Abstract
Background
Bevacizumab is a humanized antibody against vascular endothelial growth factor approved for treatment of recurrent glioblastoma. There is a need to discover imaging biomarkers that can aid in the selection of patients who will likely derive the most survival benefit from bevacizumab.
Methods
The aim of the study was to examine if pre- and posttherapy multimodal MRI features could predict progression-free survival and overall survival (OS) for patients with recurrent glioblastoma treated with bevacizumab. The patient population included 84 patients in a training cohort and 42 patients in a testing cohort, separated based on pretherapy imaging date. Tumor volumes of interest were segmented from contrast-enhanced T1-weighted and fluid attenuated inversion recovery images and were used to derive volumetric, shape, texture, parametric, and histogram features. A total of 2293 pretherapy and 9811 posttherapy features were used to generate the model.
Results
Using standard radiographic assessment criteria, the hazard ratio for predicting OS was 3.38 (P < .001). The hazard ratios for pre- and posttherapy features predicting OS were 5.10 (P < .001) and 3.64 (P < .005) for the training and testing cohorts, respectively.
Conclusion
With the use of machine learning techniques to analyze imaging features derived from pre- and posttherapy multimodal MRI, we were able to develop a predictive model for patient OS that could potentially assist clinical decision making.
Keywords: bevacizumab, glioblastoma, machine learning, recurrent, survival
Glioblastoma is the most common primary adult brain tumor and carries one of the worst prognoses amongst human cancers, with a median survival time of about 15 months after diagnosis following the best available treatment involving surgery, radiation, and chemotherapy.1–3 Patients with recurrent glioblastoma have an even more dismal prognosis, with a median survival of just 25–40 weeks.4 Bevacizumab is a humanized monoclonal antibody against vascular endothelial growth factor approved for treatment of recurrent glioblastoma. While this therapy has resulted in a significant treatment response rate in a subset of patients with recurrent glioblastoma, its overall survival benefit is modest at best.5,6 There is a need to discover imaging biomarkers that can aid in the selection of patients who will likely derive the most benefit from bevacizumab.
The radiographic appearances of glioblastomas are highly heterogeneous, often consisting of elements of high cellularity, diffuse infiltration, and necrosis.7 Glioblastomas also express pro-angiogenic cytokines, which lead to the formation of abnormal intratumoral vasculature and induce peritumoral edema.7 Tumor heterogeneity is further increased by the occurrence of complex posttreatment changes, including gliosis, necrosis, and inflammation. More advanced imaging features can provide further information about tumor heterogeneity. This is of particular interest given recent evidence that glioblastomas harbor numerous cell subpopulations.8 Previous studies have shown that imaging features extracted from conventional MRI can predict survival in patients with glioblastoma prior to treatment.9–12 Diffusion-weighted (DW) MRI can provide additional prognostic or predictive information. Specifically, apparent diffusion coefficient (ADC) values calculated from DW-MRI negatively correlate with cell density, with lower values corresponding to regions with higher cell density and higher values corresponding to areas of edema and necrosis.13 Previous studies have demonstrated that the distribution of ADC maps within tumor volume can be characterized by histograms of ADC values using Gaussian mixtures; further, the resultant ADC parameters from recurrent glioblastoma are associated with patient survival during anti-angiogenic therapy.13–15
Multivariable regression models and survival analysis of imaging features have also been developed to assess patient outcome in recurrent glioblastoma.16–18 The usefulness of these predictive models is limited by their inability to search for complex patterns. Machine learning techniques allow the analysis of large quantities of imaging features to extract highly predictive imaging phenotypes. This technique is increasingly being used to generate prognostic biomarkers for primary brain tumors.19–21 Emblem et al19 and Macyszyn et al20 applied support vector machines to MRI data to predict survival in patients with gliomas. Zacharaki et al21 used classification trees applied to imaging variables from pre- and postoperational scans to predict short versus long survival in patients with high-grade gliomas.
In this study, we retrospectively examined multimodal MRIs of 126 patients with recurrent glioblastoma prior to and after bevacizumab therapy. We hypothesized that extracted imaging features based on both conventional and DW-MRI combined with machine learning can be used to predict patient progression-free survival (PFS) and overall survival (OS).
Materials and Methods
Patients
The institutional review board approved this retrospective study with a waiver for informed consent. Using a pharmacy database, we retrospectively identified patients with pathologically confirmed glioblastoma (World Health Organization [WHO] grade IV) who had received either bevacizumab monotherapy or bevacizumab and irinotecan combination therapy for treatment of progressive or recurrent disease at our institution between December 2005 and August 2014. Recurrence was defined by new or increased size of enhancing tumor (>25% bidimensional products) based on MRI prior to bevacizumab initiation. All patients had received standard-of-care treatment, which includes surgical resection followed by radiation therapy as well as concurrent and adjuvant temozolomide. Inclusion criteria were as follows: pretreatment MRI obtained within 2 weeks prior to initiating bevacizumab; posttreatment MRI obtained within 3 months prior to initiating bevacizumab; interpretable fluid attenuated inversion recovery (FLAIR), post-gadolinium (Gd) T1-weighted and DW imaging sequences captured on either 1.5 T or 3 T MRI systems. To avoid involving cases with pseudoprogression, those with recurrence 3 months from the end of radiation therapy were excluded from our analyses. Patients without adequate imaging or who underwent complex therapy were also excluded.
Our final cohort consisted of 126 patients; 113 patients had died at the time of analysis. Previously published Response Assessment in Neuro-Oncology (RANO) criteria were used to assess disease progression for the calculation of PFS.22 OS was calculated with respect to the date of bevacizumab therapy initiation. The baseline MRI was obtained a mean of 8 days (±12 d) before bevacizumab initiation, and the follow-up MRI was obtained a mean of 37 days (±15 d) after bevacizumab initiation. The patients were then divided into a training (n = 84) and testing (n = 42) cohort based on the date of pretreatment MRI scan. Clinical variables were collected, including age, sex, mono or dual therapy, progression before or at date of postoperative MRI scan, and number of recurrences before initiation of bevacizumab therapy. The baseline clinical characteristics for the training and testing cohort included in this study are summarized in Table 1.
Table 1.
Patient characteristics in the training and validation cohorts
| Training Set (n = 84) | Testing Set (n = 42) | PFS HR | OS HR | |
|---|---|---|---|---|
| Age | 57 (49, 62) | 58 (49, 64) | 0.84 (P = .35) | 0.89 (P = .53) |
| Gender (% male) | 63% | 58% | 1.00 (P = 1.00) | 1.03 (P = .89) |
| Number of recurrences | 1 (1, 2) | 1 (1, 2) | 1.41 (P = .06) | 1.61 (P < .05) |
| Bevacizumab treatment regimen (% monotherapy) | 40% | 98% | 0.90 (P = .58) | 0.82 (P = .31) |
| PFS (days) | 147 (86, 285) | 121 (86, 224) | ||
| OS (days) | 289 (178, 506) | 297 (145, 502) |
Data presented as median (interquartile range). Hazard ratios were calculated by dichotomizing patient characteristic using median values.
Imaging Acquisition
The standard MRI protocol at our institution included nonenhanced sagittal and axial T1-weighted, axial T2-weighted fast spin echo, axial T2/FLAIR, and contrast-enhanced axial T1-weighted (T1-Gd) and 3D spoiled gradient echo imaging with coronal and sagittal reconstructions. Gadopentetate dimeglumine (Magnevist, Bayer Healthcare) was administered for contrast-enhanced imaging.
DW-MRIs were acquired before injection of contrast. They were obtained with echo time/repetition time = 80–110 ms/4–10 seconds, section thickness = 5 mm with 1-mm intersection gap, matrix size = 128 × 128, and field of view = 22–25 cm by using a monopolar spin-echo echo-planar preparation. ADC images were calculated from acquired DW images with b-value 1000 s/mm2 and b-value 0 s/mm2 images. ADC maps were generated using Advantage Workstation software (v4.3, GE Healthcare). All MR images were transferred to workstations for offline post-processing and feature extraction.23
Volumetric Tumor Segmentation
The computer-based Brain Tumor Image Analysis software (v1.2) was used to coregister and skull-strip T1, T1-Gd, T2, and T2/FLAIR images.24 Whole brain T2/FLAIR images from MRI obtained at baseline and posttherapy were used for segmentation as described previously.25 Briefly, whole tumor (WT) volume, which includes solid tumor, infiltrating tumor, and edema, was segmented from T2/FLAIR imaging. The T1-Gd images from MRI were used for enhancing tumor (ET) volume segmentation. The ET that overlapped with the WT was subtracted to obtain the nonenhancing tumor (non-ET) volume. Tumor outlines were drawn with a user-driven, manual active contour segmentation method with 3D Slicer software (v4.1). The segmented volume contours were overlaid with source T1-Gd and T2/FLAIR images and edited by the study neuroradiologist (R.Y.H.) to manually add pixels for tumor regions not included in the preliminary contour or to remove pixels for nontumor regions included in the preliminary contour.26,27
To verify imaging feature reproducibility, a second independent segmentation approach was performed. In the same way, WT volumes were segmented from T2/FLAIR imaging, and the ET volume was segmented from T1-Gd images using a semi-automatic segmentation algorithm. This algorithm combined region-based active contour and level set approach,28–30 which required an operator manually selecting a region of interest on a single slice image. To ensure correct results, computer-generated contours were superimposed onto the original images for inspection and modification by the study radiologist (M.Z.). This segmentation algorithm and a number of efficient modification tools have been integrated into the Weasis software (v2.0.3).
Imaging Registration, Normalization, and ADC Submask Generation
The whole brain ADC volume was spatially coregistered to T2/FLAIR using rigid followed by affine transformation (12 degrees of freedom) with the Matlab Imaging Processing Toolbox (v2015a). To normalize imaging intensity, the interquartile range of the normal brain regions (nontumor) was calculated for each patient and imaging modality. The intensity of all voxels for all imaging modalities was then normalized by dividing the intensity by the interquartile range of normal brain.
Regions with ADC values <1100 and 1350 10−6 mm2/sec were segmented. In addition, to characterize tumor margins, edge submasks based on margins of WT and ET were calculated by detecting the edge of WT and ET, width dilations to 5 voxel lengths outside the edge, and width dilations to 3 voxel lengths inside the edge. Each edge submask thus had a width of 8 voxel lengths that captured regions of both tumor and normal-appearing brain (Supplementary Fig. S2).
Imaging Feature Extraction
Tumor volume masks and submasks from baseline MRI were applied to both baseline and posttherapy MRIs to produce volumes of interest (VOI). Masks generated from posttreatment MRI were only applied to posttherapy MRIs to produce VOI.
For VOI from ADC images, 4-peak curve fitting was applied to corresponding ADC histograms to calculate the following histogram features: the fractional area of each peak to total histogram area and the ratio of the different peaks to each other,13 standard deviation, variance, mean, mean absolute deviation, minimum, range, root mean squared, mode, uniformity, max probability, skewness, kurtosis, energy, entropy, 10th percentile value, 25th percentile value, 50th percentile value, 75th percentile value, and 90th percentile value. For VOI from T1, T1-Gd, T2, and T2/FLAIR images, skewness and kurtosis histogram features were calculated.
The following shape features were calculated for all VOI: volume, number of centroids, surface area, compactness, solidity, max diameter, spherical disproportion, sphericity, ratio of surface area to volume, and distance of volume centroid to the centroid of the entire brain. In addition, the volume within the pretherapy but not posttherapy scan and the volume within the posttherapy scan but not pretherapy scan were calculated for all masks and submasks.
Multimodal parametric features were produced by categorizing voxel intensity of individual imaging modalities: T1 (below 25th percentiles of non-WT, between 25th and 75th percentiles of non-WT, and above 75th percentile of non-WT), T1-Gd (below and above 75th percentile of non-WT), T2 (below and above 75th percentile of non-WT), and ADC (below and above 75th percentile of non-WT). The multimodal parametric features were then defined as the fraction of the VOI within every combination of groups (24 possible combinations total).
Texture features were calculated for all VOI in the horizontal plane and the 0 degree direction by analyzing gray level run length, Laws' texture, and Haralick texture features31–33 (see Supplementary data).
Changes in shape features between baseline and posttherapy tumor masks and submasks were also included as features. Similarly, changes in histogram, multimodal parametric, and texture features between baseline MRIs (baseline volume) and posttherapy MRIs (baseline volumes) as well as between baseline MRIs (baseline volume) and posttherapy MRIs (posttherapy volumes) were included as features.
Machine Learning Algorithm
Only the training cohort (n = 84) was used to develop the learning model. The machine learning algorithm was generated using Matlab Statistics and Machine Learning Toolbox (v2015a). A random forest classifier was used to distinguish patients surviving less/more than the 50th percentile of PFS or OS in the training cohort. Random forest is an ensemble learning algorithm that combines many decision trees into a single predictive algorithm. Each decision tree was trained from data that are sampled via bagging. Random forests provide several advantages, including being able to use high dimension data (where number of features are significantly larger than the number of patients) as well as provide a method of estimation of generalization error from the calculation of an out-of-bag error.34 The out-of-bag error was defined as the mean prediction error in each patient within the training set, using only the trees that did not have that patient in the bagged sample.35 As the forest grows from the addition of more decision trees, the generalization error converges to a limit.34 Due to the high number of decision trees in the forest, the random forest method is resistant to overfitting, making the method ideal in heterogeneous datasets such as that comprising patients with recurrent glioblastoma.
All features were initially tested for their individual predictive value by calculating the area under the curve from receiver operating characteristic (ROC) curve analysis. The top 128 features were then input into the random forest algorithm. Although the threshold of trees at which performance plateaus varies depending on the application and dataset, Oshiro et al36 found from an analysis of 29 datasets that there was no significant difference between forests with 256 trees compared with those with 256, 512, 1024, 2048, and 4096 trees. For the application within this study, it was found that having more than 512 trees did not improve the performance of random forest classification.
The random forest was thus grown to 512 trees large with the number of features per decision tree set to 64. The boot fraction, which is the fraction of patients used to train each tree, was set to ½. Four random forest models were trained: pretherapy features predicting PFS, pretherapy features predicting OS, pre- and posttherapy features predicting OS, and pre- and posttherapy features predicting OS excluding the patients who showed RANO-based progression before or at the date of posttherapy scan. The random forest model for pre- and posttherapy features to predict OS excluding cases that showed RANO-based progression was performed to determine whether a quantitative imaging model provides a prognostic value for nonprogressors following treatment initiation.
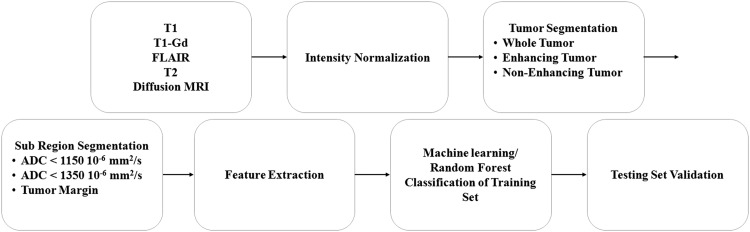
Validation of the random forest algorithm was performed by applying the trained random forest on the testing cohort. The workflow of the analysis in this study is summarized in Fig. 1.
Fig. 1.
Flow chart of analysis within this study, displaying image processing, feature extraction, machine learning, and validation steps.
Statistical Analysis
The Kaplan–Meier method was used to evaluate PFS and OS predictions. All statistical analyses were performed using Matlab v2015a. Hazard ratios (HRs) and Kaplan–Meier curves were computed for survival analysis with a significance level for a 2-sided comparison set at P < .05.
Results
Univariate ROC Analysis of Imaging Features
Pretherapy clinical features were included with features derived from pretherapy imaging. In total, there were 2293 pretherapy features. Posttherapy features include features derived from posttherapy imaging as well as difference features between features derived from pretherapy imaging and features derived from posttherapy imaging. In total, there were 9811 posttherapy features.
ROC analysis was performed on each imaging feature in the classification of both short- and long-term survivors. The top performing individual features for pretherapy features predicting PFS, pretherapy features predicting OS, pre- and posttherapy features predicting OS, and pre- and posttherapy features predicting OS excluding early progressors were shape, shape, texture, and texture, respectively.
Feature Selection and Classification
The random forest algorithm sequentially added trees until 512 trees were reached. The out-of-bag error decreased as the number of trees increased, but eventually plateaued (see Supplementary Fig. S1). For all of the random forest classifiers, the plateau was reached before 256 trees, which is consistent with what was reported by Oshiro et al.36
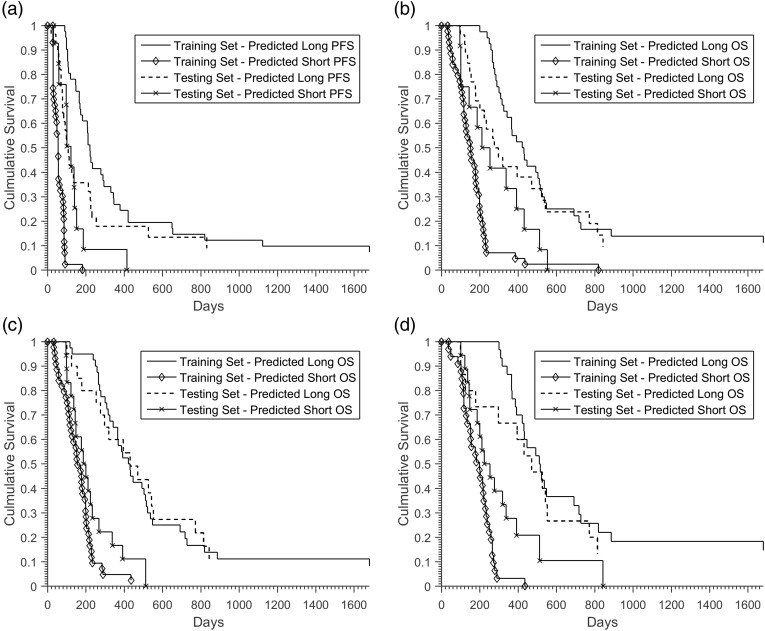
The HR for predicted short survivors compared with long survivors within the training cohort for pretherapy features predicting PFS, pretherapy features predicting OS, and pre- and posttherapy features predicting OS was 18.27 (P < .001), 6.03 (P < .001), and 5.10 (P < .001), respectively. With the exclusion of the patients who were determined to have progressive disease based on RANO criteria in the posttherapy MRI, the HR for pre- and posttherapy features predicting OS was 16.12 (P < .001). The out-of-bag accuracy for pretherapy features predicting PFS was 0.65. The out-of-bag accuracy for pretherapy features predicting OS was 0.70, which was improved through the addition of posttherapy features to 0.77. The 10 most important features, as determined by out-of-bag prediction error, for pre- and posttherapy features predicting OS are shown in Table 2. When the patients who displayed early progression based on RANO criteria in the posttherapy images were removed, the out-of-bag accuracy was 0.69 (Table 3). All predictions from the random forest classifiers within the training cohort delineated clear separation between long and short survivors in the Kaplan–Meier survival curves (Fig. 2).
Table 2.
The most important features based on random forest for pre- and posttherapy features predicting OS
| Imaging | Mask/Submask | Feature | Training Set AUC |
Testing
Set AUC |
|---|---|---|---|---|
| Pretherapy | Pretherapy
ET ADC <1350 10−6mm2/s |
Surface area to volume ratio | 0.80 | 0.56 |
| Pretherapy | Pretherapy
ET ADC <1100 10−6mm2/s |
Surface area to volume ratio | 0.78 | 0.57 |
| Posttherapy FLAIR |
Posttherapy ET | Long run low gray level emphasis | 0.81 | 0.70 |
| Posttherapy FLAIR |
Posttherapy tumor margin |
Max probability | 0.76 | 0.47 |
| Posttherapy ADC |
Posttherapy ET | Long run low gray level emphasis | 0.82 | 0.70 |
| Posttherapy ADC |
Posttherapy
WT tumor margin |
Max probability | 0.76 | 0.46 |
| Posttherapy T2 |
Posttherapy
ET ADC <1100 10−6 mm2/s |
Gray level nonuniformity | 0.86 | 0.70 |
| Posttherapy T2 |
Posttherapy ET | Long run low gray level emphasis | 0.82 | 0.69 |
| Posttherapy | Posttherapy ADC <1350 10−6 mm2/s |
Gray level nonuniformity | 0.84 | 0.72 |
| Posttherapy | Posttherapy ADC <1100 10−6 mm2/s |
Gray level nonuniformity | 0.85 | 0.71 |
Abbreviation: AUC, area under the curve.
Table 3.
Hazard ratio accuracy within the training and testing cohorts
| Training Set
(n = 84) |
Testing Set
(n = 42) |
|||
|---|---|---|---|---|
| HR | Out-of-bag Accuracy | HR | Validation Accuracy | |
| Pretherapy features predicting PFS | 18.27 (P < .001) | 0.65 | 1.37 (P = .38) | 0.57 |
| Pretherapy features predicting OS | 6.03 (P < .001) | 0.70 | 1.77 (P = .12) | 0.54 |
| Pre- and posttherapy features predicting OS | 5.10 (P < .001) | 0.77 | 3.64 (P < .005) | 0.76 |
| Pre- and posttherapy features predicting OS excluding early progressors | 16.12 (P < .001) | 0.69 | 2.02 (P = .07) | 0.69 |
Fig. 2.
Kaplan–Meier survival curves for predicted long and short survivors for (a) pretherapy features predicting PFS (training set: HR 18.27, P < .001; testing set: HR 1.37, P = .38), (b) pretherapy features predicting OS (training set: HR 6.03, P < .001; testing set: HR 1.77, P = .12), (c) pre- and posttherapy features predicting OS (training set: HR 5.10, P < .001; testing set: HR 3.64, P < .005), and (d) pre- and posttherapy features predicting OS excluding early progressors (training set: HR 16.12, P < .001; testing set: HR 2.02, P = .07).
Model Validation
The HRs for predicted short survivors compared with long survivors within the testing cohort (n = 42) are summarized in Table 3. Only pre- and posttherapy features predicting OS displayed an HR that was statistically significant (Fig. 2). The validation accuracies for pretherapy features predicting PFS, pretherapy features predicting OS, pre- and posttherapy features predicting OS, and pre- and posttherapy features predicting OS excluding early progressors were 0.57, 0.54, 0.76, and 0.69, respectively (Table 3).
Reproducibility of Imaging Features and Predictive Models
Based on tumor VOI derived from a second, independent semi-automatic segmentation approach, the imaging features were recalculated to access the reproducibility of the imaging features. For pre- and posttherapy features predicting OS, the correlation of the most important features between manual and semi-automatic segmentation was 0.85 with a standard error of 1.8% for manual segmentation and 1.5% for semi-automatic segmentation. When the random forest algorithm was applied to the resulting features, similar out-of-bag accuracies and validation accuracies were found for all applications (see Supplementary Table SI). Interestingly, semi-automatic segmentation had statistically significant HR for pretherapy features predicting PFS for both the training set (HR = 18.27, P < .001) and the testing set (HR = 2.01, P < .05).
Discussion
In this study, we examined whether a multivariable model based on pre- and early posttreatment multimodality imaging and a random forest algorithm could accurately predict survival outcome in patients with recurrent glioblastoma following anti-angiogenic therapy. The final machine learning model included clinical data as well as quantitative tumor features based on conventional MR and DW imaging. The use of out-of-bag error as well as validation on the testing cohort demonstrated the predictive potential of the model.
Previous studies have shown that imaging features such as ADC hypointensity,18 contrast material enhanced T1-weighted subtraction volume,37 change in T1 enhancing volume, residual T1 enhancing volume,25 and ratio of peaks fit in ADC histograms13,14,38 are associated with survival outcomes in recurrent glioblastoma. These studies have focused on only a few imaging features without integrating several features into a multivariate model. Here, we analyzed 2293 pretherapy features and 9811 posttherapy features simultaneously, allowing us to reveal synergistic effects of the features on prediction. In addition, through out-of-bag and testing cohort validation, we revealed distinct feature patterns predictive of survival.
Among the 4 models generated based on a manual segmentation method, statistically significant HRs in the training and testing patient cohorts were demonstrated for pre- and posttherapy features predicting OS. The results were reproduced when semi-automated segmentation was applied. It is important to note that the OS HRs for the training (5.10) and testing (3.64) cohorts were both higher than the HR (3.38) using standard radiographic evaluation (RANO). This suggests that the use of multimodal imaging features may be an improvement upon the current RANO criteria from the perspective of early posttreatment prognostication. In addition, statistically significant HRs in the training and testing sets were found for pretherapy features predicting PFS using semi-automated segmentation. This observation suggests superior performance of our predictive model when a semi-automated technique is used for feature extraction; this warrants further validation.
Several tumor volumes were calculated, including WT, ET (neoplastic and necrotic), and non-ET (neoplastic and edematous). Within these tumor volumes, further subvolumes were calculated based on ADC thresholds (high vs low cellularity) in order to increase feature specificity in the setting of tissue heterogeneity, which is well known among glioblastomas. Based on the most important features selected by the random forest algorithms, a greater proportion of the top features were derived from these subvolumes, confirming the value of subdividing tumor regions during feature extraction.
There are several limitations to our approach. Firstly, the dataset examined in this study was derived from a single-institution database; our model needs to be validated using independent or multicenter data to ensure its generalizability. In addition, manual and semi-automatic methods were used and require user input, which can be time consuming and subject to interuser variability. Although software exists for automated segmentation of glioblastoma,24,39,40 it has mostly been tested only on preoperative tumors. Despite this limitation, we have shown that our model and selected features are highly reproducible using 2 independent sets of segmentation approaches. Another limitation is that the training and testing sets differed in the percentage of patients receiving monotherapy (40% vs 98%). This difference, however, is unlikely to be significant, as the HR for mono versus dual therapy was not significant for either PFS or OS. In our study, we did not include perfusion imaging or genetic information; adding these features to our multivariable model may improve its performance, since prior studies have shown the prognostic value of these features.41–46 Nevertheless, our studies have demonstrated that features derived from conventional and DW imaging can be readily generated across different centers, since these techniques are routinely performed.
In this study, we evaluated the performance of several clinical, volumetric, shape, texture, parametric, and histogram features derived from standard MRI in the prediction of PFS and OS. In addition to analyzing the enhancing and nonenhancing components of each tumor, we improved our feature performance by subdividing tumor volumes using DW imaging. We found that several individual features were highly associated with PFS and OS; the association was even greater when the ensemble of features were integrated using a random forest algorithm. These results show that pre- and posttherapy imaging can be used to assist with clinical decision in patients with recurrent glioblastoma. Future work can investigate whether survival prediction is improved with inclusion of genotypic information and perfusion MRI.
Supplementary material
Supplementary material is available online at Neuro-Oncology (http://neuro-oncology.oxfordjournals.org/).
Funding
The work is supported by the ARRS/ASNR Scholar Award (foundation grant).
Supplementary Material
References
- 1. Stupp R, Mason WP, van den Bent MJ et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. [DOI] [PubMed] [Google Scholar]
- 3. Ostrom QT, Gittleman H, Fulop J et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015;17(Suppl 4):iv1–iv62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Norden AD, Drappatz J, Muzikansky A et al. An exploratory survival analysis of anti-angiogenic therapy for recurrent malignant glioma. J Neurooncol. 2009;92(2):149–155. [DOI] [PubMed] [Google Scholar]
- 5. Kreisl TN, Zhang W, Odia Y et al. A phase II trial of single-agent bevacizumab in patients with recurrent anaplastic glioma. Neuro Oncol. 2011;13(10):1143–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Friedman HS, Prados MD, Wen PY et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J Clin Oncol. 2009;27(28):4733–4740. [DOI] [PubMed] [Google Scholar]
- 7. Kalpathy-Cramer J, Gerstner ER, Emblem KE et al. Advanced magnetic resonance imaging of the physical processes in human glioblastoma. Cancer Res. 2014;74(17):4622–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patel AP, Tirosh I, Trombetta JJ et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Z, Jiang H, Chen X et al. Identifying the survival subtypes of glioblastoma by quantitative volumetric analysis of MRI. J Neurooncol. 2014;119(1):207–214. [DOI] [PubMed] [Google Scholar]
- 10. Mazurowski MA, Desjardins A, Malof JM. Imaging descriptors improve the predictive power of survival models for glioblastoma patients. Neuro Oncol. 2013;15(10):1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gutman DA, Cooper LAD, Hwang SN et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267(2):560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li W-B, Tang K, Chen Q et al. MRI manifestions correlate with survival of glioblastoma multiforme patients. Cancer Biol Med. 2012;9(2):120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rahman R, Hamdan A, Zweifler R et al. Histogram analysis of apparent diffusion coefficient within enhancing and nonenhancing tumor volumes in recurrent glioblastoma patients treated with bevacizumab. J Neurooncol. 2014;119(1):149–158. [DOI] [PubMed] [Google Scholar]
- 14. Pope WB, Kim HJ, Huo J et al. Recurrent glioblastoma multiforme: ADC histogram analysis predicts response to bevacizumab treatment. Radiology. 2009;252(1):182–189. [DOI] [PubMed] [Google Scholar]
- 15. Pope WB, Qiao XJ, Kim HJ et al. Apparent diffusion coefficient histogram analysis stratifies progression-free and overall survival in patients with recurrent GBM treated with bevacizumab: a multi-center study. J Neurooncol. 2012;108(3):491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ellingson BM, Lai A, Nguyen HN et al. Quantification of nonenhancing tumor burden in gliomas using effective T2 maps derived from dual-echo turbo spin-echo MRI. Clin Cancer Res. 2015;21(19):4373–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kickingereder P, Wiestler B, Burth S et al. Relative cerebral blood volume is a potential predictive imaging biomarker of bevacizumab efficacy in recurrent glioblastoma. Neuro Oncol. 2015;17(8):1139–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elson A, Paulson E, Bovi J et al. Evaluation of pre-radiotherapy apparent diffusion coefficient (ADC): patterns of recurrence and survival outcomes analysis in patients treated for glioblastoma multiforme. J Neurooncol. 2015;123(1):179–188. [DOI] [PubMed] [Google Scholar]
- 19. Emblem KE, Pinho MC, Zöllner FG et al. A generic support vector machine model for preoperative glioma survival associations. Radiology. 2015;275(1):228–234. [DOI] [PubMed] [Google Scholar]
- 20. Macyszyn L, Akbari H, Pisapia JM et al. Imaging patterns predict patient survival and molecular subtype in glioblastoma via machine learning techniques. Neuro Oncol. 2016;18(3):417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zacharaki EI, Morita N, Bhatt P et al. Survival analysis of patients with high-grade gliomas based on data mining of imaging variables. AJNR Am J Neuroradiol. 2012;33(6):1065–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wen PY, Macdonald DR, Reardon DA et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 23. Plesniak W, Rosen BR, Pieper S, Gollub RL. High throughput tools to access images from clinical archives for research. J Digit Imaging. 2015;28(2):194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Porz N, Bauer S, Pica A et al. Multi-modal glioblastoma segmentation: man versus machine. PLoS One. 2014;9(5):e96873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang RY, Rahman R, Hamdan A et al. Recurrent glioblastoma: volumetric assessment and stratification of patient survival with early posttreatment magnetic resonance imaging in patients treated with bevacizumab. Cancer. 2013;119(19):3479–3488. [DOI] [PubMed] [Google Scholar]
- 26. Pieper S, Lorensen B, Schroeder W, Kikinis R. The NA-MIC Kit: ITK, VTK, pipelines, grids and 3D slicer as an open platform for the medical image computing community. In: 3rd IEEE Int. Symp. Biomed. Imaging Nano to Macro, 2006. 698–701. [Google Scholar]
- 27. Pieper S, Halle M, Kikinis R. 3D SLICER. In: Proc. 1st IEEE Int. Symp. Biomed. Imaging Nano Macro 632–635. [Google Scholar]
- 28. Guo X, Schwartz L, Zhao B. Semi-automated segmentation of multimodal brain tumor using active contours. In: Proc. NCI-MICCAI BRATS Nagoya, Japan 17–30. [Google Scholar]
- 29. Chow DS, Qi J, Guo X et al. Semiautomated volumetric measurement on postcontrast MR imaging for analysis of recurrent and residual disease in glioblastoma multiforme. AJNR Am J Neuroradiol. 2014;35(3):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Menze BH, Jakab A, Bauer S et al. The multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging. 2015;34(10):1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Galloway MM. Texture analysis using gray level run lengths. Comput Graph Image Process. 1975;4(2):172–179. [Google Scholar]
- 32. Lee D-C, Schenk T. Image segmentation from texture measurement. In: Fritz L, Lucas J (ed). Washington, DC: ISPRS; 195–199. [Google Scholar]
- 33. Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;3(6):610–621. [Google Scholar]
- 34. Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 35. James G, Witten D, Hastie T, Tibshirani R. An Introduction to Statistical Learning. Berlin: Springer; 2013.
- 36. Oshiro TM, Perez PS, Baranauskas JA. How many trees in a random forest? Mach Learn Data Min Pattern Recognit. 2012;7376:154–168. [Google Scholar]
- 37. Ellingson BM, Kim HJ, Woodworth DC et al. Recurrent glioblastoma treated with bevacizumab: contrast-enhanced T1-weighted subtraction maps improve tumor delineation and aid prediction of survival in a multicenter clinical trial. Radiology. 2014;271(1):200–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pope WB, Lai A, Mehta R et al. Apparent diffusion coefficent histogram analysis stratifies progression-free survival in newly diagnosed bevacizumab-treated glioblastoma. AJNR Am J Neuroradiol. 2011;32(5):882–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Juan-Albarracín J, Fuster-Garcia E, Manjón JV et al. Automated glioblastoma segmentation based on a multiparametric structured unsupervised classification. PLoS One. 2015;10(5):e0125143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Steed TC, Treiber JM, Patel KS et al. Iterative probabilistic voxel labeling: automated segmentation for analysis of The Cancer Imaging Archive glioblastoma images. AJNR Am J Neuroradiol. 2015;36(4):678–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J, Jain R, Khalil K et al. Texture feature ratios from relative CBV maps of perfusion MRI are associated with patient survival in glioblastoma. AJNR Am J Neuroradiol. 2016;37(1):37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deike K, Wiestler B, Graf M et al. Prognostic value of combined visualization of MR diffusion and perfusion maps in glioblastoma. J Neurooncol. 2016;126(3):463–472. [DOI] [PubMed] [Google Scholar]
- 43. Bonekamp D, Deike K, Wiestler B et al. Association of overall survival in patients with newly diagnosed glioblastoma with contrast-enhanced perfusion MRI: comparison of intraindividually matched T1 - and T2 (*)-based bolus techniques. J Magn Reson Imaging. 2015;42(1):87–96. [DOI] [PubMed] [Google Scholar]
- 44. Di Stefano AL, Labussiere M, Lombardi G et al. VEGFA SNP rs2010963 is associated with vascular toxicity in recurrent glioblastomas and longer response to bevacizumab. J Neurooncol. 2015;121(3):499–504. [DOI] [PubMed] [Google Scholar]
- 45. D'Alessandris QG, Martini M, Cenci T et al. VEGF isoforms as outcome biomarker for anti-angiogenic therapy in recurrent glioblastoma. Neurology. 2015;84(18):1906–1908. [DOI] [PubMed] [Google Scholar]
- 46. Lv S, Teugels E, Sadones J et al. Correlation between IDH1 gene mutation status and survival of patients treated for recurrent glioma. Anticancer Res. 2011;31(12):4457–4463. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.