2.1. T Cell-Inflamed Versus Non-T Cell-Inflamed Tumor Microenvironment
Interrogation of the tumor microenvironment in metastatic melanoma was initially pursued to test the hypothesis that resistance mechanisms downstream from T cell priming in response to melanoma vaccines might dominate and enable tumor escape [1, 2]. Baseline biopsies of melanoma metastases were obtained and evaluated by gene expression profiling, to correlate with clinical outcome from vaccination. Two major subsets of tumor microenvironment could be identified that were largely characterized by the presence or absence of a transcriptional profile indicative of a pre-existing T cell infiltrate (Fig. 2.1). The T cell-inflamed subset of tumors was dominated by T cell markers and chemokines that likely mediate effector T cell recruitment [3–5]. Expression of the chemokines CCL2, −3, −4, −5 and CXCL9, −10 was observed to correlate with T cell presence, and each of these chemokines was sufficient to recruit CD8+ effector T cells in vitro [3]. Recently, CXCR3-binding chemokines (such as CXCL9 and CXCL10) were found to be critical and necessary for trafficking of activated CD8+ T cells into tumor sites [6]. Immunohistochemistry confirmed the presence of CD8+ T cells, macrophages, as well as some B cells and plasma cells in the T cell-inflamed lesions [3]. This subset of tumors was remarkably distinct from the non-T cell inflamed subset, and the biology suggests that spontaneous T cell priming and recruitment into the tumor microenvironment had occurred in those patients, even prior to any therapy. Interestingly, reflecting back onto the original goal of this analysis, the clinical responders to vaccination were seen among patients with the T cell-inflamed phenotype [4]. Thus, it appears as though tumors capable of supporting recruitment of activated CD8+ T cells are those that stand to benefit from interventions that increase the frequency of tumor antigen-specific T cells in the circulation, such as vaccination.
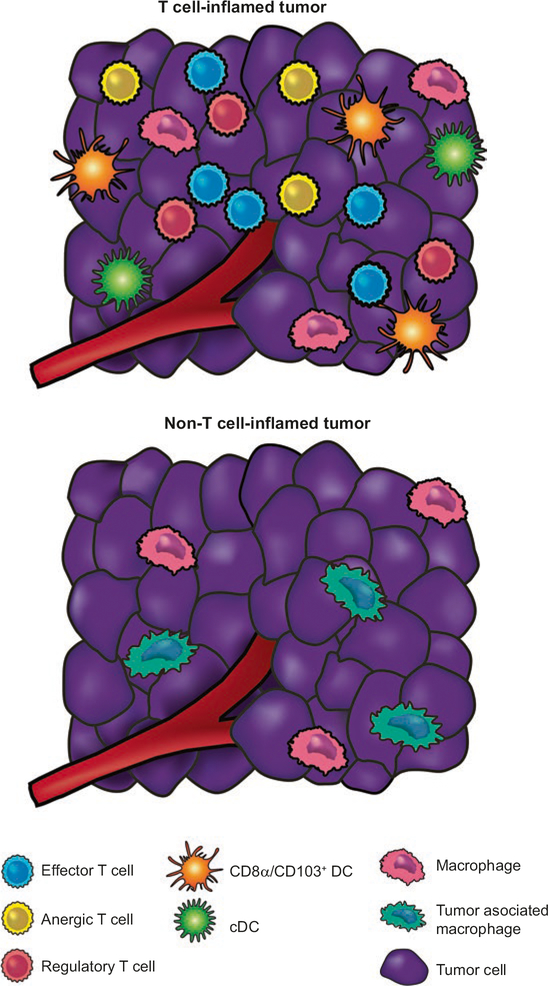
Fig. 2.1.
Immunologic composition of the T cell-inflamed versus non-T cell-inflamed tumor microenvironments. The T cell-inflamed tumors contain variable numbers of CD8+ T cells and CD8α/CD103-lineage DCs, but also possess the highest density of FoxP3+ Tregs. In addition, many of the conventional T cells have a dysfunctional anergic phenotype. In contrast, the non-T cell-inflamed tumors lack these elements but still contain blood vessels, fibroblasts, and macrophages that help support tumor growth. Recruitment of CD8+ effector cells is largely dependent on the chemokines CXCL9 and CXCL10, which engage the receptor CXCR3. Treg recruitment is primarily driven by CCL22, which is in part produced by activated CD8+ T cells
The T cell-inflamed subset of melanoma metastases is remarkably similar to the phenotype described in early stage colon cancer and other tumors in which activated T cells have been associated with favorable prognosis [7–9]. In several small studies of HLA-A2+ patients, CD8+ T cells specific for melanoma differentiation antigens such as Melan-A were identified from tumor sites using peptide-HLA-A2 tetramer analysis [10–12]. Therefore, at least a subset of T cells specific for tumor antigens is present among these infiltrates. The fact that the starting point for adoptive T cell approaches utilizing tumor-infiltrating lymphocytes (TIL), which have demonstrated approximately a 50% response rate in metastatic melanoma [13], is T cells harvested from the tumor, also supports the notion that tumor-specific T cells are present. However, the function of these T cells in situ is impaired. Various degrees of dysfunction of tumor antigen-specific T cells have been described upon analysis directly ex vivo [10–12]. Together, these results suggest that the reason for tumor progression despite the presence of specific adaptive immunity in this subset of patients is likely secondary to immune suppressive mechanisms acting at the level of the tumor microenvironment. Interestingly, in some cases the presence of memory virus-specific CD8+ T cells also has been observed in T cell-inflamed melanomas. However, their function seems to be intact [10, 14], and these probably represent non-specifically recruited activated T cells migrating along chemokine gradients but not participating in tumor recognition. These observations suggest that a component of T cell dysfunction in the tumor microenvironment is antigen-specific and restricted to tumor-reactive T cells.
In contrast to the rich set of immune genes expressed in the T cell-inflamed tumor microenvironment phenotype, the non-T cell-inflamed tumors lacked this broad signature. In particular, there was a lack of T cell markers and of chemokines that can mediate T cell recruitment [3]. These tumors still contain macrophages and vascular endothelial cells, and work from others has indicated the presence of fibroblasts and extracellular matrix, and in some cases immature dendritic cells [15–19]. It is not yet certain whether tumors that lack spontaneous T cell infiltration are defective only at the level of initial T cell priming against tumor antigens or whether there are additional mechanisms that exclude activated T cells from migrating into the tumor microenvironment, but is seems plausible that both processes may be operational. The idea that both the priming and the effector phases of the anti-tumor immune response are defective in non-T cell-inflamed tumors is supported by recent data in genetically engineered mouse models. Tumors with poor T cell infiltration appear to have higher expression of several angiogenic factors [3, 19]. Vascular endothelial cells from T cell-infiltrated versus T cell-non-infiltrated tumors have been reported to have distinct gene expression profiles, and the endothelin B receptor has been identified as a vascular target in an ovarian cancer context [19]. Thus, effector T cell trafficking into the tumor microenvironment is complex, and depends on adhesion molecules and homing receptors expressed on vascular endothelial cells in concert with chemokines produced by tumor cells and/or stromal cells within the tumor microenvironment. This process is likely necessary for clinical response to immunotherapies in most instances.
It has been argued that non-T cell-inflamed tumors might lack neoantigens for T cell recognition and therefore might not be immunogenic because they are not antigenic. A recent report has suggested that patients who failed to derive clinical benefit from the anti-CTLA-4 mAb ipilimumab might have fewer mutations and lack the antigens present in the tumors of responding patients [20]. However, we have recently analyzed exome sequencing versus germline data from the metastatic melanoma samples that are among The Cancer Genome Atlas data set. RNAseq data were used to categorize patients as having a T cell-inflamed versus a non-T cell-inflamed tumor microenvironment. The frequency of non-synonymous mutations, expression of cancer-testis antigen genes, and the expression of melanoma differentiation antigens were enumerated between these groups. No differences were observed with any of these parameters between the two cohorts of patients [21]. These data indicate that lack of antigen expression is unlikely to explain the non-T cell-inflamed tumor microenvironment phenotype in melanoma. These data are encouraging, as they suggest that strategies to overcome the barrier of T cell migration into tumor sites might ultimately enable immunotherapy efficacy in non-T cell-inflamed tumors.
2.2. Negative Regulatory Pathways Impeding Immune Efficacy in T Cell-Inflamed Tumors
Because of the presence of dysfunctional T cells in the same microenvironment as antigen-expressing tumors cells, the T cell-inflamed subset of tumors was probed for candidate immune-inhibitory mechanisms that might contribute to T cell dysfunction in situ. Gene expression profiling data revealed the presence of transcripts encoding indoleamine-2,3-dioxygenase (IDO) in these tumors, a molecule that had previously been demonstrated to contribute to peripheral tolerance [22]. Interrogation for additional candidates revealed that these tumors also expressed PD-L1 and Foxp3 transcripts [23, 24]. Quantitative analysis of individual tumors revealed that the expression level of each of these three transcripts was significantly correlated, and that the degree of expression was also associated with T cell markers. Immunohistochemistry confirmed that PD-L1 and IDO protein expression, and also nuclear Foxp3+CD4+ cells, were found within T cell-inflamed tumors in the same region as CD8+ T cells (Fig. 2.2). However, non-T cell-inflamed melanomas generally lacked these factors. These observations suggested that these immune suppressive mechanisms might not be a property of the tumor cells themselves but rather immune-intrinsic negative feedback processes that follow the recruitment of activated CD8+ T cells. Indeed, mouse mechanistic studies confirmed that CD8+ T cells were required for the upregulation of all of these three factors within the tumor microenvironment. For PD-L1 and IDO induction, the requisite factor produced by the CD8+ T cells was IFN-γ. For FoxP3+ Tregs, production of the chemokine CCL22 was identified, which mediated Treg recruitment into tumor sites [24]. Using laser capture microdissection, a correlation between IFN-γ production by TILs and local PD-L1 expression also was observed by Taube and colleagues in human tumors [25], supporting the notion that infiltrating T cells become activated by specific antigen and consequently produce IFN-γ and upregulate PD-L1 expression. The fact that these immune evasion mechanisms are part of the host response implies that targeting these pathways therapeutically should have an increased likelihood of efficacy because they are less dependent on tumor cell properties and the associated mutability that can frequently lead to therapeutic resistance.
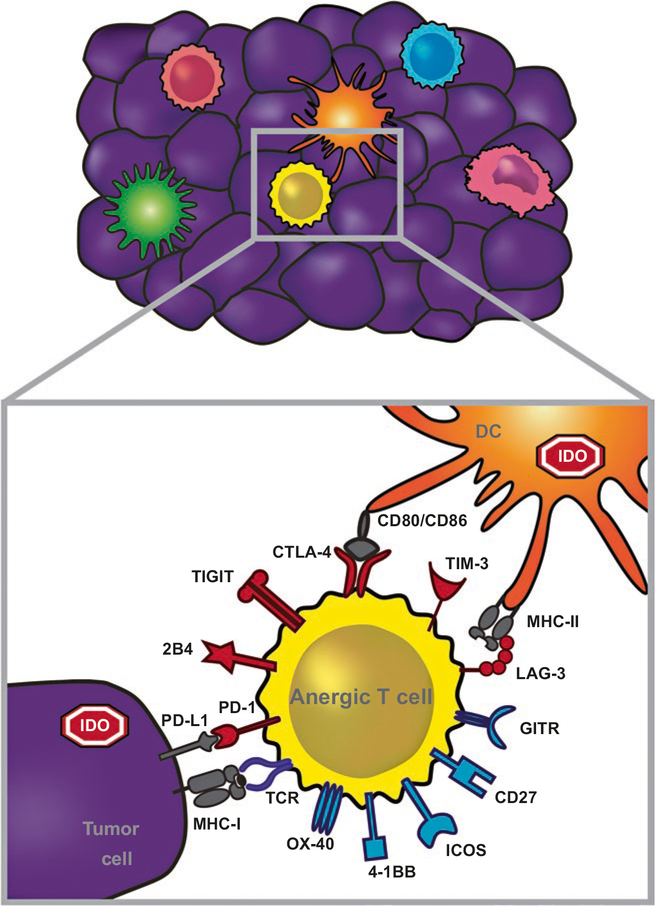
Fig. 2.2.
Immunotherapeutic targets that are preferentially relevant for the T cell-inflamed tumor microenvironment subset. T cell-inflamed tumors contain activated CD8+ T cells but also express IDO and PD-L1, which inhibit T cell function. The dysfunctional/anergic T cells in the tumor microenvironment also can express an array of additional inhibitory receptors, including LAG-3, Tigit, Tim3, and 2B4. But in addition, these T cells also para-doxically express costimulatory receptors, including 4–1BB, Ox40, ICOS, GITR, and CD27. Both blockade of inhibitory receptors and ligation of costimulatory receptors are being developed as cancer therapeutics. Additional candidate immune suppressive factors not shown here that have yet to be effectively targeted clinically include TGF-β, IL-10, iNOS, and PGE2
Preclinical studies targeting CTLA-4, PD-L1, and IDO have indicated that the therapeutic effect is associated with re-activation of CD8+ T cells directly within the tumor microenvironment [26]. The major biologic correlate that is restored with blockade of these pathways is the ability of tumor-infiltrating CD8+ T cells to produce IL-2 and to proliferate when analyzed ex vivo. In order to test whether the therapeutic effect required influx of new T cells at all, the drug FTY720 was utilized, which prevents new T cell egress from lymph nodes. In fact, both restoration of IL-2 production and proliferation of TIL as well as tumor regression were preserved despite FTY720 administration [26], arguing that the immediate functional effects of checkpoint blockade can all be explained by re-activation of T cells already present within the tumor microenvironment. Consistent with these data, clinical response with anti-PD-1 mAb in metastatic melanoma was found to occur predominantly in patients with pre-existing T cell infiltrates in the region of PD-L1 upregulation [25, 27, 28]. Following anti-PD-1 administration, these CD8+ T cells seemed to proliferate and expand, as indicated by Ki67 expression, and to penetrate deeply throughout the tumor [28]. Thus, the preponderance of clinical response with active immune-therapies also is likely mediated through restored function of pre-existing TIL.
In addition to the presence of PD-L1, IDO, and Tregs that likely mediate extrinsic suppression, a T cell-intrinsic mechanism is also likely contributing to tumor escape in the T cell-inflamed cancers. This phenomenon is similar to T cell anergy that has been characterized extensively using in vitro model systems, an observation which has provided a tool for identifying additional immune regulatory targets on dysfunctional T cells within the tumor microenvironment. In vitro experiments using CD4+ Th1 clones as a model system have identified the transcription factor EGR2 as a critical mediator of T cell dysfunction [29]. EGR2 is induced following TCR ligation alone, and leads to upregulation of the lipid phosphatase diacylglycerol kinase, which in turn inhibits TCR-mediated Ras pathway activation [30]. Conditional EGR2-knockout T cells have shown improved anti-tumor activity in vivo [29], arguing for a functional relevance of this pathway in anti-tumor immunity. With this functional importance in mind, experiments were conducted to identify the full spectrum of EGR2 target genes in anergic T cells. Gene expression profiling of wild-type versus EGR2-deleted T cells was performed, to identify EGR2-dependent genes. In parallel, a genome-wide ChIPseq study was performed, to identify genes directly bound by EGR2. Merging these two datasets revealed 50 genes that characterized the EGR2 transcriptome [31]. Interestingly, several of these target genes encode surface receptors that allow phenotyping using flow cytometry, including LAG-3 and 4–1BB. LAG-3 is an inhibitory receptor with homology to CD4 that recognizes at least class II MHC as a ligand [32]. 4–1BB is a costimulatory receptor of the TNFR superfamily that engages the NF-κB pathway [33]. Returning to the tumor microenvironment, flow cytometric analysis revealed that a major population of CD8+ TIL co-expressed LAG-3 and 4–1BB. All of these cells were also PD-1-positive. By cell sorting and stimulation in vivo, it was found that the LAG-3+4–1BB+ subset was the most dysfunctional as reflected by IL-2 production and proliferation. The majority of tumor-specific T cells were found to fall into this subset. Thus, these likely represent important markers for identifying the dysfunctional tumor antigen-specific T cell subset within the tumor microenvironment (Williams and Gajewski, manuscript in preparation). Administration of a blocking mAb against LAG-3 along with an agonistic Ab against 4–1BB showed profound anti-tumor activity in vivo. Anti-4–1BB also synergized therapeutically with either anti-CTLA-4 or anti-PD-L1 mAbs (Horton and Gajewski, unpublished data). Interestingly, all of these combination therapies also depend upon re-activation of T cells already present within the tumor microenvironment. These data suggest that thorough phenotypic analysis of dys-functional TIL should reveal the total array of immune regulatory receptors amenable to in vivo therapeutic targeting.
Because of the presence of multiple immune regulatory factors in the same T cell-inflamed tumor microenvironment, and based on preclinical evidence for synergistic efficacy, multiple phase I/II trials are underway to evaluate key combinations. These include an IDO inhibitor combined with either anti-CTLA-4 or anti-PD-1 mAbs, anti-LAG-3 + anti-PD-1, and anti-4–1BB mAb in various combinations. The potential for combination immunotherapy to have superior efficacy is supported by recent data using anti-CTLA-4 + anti-PD-1, which revealed a higher response rate than either agent alone in metastatic melanoma, albeit with a higher rate of adverse events [34]. Over time, additional combinations that have comparable efficacy and perhaps decreased toxicity will hopefully be identified, both for melanoma and for other cancer types showing a fraction of patients characterized by the T cell-inflamed tumor microenvironment.
2.3. Innate Immune Mechanisms Bridging to Spontaneous Anti-Tumor T Cell Responses
Expanding the efficacy of immunotherapies will require a better understanding of the mechanisms mediating the non-T cell-inflamed tumor microenvironment. As a first approach, possible innate immune pathways involved in generating the T cell-inflamed tumor microenvironment when it does occur have been pursued. In general, in order for adaptive T cell responses to be induced against an antigen, dendritic cells (DCs) or other involved antigen-presenting cells (APCs) need to be activated themselves for productive adaptive immunity. In the context of infectious agents, this is typically through stimulation of Toll-like receptors (TLRs) by pathogen-associated molecular patterns (PAMPs), such as endotoxin that is recognized by TLR4 [35]. However, it had not been clear what factors might provide innate immune signaling in the context of sterile tumors in which infectious agents are not implicated. Melanoma gene expression profile data were interrogated for evidence of innate immune pathway activation. A major clue came from the observation that tumors that contained a T cell infiltrate also showed evidence for a transcriptional signature known to be induced by type I IFNs [3, 36]. Armed with that information, mouse mechanistic experiments were carried out to determine whether type I IFN signaling on host cells was necessary for spontaneous priming of CD8+ T cells against tumor-associated antigens. In fact, type I IFNR−/− mice, or mice deficient in the downstream transcription factor Stat1, showed markedly reduced T cell responses against tumor-associated antigens in multiple transplantable tumor models [36]. The requirement for type I IFN signaling was mapped to the level of APCs, and indeed specific deletion of the type I IFNR in DCs was sufficient to reproduce this defect. Mixed bone marrow chimera experiments demonstrated that the specific subset of DCs involved in this effect was the Batf3-driven lineage that expresses CD8α or CD103 [36–38]. In addition, IFN-β production was found to be induced in DCs upon implantation of a tumor in vivo. These data suggest that early innate immune recognition of cancer cells in vivo involved activation of a pathway that results in IFN-β production, which in turn was necessary for complete DC activation and CD8+ T cell priming to give rise to the T cell-infiltrated tumor microenvironment phenotype [39].
These observations led to the next level question of identifying the receptor system and putative ligands that induce IFN-β production by host DCs in the context of a growing tumor in vivo. By using a series of knockout mice specifically lacking TLRs, the adaptors MyD88 or Trif, the intracellular RNA sensing pathway involving MAVS, or the extracellular ATP sensing receptor P2X7R, most of the innate immune pathways that have been implicated in various infectious disease models to promote type I IFN production were ruled out as being essential. By process of elimination, this pointed to the STING pathway as an important candidate. STING is an adapter that is activated by cyclic dinucleotides generated by cGAS, which in turn is directly activated by cytosolic DNA [40–42]. This pathway has been implicated in the sensing of DNA viruses, but also in selected autoimmune models [43, 44]. Moreover, activating mutations of STING have been identified in human patients with a vasculitis/pulmonary inflammation syndrome characterized by increased type I IFN production [45]. Indeed, the use of STING−/− mice revealed that spontaneous T cell priming against tumor antigens was markedly reduced in vivo, and rejection of immunogenic tumors was ablated [46]. Moreover, tumor-derived DNA was detected within the cytosol of a major population of tumor-infiltrating DCs, which was associated with STING pathway activation and IFN-β production. Therefore, the host STING pathway appears to be a major innate immune sensing pathway that is activated in the tumor context to drive DC activation and subsequent T cell priming against tumor-associated antigens in vivo. Several additional tumor model systems have confirmed a role for the STING pathway in antitumor immunity in vivo [47–49].
The realization of the importance of this particular innate immune pathway in the cancer context is generating new therapeutic strategies that might be utilized to activate or mimic the cGAS/STING axis for promoting immune-mediated tumor control, particularly in the non-inflamed tumor subset. Recent studies have pursued intratumoral injection of STING agonists. 5,6-Dimethylxanthenone-4-acetic acid (DMXAA) is a flavonoid compound that was previously shown to have anti-tumor activity in mouse models [50]. This drug ultimately failed in humans when combined with chemotherapy in a Phase 3 trial in non-small cell lung cancer [51]. Structure-function studies demonstrated that DMXAA is a direct ligand for mouse STING [52, 53]. However, sequence differences in human STING rendered it unable to bind DMXAA, therefore abrogating its activity in human cells and explaining the lack of clinical activity of this compound. Recent evidence has confirmed that DMXAA is a strong agonist of the mouse STING pathway in vitro and in vivo. Intratumoral injection of DMXAA markedly augmented endogenous priming of tumor antigen-specific CD8+ T cells and caused complete tumor elimination. Rejection was completely dependent on host STING, and most of the effect depended on T cells and type I IFNs. New STING agonists that stimulate all known human STING polymorphic variants have been developed that also bind mouse STING and showed similarly potent efficacy in preclinical tumor models [54]. These agents will be attractive for clinical translation as a potential strategy to initiate de novo inflammation, DC activation, and T cell priming especially in non-T cell-inflamed tumors.
An alternative approach for promoting appropriate innate immune activation in the tumor microenvironment is through targeted radiation. Directed radiation to the tumor site also appears to induce type I IFN production, augment specific T cell priming, and support T cell-mediated tumor control in vivo [55]. Based on the observation that the STING pathway was critical for spontaneous innate immune sensing of tumors in vivo, it was of interest to determine whether the STING pathway was also important for the therapeutic effect of radiation. Indeed, recent data have revealed that the therapeutic efficacy of radiation was largely ablated in STING−/− hosts. This defect was associated with blunted type I IFN induction and markedly reduced T cell priming. In contrast, no defect in the therapeutic effect of radiation was observed using mice lacking specific TLR signaling pathways [56]. Thus, radiation may facilitate the proper acquisition of tumor-derived DNA by host DCs in the tumor microenvironment, thereby leading to improved T cell priming as well as coordination of the effector phase of the anti-tumor immune response.
2.4. Reverse-Translational Research to Identify New Therapeutic Angles for Non-T Cell-Inflamed Tumors
An additional major strategy for identifying molecular mechanisms that control the presence or absence of a T cell-inflamed tumor microenvironment is to interrogate categories of genomic heterogeneity directly from patients. By clustering patients has having a T cell-inflamed versus non-T cell-inflamed tumor microenvironment using gene expression profiling as a defined phenotype, reproducible genetic or genomic patterns can be identified as a correlation. Because the T cell-inflamed tumor microenvironment is also a good predictive biomarker for response to immunotherapies such as anti-PD-1, this question can also be viewed as a pharmacogenomic analysis for mechanisms of primary resistance to these agents. Based on these notions, three potential sources of inter-patient heterogeneity could be envisioned that have the potential to influence whether a tumor in a given subject might contain or lack spontaneous T cell infiltration. These categories are differences in accessory oncogene pathways within the tumor cells based on somatic mutational events, germline polymorphisms in immunoregulatory genes that could set thresholds for immune cell activation, or environmental differences that could have global effects on immune functionality. Regarding the latter category, the major phenomenon that has recently garnered interest is the impact of the intestinal microbiome on systemic immune responses in the host. Importantly, each of these parameters is measurable in individual patients. Somatic heterogeneity in tumors can be assessed through exome sequencing and pathway analysis, germline heterogeneity in the host can be evaluated using SNP arrays on peripheral blood cells, and patterns of differences in the intestinal microbiome can be identified through 16S ribosomal RNA sequencing on stool samples. Associations between individual sequences and either the T cell-inflamed tumor microenvironment or clinical outcome to immunotherapy can then be investigated. These analyses require prospective tissue collection from patients embarking on immunotherapy treatments—fresh tumor biopsies, peripheral blood specimens, and stool samples. Broad-based tissue banking from cancer patients participating in immunotherapy clinical trials should be supported and represents a rich discovery opportunity to identify mechanisms of immunotherapy success versus resistance.
Such an analysis has been initiated in metastatic melanoma patients, focusing first on somatic differences at the level of the tumor itself. Using a series of 266 melanoma metastases, tumors were categorized based on the presence or absence of the gene signature indicative of the T cell-inflamed tumor microenvironment [3]. These same tumors were also subjected to exome sequencing, as well as pathway analysis using the Ingenuity platform based on gene expression patterns in the non-T cell-inflamed subset. Strikingly, these data indicated that nearly one-half of the non-T cell-inflamed tumors showed evidence of activation of the Wnt/β-catenin pathway. Some tumors had activating mutations in β-catenin itself, some had inactivating mutations in negative regulators of β-catenin, and some showed over-expression of Wnt7B or Frizzled 3. Using a genetically engineered mouse model in which melanomas were induced that either did or did not include conditionally expressed active β-catenin, mechanistic experiments confirmed that tumor cell-intrinsic β-catenin activation was sufficient to exclude T cell infiltrates in vivo. The molecular mechanism was narrowed down to a loss of chemokines that mediate recruitment of Batf3-lineage DCs into the tumor microenvironment, leading to defective T cell priming. The therapeutic activity of anti-CTLA-4 + anti-PD-L1 mAb normally seen was lost when tumors additionally expressed active β-catenin [57]. Thus, the Wnt/β-catenin pathway is the first identified tumor-intrinsic oncogene pathway to result in immune exclusion and resistance to immunotherapy. These data suggest that pharmacologic strategies to block β-catenin activity might not only be directly therapeutic against tumor cells, but additionally might support a positive interaction with host immunity.
Ongoing work also investigates germline polymorphisms as they relate to the presence or absence of the T cell-inflamed tumor microenvironment. There is precedent for germline genetic differences influencing response to immunotherapy. A SNP in the gene encoding the chemokine receptor CCR5 was identified that was associated with clinical response to high-dose IL-2 [58]. More recently, a polymorphism in the IRF5 gene was identified that was associated with clinical benefit in a cohort of patients treated with T cell-adoptive transfer [59]. Numerous polymorphisms have been identified in immune regulatory genes that are associated with various types of autoimmunity, including lupus [60], and many patients who are treated with immune-potentiating drugs do develop autoimmune-like adverse events. Thus, it is attractive to consider that specific germline SNPs might be associated either with clinical response or with side effects upon treatment with anti-CTLA-4 or anti-PD-1 mAbs.
The third category of biomarkers is the composition of the intestinal microbiota. The group of Trinchieri and colleagues has found in a mouse model that treatment with anti-bacterial antibiotics, which altered intestinal microbial composition, reduced the therapeutic efficacy of immunotherapy with the TLR9 agonist CpG combined with anti-IL-10R antibody in a transplantable tumor model [61]. In addition, Zitvogel and colleagues reported that the immune-potentiating effect of cyclophosphamide is associated with translocation of commensal bacteria [62]. These early data have prompted a comprehensive analysis of the intestinal microbiota using 16S rRNA sequencing from patients undergoing treatment with immunotherapeutics. Restoring the presence of specific commensal that maximize anti-tumor immunity, such as through the use of a probiotic, represents an additional future immunotherapeutic strategy.
2.5. Conclusions and Future Directions
The paradigm of the T cell-inflamed and non-T cell inflamed tumor microenvironment has provided a useful working model for identifying therapeutic targets for immunotherapy, understanding mechanisms of response versus resistance, and pursuing new strategies for overcoming resistance to expand the range of immunotherapy efficacy. Inasmuch as many of these concepts have been explored predominantly in melanoma, there is a rich opportunity to investigate these principles similarly in other tumor types. A summary of candidate interventions to improve immunotherapy efficacy to include the non-T cell-inflamed tumor microenvironment based on these principles is illustrated in Fig. 2.3. One could envision intratumoral administration of innate immune activators such as STING agonists, to trigger de novo DC activation and T cell priming and recruitment. Tumor-focused radiation also may have these effects. If specific oncogene pathways are activated such as β-catenin, then targeted inhibitors could be administered to block such pathways and restore immune cell entry. If unfavorable germline genetics are identified, specific gene products might be amenable to pharmacologic manipulation as well. Finally, if commensal bacteria are identified that might amplify host anti-tumor immunity, then probiotics could be developed to improve T cell infiltration and clinical efficacy of immunotherapies such as anti-PD-1. Ultimately, combination therapies will likely provide the broadest and deepest clinical benefit.
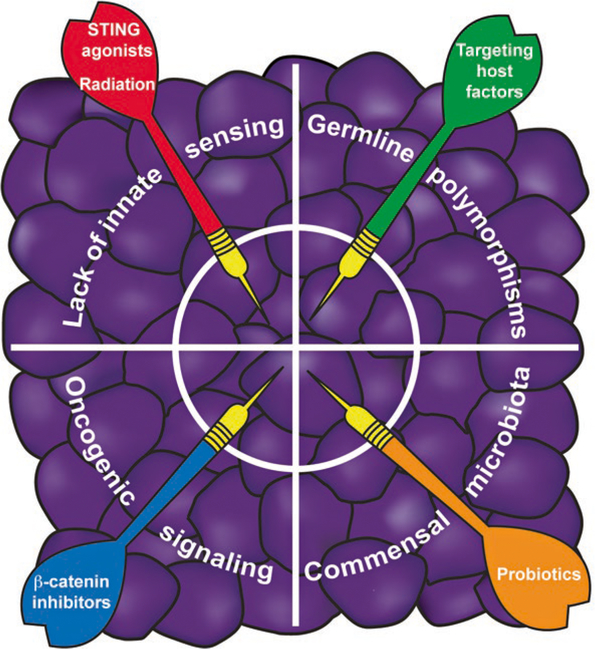
Fig. 2.3.
Summary of four types of strategies that could be considered to overcome the barrier of the non-T cell-inflamed tumor microenvironment. It is envisioned that intratumoral administration of innate immune activators or local tumor radiation, modulators of host polymorphic gene products, blockade of immune-exclusionary oncogene pathways, and delivery of probiotics that amplify anti-tumor immunity all could be considered for ultimate clinical translation
References
- 1.Peterson AC, Harlin H, Gajewski TF. Immunization with melan-A peptide-pulsed peripheral blood mono-nuclear cells plus recombinant human interleukin-12 induces clinical activity and T-cell responses in advanced melanoma. J Clin Oncol. 2003;21:2342–8. [DOI] [PubMed] [Google Scholar]
- 2.Gajewski TF, Meng Y, Blank C, Brown I, Kacha A, Kline J, Harlin H. Immune resistance orchestrated by the tumor microenvironment. Immunol Rev. 2006;213:131–45. [DOI] [PubMed] [Google Scholar]
- 3.Harlin H, Meng Y, Peterson AC, Zha Y, Tretiakova M, Slingluff C, McKee M, Gajewski TF. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69:3077–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gajewski TF, Louahed J, Brichard VG. Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer J. 2010;16:399–403. [DOI] [PubMed] [Google Scholar]
- 5.Ulloa-Montoya F, Louahed J, Dizier B, Gruselle O, Spiessens B, Lehmann FF, Suciu S, Kruit WH, Eggermont AM, Vansteenkiste J, Brichard VG. Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clin Oncol. 2013;31:2388–95. [DOI] [PubMed] [Google Scholar]
- 6.Mikucki ME, Fisher DT, Matsuzaki J, Skitzki JJ, Gaulin NB, Muhitch JB, Ku AW, Frelinger JG, Odunsi K, Gajewski TF, Luster AD, Evans SS. Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat Commun. 2015;6:7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pages F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, Meatchi T, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Galon J. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–66. [DOI] [PubMed] [Google Scholar]
- 8.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–4. [DOI] [PubMed] [Google Scholar]
- 9.Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, Bruneval P, Trajanoski Z, Fridman WH, Pages F, Galon J. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol. 2011;29:610–8. [DOI] [PubMed] [Google Scholar]
- 10.Harlin H, Kuna TV, Peterson AC, Meng Y, Gajewski TF. Tumor progression despite massive influx of activated CD8(+) T cells in a patient with malignant melanoma ascites. Cancer Immunol Immunother. 2006;55:1185–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mortarini R, Piris A, Maurichi A, Molla A, Bersani I, Bono A, Bartoli C, Santinami M, Lombardo C, Ravagnani F, Cascinelli N, Parmiani G, Anichini A. Lack of terminally differentiated tumor-specific CD8+ T cells at tumor site in spite of antitumor immunity to self-antigens in human metastatic melanoma. Cancer Res. 2003;63:2535–45. [PubMed] [Google Scholar]
- 12.Zippelius A, Batard P, Rubio-Godoy V, Bioley G, Lienard D, Lejeune F, Rimoldi D, Guillaume P, Meidenbauer N, Mackensen A, Rufer N, Lubenow N, Speiser D, Cerottini JC, Romero P, Pittet MJ. Effector function of human tumor-specific CD8 T cells in melanoma lesions: a state of local functional tolerance. Cancer Res. 2004;64:2865–73. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg SA, Dudley ME. Cancer regression in patients with metastatic melanoma after the transfer of autologous antitumor lymphocytes. Proc Natl Acad Sci U S A. 2004;101(Suppl 2):14639–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufman HL, Kim DW, DeRaffele G, Mitcham J, Coffin RS, Kim-Schulze S. Local and distant immunity induced by intralesional vaccination with an oncolytic herpes virus encoding GM-CSF in patients with stage IIIc and IV melanoma. Ann Surg Oncol. 2010;17:718–30. [DOI] [PubMed] [Google Scholar]
- 15.Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, Gopinathan A, Tuveson DA, Fearon DT. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330:827–30. [DOI] [PubMed] [Google Scholar]
- 16.Salmon H, Franciszkiewicz K, Damotte D, Dieu-Nosjean MC, Validire P, Trautmann A, Mami-Chouaib F, Donnadieu E. Matrix architecture defines the preferential localization and migration of T cells into the stroma of human lung tumors. J Clin Invest. 2012;122:899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demoulin S, Herfs M, Delvenne P, Hubert P. Tumor microenvironment converts plasmacytoid dendritic cells into immunosuppressive/tolerogenic cells: insight into the molecular mechanisms. J Leukoc Biol. 2013;93:343–52. [DOI] [PubMed] [Google Scholar]
- 18.Watkins SK, Zhu Z, Riboldi E, Shafer-Weaver KA, Stagliano KE, Sklavos MM, Ambs S, Yagita H, Hurwitz AA. FOXO3 programs tumor-associated DCs to become tolerogenic in human and murine prostate cancer. J Clin Invest. 2011;121:1361–72. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Buckanovich RJ, Facciabene A, Kim S, Benencia F, Sasaroli D, Balint K, Katsaros D, O’Brien-Jenkins A, Gimotty PA, Coukos G. Endothelin B receptor mediates the endothelial barrier to T cell homing to tumors and disables immune therapy. Nat Med. 2008;14:28–36. [DOI] [PubMed] [Google Scholar]
- 20.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gajewski TF, Zha Y-y, Hernandez K, Li Y, Bao R, Alexieff P, Andrade J, Luke JJ, Spranger S. Density of immunogenic antigens and presence or absence of the T cell-inflamed tumor microenvironment in metastatic melanomal. J Clin Oncol. 2015;33(suppl; abstr):3002. [Google Scholar]
- 22.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–3. [DOI] [PubMed] [Google Scholar]
- 23.Gajewski TF. Failure at the effector phase: immune barriers at the level of the melanoma tumor microenvironment. Clin Cancer Res. 2007;13:5256–61. [DOI] [PubMed] [Google Scholar]
- 24.Spranger S, Spaapen R, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Upregulation of PD-L1, IDO and Tregs in the melanoma tumor microenvironment is driven by CD8+ T cells. Sci Transl Med. 2013;5(200):200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, Chen L. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4:127ra137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spranger S, Koblish HK, Horton B, Scherle PA, Newton R, Gajewski TF. Mechanism of tumor rejection with doublets of CTLA-4, PD-1/PD-L1, or IDO blockade involves restored IL-2 production and proliferation of CD8(+) T cells directly within the tumor microenvironment. J Immunother Cancer. 2014;2:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, Zha Y, Driessens G, Locke F, Gajewski TF. Transcriptional regulator early growth response gene 2 (Egr2) is required for T cell anergy in vitro and in vivo. J Exp Med. 2012;209:2157–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y, Zha Y, Gajewski TF. Molecular regulation of T-cell anergy. EMBO Rep. 2008;9:50–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y, Zha Y, Spaapen RM, Mathew R, Barr K, Bendelac A, Gajewski TF. Egr2-dependent gene expression profiling and ChIP-Seq reveal novel biologic targets in T cell anergy. Mol Immunol. 2013;55:283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Triebel F, Jitsukawa S, Baixeras E, Roman-Roman S, Genevee C, Viegas-Pequignot E, Hercend T. LAG-3, a novel lymphocyte activation gene closely related to CD4. J Exp Med. 1990;171:1393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuford WW, Klussman K, Tritchler DD, Loo DT, Chalupny J, Siadak AW, Brown TJ, Emswiler J, Raecho H, Larsen CP, Pearson TC, Ledbetter JA, Aruffo A, Mittler RS. 4–1BB costimulatory signals preferentially induce CD8+ T cell proliferation and lead to the amplification in vivo of cytotoxic T cell responses. J Exp Med. 1997;186:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, Shaheen M, Ernstoff MS, Minor D, Salama AK, Taylor M, Ott PA, Rollin LM, Horak C, Gagnier P, Wolchok JD, Hodi FS. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N Engl J Med. 2015;372:2006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–95. [DOI] [PubMed] [Google Scholar]
- 36.Fuertes MB, Kacha AK, Kline J, Woo SR, Kranz DM, Murphy KM, Gajewski TF. Host type I IFN signals are required for antitumor CD8+ T cell responses through CD8{alpha}+ dendritic cells. J Exp Med. 2011;208(10):2005–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, Murphy KM. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diamond MS, Kinder M, Matsushita H, Mashayekhi M, Dunn GP, Archambault JM, Lee H, Arthur CD, White JM, Kalinke U, Murphy KM, Schreiber RD. Type I interferon is selectively required by dendritic cells for immune rejection of tumors. J Exp Med. 2011;208:1989–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuertes MB, Woo SR, Burnett B, Fu YX, Gajewski TF. Type I interferon response and innate immune sensing of cancer. Trends Immunol. 2013;34:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X, Shi H, Wu J, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol Cell. 2013;51:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahn J, Gutman D, Saijo S, Barber GN. STING manifests self DNA-dependent inflammatory disease. Proc Natl Acad Sci U S A. 2012;109:19386–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn J, Ruiz P, Barber GN. Intrinsic self-DNA triggers inflammatory disease dependent on STING. J Immunol. 2014;193:4634–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, Tenbrock K, Wittkowski H, Jones OY, Kuehn HS, Lee CC, DiMattia MA, Cowen EW, Gonzalez B, Palmer I, DiGiovanna JJ, Biancotto A, Kim H, Tsai WL, Trier AM, Huang Y, Stone DL, Hill S, Kim HJ, St Hilaire C, Gurprasad S, Plass N, Chapelle D, Horkayne-Szakaly I, Foell D, Barysenka A, Candotti F, Holland SM, Hughes JD, Mehmet H, Issekutz AC, Raffeld M, McElwee J, Fontana JR, Minniti CP, Moir S, Kastner DL, Gadina M, Steven AC, Wingfield PT, Brooks SR, Rosenzweig SD, Fleisher TA, Deng Z, Boehm M, Paller AS, Goldbach-Mansky R. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo S-R, Fuertes MB, Corrales L, Spranger S, Furdyna MJ, Leung MYK, Duggan R, Wang Y, Barber GN, Fitzgerald KA, Alegre M-L, Gajewski TF. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahn J, Xia T, Konno H, Konno K, Ruiz P, Barber GN. Inflammation-driven carcinogenesis is mediated through STING. Nat Commun. 2014;5:5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohkuri T, Ghosh A, Kosaka A, Zhu J, Ikeura M, David M, Watkins SC, Sarkar SN, Okada H. STING contributes to antiglioma immunity via triggering type I IFN signals in the tumor microenvironment. Cancer Immunol Res. 2014;2(12):1199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu Q, Man SM, Gurung P, Liu Z, Vogel P, Lamkanfi M, Kanneganti TD. Cutting edge: STING mediates protection against colorectal tumorigenesis by governing the magnitude of intestinal inflammation. J Immunol. 2014;193:4779–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baguley BC, Ching LM. Immunomodulatory actions of xanthenone anticancer agents. BioDrugs. 1997;8:119–27. [DOI] [PubMed] [Google Scholar]
- 51.Lara PN Jr, Douillard JY, Nakagawa K, von Pawel J, McKeage MJ, Albert I, Losonczy G, Reck M, Heo DS, Fan X, Fandi A, Scagliotti G. Randomized phase III placebo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent vadimezan (ASA404) in advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:2965–71. [DOI] [PubMed] [Google Scholar]
- 52.Conlon J, Burdette DL, Sharma S, Bhat N, Thompson M, Jiang Z, Rathinam VA, Monks B, Jin T, Xiao TS, Vogel SN, Vance RE, Fitzgerald KA. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J Immunol. 2013;190:5216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao P, Ascano M, Zillinger T, Wang W, Dai P, Serganov AA, Gaffney BL, Shuman S, Jones RA, Deng L, Hartmann G, Barchet W, Tuschl T, Patel DJ. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell. 2013;154:748–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corrales L, Glickman LH, McWhirter SM, Kanne DB, Sivick KE, Katibah GE, Woo SR, Lemmens E, Banda T, Leong JJ, Metchette K, Dubensky TW Jr, Gajewski TF. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 2015;11:1018–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burnette BC, Liang H, Lee Y, Chlewicki L, Khodarev NN, Weichselbaum RR, Fu YX, Auh SL. The efficacy of radiotherapy relies upon induction of type i interferon-dependent innate and adaptive immunity. Cancer Res. 2011;71:2488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng L, Liang H, Xu M, Yang X, Burnette B, Arina A, Li X-D, Mauceri H, Beckett M, Darga T, Huang X, Gajewski TF, Chen ZJ, Fu Y-X, Weichselbaum RR. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 2014;41:843–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–5. [DOI] [PubMed] [Google Scholar]
- 58.Ugurel S, Schrama D, Keller G, Schadendorf D, Brocker EB, Houben R, Zapatka M, Fink W, Kaufman HL, Becker JC. Impact of the CCR5 gene polymorphism on the survival of metastatic melanoma patients receiving immunotherapy. Cancer Immunol Immunother. 2008;57:685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uccellini L, De Giorgi V, Zhao Y, Tumaini B, Erdenebileg N, Dudley ME, Tomei S, Bedognetti D, Ascierto ML, Liu Q, Simon R, Kottyan L, Kaufman KM, Harley JB, Wang E, Rosenberg SA, Marincola FM. IRF5 gene polymorphisms in melanoma. J Transl Med. 2012;10:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kariuki SN, Franek BS, Kumar AA, Arrington J, Mikolaitis RA, Utset TO, Jolly M, Crow MK, Skol AD, Niewold TB. Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res Ther. 2010;12:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, Dai RM, Kiu H, Cardone M, Naik S, Patri AK, Wang E, Marincola FM, Frank KM, Belkaid Y, Trinchieri G, Goldszmid RS. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillere R, Hannani D, Enot DP, Pfirschke C, Engblom C, Pittet MJ, Schlitzer A, Ginhoux F, Apetoh L, Chachaty E, Woerther PL, Eberl G, Berard M, Ecobichon C, Clermont D, Bizet C, Gaboriau-Routhiau V, Cerf-Bensussan N, Opolon P, Yessaad N, Vivier E, Ryffel B, Elson CO, Dore J, Kroemer G, Lepage P, Boneca IG, Ghiringhelli F, Zitvogel L. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342:971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]