Abstract
Ionizing radiation (IR) is a ubiquitous component of our environment and an important tool in research and medical treatment. At the same time, IR is a potent genotoxic and epigenotoxic stressor, exposure to which may lead to negative health outcomes. While the genotoxocity is well described and characterized, the epigenetic effects of exposure to IR and their mechanisms remain under-investigated. In this conceptual review, we propose the IR-induced changes to one-carbon metabolism as prerequisites to alterations in the cellular epigenome. We also provide evidence from both experimental and clinical studies describing the interactions between IR and one-carbon metabolism. We further discuss the potential for the manipulation of the one-carbon metabolism in clinical applications for the purpose of normal tissue protection and for increasing the radiosensitivity of cancerous cells.
Keywords: DNA methylation, epigenetics, ionizing radiation, methionine, methyl donors
Introduction: ionizing radiation and epigenetics
Ionizing radiation (IR) is a ubiquitous environmental stressor and a widely-used tool in many spheres of human life. One of the largest sources of IR exposure comes from medical radiation when utilized as a diagnostic and treatment modality. Approximately 50% of all cancer patients receive radiotherapy and over 70 million computed tomography (CT) scans are performed annually in the US alone (1, 2), creating an ever-growing number of patients who are routinely exposed.
While it is generally accepted that the benefits of medical radiation outweigh the risks, there is considerable concern about unintended side-effects, as exposure to IR may result in a number of negative outcomes, including the development of cancer and degenerative diseases (3–5). Radiation-induced genomic instability and carcinogenesis are stochastic effects, where there appears to be no threshold dose and the risk of these effects increases with increasing dose. In addition, exposure to IR can have deterministic effects, short-term and long-term injury in normal (non-tumor) tissues for which, there appears to be a threshold dose below which, these effects do not occur (6). Normal tissue radiation injury can vary from acute radiation syndrome that is seen after large parts of the body have been exposed to relatively high doses of IR, typically within a few minutes (7, 8), to early and late injury and adverse remodeling in tissues that are exposed to IR during radiotherapy. Side effects of radiotherapy include skin erythema after breast cancer treatment (9), radiation enteropathy due to exposure of the intestinal tract in abdominal radiotherapy (10) and fibrosis in the lung and heart that may develop several years after thoracic radiotherapy (11, 12).
It is now generally accepted that both genotoxic and epigenotoxic properties of IR underline the mechanisms of those effects. At the same time, while the ability of IR to damage DNA (genotoxicity) is a well-known and well-characterized phenomenon, epigenetic effects (or those that are not related to alterations in DNA sequence) of exposure were discovered relatively recently and are not well understood.
Epigenetics is the study of heritable changes in gene expression that are not associated with alterations in the primary DNA sequence. The epigenetic mechanisms of regulation include methylation of DNA, post-translational histone modifications, nucleosome positioning along DNA and non-coding RNAs. These mechanisms are vital for normal development and maintenance of cellular homeostasis. Specifically, DNA and histone methylation regulate the expression of genetic information in a cell, tissue and sex-specific manner (13, 14). They also play critical roles in controlling the expression of repetitive elements (RE) – transposable elements and satellite DNA – that together comprise over half of the mammalian genomes (15).
Alterations in DNA and/or histone methylation may substantially affect the cellular epigenome, leading to altered gene and RE expression and resulting in genomic instability and the development of pathological states, including cancer. In fact, loss of global DNA methylation was the first epigenetic alteration reported in virtually all human cancers (16, 17). Later, DNA hypermethylation at the promoter regions of tumor-suppressor genes was also reported in various cancers (18–20). Further studies have demonstrated that epigenetic alterations, primarily changes in DNA and histone methylation, are not simply the consequences of cancer but may often serve as the drivers of carcinogenesis and can be detected in early stages of cancer development (21–25).
Detection of epigenetic alterations in tumors associated with occupational exposure to radiation suggested that epigenetics may also contribute to IR-induced carcinogenesis. For instance, hypermethylation of p16INK4a (26) and GATA5 (27) genes was observed in lung adenocarcinomas of occupationally exposed workers in comparison with adenocarcinomas from the cohort of non-exposed patients. Further studies using experimental rodent models have convincingly demonstrated that IR affects DNA and histone methylation in the target organs, such as bone marrow, thymus and spleen (28). The majority of the existing literature in the field indicates that exposures to doses of IR 1 Gy and above, are usually characterized by the loss of global DNA methylation in these organs (29–32). Subsequent studies demonstrated that the observed changes in DNA methylation stem primarily from RE, while the gene-specific alterations seem to be less obvious (33–36).
Less is known about the effects of IR on histone methylation, however, these changes are primarily characterized by the loss of histone methylation from hours to several days after irradiation, similar to the effects on DNA methylation. For instance, histone marks that are responsible for the formation of transcriptionally silent heterochromatin structure – histone H3 lysine 9 (H3K9me3) and histone H4 lysine 20 (H4K20me3) trimethylation – are negatively regulated after exposure to both low and high-dose IR (30, 37). This relaxed chromatin structure may allow for easier access of repair complexes to the sites of DNA damage. At the same time, histone methylation is generally more labile than methylation of DNA and often changes observed shortly after irradiation are not detectable at later time-points (37).
Radiation-induced alterations in DNA and histone methylation
Despite significant progress in radiation epigenetics in the past decade, the mechanisms of radiation-induced changes in DNA and histone methylation remain largely unknown. A number of hypotheses have been proposed, suggesting different mechanisms including the affected function of DNA and histone methyltransferases, interference of DNA damage with the ability of DNA methyltransferases to methylate DNA, DNA damage and repair and radiation-induced proliferation, to name a few (reviewed in ref. 28).
DNA and histone methyltransferases are the key enzymes needed for methylation of the two abovementioned substrates. While there are a limited number of DNA methyltransferases (DNMT) which are primarily represented by the predominantly maintenance DNA methyl-transferase DNMT1 and de novo methyltransferases Dnmt3a and Dnmt3b, histone methylation at different residues are facilitated by specific histone methyltransferases.
IR has been shown to affect mRNA and protein levels of DNA methyltransferases, as well as their enzymatic activity. In particular, the levels of the de novo DNA methyltransferases Dnmt3a and Dnmt3b were found to be decreased 3 months after total body irradiation to low absorbed mean doses of either heavy irons (56Fe) or protons in the mouse model (38). Similarly, exposure to low-dose X-rays resulted in simultaneous decreased protein levels of Dnmt1, Dnmt3a and Dnmt3b in the murine thymus (30). Interestingly, in the same study, the authors have also demonstrated the IR-induced loss in histone H4 lysine 20 trimethylation (H4K20me3), however, the status of methylases and demethylases specific to this histone mark was not assessed.
In cell lines, nuclear DNA methyltransferase activity was found to be decreased up to 3 days after exposure to 10 Gy of γ-rays (39). At the same time, there was a parallel increase in DNA methyltransferase cytoplasmic activity. Sequestration of Dnmt1 in the cytoplasm in its active form may substantially contribute to IR-induced DNA hypomethylation (40).
In addition to affecting methyltransferases, accumulating evidence suggests that IR also affects the availability of methyl donors (38, 41). Both DNA and histone methylation require the donation of a CH3 group from the universal methyl donor S-adenosylmethionine (SAM). Addition of those methyl groups modifies how proteins interact with a section of DNA or chromatin and in turn, influence the level of expression of that region. Any impact on the availability of methyl donors and in the enzymes responsible for their metabolism will consequently affect the level of DNA and histone methylation. Those methyl groups used for DNA and histone methylation originate from one-carbon metabolism.
One-carbon metabolism
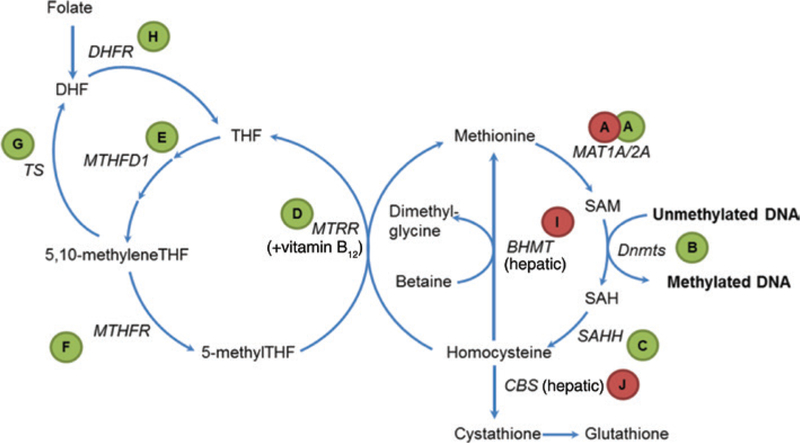
The reactions surrounding the transfer of the methyl group from SAM to the acceptor molecules and the regeneration of SAM are the key components of folate-dependent one-carbon metabolism (Figure 1). The latter ties together gene regulation, amino acid synthesis, purine and pyrimidine synthesis, four vitamins and antioxidants, to name a few and over a hundred of biomethylation reactions (42). The ramifications of changes in that pathway affects nearly all cellular functions.
Figure 1:
Overview of one-carbon metabolism.
The direct precursor of SAM is the essential amino acid methionine. The adenylation reaction is catalyzed by the enzyme methionine adenosyltransferase (MAT, Figure 1A). There are two forms of this enzyme: the hepatic form MAT1A and the ubiquitously expressed extrahepatic form MAT2A. SAM is utilized as a substrate for dozens of methyltransferases, including DNA methyltransferases (Figure 1B), yielding S-adenosylhomocysteine (SAH). In turn, SAH exerts substrate inhibition on methyltransferases because they have higher affinity to SAH compared to SAM (43, 44). SAH is further converted to homocysteine through the action of the enzyme S-adenosylhomocysteine hydrolase (SAHH, Figure 1C). Approximately, half of homocysteine can then be remethylated back to methionine using the 5-methyltetrahydrofolate through the enzyme methionine synthase (MTRR, Figure 1D) and another half converted to cystathione by cystathione beta synthase (CBS, Figure 1J) (45). Methionine synthase is ubiquitously expressed and necessitates the contribution of vitamin B12, also known as cobalamin, in the form of methylcobalamin as a cofactor. The origin of the methyl group of SAM derives from 5-methylfolate which links methionine and SAM metabolism to folate-dependent one-carbon metabolism.
Homocysteine is an amino acid that is not used in protein synthesis (non-protein-forming). When elevated in blood, homocysteine level is an independent risk factor for neurological disorders, bone tissue damage and cardiovascular disease (46–48). While administration of B vitamins (see Figure 1) successfully lowers plasma homocysteine levels, it is unclear if this leads to any health benefits. It also remains unclear whether homocysteine itself is the cause of disease, or if it merely reflects other underlying health problems (49).
Through 5-methyltetrahydrofolate, the folic acid cycle provides another source of methyl groups for DNA methylation. Folic acid in the diet is reduced to dihydrofolate and tetrahydrofolate (THF) by the enzyme dihydrofolate reductase. THF is metabolized by the enzyme serine hydroxymethyltransferase (SHMT) to 5,10 methyleneTHF which is then metabolized to 5-methylTHF by methyltetrahydrofolate reductase, MTHFR (Figure 1F). MTHFR catalyzes the rate-limiting reaction to 5-methylTHF in the methyl cycle.
Alterations in one carbon metabolism and human disease
Two polymorphisms have been identified in human populations that influence MTHFR enzymatic activity. One is A1298C (Glu429Ala) and the other one is C677T (Ala-222Val). This second is by far the more studied of the two. Compared to the ancestral C allele, the homozygous form of the T allele has 30% residual activity (50). It has been associated not only with variation in the blood levels of homocysteine (51), but also with elevated risks of vascular disease (52) and different types of cancers (53). However, publication bias is suspected and the effect may be significantly less than previously thought (54). A few more enzymes such as thymidylate synthase (TS, Figure 1G) and dihydrofolate reductase (DHFR, Figure 1H) serve additional roles in folate-dependent one-carbon metabolism.
In addition to common methionine-folate cycles in somatic cells, the liver expresses two enzymes that provide alternative pathways for the degradation of homocysteine. These are betaine homocysteine methyltransferase (BHMT, Figure 1I) and CBS (Figure 1J). BHMT utilizes trimethylglycine – also known as betaine and derived from the dietary precursor choline – to remethylate homocysteine to methionine. Conversely, CBS utilizes a transulfuration reaction to produce cystathionine, which is later converted to cysteine and the antioxidant glutathione. A genetic defect in this enzyme is the cause for a rare inborn disorder called cystathionine β synthase deficiency that leads to abnormally high levels of homocysteine. Those levels are 5 to 10 times higher than those seen in patients with hyperhomocysteinemia associated with vascular disease.
Effect of radiation on one-carbon metabolism
Folate is susceptible to oxidative degradation and this process is exacerbated by exposure to IR (55). Irradiation with γ rays led to reduced total folate levels by about half in the mouse liver 4 days after exposure (56). Folate was also found to be decreased in plasma (5 to 120 h) and in bone marrow (24 h) after total body irradiation with 3 Gy X-ray (57). As a consequence, enzymes in the folic acid cycle were also affected after exposure to IR. There was a dose-dependent increase in the gene expression for the enzymes TS, DHFR and methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1 (MTHFD1) that peaked 2 h after lymphoblastoid cell lines were exposed to 10 Gy of γ-rays (58). This is supported by the observation of an increase in enzymatic activity for TS and DHFR up to 96 h after total body irradiation with 2–7 Gy of γ-rays (56). Interestingly, the effect was opposite for the rate-limiting MTHFR, where there was a reduction in activity that was sustained over a week after exposure.
In addition to these observations in folate, cobalamin levels were also reported to be affected by IR. At the end of 5 weeks of radiotherapy to the pelvic area, patients showed significantly reduced mean serum levels of cobalamin (59). The levels continued to decrease up to 6 weeks after the end of the treatment period and remained significantly decreased a year later. Similar findings were identified after radiotherapy for cervical and bladder cancer (60–62). In the case of cobalamin, reduced absorption due to radiation damage to the intestinal lumen is the most likely reason for the changes in serum levels (61, 62).
Exposure to IR also seems to affect the distribution of methyl groups from methionine (63). The level of tracer for the methyl carbon was decreased in heart, brain, kidney and muscle and increased in the gastrointestinal tract, skin, thymus and thyroid. Concentrations of methionine, SAM, SAH and the ratio of SAM and SAH were also shown to be affected by exposure to protons and 56Fe radiation in the mouse heart (38). It has been proposed that alterations in one-carbon metabolism may serve as driving mechanisms of epigenetic alterations, DNA and histone methylation, in particular (28). Indeed, decreases in methionine or synthesis of SAM would substantially affect the availability of universal methyl donor (SAM), needed for the maintenance of DNA and histone methylation. To our knowledge, there is only one study that simultaneously addressed, the one-carbon metabolism and methylation of DNA in response to IR. Interestingly, whole-body exposure to low absorbed mean doses of protons and 56Fe resulted in increased cardiac tissue levels of methionine and SAM and was paralleled by the increase in DNA methylation of RE, suggesting an overall increase in global DNA methylation in the heart (38). It must be mentioned that both protons and 56Fe are sources of high linear energy transfer (LET) radiation, biological effects of exposure (including epigenetic effects), which are often very different from the exposure to terrestrial low-LET radiation (31, 64, 65). Some studies investigated the potential of methionine supplementation on DNA methylation in response to exposure to IR; these studies are discussed further in this review.
Methyl donor deficiency
Given the effects exerted by IR on one-carbon metabolism, it is not surprising that the response to IR can be modified by altering the supply of methyl group donors. Manipulations of folate, choline, cobalamin and methionine intake have been demonstrated to have impacts on birth defects, cognition and risk of cancer.
In in vitro models, folate deficiency was shown to sensitize cells to radiation-induced chromosomal instability (66, 67). The incidence of micronuclei formed, as a result of exposure to IR was inversely proportional to the concentration of folic acid in the culture medium. Homozygosity for the MTHFR C677T polymorphism further amplified the damaging effect of low folic acid concentration (67). In vivo, folate deficiency induced a change in the pattern of histone methylation (68). There was a suppression of H3K27 histone methyltransferase activity (a suppressive mark) with a parallel increase in H3K4 histone methyltransferase activity (an activating mark). IR also induced a decrease in the liver folate levels and depletion in the universal methyl donor SAM.
As suggested previously, IR leads to a decrease in liver folate levels (56). Combined with a folate-deficient diet, levels can be driven down even further (68). However, folate deficiency combined with IR also decreased liver and kidney choline levels (69). In parallel, exposure to IR led to an increase of choline into the circulation that peaked around 48 h and slowly decreased afterwards. Interestingly, the levels of choline in the brain were also elevated after the combination of folate-deficient diet and IR (69). A similar phenomenon was observed with a choline-free diet in combination with IR. The levels of choline in the liver were decreased while they were increased in the serum and brain (70). The combined insult is presumed to divert the choline reserves away from the liver, their storage point and into the serum to be directed towards other utilizing organs (69).
Methyl donor supplementation
Given the amplifying effect of a folate or choline-deficient diet on the effects exerted by exposure to IR, several groups have explored the protective potential of methyl donor supplements. Indeed, betaine showed radioprotective properties on several fronts. Supplementation with betaine reduced the number of chromosomal aberrations in human lymphocytes exposed to either γ-rays or carbon ions (71). It also showed protection for the intestinal lumen by increasing crypt survival after irradiation with γ-rays, although this effect was not seen after irradiation with carbon ions (72). Overall, treatment with betaine supplementation also increased survival in animal models (71, 72).
Similar beneficial effects were observed using a supplementation mix of folic acid, choline and cobalamin. This treatment was able to preserve liver and serum folate levels after exposures up to 6 Gy (73). Homocysteine levels were also near baseline levels in the serum. In the liver, an IR-induced increase in homocysteine (2–2.5-fold) was observed at a 48 h time-point even on the supplemented diet, however this increase was ~4-fold on the normal diet. The supplemented diet also rescued DNA methylation and DNMT levels at this time-point.
The D isomer of methionine also shows promises at mitigating radiation effects. Survival of animals after irradiation was improved and there was an improvement in the mouth and tongue mucosal epithelial cells seen after D-methionine treatment (74, 75). To our knowledge, no studies have evaluated the radioprotective effects of L-methionine, which is the form used by cells. Although L-methionine is a methyl donor, it also displays toxicity at high levels. When given in excess, it interferes with ureanitrogen balance (76). High dietary methionine intake also leads to increased levels of serum homocysteine (77) and may result in the formation of highly toxic methanethiol-cysteine disulfides (78).
Methionine restriction
While long-term complete methionine deficiency leads to liver steatosis (79), methionine restriction has been associated with a number of health benefits in animal models. In a way that is postulated to be similar to caloric restriction, a reduction of 40%–80% of the levels of methionine in the diet leads to an increase in longevity in both rats and mice (80, 81).
Methionine restriction improves barrier function after exposure to IR, including in the gut (82–84). As irradiation results in reduced intestinal barrier integrity (85), methionine restriction may provide certain protection against gastrointestinal toxicity as a result of radiotherapy or accidental exposure. It is possible that the positive effect of the D isomer of methionine after IR may actually be due to a competition with the protein-building form of the amino acid.
Cancerous cells are known for high sensitivity to methionine restriction. While normal cells tolerate methionine deficiency, the vast majority of cancer cells are methionine auxotrophs, meaning they exhibit irreplaceable nutritional requirements for methionine (86). A number of studies convincingly demonstrated that methionine deprivation holds substantial potential to increase the effects of chemotherapeutic agents both in vitro and in experimental models (86–90). Several clinical studies have also investigated the potential of the methionine-deprived diet in cancer therapy (91, 92). To our knowledge, however, no studies have addressed the effects of methionine deprivation combined with radiotherapy.
Expert opinion
It is increasingly recognized that exposure to IR affects the one-carbon metabolism. Alterations to methionine, one of the central molecules in the one-carbon metabolism and major substrate for methyl donors, may further impact normal DNA and histone methylation. Some experimental evidence supports this hypothesis, however further studies examining the link between IR-induced changes to the methionine cycle and epigenetic alterations are clearly needed.
Besides the numerous impacts IR exerts on one-carbon metabolism, it is evident that alterations to one-carbon metabolism influence the response to IR. This new understanding of the interaction between these two previously separated fields brings the possibility of using interventions on one-carbon metabolism as a strategy for modulation of both tumor and normal tissue responses to IR exposure. These applications are especially relevant for patients receiving radiotherapy to abdominal and pelvic areas, where gastrointestinal toxicity often appears to be the most frequent dose-limiting limiting factor (10, 93). For instance, methionine deprivation that may exert protective effects on the gut (i.e. by regulating the tight junction-related proteins and suppressing microflora) combined with extreme sensitivity of cancer cells to methionine starvation, may substantially increase efficacy of radiotherapy.
On the other hand, methionine has been proposed as a potential radiomitigator as methionine dietary supplementation may increase the availability of methyl donors for DNA and histone methylation, as well as provide additional substrates for the synthesis of glutathione. It must be kept in mind, however, that despite its importance for normal homeostasis, methionine is also considered the most toxic amino acid that may potentiate IR-induced gastrointestinal toxicity – either via formation of toxic methanethiol-cysteine disulfides, or by boosting the gut microbiome growth (development of the small intestine bacterial out-growth syndrome) (reviewed in refs. 28, 94). Therefore, new research approaches must be established in order to learn how to benefit normal cells and cause maximal harm to cancer cells by this delicate methionine manipulation.
In these regards, metabolomics is one of the most promising and rapidly developing avenues. Metabolomics, an emerging technology, is the comprehensive study of the metabolome, the repertoire of small molecules or biochemicals present in cells, tissue and body fluids. Recent advances in mass spectrometry-based metabolomics technology have augmented the targeted quantification of metabolites with remarkable specificity and sensitivity. This is advantageous for rapid and reliable quantification of metabolites of one-carbon metabolism. The metabolites are quantified using stable isotope dilution – multiple reaction monitoring – mass spectrometry (SID-MRM-MS) using a triple quadrupole mass spectrometer wherein the quantitation is based on transitions (daughter ions) specific to the metabolite of interest. The ability to interrogate precise changes in one-carbon metabolism and related pathways is valuable in the context of biochemical alterations, as well as promises to gain more insights into the normal and cancerous cells response to IR.
Outlook
The last 10 years have seen significant progress in our understanding of the metabolic and epigenetic effects of exposure to IR. We predict that the next 10 years will be dedicated to using one-carbon metabolism to manipulate the cellular epigenome, as well as to modulate normal and cancerous tissues’ response to IR.
The potential modulation of dietary methionine intake is of particular interest as it seems to be the most confronting. Some data suggests that methionine supplementation may have a radioprotective effect on the normal tissue, while other data suggests the potentiation of IR-induced normal tissue toxicity. Interestingly, beyond the generally accepted cancer cell-specific toxicity due to the methionine starvation, accumulating evidence also indicates that methionine overload can be equally toxic to cancer cells (95–97). This duality will certainly require further investigations that we foresee to occur in the next decade.
One of the obstacles in the clinical application of methionine deprivation is its toxicity during the long duration of cancer therapy. Indeed, administration of a methionine-deficient diet causes substantial and often unaffordable weight loss in cancer patients (0.5 kg/week) (98). Furthermore, studies in cancer patients and in vivo models report methionine deprivation-induced thrombocytopenia and neutropenia, as well as the development of hepatosteatosis (91, 92, 99). However, recent advances in tumor imaging and irradiation techniques, along with the development of stereotactic body radiation therapy (SBRT), have begun to reveal increased local tumor control within the substantially decreased duration of radiation treatments compared to conventional fractionation (100–102). This opens a new horizon for methionine deprivation utilization in clinical practice, since a short-term methionine starvation can be expected to have significantly lower potential for toxicity and negative health outcomes. We anticipate the studies investigating the combined effects of methionine deprivation with radiotherapy in the near future.
Highlights.
Exposure to IR leads to epigenetic alterations, often exhibited as a loss of global and RE-specific DNA and histone methylation.
Exposure to IR affects one-carbon metabolism and the tissue concentrations of methyl donors, suggesting a plausible mechanism for IR-induced epigenetic alterations.
Supplementation in methyl donors can mitigate the genetic, epigenetic and apical effects of IR.
Methionine dietary supplementation and deprivation may – positively and negatively – regulate both normal and cancerous tissue sensitivity to IR.
Acknowledgments:
The authors are thankful to Dr. Kristy Kutanzi for critical reading and to Christopher Fettes for editing the manuscript. We apologize to our colleagues whose research was not mentioned here owing to the focus on specific models and space limitations. Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number 1P20GM109005 and the National Space Biomedical Research Institute through the National Aeronautics and Space Administration NCC 9-58, grant number RE03701. J.T. was supported by T32 GM106999.
List of abbreviations
- BHMT
betaine-homocysteine methyltransferase
- CBS
cystathionine beta synthase
- DHFR
dihydrofolate reductase
- DNMT
DNA methyltransferase
- IR
ionizing radiation
- MAT
methionine adenosyltransferase
- MTHFD1
methylenetetrahydrofolate dehydrogenase, cyclohydrolase and formyltetrahydrofolate synthetase 1
- MTHFR
methyltetrahydrofolate reductase
- RE
repetitive elements
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- THF
tetrahydrofolate
- TS
thymidylate synthase
Contributor Information
Isabelle R. Miousse, Department of Environmental and Occupational Health, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA
Julia Tobacyk, Departments of Environmental and Occupational Health, and Pharmacology and Toxicology, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Stepan Melnyk, Department of Pediatrics, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
S. Jill James, Department of Pediatrics, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Amrita K. Cheema, Departments of Oncology and Biochemistry, Molecular and Cellular Biology, Georgetown University Medical Center, Washington DC 20057, USA
Marjan Boerma, Division of Radiation Health, Department of Pharmaceutical Sciences, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Martin Hauer-Jensen, Division of Radiation Health, Department of Pharmaceutical Sciences, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
Igor Koturbash, Department of Environmental and Occupational Health, University of Arkansas for Medical Sciences, Little Rock, AR 72205, USA.
References
- 1.Brenner DJ. Slowing the increase in the population dose resulting from CT scans. Radiat Res 2010; 174: 809–15. [DOI] [PubMed] [Google Scholar]
- 2.Moding EJ, Kastan MB, Kirsch DG. Strategies for optimizing the response of cancer and normal tissues to radiation. Nat Rev Drug Discov 2013; 12: 526–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kutanzi KR, Lumen A, Koturbash I, Miousse IR. Pediatric exposures to ionizing radiation: carcinogenic considerations. Int J Env Res Pub He 2016; 13: pii: E1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boerma M, Sridharan V, Mao XW, Nelson GA, Cheema AK, Koturbash I, Singh SP, Tackett AJ, Hauer-Jensen M. Effects of ionizing radiation on the heart. Mutat Res-Rev Mutat 2016; 770: 319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart FA, Akleyev AV, Hauer Jensen M, Hendry JH, Kleiman NJ, Macvittie TJ, Aleman BM, Edgar AB, Mabuchi K, Muirhead CR, Shore RE, Wallace WH. ICRP publication 118: ICRP statement on tissue reactions and early and late effects of radiation in normal tissues and organs–threshold doses for tissue reactions in a radiation protection context. Annal ICRP 2012; 41: 1–322. [DOI] [PubMed] [Google Scholar]
- 6.Hall E, Giaccia A. Radiobiology for the radiologist, 7th ed., Philadelphia: Lippincott Williams and Wilkins, 2011. [Google Scholar]
- 7.Dorr H, Meineke V. Acute radiation syndrome caused by accidental radiation exposure – therapeutic principles. BMC Med 2011; 9: 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mettler FA, Gus’kova AK, Gusev I. Health effects in those with acute radiation sickness from the Chernobyl accident. Health Phys 2007; 93: 462–9. [DOI] [PubMed] [Google Scholar]
- 9.Harris S Breast cancer management. 6. Radiotherapy for early and advanced breast cancer. Int J Clin Pract 2001; 55: 609–12. [PubMed] [Google Scholar]
- 10.Hauer-Jensen M, Denham JW, Andreyev HJN. Radiation enteropathy-pathogenesis, treatment and prevention. Nat Rev Gastro Hepat 2014; 11: 470–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Versluys AB, Bresters D. Pulmonary Complications of Childhood Cancer Treatment. Paediatr Respir Rev 2016; 17: 63–70. [DOI] [PubMed] [Google Scholar]
- 12.Umezawa R, Ota H, Takanami K, Ichinose A, Matsushita H, Saito H, Takase K, Jingu K. MRI findings of-radiation-induced myocardial damage in patients with oesophageal cancer. Clin Radiol 2014; 69: 1273–9. [DOI] [PubMed] [Google Scholar]
- 13.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012; 13: 484–92. [DOI] [PubMed] [Google Scholar]
- 14.Espada J, Esteller M. Epigenetic control of nuclear architecture. Cell Mol Life Sci 2007; 64: 449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miousse IR, Chalbot MCG, Lumen A, Ferguson A, Kavouras IG, Koturbash I. Response of transposable elements to environmental stressors. Mutat Res-Rev Mutat 2015; 765: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gama-sosa MA, Slagel VA, Trewyn RW, Oxenhandler R, Kuo KC, Gehrke CW, Ehrlich M. The 5-methylcytosine content of DNA from human-tumors. Nucleic Acids Res 1983; 11: 6883–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983; 301: 89–92. [DOI] [PubMed] [Google Scholar]
- 18.Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res 1995; 55: 5195–9. [PubMed] [Google Scholar]
- 19.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res 1999; 59: 3730–40. [PubMed] [Google Scholar]
- 20.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol 2009; 19: 181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koturbash I, Beland FA, Pogribny IP. Role of epigenetic events in chemical carcinogenesis – a justification for incorporating epigenetic evaluations in cancer risk assessment. Toxicol Mech Method 2011; 21: 289–97. [DOI] [PubMed] [Google Scholar]
- 22.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis 2000; 21: 461–7. [DOI] [PubMed] [Google Scholar]
- 23.Koturbash I, Simpson NE, Beland FA, Pogribny IP. Alterations in histone H4 lysine 20 methylation: implications for cancer detection and prevention. Antioxid Redox Sign 2012; 17: 365–74. [DOI] [PubMed] [Google Scholar]
- 24.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev 2012; 22: 50–5. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med 2011; 17: 330–9. [DOI] [PubMed] [Google Scholar]
- 26.Belinsky SA, Klinge DM, Liechty KC, March TH, Kang T, Gilliland FD, Sotnic N, Adamova G, Rusinova G, Telnov V. Plutonium targets the p16 gene for inactivation by promoter hypermethylation in human lung adenocarcinoma. Carcinogenesis 2004; 25: 1063–7. [DOI] [PubMed] [Google Scholar]
- 27.Lyon CM, Klinge DM, Liechty KC, Gentry FD, March TH, Kang T, Gilliland FD, Adamova G, Rusinova G, Telnov V, Belinsky SA. Radiation-induced lung adenocarcinoma is associated with increased frequency of genes, inactivated by promoter hypermethylation. Radiat Res 2007; 168: 409–14. [DOI] [PubMed] [Google Scholar]
- 28.Miousse IR, Kutanzi KR, Koturbash I. Effects of ionizing radiation on DNA methylation: from experimental biology to clinical applications. Int J Radiat Biol 2017; 93: 457–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koturbash I, Pogribny I, Kovalchuk O. Stable loss of global DNA methylation in the radiation-target tissue – a possible mechanism contributing to radiation carcinogenesis? Biochem Bioph Res Co 2005; 337: 526–33. [DOI] [PubMed] [Google Scholar]
- 30.Pogrlbny I, Koturbash I, Tryndyak V, Hudson D, Stevenson SML, Sedelnikova O, Bonner W, Kovalchuk O. Fractionated low-dose radiation exposure leads to accumulation of DNA damage and profound alterations in DNA and histone methylation in the murine thymus. Mol Cancer Res 2005; 3: 553–61. [DOI] [PubMed] [Google Scholar]
- 31.Miousse IR, Shao L, Chang J, Feng W, Wang Y, Allen AR, Turner J, Stewart B, Raber J, Zhou D, Koturbash I. Exposure to low-dose Fe-56-ion radiation induces long-term epigenetic alterations in mouse bone marrow hematopoietic progenitor and stem cells. Radiat Res 2014; 182: 92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giotopoulos G, McCormick C, Cole C, Zanker A, Jawad M, Brown R, Plumb M. DNA methylation during mouse hemopoietic differentiation and radiation-induced leukemia. Exp Hematol 2006; 34: 1462–70. [DOI] [PubMed] [Google Scholar]
- 33.Lahtz C, Bates SE, Jiang Y, Li AX, Wu X, Hahn MA, Pfeifer GP. Gamma irradiation does not induce detectable changes in DNA methylation directly following exposure of human cells. PLoS One 2012; 7: e44858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prior S, Miousse IR, Nzabarushimana E, Pathak R, Skinner C, Kutanzi KR, Allen AR, Raber J, Tackett AJ, Hauer-Jensen M, Nelson GA, Koturbash I. Densely ionizing radiation affects DNA methylation of selective LINE-1 elements. Environ Res 2016; 150: 470–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nzabarushimana E, Miousse IR, Shao L, Chang J, Allen AR, Turner J, Stewart B, Raber J, Koturbash I. Long-term epigenetic effects of exposure to low doses of Fe-56 in the mouse lung. J Radiat Res 2014; 55: 823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antwih DA, Gabbara KM, Lancaster WD, Ruden DM, Zielske SP. Radiation-induced epigenetic DNA methylation modification of radiation-response pathways. Epigenetics 2013; 8: 839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koturbash I, Boyko A, Rodriguez-Juarez R, McDonald RJ, Tryndyak VP, Kovalchuk I, Pogribny IP, Kovalchuk O. Role of epigenetic effectors in maintenance of the long-term persistent bystander effect in spleen in vivo. Carcinogenesis 2007; 28: 1831–8. [DOI] [PubMed] [Google Scholar]
- 38.Koturbash I, Miousse IR, Sridharan V, Nzabarushimana E, Skinner CM, Melnyk SB, Pavliv O, Hauer-Jensen M, Nelson GA, Boerma M. Radiation-induced changes in DNA methylation of repetitive elements in the mouse heart. Mutat Res-Fund Mol M 2016; 787: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalinich JF, Catravas GN, Snyder SL. The effect of γ-radiation on DNA methylation. Radiat Res 1989; 117: 185–97. [PubMed] [Google Scholar]
- 40.Inano K, Suetake I, Ueda T, Miyake Y, Nakamura M, Okada M, Tajima S. Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J Biochem (Tokyo) 2000; 128: 315–21. [DOI] [PubMed] [Google Scholar]
- 41.Ghosh SP, Singh R, Chakraborty K, Kulkarni S, Uppal A, Luo Y, Kaur P, Pathak R, Kumar KS, Hauer-Jensen M, Cheema AK. Metabolomic changes in gastrointestinal tissues after whole body radiation in a murine model. Mol Biosyst 2013; 9: 723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Selhub J. Folate, vitamin B12 and vitamin B6 and one carbon metabolism. J Nutrit Health Aging 2001; 6: 39–42. [PubMed] [Google Scholar]
- 43.Nazki FH, Sameer AS, Ganaie BA. Folate: Metabolism, genes, polymorphisms and the associated diseases. Gene 2014; 533: 11–20. [DOI] [PubMed] [Google Scholar]
- 44.Yi P, Melnyk S, Pogribna M, Pogribny IP, Hines RJ, James SJ. Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J Biol Chem 2000; 275: 29318–23. [DOI] [PubMed] [Google Scholar]
- 45.Frye RE, James SJ. Metabolic pathology of autism in relation to redox metabolism. Biomark Med 2014; 8: 321–30. [DOI] [PubMed] [Google Scholar]
- 46.Robinson K. Homocysteine and vascular disease. Eur Heart J 2002; 23: 1482–4. [DOI] [PubMed] [Google Scholar]
- 47.Polito L, Poloni TE, Vaccaro R, Abbondanza S, Mangieri M, Davin A, Villani S, Guaita A. High homocysteine and epistasis between MTHFR and APOE: association with cognitive performance in the elderly. Exp Gerontol 2016; 76: 9–16. [DOI] [PubMed] [Google Scholar]
- 48.Behera J, Bala J, Nuru M, Tyagi SC, Tyagi N. Homocysteine as a Pathological Biomarker for Bone Disease. J Cell Physiol 2016. [DOI] [PMC free article] [PubMed]
- 49.Hannibal L, Blom HJ. Homocysteine and disease: causal associations or epiphenomenons? Mol Aspects Med 2017; 53: 36–42. [DOI] [PubMed] [Google Scholar]
- 50.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP, Rozen R. A Candidate genetic risk factor for vascular-disease – a common mutation in methylenetetrahydrofolate reductase. Nat Genet 1995; 10: 111–3. [DOI] [PubMed] [Google Scholar]
- 51.Jacques PF, Bostom AG, Williams RR, Ellison RC, Eckfeldt JH, Rosenberg IH, Selhub J, Rozen R. Relation between folate status, a common mutation in methylenetetrahydrofolate reductase, and plasma homocysteine concentrations. Circulation 1996; 93: 7–9. [DOI] [PubMed] [Google Scholar]
- 52.Wald DS, Morris JK, Wald NJ. Reconciling the evidence on serum homocysteine and ischaemic heart disease: a meta-analysis. PLoS One 2011; 6: e16473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Izmirli M A literature review of MTHFR (C677T and A1298C polymorphisms) and cancer risk. Mol Biol Rep 2013; 40: 625–37. [DOI] [PubMed] [Google Scholar]
- 54.Clarke R, Bennett DA, Parish S, Verhoef P, Dotsch-Klerk M, Lathrop M, Xu P, Nordestgaard BG, Holm H, Hopewell JC, Saleheen D, Tanaka T, Anand SS, Chambers JC, Kleber ME, Ouwehand WH, Yamada Y, Elbers C, Peters B, Stewart AF, Reilly MM, Thorand B, Yusuf S, Engert JC, Assimes TL, Kooner J, Danesh J, Watkins H, Samani NJ, Collins R, Peto R; MTHFR Studies Collaborative Group. Homocysteine and coronary heart disease: meta-analysis of MTHFR case-control studies, avoiding publication bias. PLoS Med 2012; 9: e1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kesavan V, Pote MS, Batra V, Viswanathan G. Increased folate catabolism following total body γ-irradiation in mice. J Radiat Res 2003; 44: 141–4. [DOI] [PubMed] [Google Scholar]
- 56.Batra V, Kesavan V, Mishra KP. Modulation of enzymes involved in folate dependent one-carbon metabolism by γ-radiation stress in mice. J Radiat Res 2004; 45: 527–33. [DOI] [PubMed] [Google Scholar]
- 57.Endoh K, Murakami M, Umegaki K. Vulnerability of folate in plasma and bone marrow to total body irradiation in mice. Int J Radiat Biol 2007; 83: 65–71. [DOI] [PubMed] [Google Scholar]
- 58.Radivoyevitch T Folate system correlations in DNA microarray data. BMC Cancer 2005; 5: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guren MG, Schneede J, Tveit KM, Ueland PM, Nexo E, Dueland S. Biochemical signs of impaired cobalamin status during and after radiotherapy for rectal cancer. Int J Radiat Oncol 2004; 60: 807–13. [DOI] [PubMed] [Google Scholar]
- 60.Kinn AC, Lantz B. Vitamin-B12 deficiency after irradiation for bladder-carcinoma. J Urol 1984; 131: 888–90. [DOI] [PubMed] [Google Scholar]
- 61.Vistad I, Kristensen GB, Fossa SD, Dahl AA, Morkrid L. Intestinal Malabsorption in Long-Term Survivors of Cervical Cancer Treated with Radiotherapy. Int J Radiat Oncol 2009; 73: 1141–7. [DOI] [PubMed] [Google Scholar]
- 62.Anderson CG, Walton KR, Chanarin I. Megaloblastic-anemia after pelvic radiotherapy for carcinoma of the cervix. J Clin Pathol 1981; 34: 151–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edwards CH, Edwards GA, Gadsden EL. Effect of irradiation on tissue uptake of methionine-2–14C + methionine methyl-14C. Radiat Res 1964; 22: 116. [PubMed] [Google Scholar]
- 64.Aypar U, Morgan WF, Baulch JE. Radiation-induced epigenetic alterations after low and high LET irradiations. Mutat Res Fund Mol M 2011; 707: 24–33. [DOI] [PubMed] [Google Scholar]
- 65.Goetz W, Morgan MNM, Baulch JE. The effect of radiation quality on genomic DNA methylation profiles in irradiated human cell lines. Radiat Res 2011; 175: 575–87. [DOI] [PubMed] [Google Scholar]
- 66.Beetstra S, Thomas P, Salisbury C, Turner J, Fenech M. Folic acid deficiency increases chromosomal instability, chromosome 21 aneuploidy and sensitivity to radiation-induced micronuclei. Mutat Res-Fund Mol M 2005; 578: 317–26. [DOI] [PubMed] [Google Scholar]
- 67.Leopardi P, Marcon F, Caiola S, Cafolla A, Siniscalchi E, Zijno A, Crebelli R. Effects of folic acid deficiency and MTHFR C677T polymorphism on spontaneous and radiation-induced micronuclei in human lymphocytes. Mutagenesis 2006; 21: 327–33. [DOI] [PubMed] [Google Scholar]
- 68.Batra V, Devasagayam TPA. Interaction between γ-radiation and dietary folate starvation metabolically reprograms global hepatic histone H3 methylation at lysine 4 and lysine 27 residues. Food Chem Toxicol 2012; 50: 464–72. [DOI] [PubMed] [Google Scholar]
- 69.Batra V, Devasagayam TPA. Interaction between cytotoxic effects of γ-radiation and folate deficiency in relation to choline reserves. Toxicology 2009; 255: 91–9. [DOI] [PubMed] [Google Scholar]
- 70.Batra V, Kislay B, Devasagayam TPA. Interaction between total body γ-irradiation and choline deficiency triggers immediate modulation of choline and choline-containing moieties. Int J Radiat Biol 2011; 87: 1196–207. [DOI] [PubMed] [Google Scholar]
- 71.Monobe M, Uzawa A, Hino M, Ando K, Kojima S. Glycine betaine, a beer component, protects radiation-induced injury. J Radiat Res 2005; 46: 117–21. [DOI] [PubMed] [Google Scholar]
- 72.Monobe M, Hamano N, Sumi M, Mukai K, Moritake T, Anzai K, Uzawa A, Ando K. Effects of glycine betaine on bone marrow death and intestinal damage by γ rays and carbon ions. Radiat Prot Dosim 2006; 122: 494–7. [DOI] [PubMed] [Google Scholar]
- 73.Batra V, Sridhar S, Devasagayam TPA. Enhanced one-carbon flux towards DNA methylation: effect of dietary methyl supplements against γ-radiation-induced epigenetic modifications. Chem-Biol Interact 2010; 183: 425–33. [DOI] [PubMed] [Google Scholar]
- 74.Yoshikawa M D-methionine as a protector for irradiation-induced oral mucositis and xerostomia. Nihon yakurigaku zasshi Folia pharmacologica Japonica 2013; 141: 310. [DOI] [PubMed] [Google Scholar]
- 75.Vuyyuri SB, Hamstra DA, Khanna D, Hamilton CA, Markwart SM, Campbell KC, Sunkara P, Ross BD, Rehemtulla A. Evaluation of D-methionine as a novel oral radiation protector for prevention of mucositis. Clin Cancer Res 2008; 14: 2161–70. [DOI] [PubMed] [Google Scholar]
- 76.Meakins TS, Persaud C, Jackson AA. Dietary supplementation with L-methionine impairs the utilization of urea-nitrogen and increases 5-L-oxoprolinuria in normal women consuming a low protein diet. J Nutr 1998; 128: 720–7. [DOI] [PubMed] [Google Scholar]
- 77.Chambers JC, McGregor A, Jean-Marie J, Obeid OA, Kooner JS. Demonstration of rapid onset vascular endothelial dysfunction after hyperhomocysteinemia - an effect reversible with vitamin C therapy. Circulation 1999; 99: 1156–60. [DOI] [PubMed] [Google Scholar]
- 78.Benevenga NJ, Steele RD. Adverse-effects of excessive consumption of amino-acids. Annu Rev Nutr 1984; 4: 157–81. [DOI] [PubMed] [Google Scholar]
- 79.Weltman MD, Farrell GC, Liddle C. Increased hepatocyte CYP2E1 expression in a rat nutritional model of hepatic steatosis with inflammation. Gastroenterology 1996; 111: 1645–53. [DOI] [PubMed] [Google Scholar]
- 80.Orentreich N, Matias JR, Defelice A, Zimmerman JA. Low methionine ingestion by rats extends life-span. J Nutr 1993; 123: 269–74. [DOI] [PubMed] [Google Scholar]
- 81.Miller RA, Buehner G, Chang Y, Harper JM, Sigler R, Smith-Wheelock M. Methionine-deficient diet extends mouse lifespan, slows immune and lens aging, alters glucose, T4, IGF-I and insulin levels, and increases hepatocyte MIF levels and stress resistance. Aging Cell 2005; 4: 119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mullin JM, Skrovanek SM, Ramalingam A, DiGuilio KM, Valenzano MC. Methionine restriction fundamentally supports health by tightening epithelial barriers. Ann Ny Acad Sci 2016; 1363: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramalingam A, Wang X, Gabello M, Valenzano MC, Soler AP, Ko A, Morin PJ, Mullin JM. Dietary methionine restriction improves colon tight junction barrier function and alters claudin expression pattern. Am J Physiol-Cell Ph 2010; 299: C1028–C1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skrovanek S, Valenzano MC, Mullin JM. Restriction of sulfur-containing amino acids alters claudin composition and improves tight junction barrier function. Am J Physiol-Reg I 2007; 293: R1046–55. [DOI] [PubMed] [Google Scholar]
- 85.Monti P, Wysocki J, Van der Meeren A, Griffiths N. The contribution of radiation-induced injury to the gastrointestinal tract in the development of multi-organ dysfunction syndrome or failure. Br J Radiol 2014; 1(Suppl 27): 89–94. [Google Scholar]
- 86.Hoffman RM. Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: a 40-year odyssey. Expert Opin Biol Th 2015; 15: 21–31. [DOI] [PubMed] [Google Scholar]
- 87.Agrawal V, Alpini SEJ, Stone EM, Frenkel EP, Frankel AE. Targeting methionine auxotrophy in cancer: discovery & exploration. Expert Opin Biol Th 2012; 12: 53–61. [DOI] [PubMed] [Google Scholar]
- 88.Guenin S, Morvan D, Thivat E, Stepien G, Demidem A. Combined methionine deprivation and chloroethylnitrosourea have time-dependent therapeutic synergy on melanoma tumors that NMR spectroscopy-based metabolomics explains by methionine and phospholipid metabolism reprogramming. Nutr Cancer 2009; 61: 518–29. [DOI] [PubMed] [Google Scholar]
- 89.Strekalova E, Malin D, Good DM, Cryns VL. Methionine deprivation induces a targetable vulnerability in triple-negative breast cancer cells by enhancing TRAIL receptor-2 expression. Clin Cancer Res 2015; 21: 2780–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kokkinakis DM, Brickner AG, Kirkwood JM, Liu XY, Goldwasser JE, Kastrama A, Sander C, Bocangel D, Chada S. Mitotic arrest, apoptosis, and sensitization to chemotherapy of melanomas by methionine deprivation stress. Mol Cancer Res 2006; 4: 575–89. [DOI] [PubMed] [Google Scholar]
- 91.Durando X, Thivat E, Farges MC, Cellarier E, D’Incan M, Demidem A, Vasson MP, Barthomeuf C, Chollet P. Optimal methionine-free diet duration for nitrourea treatment: a phase I clinical trial. Nutr Cancer 2008. ’; 60: 23–30. [DOI] [PubMed] [Google Scholar]
- 92.Thivat E, Durando X, Demidem A, Farges MC, Rapp M, Cellarier E, Guenin S, D’Incan M, Vasson MP, Chollet P. A methionine-free diet associated with nitrosourea treatment down-regulates methylguanine-DNA methyl transferase activity in patients with metastatic cancer. Anticancer Res 2007; 27: 2779–83. [PubMed] [Google Scholar]
- 93.Hauer-Jensen M, Wang J, Boerma M, Fu Q, Denham JW. Radiation damage to the gastrointestinal tract: mechanisms, diagnosis, and management. Curr Opin Support Palliative Care 2007; 1: 23–9. [DOI] [PubMed] [Google Scholar]
- 94.Ferreira MR, Muls A, Dearnaley DP, Andreyev HJN. Microbiota and radiation-induced bowel toxicity: lessons from inflammatory bowel disease for the radiation oncologist. Lancet Oncol 2014; 15: E139–47. [DOI] [PubMed] [Google Scholar]
- 95.Benavides MA, Bosland MC, da Silva CP, Sares CTG, de Oliveira AMC, Kemp R, dos Reis RB, Martins VR, Sampaio SV, Bland KI, Grizzle WE, dos Santos JS. L-Methionine inhibits growth of human pancreatic cancer cells. Anti-Cancer Drug 2014; 25: 200–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Benavides MA, Hagen KL, Fang W, Du P, Lin S, Moyer MP, Yang W, Bland KI, Grizzle WE, Bosland MC. Suppression by L-methionine of cell cycle progression in LNCaP and MCF-7 cells but not benign cells. Anticancer Res 2010; 30: 1881–5. [PMC free article] [PubMed] [Google Scholar]
- 97.Benavides MA, Hu D, Baraoidan MK, Bruno A, Du P, Lin S, Yang W, Bland KI, Grizzle WE, Bosland MC. L-methionine-induced alterations in molecular signatures in MCF-7 and LNCaP cancer cells. J Cancer Res Clin 2011; 137: 441–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Epner DE, Morrow S, Wilcox M, Houghton JL. Nutrient intake and nutritional indexes in adults with metastatic cancer on a phase I clinical trial of dietary methionine restriction. Nutr Cancer 2002; 42: 158–66. [DOI] [PubMed] [Google Scholar]
- 99.Koteish A, Diehl AM. Animal models of steatosis. Semin Liver Dis 2001; 21: 89–104. [DOI] [PubMed] [Google Scholar]
- 100.Brown JM, Brenner DJ, Carlson DJ. Dose Escalation, not “new biology,” can account for the efficacy of stereotactic body radiation therapy with non-small cell lung cancer. Int J Radiat Oncol 2013; 85: 1159–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Song CW, Lee YJ, Griffin RJ, Park I, Koonce NA, Hui S, Kim MS, Dusenbery KE, Sperduto PW, Cho LC. Indirect tumor cell death after high-dose hypofractionated irradiation: implications for stereotactic body radiation therapy and stereotactic radiation surgery. Int J Radiat Oncol 2015; 93: 166–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Song CW, Cho LC, Yuan JL, Dusenbery KE, Griffin RJ, Levitt SH. Radiobiology of stereotactic body radiation therapy/stereotactic radiosurgery and the linear-quadratic model. Int J Radiat Oncol 2013; 87: 18–9. [DOI] [PubMed] [Google Scholar]