units of red blood cells induce bone marrow transplant rejection in a mouse model. Blood. 2009;114(11):2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peterson KR, Fedosyuk H, Zelenchuk L, et al. Transgenic Cre expression mice for generation of erythroid-specific gene alterations. Genesis. 2004;39(1):1-9. [DOI] [PubMed] [Google Scholar]
- 13.D’Alessandro A, Culp-Hill R, Reisz JA, et al. ; Recipient Epidemiology and Donor Evaluation Study-III (REDS-III). Heterogeneity of blood processing and storage additives in different centers impacts stored red blood cell metabolism as much as storage time: lessons from REDS-III-Omics. Transfusion. 2019;59(1):89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohgami RS, Campagna DR, Greer EL, et al. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37(11):1264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith NH, Henry KL, Cadwell CM, et al. Generation of transgenic mice with antithetical KEL1 and KEL2 human blood group antigens on red blood cells. Transfusion. 2012;52(12):2620-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue F, Cheng Y, Breschi A, et al. ; Mouse ENCODE Consortium. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gibb DR, Liu J, Natarajan P, et al. Type I IFN is necessary and sufficient for inflammation-induced red blood cell alloimmunization in mice. J Immunol. 2017;199(3):1041-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nemkov T, Sun K, Reisz JA, et al. Hypoxia modulates the purine salvage pathway and decreases red blood cell and supernatant levels of hypoxanthine during refrigerated storage. Haematologica. 2018;103(2):361-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohgami RS, Campagna DR, Antiochos B, et al. nm1054: a spontaneous, recessive, hypochromic, microcytic anemia mutation in the mouse. Blood. 2005;106(10):3625-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanc L, Papoin J, Debnath G, et al. Abnormal erythroid maturation leads to microcytic anemia in the TSAP6/Steap3 null mouse model. Am J Hematol. 2015;90(3):235-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grandchamp B, Hetet G, Kannengiesser C, et al. A novel type of congenital hypochromic anemia associated with a nonsense mutation in the STEAP3/TSAP6 gene. Blood. 2011;118(25):6660-6666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.