Abstract
Objectives
The discovery of mobile colistin resistance mcr-1, a plasmid-borne polymyxin resistance gene, highlights the potential for widespread resistance to the last-line polymyxins. In the present study, we investigated the impact of mcr-1 acquisition on polymyxin resistance and biological fitness in Klebsiella pneumoniae.
Methods
K. pneumoniae B5055 was used as the parental strain for the construction of strains carrying vector only (pBBR1MCS-5) and mcr-1 recombinant plasmids (pmcr-1). Plasmid stability was determined by serial passaging for 10 consecutive days in antibiotic-free LB broth, followed by patching on gentamicin-containing and antibiotic-free LB agar plates. Lipid A was analysed using LC–MS. The biological fitness was examined using an in vitro competition assay analysed with flow cytometry. The in vivo fitness cost of mcr-1 was evaluated in a neutropenic mouse thigh infection model.
Results
Increased polymyxin resistance was observed following acquisition of mcr-1 in K. pneumoniae B5055. The modification of lipid A with phosphoethanolamine following mcr-1 addition was demonstrated by lipid A profiling. The plasmid stability assay revealed the instability of the plasmid after acquiring mcr-1. Reduced in vitro biological fitness and in vivo growth were observed with the mcr-1-carrying K. pneumoniae strain.
Conclusions
Although mcr-1 confers a moderate level of polymyxin resistance, it is associated with a significant biological fitness cost in K. pneumoniae. This indicates that mcr-1-mediated resistance in K. pneumoniae could be attenuated by limiting the usage of polymyxins.
Introduction
The spread of MDR amongst Gram-negative bacteria has emerged as one of the most serious global public health threats.1 Owing to the paucity of newly approved antibiotics with novel modes of action, polymyxins (i.e. colistin and polymyxin B) have been increasingly used for treating MDR Gram-negative bacterial infections.2 However, in recent times, there have been increasing reports of polymyxin-resistant Gram-negative bacterial infections in patients and animals.3–8 The primary mechanism of polymyxin resistance in Gram-negative bacteria involves modification of lipid A with positively charged residues, such as aminoarabinose or phosphoethanolamine (pEtN).9,10 These modifications diminish the interaction of polymyxin molecules with the outer membrane, leading to resistance.11 Modifications to the glucosamine phosphate groups of lipid A are usually mediated by chromosomal mutation, resulting in up-regulation of two-component systems, PhoP/PhoQ and PmrA/PmrB.12,13 The inactivation of the mgrB gene, a negative regulator of the PhoP/PhoQ system, has been shown to play a key role in polymyxin resistance in Klebsiella pneumoniae.14,15
The first plasmid-borne polymyxin resistance gene, mcr-1, was detected in a livestock isolate in China in November 2015.16 Subsequently, several variants of the mcr genes (mcr-1 to -5) have been reported in a number of countries.17–23 The mcr gene has been identified globally in various Gram-negative bacteria isolated from animals, meat, processed meat products, vegetables and humans.24,25 The mcr gene encodes a pEtN transferase that modifies the negative phosphate groups of lipid A with positively charged pEtN residues.16 This readily transferable polymyxin resistance gene holds the potential to rapidly spread resistance. Escherichia coli remains as the most prevalent Gram-negative bacterium harbouring the mcr gene, whereas lower prevalence is observed in K. pneumoniae; however, the latter pathogen is a leading cause of nosocomial infections globally.26,27
The mcr-1 gene is believed to have originated from animal-associated bacteria. It has been frequently isolated from livestock and is supposedly sustained by the heavy usage of colistin in the agricultural and veterinary sectors.16,28 This hypothesis is supported by the identification of the earliest mcr-1-harbouring E. coli strain isolated from farm chickens in the 1980s, which coincides with the onset of colistin use in poultry farming in China.29 Purportedly, the heavy usage of colistin in Chinese agriculture may have acted as the selective pressure that promoted the spread and evolution of mcr-1-mediated resistance. Although the prevalence of mcr-1 in bacteria isolated from human sources is not as high as that from animals, the zoonotic potential of mcr-1-carrying bacteria has been indicated by a number of studies.28,30,31 Zoonotic infections by bacteria carrying mcr-1 highlight the urgent need to limit the use of colistin in agriculture and veterinary practices. Fortunately, the use of colistin in animal feed was banned in China soon after the increasing reports of mcr-1.32
The development of antibiotic resistance often comes with a cost to biological fitness, defined by reduced competitive ability in an antibiotic-free environment, a phenomenon that usually allows the fitter, often susceptible, strain to outcompete the resistant strain.33 The primary aim of the present study was to investigate the potential fitness cost incurred by mcr-1 in K. pneumoniae.
Materials and methods
Bacterial strains and plasmids
K. pneumoniae B5055 (K2:O1) was employed as the parental strain for this study and E. coli JW1 was used as a reference strain for the in vitro biological fitness assays.34 The codon-optimized mcr-1 gene was synthesized by Invitrogen™ GeneArt™. An mcr-1 recombinant plasmid (pmcr-1) was constructed by ligating mcr-1 into the low-copy, broad-host-range vector pBBR1MCS-5 at the XhoI and HindIII sites using T4 DNA ligase (New England BioLabs).35 The vector (pBBR1MCS-5) and mcr-1 recombinant plasmid (pmcr-1) were isolated using a QIAprep® Spin Miniprep kit and transformed into K. pneumoniae B5055 by electroporation conducted using a 0.1 cm electroporation cuvette with MicroPulser (Bio-Rad) at the EC1 setting (1.8 kV). Cells were recovered in LB broth for 1 h, followed by plating on LB agar with 20 mg/L gentamicin (Sigma-Aldrich). Successful transformants were confirmed by PCR and Sanger sequencing using M13 Fw (5′-GTAAAACGACGGCCAGT-3′) and M13 Rv (5′-AACAGCTATGACCATG-3′) primers (Micromon, Monash University). Unless otherwise stated, K. pneumoniae B5055pBBR1MCS-5 and K. pneumoniae B5055pmcr-1 were grown in LB broth supplemented with 20 mg/L gentamicin and E. coli JW1 was supplemented with 50 mg/L ampicillin (Sigma-Aldrich).
In vitro polymyxin B susceptibility
MICs of polymyxin B were determined using broth microdilution in CAMHB (Oxoid).36 Polymyxin B (Beta Pharma) was tested over 0.125–128 mg/L and gentamicin (final concentration of 20 mg/L) was added to the growth medium of the plasmid-carrying strains as required.
Plasmid stability
A conventional plate count method was employed to examine the stability of the vector and mcr-1 recombinant plasmid in K. pneumoniae strain B5055. Briefly, cultures were grown at 37°C in a shaking water bath (200 rpm) and serially passaged for 10 consecutive days with 1:1000 dilution in antibiotic-free LB broth, allowing ∼10 cell generations per day. Samples were taken each day, serially diluted and plated on antibiotic-free LB agar. After incubation overnight, 100 colonies were randomly selected and patched onto gentamicin-containing and antibiotic-free LB agar plates. The percentage of plasmid retention was calculated by dividing the number of colonies on gentamicin-containing LB agar by the total number of colonies patched. Three biological replicates were included for each group.
Extraction and structural analysis of lipid A
Lipid A was extracted from K. pneumoniae B5055 using a mild acid hydrolysis method.37 In brief, bacterial culture at an OD600 of 0.8 was collected by centrifugation at 9600 g and washed twice with PBS (pH 7.4). The cells were resuspended in PBS, followed by addition of chloroform and methanol to form a single-phase Bligh-Dyer mixture (water/chloroform/methanol, 0.8:1:2, v/v). LPS was pelleted by centrifugation at 3220 g and washed once with single-phase Bligh-Dyer mixture. The pellet was then resuspended in hydrolysis buffer [50 mM sodium acetate (pH 4.5)/1% SDS], followed by sonication and incubation in boiling water for 45 min to ensure complete hydrolysis. The solution was converted into a two-phase Bligh-Dyer mixture with addition of chloroform and methanol to form a final mixture of water/chloroform/methanol (0.9:1:1, v/v). Following centrifugation at 3220 g, the lower phase containing lipid A was extracted and allowed to dry overnight. For LC–MS analysis, the dried lipid A was reconstituted in chloroform/methanol (1:1, v/v) and the supernatant was collected into a glass LC–MS vial after centrifugation at 14 000 g. The structure of lipid A was analysed using a Q-Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific).
In vitro biological fitness assay
The fitness of K. pneumoniae strains was measured in competition against the reference strain E. coli JW1. Bacteria were inoculated into sterile filtered M9 minimal medium supplemented with antibiotics as appropriate.38 Overnight cultures were pelleted and washed with PBS prior to resuspension in fresh M9 minimal medium. An equal volume (1 mL) of the experimental strain and reference strain at ∼109 cfu/mL was mixed by vortexing. The mixed bacterial culture was diluted 1:100 in fresh M9 minimal medium and incubated for 24 h, which permitted ∼6.6 cell generations. Samples at 0 and 24 h were diluted 1:100 in PBS and analysed using flow cytometry. The experimental and reference strains were analysed using the dot plot of side scatter (SSC) versus forward scatter (FSC), in which gates were placed around the two populations, which were distinguishable based on morphological differences between the experimental and reference strains. The strains belonging to the populations gated were identified by referring to the single experimental and reference strain control. The selection coefficients were determined based on the following regression model: selection coefficient = [ln(E/R)t − ln(E/R)0]/T, where E/R is the ratio of cell numbers of the experimental to the reference strain and T is the generation number.39 All calculated selection coefficients were normalized to set the K. pneumoniae B5055 parental strain as zero by subtracting the mean selection coefficient of the parental strain. Four biological replicates were included for each sample. Data were analysed using the non-parametric Mann–Whitney U-test (GraphPad Prism software).
In vivo effect of mcr-1 in a neutropenic mouse thigh infection model
Ethical approval was obtained from the Monash University Animal Ethics Committee and the experiment was conducted in accordance with the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes. The 8-week-old female Swiss mice (Monash University, Clayton, Victoria, Australia) were housed with food and water available ad libitum. Neutropenia was induced by administrating two doses of cyclophosphamide intraperitoneally at 150 mg/kg for the first dose and 100 mg/kg for the second dose.40 Early logarithmic-phase K. pneumoniae strains (1 × 105 cfu/mL) were introduced intramuscularly into each posterior thigh muscle to establish thigh infection and two mice were included in each group (i.e. four thighs). At 2 h after inoculation, polymyxin B was administered subcutaneously at 10 mg/kg every 8 h (total dose of 30 mg/kg/day) to the treatment group. Mice were euthanized at 24 h and both thighs were harvested, followed by homogenization with saline and filtration using a sterile filter bag (pore size 280 μm) for viable counting. The agar plates were incubated overnight at 37°C and the number of cfu of bacteria per thigh was determined. Data were analysed using the non-parametric Mann–Whitney U-test.
Results
Prior to commencing our phenotypic investigation, we confirmed the presence of the vector (pBBR1MCS-5) and mcr-1 recombinant plasmid (pmcr-1) in K. pneumoniae B5055 with PCR using M13-specific forward and reverse primers. Comparison of the vector and mcr-1 recombinant plasmid confirmed the presence of a 1635 bp insertion in pBBR1MCS-5 and the insert was confirmed with Sanger sequencing.
Contribution of mcr-1 to polymyxin resistance and plasmid stability
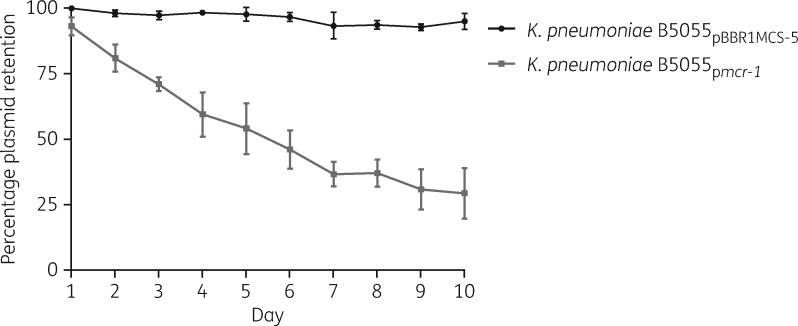
To evaluate the intrinsic resistance of the K. pneumoniae B5055 parental strain and the contribution of mcr-1 to polymyxin resistance, we first sought to determine their respective MICs. The K. pneumoniae B5055 parental strain, vector-only K. pneumoniae B5055pBBR1MCS-5 strain and mcr-1-expressing K. pneumoniae B5055pmcr-1 strain displayed MICs of 1, 0.5 and 4 mg/L, respectively. The stabilities of the mcr-1 recombinant plasmid and the vector in K. pneumoniae B5055 were determined by serial passaging for 10 days in the absence of gentamicin (Figure 1). K. pneumoniae B5055pBBR1MCS-5 showed high plasmid stability across the 10 day passages (i.e. ∼100 generations), whereas a gradual loss of the mcr-1 recombinant plasmid was observed, with only 50% of the population maintaining the mcr-1 recombinant plasmid after 6 day passage (i.e. ∼60 generations). This result suggests that in the absence of antibiotic selective pressure the maintenance of mcr-1 in the population was decreased.
Figure 1.
Stability of the empty vector (pBBR1MCS-5) and mcr-1 recombinant plasmid (pmcr-1) in K. pneumoniae B5055 strains over 10 days (∼10 generations per day) in the absence of antibiotic. Error bars represent the standard deviation of the mean (n = 3).
Lipid A profiling
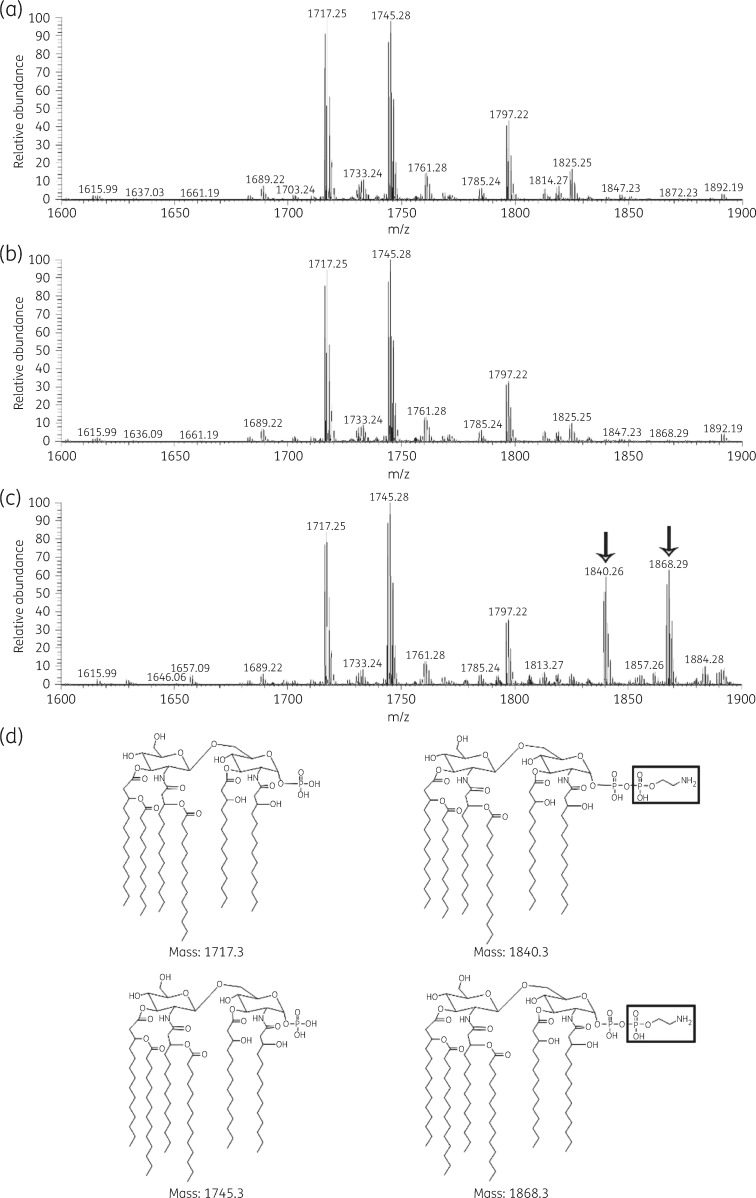
To confirm the role of mcr-1 in pEtN modification of lipid A, we next sought to investigate the lipid A profile of each K. pneumoniae strain by LC–MS (Figure 2). The MS analyses from all three strains showed two predominant peaks at m/z 1717.25 and 1745.28. The peak at m/z 1717.25 represents hexa-acylated lipid A species with four primary 3-hydroxymyristate (C14 [3-OH]) fatty acyls, two secondary fatty acyls (one laurate [C12]acyl and one myristate [C14]acyl), and the loss of one phosphate group from the glucosamine. The second peak at m/z 1745.28 represents hexa-acylated lipid A with four primary 3-hydroxymyristate (C14[3-OH]) fatty acyls, two secondary fatty acyls (myristate [C14]acyls), and the loss of one phosphate group from the glucosamine. MS analysis of the lipid A structure of K. pneumoniae B5055pmcr-1 revealed two additional peaks at m/z 1840.26 and 1868.29, representing the addition of a pEtN residue (Δm/z = +123) to the hexa-acylated lipid A molecules described earlier at m/z 1717.25 and 1745.28, respectively.41
Figure 2.
Mass spectra of lipid A from the (a) K. pneumoniae B5055 parental strain, (b) vector-only K. pneumoniae B5055pBBR1MCS-5 strain and (c) mcr-1-expressing K. pneumoniae B5055pmcr-1 strain. The pEtN-modified lipid A species are shown by the arrows. (d) Structures of predominant hexa-acylated lipid A species from each strain, with the corresponding pEtN-modified species on the right with pEtN shown in the boxes.
Biological cost of mcr-1 in vitro and in vivo
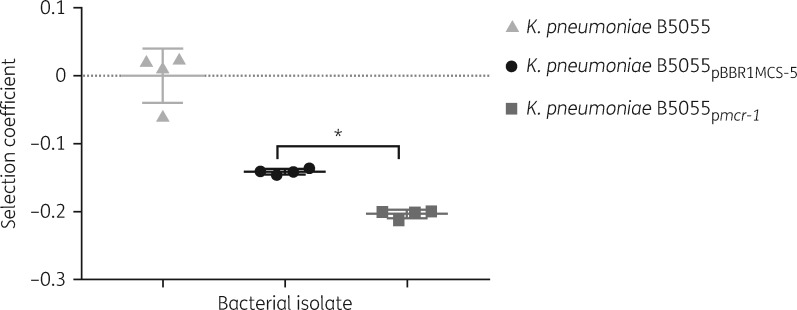
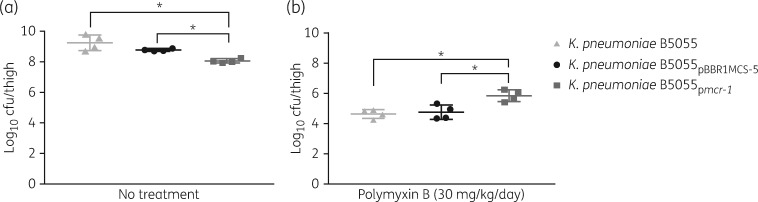
Competition assays were performed to determine whether mcr-1 is associated with biological cost in K. pneumoniae (Figure S1, available as Supplementary data at JAC Online). In comparison with the parent strain, lower selection coefficients were observed in vitro for both strains carrying the vector alone and mcr-1. However, a significantly lower selection coefficient (P < 0.05) was observed for K. pneumoniae B5055pmcr-1 in the in vitro study, compared with the strain carrying the vector alone (Figure 3). This result showed a biological fitness cost incurred in vitro by harbouring mcr-1. It appeared that in the absence of polymyxin B the strain carrying the vector only showed a minor reduction in growth in the mouse thigh infection model (P = 0.34). In comparison, it is clearly evident that the growth of the strain carrying the plasmid harbouring mcr-1 was significantly reduced in the infected mouse thighs (Figure 4a; P < 0.05). The impact of mcr-1 on polymyxin resistance in vivo was also illustrated using the same infection model but in the presence of polymyxin. After polymyxin B treatment (30 mg/kg/day), ∼10 000-fold reduction in the bacterial viability was observed at 24 h for K. pneumoniae B5055 and K. pneumoniae B5055pBBR1MCS-5, compared with the untreated group, but only a 100-fold reduction for K. pneumoniae B5055pmcr-1 (Figure 4b; P < 0.05).
Figure 3.
Selection coefficient for K. pneumoniae B5055 parental strain, vector-only K. pneumoniae B5055pBBR1MCS-5 strain and mcr-1-expressing K. pneumoniae B5055pmcr-1strain against the reference strain E. coli JW1. Error bars represent the standard deviation of the mean (n = 4). *P ≤ 0.05.
Figure 4.
In vivo effect of mcr-1 in a neutropenic mouse thigh infection model. Recovered bacterial burdens at 24 h (a) without any treatment and (b) treated with polymyxin B (30 mg/kg/day). Error bars represent the standard deviation of the mean (n = 4). *P ≤ 0.05.
Discussion
The identification of mcr-1 as the first plasmid-borne polymyxin resistance gene prompted extensive and retrospective surveillance investigations across a range of Gram-negative species.7,16mcr-1 is commonly found in Gram-negative bacteria isolated from animals, humans and the environment globally.7,24 The primary aims of the present study were to investigate the impact of mcr-1 on polymyxin resistance and the fitness of mcr-1-carrying K. pneumoniae.
According to EUCAST, the breakpoint for polymyxin susceptibility in Enterobacteriaceae is defined as ≤2 mg/L.42 When mcr-1 was introduced into K. pneumoniae B5055 via transformation with the recombinant plasmid (pmcr-1), the polymyxin B MIC increased from 1 to 4 mg/L (i.e. the polymyxin breakpoint for resistance). This moderate level of resistance has been demonstrated by other reports with bacterial isolates having MICs of 4–8 mg/L following experimental acquisition of mcr-1-carrying plasmids by polymyxin-susceptible E. coli strains via conjugation and transformation.16,43–49 Although the level of mcr-1-conferred polymyxin resistance is moderate, the ability of mcr-1 to improve bacterial survival in the presence of polymyxins was demonstrated in vivo using a neutropenic mouse thigh infection model. Similar findings were reported by Liu et al.,16 who showed that the presence of mcr-1 provided E. coli with adequate protection against colistin treatment in vivo. In order to exclude the possibility that the presence of other genetic elements in the plasmid contributed to the observed increase in polymyxin resistance, we also demonstrated that K. pneumoniae B5055pBBR1MCS-5 (which carries only the vector) was susceptible to polymyxin treatment.
The evolution and persistence of antibiotic resistance in the bacterial population depends on a complex calculus factoring the biological fitness cost associated with the resistance and the impact of the resistance pressure; any significant fitness cost will allow a susceptible strain to outcompete the resistant strain in the absence of an antibiotic.50 Variability in the fitness cost associated with antibiotic resistance can be attributed to various factors, including the mechanisms of resistance, bacterial species and antibiotic. A fitness cost has been shown to be associated with the acquisition of colistin resistance in Acinetobacter baumannii.51,52 Notably, the loss of LPS imposes a greater biological fitness cost in A. baumannii compared with modification of lipid A with pEtN.53 However, not all mechanisms of polymyxin resistance carry a fitness cost; for example, no significant biological cost is associated with polymyxin resistance due to chromosomal inactivation of mgrB in K. pneumoniae.54 The in vitro competition fitness assay and in vivo model from our study demonstrated that a significant biological cost was incurred when mcr-1 was introduced into K. pneumoniae in the absence of polymyxin treatment (Figures 3 and 4a). It has been reported that travellers colonized by mcr-1-carrying bacteria were able to completely eliminate these bacteria a month after of returning to their home country.55 This could suggest that mcr-1-carrying bacteria have reduced fitness in the antibiotic-free environment. Another possibility is that loss of the mcr-1 gene from the bacteria is due to the instability of the mcr-1-harbouring plasmids in the absence of selective pressure. Our study found that plasmid stability in the absence of selective pressure was indeed reduced with the acquisition of mcr-1. The heavy usage of polymyxins in the veterinary and agriculture environment could act as the selective pressure that promotes the dissemination of mcr-1.
The use of an artificial vector, pBBR1MCS-5, was a limitation of the present study. The vector itself showed a minor detrimental effect on the host bacteria in the absence of polymyxin treatment, but the mechanism is unknown. A recent study published after the submission of our manuscript showed that a negative fitness effect was correlated with both an artificial vector (pCR-Blunt II-TOPO) carrying mcr-1 and a native mcr-1 plasmid (pKP2442) in K. pneumoniae.56 Unfortunately, a Galleria mellonella model, rather than rodents, was employed for measuring the in vivo fitness cost in this recent study.56 Our present study employed a widely used mouse infection model, which is more clinically relevant, and demonstrated the negative influence of mcr-1 on in vivo fitness in K. pneumoniae.
In conclusion, our study demonstrates the beneficial effect of mcr-1 on the survival of K. pneumoniae in the presence of polymyxins. In the absence of selective pressure, mcr-1 negatively impacts the biological fitness of K. pneumoniae. It is likely that the spread of mcr-1 might be potentially attenuated by reducing polymyxin usage in both agriculture and healthcare sectors.
Supplementary Material
Acknowledgements
We wish to acknowledge the technical assistance provided by Ms Jesmin Akter, Mr Ke Chen and Mr Yang Hu.
Funding
This study was supported by a grant from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (R01 AI111965) and an Australian National Health and Medical Research Council (NHMRC) project grant (1064896). J. L. is an NHMRC Senior Research Fellow. T. V. is an Australian NHMRC Career Development Research Fellow.
Transparency declarations
None to declare.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Supplementary data
Figure S1 is available as Supplementary data at JAC Online.
References
- 1. WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf? ua=1.
- 2. Velkov T, Roberts KD, Nation RL. et al. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol 2013; 8: 711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Olaitan AO, Diene SM, Kempf M. et al. Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int J Antimicrob Agents 2014; 44: 500–7. [DOI] [PubMed] [Google Scholar]
- 4. Rossi F, Girardello R, Cury AP. et al. Emergence of colistin resistance in the largest university hospital complex of Sao Paulo, Brazil, over five years. Braz J Infect Dis 2017; 21: 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahmed SS, Alp E, Hopman J. et al. Global epidemiology on colistin resistant Acinetobacter baumannii. J Infect Dis Ther 2016; 4: 287. [Google Scholar]
- 6. Huang X, Yu L, Chen X. et al. High prevalence of colistin resistance and mcr-1 gene in Escherichia coli isolated from food animals in China. Front Microbiol 2017; 8: 562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwarz S, Johnson AP.. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 2016; 71: 2066–70. [DOI] [PubMed] [Google Scholar]
- 8. Liakopoulos A, Mevius DJ, Olsen B. et al. The colistin resistance mcr-1 gene is going wild. J Antimicrob Chemother 2016; 71: 2335–6. [DOI] [PubMed] [Google Scholar]
- 9. Baron S, Hadjadj L, Rolain JM. et al. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents 2016; 48: 583–91. [DOI] [PubMed] [Google Scholar]
- 10. Olaitan AO, Morand S, Rolain JM.. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 2014; 5: 643.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trimble MJ, Mlynárčik P, Kolář M. et al. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb Perspect Med 2016; 6: a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim SY, Choi HJ, Ko KS.. Differential expression of two-component systems, pmrAB and phoPQ, with different growth phases of Klebsiella pneumoniae in the presence or absence of colistin. Curr Microbiol 2014; 69: 37–41. [DOI] [PubMed] [Google Scholar]
- 13. Cheng HY, Chen YF, Peng HL.. Molecular characterization of the PhoPQ-PmrD-PmrAB mediated pathway regulating polymyxin B resistance in Klebsiella pneumoniae CG43. J Biomed Sci 2010; 17: 60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cannatelli A, Giani T, D’Andrea MM. et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 2014; 58: 5696–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poirel L, Jayol A, Bontron S. et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 2015; 70: 75–80. [DOI] [PubMed] [Google Scholar]
- 16. Liu YY, Wang Y, Walsh TR. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016; 16: 161–8. [DOI] [PubMed] [Google Scholar]
- 17. Di Pilato V, Arena F, Tascini C. et al. mcr-1.2: a new mcr variant carried on a transferable plasmid from a colistin-resistant KPC carbapenemase-producing Klebsiella pneumoniae of sequence type 512. Antimicrob Agents Chemother 2016; 60: 5612–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tijet N, Faccone D, Rapoport M. et al. Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS One 2017; 12: e0180347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang YQ, Li YX, Song T. et al. Colistin resistance gene mcr-1 and its variant in Escherichia coli isolates from chickens in China. Antimicrob Agents Chemother 2017; 61: e01204-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xavier BB, Lammens C, Ruhal R. et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 2016; 21: pii=30280. [DOI] [PubMed] [Google Scholar]
- 21. Borowiak M, Fischer J, Hammerl JA. et al. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 2017; 72: 3317–24. [DOI] [PubMed] [Google Scholar]
- 22. Carattoli A, Villa L, Feudi C. et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 2017; 22: pii=30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yin W, Li H, Shen Y. et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 2017; 8: e00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Skov RL, Monnet DL.. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill 2016; 21: pii=30155. [DOI] [PubMed] [Google Scholar]
- 25. Luo J, Yao X, Lv L. et al. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli from retail vegetables, China. Antimicrob Agents Chemother 2017; 61: e01139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Onori R, Gaiarsa S, Comandatore F. et al. Tracking nosocomial Klebsiella pneumoniae infections and outbreaks by whole-genome analysis: small-scale Italian scenario within a single hospital. J Clin Microbiol 2015; 53: 2861–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Podschun R, Ullmann U.. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998; 11: 589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poirel L, Nordmann P.. Emerging plasmid-encoded colistin resistance: the animal world as the culprit? J Antimicrob Chemother 2016; 71: 2326–7. [DOI] [PubMed] [Google Scholar]
- 29. Shen Z, Wang Y, Shen Y. et al. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis 2016; 16: 293. [DOI] [PubMed] [Google Scholar]
- 30. Poirel L, Kieffer N, Liassine N. et al. Plasmid-mediated carbapenem and colistin resistance in a clinical isolate of Escherichia coli. Lancet Infect Dis 2016; 16: 281. [DOI] [PubMed] [Google Scholar]
- 31. Elnahriry SS, Khalifa HO, Soliman AM. et al. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob Agents Chemother 2016; 60: 3249–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walsh TR, Wu Y.. China bans colistin as a feed additive for animals. Lancet Infect Dis 2016; 16: 1102–3. [DOI] [PubMed] [Google Scholar]
- 33. Andersson DI, Levin BR.. The biological cost of antibiotic resistance. Curr Opin Microbiol 1999; 2: 489–93. [DOI] [PubMed] [Google Scholar]
- 34. Clements A, Tull D, Jenney AW. et al. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J Biol Chem 2007; 282: 15569–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kovach ME, Elzer PH, Hill DS. et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 1995; 166: 175–6. [DOI] [PubMed] [Google Scholar]
- 36. Lin YW, Zhou Q, Onufrak NJ. et al. Aerosolized polymyxin B for treatment of respiratory tract infections: determination of pharmacokinetic-pharmacodynamic indices for aerosolized polymyxin B against Pseudomonas aeruginosa in a mouse lung infection model. Antimicrob Agents Chemother 2017; 61: e00211–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Que NLS, Lin S, Cotter RJ. et al. Purification and mass spectrometry of six lipid A species from the bacterial endosymbiont Rhizobium etli. Demonstration of a conserved distal unit and a variable proximal portion. J Biol Chem 2000; 275: 28006–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cold Spring Harbor Protocols. M9 Recipe 2006. 10.1101/pdb.rec8146. [DOI]
- 39. Dykhuizen DE. Experimental studies of natural selection in bacteria. Annu Rev Ecol Syst 1990; 21: 373–98. [Google Scholar]
- 40. Cheah SE, Wang J, Nguyen VT. et al. New pharmacokinetic/pharmacodynamic studies of systemically administered colistin against Pseudomonas aeruginosa and Acinetobacter baumannii in mouse thigh and lung infection models: smaller response in lung infection. J Antimicrob Chemother 2015; 70: 3291–7. [DOI] [PubMed] [Google Scholar]
- 41. Kim S-H, Jia W, Parreira VR. et al. Phosphoethanolamine substitution in the lipid A of Escherichia coli O157: H7 and its association with PmrC. Microbiology 2006; 152: 657–66. [DOI] [PubMed] [Google Scholar]
- 42. EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 7.1, 2017. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_?>files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf.
- 43. Yang YQ, Zhang AY, Ma SZ. et al. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J Antimicrob Chemother 2016; 71: 2336–8. [DOI] [PubMed] [Google Scholar]
- 44. Sonnevend Á, Ghazawi A, Alqahtani M. et al. Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int J Infect Dis 2016; 50: 85–90. [DOI] [PubMed] [Google Scholar]
- 45. Berrazeg M, Hadjadj L, Ayad A. et al. First detected human case in Algeria of mcr-1 plasmid mediated colistin resistance: a 2011 Escherichia coli isolate. Antimicrob Agents Chemother 2016; 60: 6996–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meinersmann RJ, Ladely SR, Bono JL. et al. Complete genome sequence of a colistin resistance gene (mcr-1)-bearing isolate of Escherichia coli from the United States. Genome Announc 2016; 4: e01283-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zheng B, Dong H, Xu H. et al. Coexistence of MCR-1 and NDM-1 in clinical Escherichia coli isolates. Clin Infect Dis 2016; 63: 1393–5. [DOI] [PubMed] [Google Scholar]
- 48. Ortiz de la Tabla V, Ortega A, Buñuel F. et al. Detection of the high-risk clone ST131 of Escherichia coli carrying the colistin resistance gene mcr-1 and causing acute peritonitis. Int J Antimicrob Agents 2016; 49: 115–6. [DOI] [PubMed] [Google Scholar]
- 49. Lu X, Hu Y, Luo M. et al. MCR-1.6: a new MCR variant carried by an IncP plasmid in a colistin-resistant Salmonella enterica serovar Typhimurium isolated from a healthy individual. Antimicrob Agents Chemother 2017; 61: e02632-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andersson DI. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr Opin Microbiol 2006; 9: 461–5. [DOI] [PubMed] [Google Scholar]
- 51. López-Rojas R, Dominguez-Herrera J, McConnell MJ. et al. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J Infect Dis 2011; 203: 545–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pournaras S, Poulou A, Dafopoulou K. et al. Growth retardation, reduced invasiveness, and impaired colistin-mediated cell death associated with colistin resistance development in Acinetobacter baumannii. Antimicrob Agents Chemother 2014; 58: 828–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Beceiro A, Moreno A, Fernández N. et al. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 2014; 58: 518–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cannatelli A, Santos-Lopez A, Giani T. et al. Polymyxin resistance caused by mgrB inactivation is not associated with significant biological cost in Klebsiella pneumoniae. Antimicrob Agents Chemother 2015; 59: 2898–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arcilla MS, van Hattem JM, Matamoros S. et al. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis 2016; 16: 147–9. [DOI] [PubMed] [Google Scholar]
- 56. Tietgen M, Semmler T, Riedel-Christ S. et al. Impact of the colistin resistance gene mcr-1 on bacterial fitness. Int J Antimicrob Agents 2017; doi:10.1016/j.ijantimicag.2017.11.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.