Abstract
As are most non-European populations, the Han Chinese are relatively understudied in population and medical genetics studies. From low-coverage whole-genome sequencing of 11,670 Han Chinese women we present a catalog of 25,057,223 variants, including 548,401 novel variants that are seen at least 10 times in our data set. Individuals from this data set came from 24 out of 33 administrative divisions across China (including 19 provinces, 4 municipalities, and 1 autonomous region), thus allowing us to study population structure, genetic ancestry, and local adaptation in Han Chinese. We identified previously unrecognized population structure along the East–West axis of China, demonstrated a general pattern of isolation-by-distance among Han Chinese, and reported unique regional signals of admixture, such as European influences among the Northwestern provinces of China. Furthermore, we identified a number of highly differentiated, putatively adaptive, loci (e.g., MTHFR, ADH7, and FADS, among others) that may be driven by immune response, climate, and diet in the Han Chinese. Finally, we have made available allele frequency estimates stratified by administrative divisions across China in the Geography of Genetic Variant browser for the broader community. By leveraging the largest currently available genetic data set for Han Chinese, we have gained insights into the history and population structure of the world’s largest ethnic group.
Introduction
To date, a range of strategies has been employed to characterize populations. In Genomes of the Netherlands (GoNL) (Francioli et al. 2014), the trio design allowed estimation of high-quality genotypes for both single nucleotide and structural variations with intermediate (∼13×) sequencing coverage and enabled the investigation of de novo mutations. In Sardinian (Sidore et al. 2015) and Icelandic (Gudbjartsson et al. 2015) population cohorts, extensive haplotype sharing within populations was used to inform accurate genotype calling among low- (∼4–6×) and intermediate- (∼20×) coverage sequencing of ∼2,000–3,000 individuals. In the UK10K project (Walter et al. 2015), low (∼7×) whole-genome sequencing in 3,781 healthy samples from two British cohorts was combined with deep (80×) exome sequencing in three disease cohorts to accurately detect low frequency and rare variants associated with quantitative traits. However, much like the genome-wide association studies preceding the current era of sequencing studies, most sequencing efforts are biased toward European populations. To address the need to comprehensively characterize non-European populations, we describe a resource of genetic variants in the world’s largest ethnic group, Han Chinese.
The whole-genome sequencing data set of the Han Chinese analyzed here adopted a different approach. We analyzed genetic variants from very low-coverage whole-genome sequencing data of 11,670 Han Chinese women, previously generated to study major depressive disorder (MDD) (Cai et al. 2015, 2017). With a median coverage of 1.7×, this data set is predicted to identify rare (<0.5%) single nucleotide polymorphisms (SNPs) with high confidence (Li, Sidore, et al. 2011) and obtain accurate estimates of allele frequencies in a large sample. Using this genomic resource, we catalog variants, and use the data to characterize the genetic structure of the Chinese population.
Our understanding of the population structure and history of the Han Chinese has been relatively limited. Historical records suggest that the Han Chinese originated from the Central Plain region of China during the early historic and prehistoric era. Aided by their advantage in agriculture and technology, the population expanded both northward and southward to become the largest ethnic group today in China (Zhao et al. 2015). This historical movement is corroborated genetically based on uniparental markers from both modern and ancient DNA samples (Wen et al. 2004; Zhao et al. 2015). A North-to-South structure is also evident from array-based genome-wide data (Chen et al. 2009, 2016; Xu et al. 2009). However, very little structure has been observed beyond this North–South cline. This could be due to a lack of representative samples across China, and/or small sample sizes that reduced power to detect less prominent features of population structure. Furthermore, because typical representatives of Han Chinese are discrete Northern or Southern Chinese populations from the 1000 Genomes (1KG) or Human Genome Diversity Project (HGDP) data, little has been discussed of the relationship between Han Chinese and other non-East Asian populations. Nonetheless, in these relatively limited reference samples of Han Chinese, elevated ancestry from Western Eurasians had been detected among Northern Han Chinese, and speculated to be due to trades along the Silk Road (Hellenthal et al. 2014).
Taking advantage of the diverse geographical sampling of Han Chinese across China in this data set, covering 19 out of 22 provinces in China (along with four municipalities and one autonomous region), we sought to investigate the genetic structure of modern Han Chinese. First, using largely allele-frequency-based population genetic analyses, we examined patterns of ancestry and admixture across China. Next, we investigated regions of the genome undergoing extreme differentiation in Han Chinese. We also investigated the relationship of modern Han Chinese with ancient and archaic samples followed by examining the influence of archaic admixture on depressive symptoms to contrast with previous observations made in European populations (Simonti et al. 2016). Findings of our study provide additional insights to the genetic history of the Han Chinese, and our analytical framework would also be broadly applicable to future larger-scale whole-genome sequencing studies that, in the short term, will come from ultra-low-coverage sequencing.
Results
A total of 11,670 Han Chinese women were previously sequenced at a median coverage of 1.7× per individual, of which 10,640 individuals and 25,057,223 SNPs remained after quality control (QC) (Cai et al. 2015, 2017). Despite the low median coverage genome-wide, individual level genotype calls showed high concordance (>96–97%) in validation experiments (Cai et al. 2017). Furthermore, the allele frequencies in this data set are highly correlated (mean r = 0.995 across chromosomes) to those from the East Asian sample in Exome Aggregation Consortium (ExAC; Lek et al. 2016). This observation suggests that any batch effect due to genotype calling in very low-coverage sequencing should not impact allele frequency estimates and their use in downstream analyses.
Restricting analysis to variants with minor allele counts (MAC) ≥10 (9,888,655 variants), we found that the alternate alleles of 477,792 (4.8%), 567,731 (5.7%), and 868,251 (8.8%) variants are not seen in 1KG (phase 3), 1KG East Asians, and 1KG CHB+CHS panels, respectively (supplementary fig. S1 and table S1, Supplementary Material online). We defined three minor allele frequency (MAF) categories: Common (MAF ≥ 0.05), low frequency (0.005 ≤ MAF < 0.05), and rare (MAF < 0.005). As expected, a large proportion (66–79%) of novel alleles are rare in the population, and an additional 11–17% of them are of low frequency (supplementary table S1, Supplementary Material online). We also identified ∼82,000 variants with MAF ≥ 0.05 in our data set that are not seen in the 1KG CHB+CHS populations. Even though this class of variants is likely enriched for sequencing errors, a subset of these variants were identified in limited number of East Asians included in other recent large-scale sequencing efforts (Lek et al. 2016) and the frequency estimates are highly concordant (r = 0.78 and 0.90 when compared with ExAC and GNOMAD databases, supplementary fig. S1, Supplementary Material online). Taken together, these observations suggest that our data set currently consists of the largest variant map in Han Chinese both in terms of span of the genome and in sample size.
Variant Function and ClinVar Annotation
We performed variant annotation on the 9.89 million variants with MAC ≥ 10 using Variant Effect Predictor (VEP) (McLaren et al. 2016). We observed 2,312 loss-of-function variants, 12,458 damaging variants predicted by PolyPhen, and 10,277 damaging variants predicted by SIFT. In total, 17,889 variants are annotated as functionally deleterious by at least one method; among them, 5,695 (31.8%) are common, 4,268 (23.9%) are low frequency, and 7,926 (44.3%) are rare (supplementary fig. S2, Supplementary Material online). In comparison, we did not see the same degree of enrichment of rare variants among those annotated with less severe or deleterious effects (missense or synonymous variants in VEP, benign and tolerated variants predicted by PolyPhen and SIFT, respectively), consistent with stronger negative selection experienced by the functionally deleterious alleles (supplementary fig. S2, Supplementary Material online). Per individual, we found an average of 3,708 functionally deleterious alleles. Most of these alleles are common in the population (91%, compared with 7.6% low frequency and 1.6% rare), as most of the deleterious load per person is due to the common variants. Similarly, we observed a depletion of rare deleterious alleles per individual, compared with the less deleterious variant classes such as synonymous variants (supplementary table S2, Supplementary Material online).
We further investigated the functional consequences of the Han Chinese variant map by cross-referencing to ClinVar. A number of entries in ClinVar (and other similar database) are likely erroneous as these variants appear to be too frequent in a large unascertained population of predominantly European ancestry (Lek et al. 2016). Focusing on 15,157 variants reported to be pathogenic or likely pathogenic in ClinVar with frequencies estimated in ExAC, we identified three variants that are rare in non-Finnish Europeans (NFE) in ExAC but common in our Chinese data set (table 1). An additional variant, rs2276717 in SLC7A14 for retinitis pigmentosa, was just below the common frequency cutoff (4.8% in Han Chinese). The frequency for rs2276717 is likely too high in the Han Chinese populations to be truly pathogenic, considering the prevalence of the disorder in Han Chinese (see Discussion).
Table 1.
List of Rare Pathogenic Variants in Europeans but Common in Han Chinese.
| Chr | Pos | rsID | Ref | Alt | ClinVar Annotations |
Frequency Information |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant Type | Affected Gene | Clinical Significance | Syndrome Name | 1000G CEU | ExAC NFE | ExAC EAS | Han Chinese | |||||
| 3 | 170201230 | rs2276717 | C | T | Missense | SLC7A14 | Pathogenic | Retinitis pigmentosa 68 | 0 | 1.50E-05 | 0.026 | 0.048 |
| 6 | 46677098 | rs76863441 | C | A | Missense | PLA2G7 | Pathogenic | Platelet-activating factor acetylhydrolase deficiency | 0 | 3.50E-04 | 0.064 | 0.063 |
| 12 | 112241766 | rs671 | G | A | Missense | ALDH2 | Pathogenic | Acute alcohol sensitivity | 0 | 6.20E-05 | 0.266 | 0.051 |
| 13 | 20763612 | rs72474224 | C | T | Missense | GJB2 | Likely pathogenic | Nonsyndromic hearing loss and deafness | 0 | 0.002 | 0.072 | 0.056 |
Note.—Of the 15,157 sites in CLINVAR reported as Pathogenic or Likely Pathogenic and found in ExAC, we filtered by frequency of ≤0.01 in ExAC NFE and ≥0.05 in Han Chinese. Frequencies in ExAC East Asians (EAS) are also given for comparisons.
Population Structure of Han Chinese
Compared with previous genetic investigations of the Chinese population, our data set has one of the broadest sampling of Han Chinese across China. In total, self-reported birth locations of recruited individuals included all 4 metropolis municipalities in China (Beijing, Tianjin, Shanghai, and Chongqing) as well as 19 out of 22 provinces, and 1 (Guangxi) out of 5 autonomous regions in China (fig. 1 and supplementary table S3, Supplementary Material online). (No one was recruited from either of the two Special Administrative Regions, Hong Kong and Macau, in the original study.) We have made available the allele frequency estimates of the 9.89M variants with MAC ≥ 10 across the 24 administrative regions in the Geography of Genetic Variants browser (Marcus and Novembre 2017) for public use.
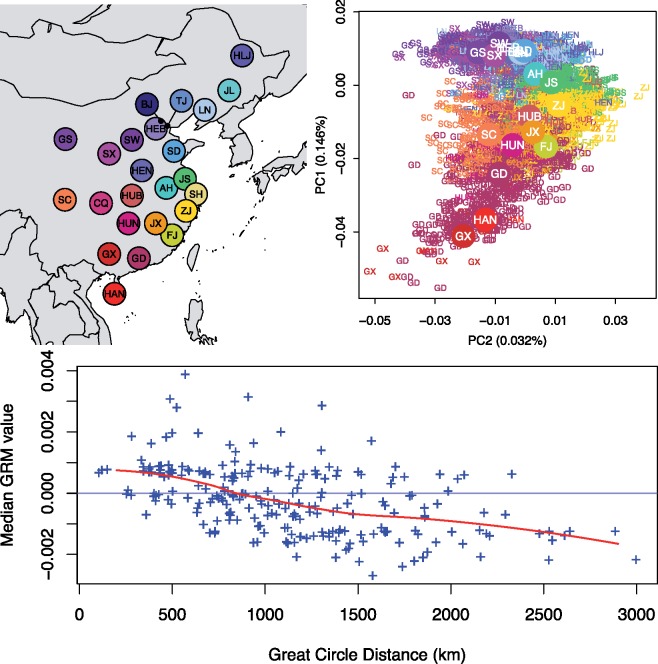
Fig. 1.
Population structure of Han Chinese. (Top Left) The geographical locations of each province of China represented in the data set, with the following abbreviations: AH, Anhui; BJ, Beijing; CQ, Chongqing; FJ, Fujian; GS, Gansu; GD, Guangdong; GX, Guangxi; HAN, Hainan; HEB, Hebei; HLJ, Heilongjiang; HEN, Henan; HUB, Hubei; HUN, Hunan; JS, Jiangsu; JX, Jiangxi; JL, Jilin; LN, Liaoning; SX, Shaanxi; SD, Shandong; SH, Shanghai; SW, Shanxi; SC, Sichuan; TJ, Tianjin; ZJ, Zhejiang. Beijing, Chongqing, Shanghai, and Tianjin are the four metropolitan municipalities of China; the remaining represents 19 out of the 22 provinces and 1 out of 5 autonomous regions of China. (Top Right) The first two PCs showing the North–South structure on PC1 and East–West structure on PC2. Individual labels were assigned based on their self-reported birth locations and colored according to the map on the left. Individuals from the four municipalities were not plotted. (Bottom) Observed decay of genetic relatedness as function of geographical distance, which is consistent with an isolation-by-distance model.
A well-known feature of Han Chinese population structure is the North–South differentiation, which we observed clearly in pairwise Fst comparisons between provinces. We found that Northern Han Chinese (Heilongjiang, Jilin, Liaoning, Beijing, Tianjin, Hebei, Gansu, Shanxi, Shandong, Shaanxi, Henan) showed relatively little genetic differentiation by Fst (supplementary fig. S3, Supplementary Material online), while the strongest signal of differentiation based on Fst was observed between Southern and Northern Han Chinese. For example, the Fst is 1.8 × 10−4 between Fujian and Beijing, and 2.5 × 10−4 between Guangdong and Beijing (supplementary fig. S3, Supplementary Material online). For comparison, this is roughly the same level of differentiation between Spanish and English or Spanish and French (Haak et al. 2015).
We also conducted a principal components analysis (PCA) (see Materials and Methods) and found the first two principal components (PCs) recapitulate the geography of China (fig. 1). Although the second PC explained only approximately a fifth as much variance compared with the first PC, both were significantly correlated with geography: The first PC corresponded to the North–South axis of China (r = 0.882, P < 1 × 10−5 for PC1 vs. latitude), while the second PC corresponded to the East–West axis of China (r = 0.701, P < 1 × 10−5 for PC2 vs. longitude) (fig. 1). The Procrustes similarity statistic between the first two PCs and geography was 0.749 (P < 1 × 10−5), with the optimal projection to latitude and longitude achieved with a 19.9° counterclockwise rotation, close to the value reported for Europeans (Novembre et al. 2008; Wang et al. 2010). Individuals from the metropolitan municipalities were more or less confined to the particular geographical locale (supplementary fig. S4, Supplementary Material online), while individuals from Guangdong exhibited exceptional dispersion, compared with other populations (supplementary fig. S5, Supplementary Material online).
Beyond the first two PCs, we observed no discernable geographical structure (although the Tracy–Widom statistics remained significant for lower PCs) (supplementary fig. S6, Supplementary Material online), contrary to the expectation if there were discrete well-differentiated subpopulations. Indeed, we found genetic relatedness for the geographically closest 20% pairs of populations to be significantly greater than that among the median 20% pairs of the populations (P = 0.00126). In general, we found genetic relatedness decays over geographical distances between pairs (Mantel’s correlation = −0.49, P < 1 × 10−5 by permutations, fig. 1). Together, these results suggest that the migration history of the Han Chinese, like that for the Europeans, followed sufficiently an isolation-by-distance model where genetic similarity decays with distance in a 2D geographical space.
Signature of Admixture with Western Eurasia, Siberia, and Neighboring East Asia Populations
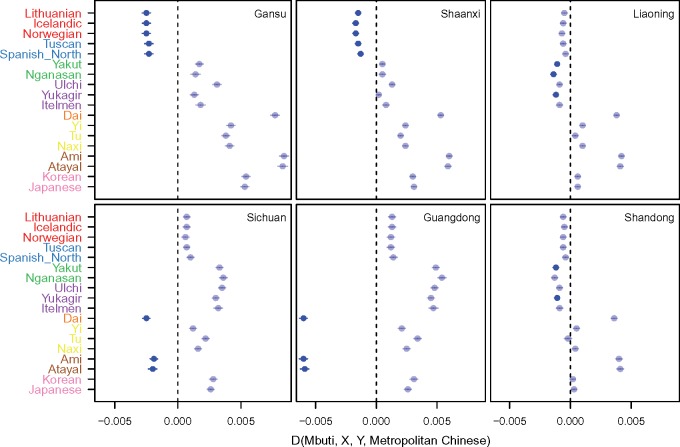
To examine evidence of admixture, we first merged our data set with the 1KG data set and a global reference data set genotyped on the Human Origins array (Patterson et al. 2012). For each Chinese province in our data set, we then used the f3-statistics to test for admixture. We used all possible pairs of other populations in the merged data set as potential ancestral populations (see Materials and Methods). In general, we found geographically localized signals of admixture across China (supplementary table S4, Supplementary Material online). For example, we identified an affinity toward Western Eurasia, particularly from Northern and Eastern Europe (e.g., English, Finnish, Lithuanian, etc.) among the Northwestern provinces of China (Gansu, Shaanxi, and Shanxi), and an affinity toward Siberia among the Northeastern provinces of China (Liaoning, Jilin, and Heilongjiang, and to a lesser extent, Shandong). In contrast, Southern provinces, such as the Southwestern province Sichuan and Southern coastal province Guangdong, showed little influence from Siberia or Western Eurasia, but instead exhibited influences from the ethnic minorities geographically situated in the Southwest and Southeast of China, such as the Ami and Atayal from Taiwan and the Dai. To better visualize these regional patterns, we corroborated these general trends with the D-statistic to compare the extent of allele sharing between selected global populations and Han Chinese of different regions, using as a baseline an average metropolitan Chinese population (see Materials and Methods, fig. 2).
Fig. 2.
Allelic sharing between selected Chinese provinces and non-Han Chinese populations in the Human Origins Array data. The allelic sharing between Han Chinese from Gansu, Shaanxi, Liaoning, Sichuan, Guangdong, and Shandong with example non-Han Chinese populations was evaluated by computing the D-statistics D(Mbuti, X, Y, Metropolitan Chinese), where population X is a population shown on the ordinate, and population Y is one of the Chinese provinces. Populations on the ordinate are color-coded by subcontinental origins: red, Northern Europeans; blue, Southern Europeans; green, Central Siberia; purple, Eastern Siberia; yellow, Western Chinese minorities; brown, Aboriginal Taiwanese; pink, Korean and Japanese. Metropolitan Chinese is a random sample of 200 individuals from one of the four metropolis municipalities, taken as representative Chinese. A significantly negative value (bolded, Z ≤ −5) suggests significant sharing between populations X and Y, relative to the representative Chinese, thus is consistent with a regional signal of admixture.
Note that an admixing source population implicated in admixture in this analysis may not be the actual source population, but a related one. Therefore, it is conceivable that some of these admixture signals of West Eurasians or Siberians in Han Chinese may be mediated by intermixing with nearby ethnic-minority Chinese populations such as Hezhen, Tu, Uygur, or others. However, we found that while these ethnic-minority Chinese could serve as one of the potential admixing populations for Han Chinese, the admixture signals were usually weaker than when a Western Eurasian or Siberian population was used as one of the admixing sources (supplementary table S5, Supplementary Material online). Similarly, the West Eurasian influence in Gansu may be mediated by the Mongolians, but the signals were also weaker when using Mongolians as one of the source populations (supplementary table S5, Supplementary Material online). Nevertheless, the best admixing source populations implicated in our analysis were often closely situated, suggesting a generally continuous admixing process rather than a discrete pulse-like admixture process.
We also examined whether Han Chinese could be the admixing source population for neighboring East Asian populations, such as Koreans, Japanese, and Native Taiwanese (supplementary table S4, Supplementary Material online). Koreans have a history of political and demographic relationship with mainland China, and are one of the officially recognized ethnic minority in China, as are Chinese in Korea. Our results were consistent with the Koreans receiving gene flows from China, Siberia, and Japan (supplementary table S4, Supplementary Material online), but curiously the Chinese provinces, even those close to the Korean peninsula, generally did not show strong signals with Korean as a plausible admixing source, suggesting a largely unidirectional gene flow to Korea or diffusion of Korean gene flow into the much larger Chinese population. We detected no discernable signal of admixture among the island populations of Hainan, Ami and Atayal ethnic minorities from Taiwan, and Japan. This could be due to small sample size, or long-term isolation among the island ethnic minorities that decreased the power of the f3-test for admixture (Patterson et al. 2012).
Genetic Relationship with Ancient Human Populations
The array of ancient DNA data now publicly available has tremendously informed the genetic ancestry of modern day Europeans. We therefore assessed the genetic relationship between Han Chinese and publicly available prehistoric human samples, mainly of European origins (Lazaridis et al. 2014, 2016; Fu et al. 2016; Siska et al. 2017; Yang, Gao, et al. 2017). We assembled 322 ancient samples that geographically span three broadly defined regions (Europe/Anatolia, Central Asia, and Siberia/East Asia) (see Materials and Methods). Temporally, the ancient human samples span a period from ∼2,300 to ∼45,000 years before present (BP), with most samples originating since the Mesolithic times ∼9,000 years ago.
We measured allele frequency covariances between ancient populations and individuals from various Chinese provinces in our data set using the outgroup f3-statistics (Raghavan et al. 2014). This measure estimates the amount of shared drift between the two populations related through a tree rooted by the outgroup population. In general, outside of the two ancient populations in East Asia, we found that shared drift with Han Chinese was greater among Siberian, Central Asian, and Northern European ancient individuals than individuals from Southern Europe, Anatolia, Levant, and the Caucasus (fig. 3 and supplementary fig. S7 and table S6, Supplementary Material online). Across China, the amount of shared drift was often significantly correlated with latitude, with Northern provinces showing higher values than Southern provinces (supplementary table S7, Supplementary Material online). This likely reflects the enrichment of European/Siberian ancient samples currently available. The exception is with some of the oldest samples (e.g., Ust Ishim, Oase1, Kostenki14, and the archaic individuals), which showed very little variation in shared drift across our Chinese sample. This is likely due to these oldest samples being a lineage basal to the European–East Asian predivergence lineage, while more recent samples showed greater variability across geography (supplementary fig. S8, Supplementary Material online).
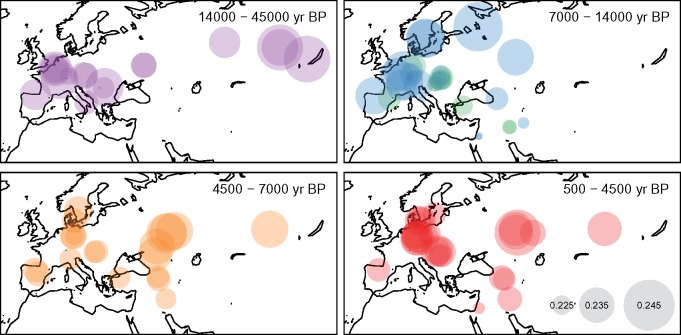
Fig. 3.
Shared drift between Han Chinese and ancient DNA samples from Eurasia. Using Shanghai individuals as representatives, shared drift between Chinese and ancient humans is computed by calculating the outgroup f3-statistics. Ancient individuals are separated into roughly four temporal categories spanning from 500 to 45,000 years BP. In the time period 7,000–14,000 years BP, ancient individuals are further divided based on their cultural context: pre-Neolithic hunter-gatherers in blue, Neolithic farmers in green. During this period, we found modern Chinese individuals showing greater shared drift with pre-Neolithic hunter-gatherers rather than Neolithic farmers.
Focusing on central and west Eurasia from where vast majority of the ancient samples originated, before the major warming period (∼14,000 years BP) some of the largest shared drift values observed were between the Siberian ancient individuals (MA1 and AG2, shared drift = 0.243 and 0.242, respectively) and Han Chinese (fig. 3). During the transition period from Paleolithic/Mesolithic to early Neolithic (∼7,000–14,000 years BP), Europe was inhabited by a mixture of both hunter-gatherers and Neolithic farmers. Between the two roughly contemporaneous cultures, Han Chinese showed greater shared drift with the pre-Neolithic hunter-gatherers (blue in fig. 3) than with the Neolithic farmers (green in fig. 3). In particular, the hunter-gatherers from Karelia, Loschbour, and Motala across the Eastern, Western, and Northern Europe, respectively, had the highest values of shared drift during this time period (outgroup f3 = 0.245, 0.238, and 0.238; standard errors = 0.0029, 0.0031, and 0.0028, respectively), compared with Neolithic individuals (outgroup f3 ranged from 0.222 to 0.229; standard errors of 0.002 to 0.003). The more intermediate values observed for later time periods might reflect the documented mixing between hunter-gatherers, Eastern pastoralists, and Neolithic farmers in Europe and their subsequent dispersal (Lazaridis et al. 2014; Haak et al. 2015; Mathieson et al. 2015).
Genetic Relationship with Archaic Hominin Individuals
Past studies of Neandertal genomes have shown that the East Asians have inherited ∼20% more Neandertal ancestry than Europeans and that this excess ancestry may reflect a second pulse of admixture in East Asians or a dilution of Neandertal admixture in Europeans (Prufer et al. 2014; Sankararaman et al. 2014, 2016; Vernot and Akey 2014, 2015; Kim and Lohmueller 2015). We largely recapitulated the relationship of a number of Neandertal samples and Denisovan to the Han Chinese as previously reported (supplementary fig. S9, Supplementary Material online). We observed subtle differences in allele-sharing pattern and estimated Neandertal ancestry (∼1.8–2%) across China, though the difference is not significant after correcting for multiple testing (supplementary table S8, Supplementary Material online).
Previous analyses of the locations of Neandertal segments within the genomes of non-African individuals indicated that some of the Neandertal variants were adaptively beneficial while the bulk of Neanderthal variants were deleterious in the modern human genetic background (Harris and Nielsen 2016; Juric et al. 2016). Specifically, a recent examination of Neandertal-informative markers (NIMs) among large cohort of Europeans showed that these markers explained some proportions of the phenotypic risk of a number of diseases in the electronic health record (Simonti et al. 2016), including MDD. We sought to replicate this finding in East Asians as our data set was originally ascertained as a case–control study of MDD in Han Chinese women (Cai et al. 2015).
We extracted 75,539 SNPs that were previously identified to tag Neandertal haplotypes in East Asian individuals in the 1KG project (Sankararaman et al. 2014), and assessed the contribution of these NIMs to depression in our cohort consisting 5,224 cases of MDD and 5,218 controls. The allele frequencies of these NIMs are highly correlated (r = 0.951) between our cohort and 1KG, suggesting that the NIMs are not overt outliers from the rest of the variants in our data set in terms of data quality. We tested the association between the NIMs and depression by performing a logistic regression of depression, controlling for age and the first ten PCs, for MDD and Melancholia. Using the current sample size and sequence data, we found no association surviving the Bonferroni correction (supplementary fig. S10, Supplementary Material online) and the QQ plots did not reveal any systematic inflation nor significant enrichment among top associated SNPs (data were not shown).
We also calculated the proportion of phenotypic variance explained by these NIMs using GCTA (Yang et al. 2011) for MDD. We used a prevalence of 7.5% to transform the heritability to the liability scale. We found that the variance explained by the NIMs is ∼1%, which is different from that reported in Simonti et al. (∼2%) and is not significantly different from 0 (P = 0.12). Repeating the analysis with NIMs with MAF >0.01 as well as with no covariates did not qualitatively alter the results (supplementary table S9, Supplementary Material online). Finally, we found that the heritability explained by NIMs is not significantly different from that of a background set of SNPs chosen at random to match the NIMs by derived allele frequency decile and by Linkage Disequilibrium (LD) scores (P > 0.4). Our analysis may be underpowered given the smaller sample size and low coverage, but the results could suggest that the impact of Neandertal ancestry on MDD differs between European and Han Chinese. Future investigation in larger cohorts will be informative.
PCA-Based Signal of Selection
Taking advantage of the greater geographical resolution of our data set, we conducted a genome scan for loci under directional selection within the Han Chinese populations. We used a recently published PCA-based method (Galinsky et al. 2016) and identified 24 loci showing genome-wide significant allele frequency differentiations in the top two PCs (supplementary figs. S11 and S12, Supplementary Material online). Some of the loci appeared significant in both PCs, and a number of them had been previously reported to lie in the tail end of haplotype-based selection statistics in East Asian populations (tables 2 and 3), thereby supporting the robustness of our approach here. A number of our identified loci are related to diet, UV radiation, and immune responses, the major selective forces known to impact global human populations. However, previous studies are often based in non-East Asian populations, or compared East Asian populations as a whole to non-East Asian populations; our results here suggest that these loci show within-China signal of selection.
Table 2.
Putative Loci under Selection on PC1.
| Chr | Start (Mb) | Stop (Mb) | Locus Size (Mb) | Lead SNP | P-Value | Previously Reported? | Notable Genes | Notable Phenotype Associations |
|---|---|---|---|---|---|---|---|---|
| 1 | 11.84 | 12.00 | 0.161 | 1:11856378 | 4.45E-11 | C1orf167, MTHFR, CLCN6 | Homocysteine levels | |
| 1 | 207.66 | 207.80 | 0.143 | 1:207694357 | 2.07E-08 | CR1 | ||
| 3 | 75.59 | 75.76 | 0.163 | 3:75632610 | 9.59E-12 | |||
| 3 | 162.37 | 163.25 | 0.881 | 3:162732731 | 1.41E-14 | |||
| 6 | 29.72 | 30.47 | 0.747 | 6:29878687 | 5.26E-13 | MHC, Liu et al.a, Suo et al.a | MHC region | |
| 6 | 31.31 | 31.35 | 0.04 | 6:31313972 | 1.53E-10 | MHC, Liu et al.a, Suo et al.a | MHC region | |
| 6 | 32.68 | 32.68 | 0.001 | 6:32682207 | 1.16E-08 | MHC, Liu et al.a, Suo et al.a | MHC region | |
| 11 | 61.52 | 61.69 | 0.176 | 11:61579463 | 7.80E-14 | Suo et al.a, Kothapalli et al.a | MYRF, TMEM258, FEN1, FADS1, FADS2, FADS3, RAB3IL1 | |
| 13 | 99.60 | 99.77 | 0.17 | 13:99759875 | 2.02E-11 | Suo et al.a | DOCK9, DOCK9-AS2 | |
| 14 | 105.91 | 106.38 | 0.479 | 14:106134635 | 9.34E-32 | IGH cluster | ||
| 15 | 24.34 | 24.97 | 0.639 | 15:24341809 | 1.26E-22 | PWRN2, PWRN3, PWRN1, NPAP1 | ||
| 19 | 54.74 | 54.80 | 0.057 | 19:54800371 | 2.80E-26 | Hirayasu et al. | LILRB5, LILRB2, LILRA3 | |
| 20 | 0.77 | 0.78 | 0.011 | 20:773680 | 1.09E-08 |
Note.—Locus intervals are defined as the interval at which the P-value for evidence of selection drops below 0.05. Lead SNP is the SNP with the best P-value. If the interval overlapped with previous selection scans involving East Asian samples, the references are given (Hirayasu et al. 2008; Suo et al. 2012; Liu et al. 2013; Kothapalli et al. 2016). Notable genes column lists only protein coding genes for which a variant with P < 1e-7 was mapped and annotated by VEP to its canonical transcript. Notable phenotype associations are phenotypes reported to be associated with the genes in the GWAS catalog (Welter et al. 2014).
Corroboration by a haplotype-based statistic.
Table 3.
Putative Loci under Selection on PC2.
| Chr | Start (Mb) | Stop (Mb) | Locus Size (Mb) | Lead SNP | P-Value | Previously Reported? | Notable Genes | Notable Phenotype Associations |
|---|---|---|---|---|---|---|---|---|
| 3 | 75.45 | 75.95 | 0.504 | 3:75599989 | 3.11E-32 | ALG1L6P, FAM86DP, ENPP7P2, ZNF717 | ||
| 3 | 162.12 | 163.26 | 1.139 | 3:162446617 | 2.45E-40 | |||
| 4 | 9.77 | 10.42 | 0.651 | 4:10065873 | 3.33E-12 | SLC2A9, WDR1 | ||
| 4 | 100.32 | 100.45 | 0.125 | 4:100332865 | 2.37E-08 | Suo et al.a, Grossman et al.a, Higasa et al.a | ADH7 | Upper aerodigestive tract cancer |
| 5 | 144.22 | 144.29 | 0.068 | 5:144244236 | 2.15E-08 | |||
| 6 | 32.50 | 33.18 | 0.678 | 6:32632189 | 2.66E-17 | HLA region | ||
| 7 | 158.27 | 158.59 | 0.322 | 7:158366586 | 1.76E-12 | PTPRN2, THAP5P1, NCAPG2, ESYT2 | ||
| 8 | 3.16 | 3.32 | 0.164 | 8:3177530 | 5.96E-13 | CSMD1 | Age of menarche; schizoprenia | |
| 9 | 1.54 | 1.84 | 0.299 | 9:1626977 | 1.45E-15 | |||
| 12 | 40.49 | 40.82 | 0.328 | 12:40586433 | 8.15E-09 | LRRK2b, SLC2A13b | Inflammatory bowel disease, Parkinson Disease, Crohn’s Disease | |
| 14 | 106.07 | 106.16 | 0.088 | 14:106092003 | 7.79E-09 | IGH cluster | ||
| 15 | 24.35 | 24.82 | 0.472 | 15:24341825 | 5.48E-15 | PWRN2, PWRN3, PWRN1, NPAP1 | ||
| 16 | 20.55 | 20.61 | 0.056 | 16:20599947 | 3.01E-08 | ACSM2B | ||
| 16 | 69.54 | 70.08 | 0.538 | 16:70070508 | 1.62E-09 | NFAT5, NOB1, WWP2, SNORA62, CLEC18A, PDXDC2P | Age of menarche | |
| 18 | 14.68 | 15.02 | 0.341 | 18:14696035 | 1.90E-10 | ANKRD30B | ||
| 19 | 54.74 | 54.80 | 0.057 | 19:54800371 | 1.19E-14 | Hirayasu et al. | LILRB2, LILRA3 | |
| 22 | 30.45 | 30.60 | 0.157 | 22:30599596 | 3.41E-10 | HORMAD2 | T1D, inflammatory bowel disease, Crohn’s Disease |
Note.—Locus intervals are defined as the interval at which the P-value for evidence of selection drops below 0.05. Lead SNP is the SNP with the best P-value. If the interval overlapped with previous selection scans involving East Asian samples, the references are given (Hirayasu et al. 2008; Higasa et al. 2009; Suo et al. 2012; Grossman et al. 2013). Notable genes column lists only protein-coding genes for which a variant with P < 1e-7 was mapped and annotated by VEP to its canonical transcript. Notable phenotype associations are phenotypes reported to be associated with the genes in the GWAS catalog (Welter et al. 2014).
Corroboration by a haplotype-based statistic.
Cases where no genes were identified based on the above criteria, and thus the closest genes are given.
A well-known selective pressure in human evolution is the interaction with pathogens. We identified a number of signals related to immune response. In addition to the expected signal from the human leukocyte antigen (HLA) region, we also identified the LILRA3/LILRB2 locus on chr19, which encodes a family of HLA class-I recognizing receptors, the CR1 locus on chr1, which encodes the human complement receptor 1 gene, and the IGH locus on chr14, the immunoglobulin heavy cluster (supplementary figs. S13–S15, Supplementary Material online) (Cockburn et al. 2004; Barreiro et al. 2008; Hirayasu et al. 2008; Galinsky et al. 2016). Adaptation to different diets is another well-known selective force of human evolution. We identified two loci related to diet: The FADS1, 2, 3 locus on chr11 and the ADH gene cluster on chr4. The FADS locus encodes a cluster of fatty acid desaturases, which modulate omega-3 polyunsaturated fatty acids levels, and is known to be selected in multiple populations around the globe (Fumagalli et al. 2015; Mathieson et al. 2015; Kothapalli et al. 2016; Ye et al. 2017). The ADH locus encodes a cluster of alcohol dehydrogenases involved in alcohol metabolism. Our lead SNP resides in ADH7 and is in poor LD (r2 ∼ 0.18) with the previously identified variant in ADH1B that was shown to be under selection in East Asians and Europeans (Han et al. 2007; Li, Gu, et al. 2011; Galinsky et al. 2016). The 99% credible set of variants responsible for this signal from fine-mapping analysis excluded previously known functional variants in ADH1B (supplementary fig. S16, Supplementary Material online), thus suggesting a different Han-Chinese-specific signal of selection. Lastly, we noted a signal on chr1 in the MTHFR gene encoding a methylenetetrahydrofolate reductase involved in folate metabolism (table 2). This signal may represent a complex interaction between UV radiation (to prevent photolysis of folate) and dietary folate intake (Rosenberg et al. 2002; Jablonski and Chaplin 2010).
Discussion
By analyzing the low-coverage whole-genome sequences of 11,670 Han Chinese individuals, we cataloged the most comprehensive map of genetic variation currently available in the Han Chinese population. We used this catalog to characterize population structure, admixture history, and infer signals of natural selection. Because of the very low coverage in generating this data set, we focused on using allele-frequency-based methods to address these questions. Despite this potential limitation, our results have improved our understanding of one of the largest, yet understudied, populations in the world, which will pave the way for future medical and population genetic studies of Han Chinese.
Analyses of exome sequencing studies, primarily in Europeans, have noted a number of erroneous entries in clinical genetics databases, such as ClinVar, where reported pathogenic variants appear to be too common in the population to be truly pathogenic. We cross-referenced the catalog of Han Chinese variation with the ClinVar database, looking for incidences where reported pathogenic variants are extremely rare in European populations (hence likely to be still considered pathogenic based on ExAC), but are more common in Han Chinese. We identified four such pathogenic or likely pathogenic variants in the ClinVar database (table 1). Among them, the missense variant in SLC7A14 (rs2276717) is a singleton in ExAC NFE (MAF ∼1.5 × 10−5), but has a frequency of 0.048 in Han Chinese. The variant is reported to cause an autosomal recessive form of retinitis pigmentosa (Jin et al. 2014). However, given the prevalence of ∼1 in 4,000 in China (You et al. 2013), which is on par with the estimates in the United States. (https://nei.nih.gov/health/pigmentosa/pigmentosa_facts), the maximum credible population allele frequency would be ∼0.022 in Han Chinese even under relatively relaxed assumptions of genetic architecture (see Materials and Methods). Therefore, this variant appears to be too common and unlikely to be pathogenic in the Han Chinese population, or the disease is underdiagnosed in China. Incidentally, the observed frequency in ExAC East Asians is closer to the maximum credible population frequency (0.027, but likely a mixture of Han Chinese and other East Asians). Thus, the ExAC frequency would not be as confidently filtered in a screen for pathogenic variants.
The example of rs2276717 notwithstanding, interpretation of the variants listed in Table 1 should be taken with caution. For example, rs671 in ALDH2 for alcohol dependence is a well-characterized functional variant in the alcohol metabolism pathway. It appears to be selected in East Asian populations (Oota et al. 2004) and thus it would not be surprising that the frequency is uniquely high in East Asians while being truly causal. Another example, a missense variant in GJB2, rs72474224, is reported as likely pathogenic for autosomal recessive form of nonsyndromic sensorineural hearing loss and deafness by ClinVar. The variant is rare in non-East Asian populations in ExAC (<0.001), but is common in ExAC East Asian (0.072) and Han Chinese in our data set (0.055). However, the prevalence of nonsyndromic hearing loss is also known to be higher in East Asian populations (Naeem and Newton 1996), and thus the observed frequency in Han Chinese may still be consistent with its pathogenicity. Taken together, these findings suggested that population-specific prevalence and disease risk should be taken into account when screening clinically sequenced genomes for pathogenic variants.
In terms of population structure, the North-to-South cline among the Han Chinese has been described in both studies of uniparental markers (Wen et al. 2004; Zhao et al. 2015) and genome-wide array data (Chen et al. 2009, 2016; Xu et al. 2009). We additionally observed, for the first time to our knowledge, population structure in the East-to-West axis among the Han Chinese. Moreover, we demonstrated that the general migration pattern sufficiently followed an isolation-by-distance model (fig. 1). Despite this apparent structure among the Han Chinese, however, it was also apparent that as a whole that Han Chinese exhibited relatively low levels of differentiation. This suggests continuous gene flows across China, consistent with a number of recorded large migration waves in Han Chinese history due to wars, invasions, or expansions. For example, the Northeastern part of China today, also known as Manchuria, has not always been under the direct control of Han Chinese dynasties and was closed to settlement by Han Chinese during the Qing dynasty. Han Chinese migrated into the region only starting in the late 19th century, toward the end of the Qing dynasty (Reardon-Anderson 2005). This may explain the very low genetic differentiation observed in Fst estimates and PCA among the Northern and Northeastern Han Chinese (fig. 1 and supplementary fig. S3, Supplementary Material online).
When examining patterns of admixture among Han Chinese, we found signals that were geographically localized to certain parts of China (supplementary table S4, Supplementary Material online, and fig. 2). These signals typically involved nearby non-Han Chinese populations. Note that a significant negative f3-statistic indicates that the target population is admixed, but the admixing source populations may not be the actual parental populations (Patterson et al. 2012). This would explain the large number of possible source population pairs producing significant admixture signal in our analysis (supplementary table S4, Supplementary Material online). Taken together, the signals we have detected through f3-statistics were also consistent with a model of continuous and frequent exchange of migrants with neighboring populations throughout history, rather than a single-pulse admixture model typically invoked for populations such as the African Americans. Indeed, one should be cautious when interpreting the admixture results via f3-statistics (Peter 2016).
One finding from our admixture analysis that may fit a one-pulse model was our observation of admixture from Northern/Western European populations to the Northwestern provinces of China (Gansu, Shaanxi, and Shanxi). Previous analysis of the HGDP data, based on patterns of haplotype sharing among ten individuals, estimated a single pulse of ∼6% West Eurasian ancestry among the Northern Han Chinese. The estimated date of admixture was around 1200 CE. This signal was also observed among the Tu people, an ethnic minority from Northwestern China; the authors attributed this signal to contact through the Silk Road (Hellenthal et al. 2014). We estimate a lower bound of admixture proportion due to Northern Europeans at ∼2–5%, with an admixture date of about 26 ± 3 generations for Gansu, and 47 ± 3 generations for Shaanxi (supplementary table S10, Supplementary Material online). Using a generation time of about 26–30 years, these estimates suggest admixture events occurred at around 1300 CE and 700 CE, respectively, corresponding roughly to the Yuan and Tang dynasties in China. However, these estimated dates should be interpreted with care, as both the violation of a single-pulse admixture model and the additional noise in intermarker LD estimates due to low-coverage data could impact the estimates. Moreover, an alternative model should also be considered, in that the admixture signal is induced by ancestries due to ancient, not-yet-sampled, Central Asians that are shared by both Northern Europeans and Northwestern Han Chinese.
Ancient DNA studies have shed light on the origin of agriculture in Europe. Agriculture likely arrived in Europe due to demic diffusion from Anatolia (Mathieson et al. 2015), and arrived in South Asia due to demic diffusion from Iran (Lazaridis et al. 2016). The prevailing view in the origin of agriculture in China is that domestication of crops and animals occurred independently from other agricultural centers around the world (Ho 1976; Jones and Liu 2009; Zhao 2011). Consistent with these views, we found that present-day Han Chinese showed greater genetic affinity to Mesolithic European hunter-gatherers than Neolithic European farmers from the same era (fig. 3), and showed very low affinity with the Iranian Farmers (fig. 3). Currently available ancient DNA data are largely of European origin, thus provide limited information on the spread of the Han Chinese people that may be coupled with spread of agriculture (Zhao et al. 2015). Furthermore, within China there may be two or more independent centers of domestication: One near the southern Yangtze River where rice was first domesticated, and another one near the northern Yellow River where millets were found (Zhao 2011). Therefore, more ancient DNA data from East Asia will be needed to better understand the extent to which the Han Chinese people replaced or integrated during their expansion and how that coupled with the spread of agricultural practices.
Taking advantage of this geographically diverse data set of Han Chinese, we identified a number of loci in the genome showing extreme frequency differentiation across China. A subset of these loci are likely due to selective pressures such as pathogen, diet, and variation in environmental exposures such as UV radiation. Below we highlight a few of these loci.
Presumably driven by diets, the FADS locus appeared adaptive in a number of other populations around the globe, including the Greenlandic Inuits, Europeans, and some East Asian populations (Fumagalli et al. 2015; Mathieson et al. 2015; Kothapalli et al. 2016; Ye et al. 2017). Our top variants in the FADS locus, two intronic variants in FADS1 (chr11:61571478 and chr11:61579463, r2 = 0.99 between the two), are in high LD with one of the top hits reported in Fumagalli et al. (chr11:61597212; r2 = 0.99), while the other five variants reported in Fumagalli et al. showed modest to poor LD with our lead variant here (supplementary fig. S17 and table S11, Supplementary Material online). Because the presumed selected (derived) allele in Inuits, chr11:61597212, is actually higher in frequency among Southern Han Chinese rather than Northern Chinese, yet previous haplotype-based analysis suggested that the ancestral allele is under selection in Northern Chinese and Japanese (Kothapalli et al. 2016), it remains possible that opposite forces are favoring the two alternative alleles to be selected in Northern versus Southern Han Chinese. The opposite forces may be diet-driven, reminiscent to varying selection on the two alleles in FADS locus depending on different dietary practices in Europe (Ye et al. 2017).
Another well-studied locus is in MTHFR. Our top differentiated SNP is a missense variant (rs1801133, 1:11856378), which modulates the activity of the enzyme by producing a thermolabile derivative with reduced activity (Frosst et al. 1995) and is associated with plasma homocysteine levels (Pare et al. 2009; van Meurs et al. 2013). The thermolabile (derived) allele frequency ranges from ∼30% to ∼60% in our samples from Southern and Northern China, respectively, consistent with previously reported South-to-North increasing frequency gradient in East Asia. This is in opposite direction to the observed North-to-South increasing gradient in Europe (Yafei et al. 2012). The selection has been hypothesized to be driven by protection against photolysis of folate (Jablonski and Chaplin 2010), and the opposite frequency gradient between Europe and China could be explained by the relative difference in latitude between China and Europe (Yafei et al. 2012). It should also be noted that dietary supplementation of folate could stabilize the thermolabile protein (Frosst et al. 1995). In cases where folate is plentiful in the diet, the impact of the thermolabile allele on recurrent pregnancy loss is reduced (Munoz-Moran et al. 1998). Thus, in principle, differential dietary practices across Han-Chinese could lead to the latitudinal gradient we observe here, and interactions between UV exposure and diet cannot be ruled out.
We also detected a signal at the ADH locus, which is near but different from the well-characterized variant in ADH1B locus (rs1229984, chr4:100239319, Arg48His allele) for alcohol metabolism. The ADH1B Arg48His variant is suggested to be positively selected based on comparisons between East Asians and Europeans (Han et al. 2007), as well as within Europeans (Galinsky et al. 2016). Our lead variant (rs422143, chr4:100332865) is downstream of ADH7, ∼90 kb away from ADH1B, and showed much stronger signal (P ∼ 2.4 × 10−8) than any variant in the ADH1B locus. It is in poor LD (r2 ∼ 0.18) with the rs1229984, which only showed marginal evidence of selection among our Han Chinese samples (P ∼ 0.00096). Fine-mapping analysis also excluded rs1229984 from the 99% credible set of causal variant in this locus (supplementary fig. S16, Supplementary Material online). A separate regulatory variant in the ADH1B locus, rs3811801 (chr4:100244319), was previously suggested to be a younger variant showing stronger differentiation within East Asia than rs1229984 (Li, Gu, et al. 2011; Galinsky et al. 2016). This variant was in slightly higher LD to our lead variant in ADH7 (r2 ∼ 0.33) and exhibited better evidence of selection (P ∼ 6.1 × 10−6). However, in fine-mapping analysis this variant was also excluded from the 99% credible set unless one assumes two causal variants in the locus (supplementary fig. S16, Supplementary Material online).
Additionally, there are known functional differences between ADH7 and ADH1B. Unlike ADH1B, ADH7 is mainly expressed in the upper digestive tract where it oxidizes ethanol at high concentrations early in the timeline of alcohol metabolism (Park et al. 2013). The mechanistic differences explain why the associations of variants in ADH1B with UADT cancer (squamous cell carcinoma of the upper aerodigestive tract, encompassing oral cavity, pharynx, larynx, and esophagus) were mediated through alcohol consumption behaviors, while the associations of variants in ADH7 with UADT were not (McKay et al. 2011). Therefore, it is conceivable the ADH7 selective signal is specific to Han Chinese and is independent from previously reported signal in ADH1B. The potential selective pressures for alcohol dehydrogenase variants in East Asia include protection against infectious agents (due to increased acetaldehyde levels; Han et al. 2007) and rice domestication (Peng et al. 2010), which spread along the East–West axis of China. Consistently, our detected signal in ADH7 is found on PC2.
Notably, ADH7 and MTHFR have been recently implicated as under selection in Tibetans when comparing them with Han Chinese (Yang, Jin, et al. 2017). Our results suggest a more general signal among Han Chinese that might reflect a continuously varying process across geography among the Han Chinese; this signal could be missed if one only compares discrete populations. Beyond the loci described here, there are a number of potentially novel loci found with little recognizable insights to the biological mechanisms or selective pressure behind the extreme differentiation. Some of the genes within the selection peak or nearby have been implicated in GWAS for life-history traits such as age of menarche, or immune-related traits such as inflammatory bowel disease or Crohn’s disease. However, we note that there may be Han Chinese-specific, yet uncharacterized, regions of long-range LD that could appear like a region under selection in this analysis.
A major limitation to our study is the reliance on allele frequency-based methods for analysis, as the very low-coverage data obscured the haplotypic patterns at the individual level and precluded direct merging and comparison to other reference data sets. Haplotype-based analysis may reveal additional structure beyond the two geographical axes found here. Nevertheless, our analysis framework relying heavily on allele frequency estimates is applicable to future very large-scale (>100,000 individuals) whole-genome sequencing studies, which in the short term will inevitably focus on ultra-low sequencing data such as those obtained through noninvasive prenatal testing. Despite these ongoing challenges to develop and employ appropriate analysis methods, our results collectively demonstrate the existence of significant variations in demographic and adaptive histories across Han Chinese populations. We demonstrated how the impact due to Neandertal ancestry on one type of trait, MDD and Melancholia, appears to differ between Han Chinese and Europeans. In general, these unique histories undoubtedly contributed to the variation of phenotype within Han Chinese as well as between Han Chinese and other global populations. Therefore, a better understanding of Han Chinese history will help in conducting and interpreting future medical genetic studies within the largest ethnic group of mankind.
Materials and Methods
Sample Collection, DNA Sequencing, and Variant Calling
The data set we analyzed consists of previously collected cases of recurrent major depression from 58 provincial mental health centers and psychiatric departments of general medical hospitals in 45 cities of China, and previously collected controls from patients undergoing minor surgical procedures at general hospitals or from local community centers. All subjects were self-reported Han Chinese women born in China with four Han Chinese grandparents. The descriptions of DNA sequencing, variant calling, and data access have been previously published (Cai et al. 2015, 2017). All coordinates are reported with respect to human reference build GRCh37/hg19.
Evaluating ClinVar Variants
The curated ClinVar database was downloaded from https://github.com/macarthur-lab/clinvar on April 27, 2017. We examined variants annotated as pathogenic or likely pathogenic, with no conflicting reports, with low frequency (≤0.01) in ExAC NFE, but common (≥0.05) in Han Chinese. Because of its near-absence in non-East Asian populations in ExAC, we used an online application (http://cardiodb.org/allelefrequencyapp/; Whiffin et al. 2017) to further examine rs2276717 reported in ClinVar to cause autosomal recessive retinitis pigmentosa. We assumed an autosomal recessive model, with a prevalence of ∼1 in 4,000, genetic heterogeneity of 0.02 (Jin et al. 2014), and a range of possible parameters for the allelic heterogeneity (from 0.01 to 1) and the penetrance (from 0.01 to 0.1). Under the most liberal setting (allelic heterogeneity = 1 and penetrance = 0.01), we find the maximum credible population allele frequency is 0.0224.
Frequency-Based Population Genetic Analysis
For analyses investigating the genetic relationship between Han Chinese in our data set and external reference populations, including ancient and archaic individuals, we merged our data set with the Human Origins Array data (N = 2,583; Lazaridis et al. 2016) and the 1KG phase 3 data (N = 2,504; 1000 Genomes Project Consortium et al. 2015). We further supplemented this merged data set with ancient samples not found in the public release of Human Origins Array, including ancient Europeans from the Ice Age (Fu et al. 2016), two ancient East Asian populations (Siska et al. 2017; Yang, Gao, et al. 2017), and the high-coverage Vindija 33.19 individual (Prufer et al. 2017) (http://cdna.eva.mpg.de/neandertal/Vindija/VCF/Vindija33.19/, accessed prepublication on July 15, 2016). SNP intersections of merging data sets were generated after removing apparent triallelic SNPs, SNPs with inconsistent alleles, and A/T or C/G transversion SNPs. In total, we analyzed 449,336 (out of 592,146 available) SNPs shared between Human Origins Array data and our data set, across 15,876 individuals.
Given the exceedingly high correlation of allele frequency estimates of this data set and ExAC (r > 0.99), we relied heavily on frequency-based methods to infer signals of admixture, allelic sharing, and relationships with other modern and ancient populations. Comparisons of f3-statistics with data from worldwide populations from Human Origins Array data showed no difference whether we used CHB population from 1KG or individuals from Beijing in our data set (data were not shown), suggesting that these analyses are robust to the impact of very low-coverage sequencing.
To test for admixture, we computed f3(Han Chinese; reference1, reference2), iterating through all possible pairs of reference populations found in the merged data. A significantly negative value of f3 is suggestive of admixture. For specific illustrations of regional signals, we further corroborated the observed evidence of admixture by computing D(Mbuti, X, Y, Metropolitan Chinese). Here, X was a potential admixing source population of interest from the Human Origin Array data, while Y was a specific Chinese province in our data set. The Metropolitan Chinese population was based on a random sample of 200 individuals from the four municipalities in China (Tianjin, Beijing, Shanghai, and Chongqing), used as representative of average Chinese (these four populations were not tested for admixture). Therefore, this D-statistic tested localized signals of admixture by excess of allele sharing between non-Han population X and Han Chinese province Y, compared with the metropolitan Chinese individuals. To evaluate shared history between ancient populations and Han Chinese as measured by allele frequency covariances, we used f3(Outgroup; Y, Ancient), where the Human Origin Array Mbuti population was used as the outgroup. We used qp3Pop and qpDstat package implemented in ADMIXTOOLS (v3.0). Statistical significance was assessed using the default blocked jackknife approach implemented in ADMIXTOOLS.
We estimated Neandertal ancestry in a population using a statistic analogous to one previously proposed (Mallick et al. 2016). For a target population t, we defined a set of SNPs, , where all 43 African genomes from 17 population in the Simons Genome Diversity Project are ancestral, a randomly picked allele from the Altai Neanderthal (Prufer et al. 2014) is derived while a randomly picked allele from the high-coverage Denisovan individual (Meyer et al. 2012) is ancestral and not all genotypes in t are missing. We then estimated Neanderthal ancestry in population t as:
Here, is the derived allele frequency at SNP i in population t. We converted this into an estimator of Neanderthal ancestry, , by rescaling the statistics so that population from Beijing was assigned an ancestry of 1.89% to match previous estimates (Prufer et al. 2014). The estimator is not an unbiased estimate of Neanderthal ancestry, but differences in these statistics (using Beijing as the baseline) are informative of differential relationships to the Neanderthals between populations. We assessed statistical significance using a block jackknife with 10-Mb blocks.
PCA-Based Analysis of Population Structure and Natural Selection
While our frequency-based analyses were robust to the impact of very low-coverage sequencing because only aggregate data were used, exploratory PCA using individual level genotypes showed a number of individuals clustered along PC2 due to extremely low-coverage and consequently poorer genotype/imputation qualities. Therefore, for PCA-based analysis we restricted our analysis to genotypes of 429,935 SNPs, representing a random selection of a tenth of all SNPs with MAF ≥ 10%; common SNPs have much higher genotyping accuracy at the individual level in a very low-coverage sequencing design (Cai et al. 2017).
We then constructed a classifier to select a subset of individuals least biased by poorly imputed genotypes for analysis. Specifically, we first randomly selected 500 samples of apparent poor quality (outliers on PC2) and 2,000 samples of apparent good quality (nonoutliers). We trained a univariate logistic model for each of 26 individual-level QC features of the sequence data generation process, and evaluated the classifier in 10-fold cross-validation. Because the 26 QC features were substantially correlated with each other, we greedily pruned the features such that no feature was correlated with any other feature with r2 ≥ 0.2, while preferentially retained the feature with best accuracy in classification. In the end, we retained six features (proportion of genotypes with maximum posterior genotype probability ≤ 0.9, proportion of reads passing sequencing machine default QC, rate of reads mapped by Stampy, proportion of reads with low quality scores, raw number of mitochondrial reads, and number of transversion singletons) to build a multivariate logistic model. The model estimates a classification probability for each individual in our reference sample, and we chose a threshold based on this probability that would achieve 90% sensitivity of selecting poor quality sample in 10-fold cross-validation. We then used this model and threshold to classify the remaining 8,140 individuals not used in the training set. We additionally filtered individuals with inconsistent or missing demographic labels, individuals due to extended relatedness (removing third-degree relationships by KING v1.4; Manichaikul et al. 2010), and population outliers in a second round of PCA analysis (±4 SD in any of the top ten PCs). In total, we removed 3,183 individuals and retained 7,457 individuals for PCA-based analysis (supplementary table S4, Supplementary Material online).
PCA was conducted using EIGENSTRAT (v6.1). Following previous practice (Novembre et al. 2008), we removed one SNP of each pair of SNPs with r2 ≥ 0.8 (in windows of 50 SNPs and steps of 5 SNPs) as well as SNPs in regions known to exhibit extended long-range LD (Price et al. 2008) prior to analysis. We used each person’s self-reported birthplace at the province level as labels. To quantify the correlation between PCs and geography, we computed Pearson’s correlation coefficient between the median PC value for each population with their latitude and longitude. Furthermore, we computed the Procrustes similarity statistics (Wang et al. 2010) by superimposing the population-level coordinates from the first two PCs onto the latitude and longitude coordinates after applying Gall–Peters projection to the geographical coordinates. Statistical significance was assessed by 100,000 permutations of the population label associated with each individual.
To test an isolation-by-distance model, we computed for each pair of populations in our data set (except for Guangxi and Hainan, due to their small sample sizes) the median genetic relatedness based on the genetic relationship matrix used in PCA. We correlated the pairwise relatedness with the great circle distance between the pair. We used the t-test to examine whether the genetic relatedness among the 20% most geographically proximal pairs were larger than the middle 20% pairs of populations. Moreover, we used the Mantel test to compare the genetic relationship and the geographical distance matrices. Significance was assessed by 10,000 rounds of permutation as implemented in the “vegan” package in R.
To identify loci under selection, we considered only the top two PCs as these reflected geographical structure. We initially screened 429,935 SNPs used in the PCA, using a genome-wide significance threshold of 5.8 × 10−8 (corresponding to Bonferroni correction for testing 430K SNPs and 2 PCs), and then followed up each significant locus using all SNPs available in our data with MAF ≥1% in order to narrow the list of possible causal variants. PCA-based selection statistics was calculated using the “pcaselection” program in Eigenstrat (v6.1.1) (Galinsky et al. 2016). We further fine-mapped the ADH locus using CAVIARBF (Chen et al. 2015) to delineate the selection signal attributable to ADH7 and ADH1B. We included all variants (MAF ≥ 1%) spanning ADH locus plus 100 kb upstream and downstream (chr4:99892130–100456291, which includes other genes). Signed Z-scores were computed from the Eigenstrat output. We applied CAVIARBF using in-sample signed variant correlation matrix, assuming either one or two causal variants in the region, and setting σa to be 0.1 per user manual. (Repeating analysis with σa = 0.2 did not qualitatively change the results.) Default priors were used in the CAVIARBF model selection step.
Estimating Admixture Dates and Proportions
We used ALDER v1.03 (Loh et al. 2013) to estimate the date of the admixture and an f4 ratio to estimate the admixture proportions (qpF4ratio in ADMIXTOOLS) (Patterson et al. 2012). For ALDER, we looked for successful fit of LD-decay curves corresponding to a pair of Northern European and Southern Chinese population serving as the proxies of admixing source populations to the Northwestern provinces (Gansu, Shaanxi, and Shanxi) where we detected admixture signals. We report the fit with largest amplitude from fitting of exponential curves from two source populations that corresponded to similar geographical regions as the two source populations reported by f3-test (as the exact two source populations may not produce a successful fit). We used the same set of individuals used in PCA analysis to reduce the noise in LD estimates. For f4-ratio test, we used the two source populations inferred from f3-test or from ALDER, and the Sardinians and Chimp sequences as the outgroups to European source population and human populations, respectively. The Sardinians were used as an outgroup of choice because Southern Europeans in general did not contribute to the admixture signals we detected among Han Chinese, and that the isolated Sardinians more likely reflect the genetic ancestry of ancient Southern Europe or Near East Anatolia (Chiang et al. 2018). Replacing Sardinians with Sicilians or Early Neolithic Farmer samples did not significantly change the results.
Supplementary Material
Acknowledgments
The authors would like to thank Na Cai, Warren W. Kretzschmar, Kenneth S. Kendler, Jonathan Flint, and members of the entire CONVERGE consortium for discussion, clarifications and assistance in processing the whole genome sequencing data, Joseph H. Marcus, Daniel Cook, and John Novembre for assistance in posting the allele frequency estimates to the Geography of Genetic Variants browser, and Linda Polfus for comments of the manuscript. C.W.K.C. was supported in part by National Institute of Health grant T32NS048004. C.R.R was supported in part by National Institute of Health grant T32HG002536. S.S. was supported in part by National Institute of Health grants R00GM111744 and R35GM125055. S.M. was supported by a Quantitative Computational Biology (QCB) Collaboratory Postdoctoral Fellowship and the QCB Collaboratory community. S.M. is also supported by National Science Foundation grants 0513612, 0731455, 0729049, 0916676, 1065276, 1302448, 1320589 and 1331176, and National Institute of Health grants K25HL080079, U01DA024417, P01HL30568, P01HL28481, R01GM083198, R01ES021801, R01MH101782 and R01ES022282.
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. 2015. A global reference for human genetic variation. Nature 526(7571): 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L.. 2008. Natural selection has driven population differentiation in modern humans. Nat Genet. 40(3): 340–345. [DOI] [PubMed] [Google Scholar]
- Cai N, Bigdeli TB, Kretzschmar WW, Li Y, Liang J, Hu J, Peterson RE, Bacanu S, Webb BT, Riley B, et al. 2017. 11,670 whole-genome sequences representative of the Han Chinese population from the CONVERGE project. Sci Data. 4:170011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cai N, Bigdeli TB, Kretzschmar WW, Li Y, Liang J, Song L, Hu J, Li Q, Jin W, Hu Z, et al. 2015. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523(7562): 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Yang JH, Chiang CWK, Hsiung CN, Wu PE, Chang LC, Chu HW, Chang J, Song IW, Yang SL, et al. 2016. Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum Mol Genet. 25(24): 5321–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zheng H, Bei JX, Sun L, Jia WH, Li T, Zhang F, Seielstad M, Zeng YX, Zhang X, et al. 2009. Genetic structure of the Han Chinese population revealed by genome-wide SNP variation. Am J Hum Genet. 85(6): 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Larrabee BR, Ovsyannikova IG, Kennedy RB, Haralambieva IH, Poland GA, Schaid DJ.. 2015. Fine mapping causal variants with an approximate Bayesian method using marginal test statistics. Genetics 200(3): 719–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CW, Marcus JH, Sidore C, ABiddanda A, Al-Asadi H, Zoledziewska M, Pitzalis M, Busonero F, Maschio A, Pistis G, et al. 2018. Genomic history of the Sardinian population. Nat Genet. 50(10): 1426–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn IA, Mackinnon MJ, O'Donnell A, Allen SJ, Moulds JM, Baisor M, Bockarie M, Reeder JC, Rowe JA.. 2004. A human complement receptor 1 polymorphism that reduces Plasmodium falciparum rosetting confers protection against severe malaria. Proc Natl Acad Sci U S A. 101(1): 272–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francioli LC, Menelaou A, Pulit SL, van Dijk F, Palamara PF, Elbers CC, Neerincx PBT, Ye K, Guryev V, Kloosterman WP, et al. 2014. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 46(8): 818–825., [DOI] [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP.. 1995. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 10(1): 111–113. [DOI] [PubMed] [Google Scholar]
- Fu Q, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes D, Furtwangler A, Haak W, Meyer M, Mittnik A, et al. 2016. The genetic history of Ice Age Europe. Nature 534(7606): 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, et al. 2015. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349(6254): 1343–1347. [DOI] [PubMed] [Google Scholar]
- Galinsky KJ, Bhatia G, Loh PR, Georgiev S, Mukherjee S, Patterson NJ, Price AL.. 2016. Fast principal-component analysis reveals convergent evolution of ADH1B in Europe and East Asia. Am J Hum Genet. 98(3): 456–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152(4): 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, Besenbacher S, Magnusson G, Halldorsson BV, Hjartarson E, et al. 2015. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 47(5): 435–444. [DOI] [PubMed] [Google Scholar]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522(7555): 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Gu S, Oota H, Osier MV, Pakstis AJ, Speed WC, Kidd JR, Kidd KK.. 2007. Evidence of positive selection on a class I ADH locus. Am J Hum Genet. 80(3): 441–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K, Nielsen R.. 2016. The genetic cost of Neanderthal introgression. Genetics 203(2): 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellenthal G, Busby GB, Band G, Wilson JF, Capelli C, Falush D, Myers S.. 2014. A genetic atlas of human admixture history. Science 343(6172): 747–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higasa K, Kukita Y, Kato K, Wake N, Tahira T, Hayashi K.. 2009. Evaluation of haplotype inference using definitive haplotype data obtained from complete hydatidiform moles, and its significance for the analyses of positively selected regions. PLoS Genet. 5(5): e1000468.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayasu K, Ohashi J, Tanaka H, Kashiwase K, Ogawa A, Takanashi M, Satake M, Jia GJ, Chimge NO, Sideltseva EW, et al. 2008. Evidence for natural selection on leukocyte immunoglobulin-like receptors for HLA class I in Northeast Asians. Am J Hum Genet. 82(5): 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P-T. 1976. The cradle of the East: an Inquiry into the indigenous origins of techniques and ideas of Neolithic and early historic China, 5000-1000 B.C. Chicago: The University of Chicago Press. [Google Scholar]
- Jablonski NG, Chaplin G.. 2010. Colloquium paper: human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci U S A. 107(Suppl 2):8962–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin ZB, Huang XF, Lv JN, Xiang L, Li DQ, Chen J, Huang C, Wu J, Lu F, Qu J.. 2014. SLC7A14 linked to autosomal recessive retinitis pigmentosa. Nat Commun. 5:3517.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Liu X.. 2009. Archaeology. Origins of agriculture in East Asia. Science 324(5928): 730–731. [DOI] [PubMed] [Google Scholar]
- Juric I, Aeschbacher S, Coop G.. 2016. The strength of selection against Neanderthal introgression. PLoS Genet. 12(11): e1006340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Lohmueller KE.. 2015. Selection and reduced population size cannot explain higher amounts of Neandertal ancestry in East Asian than in European human populations. Am J Hum Genet. 96(3): 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli KSD, Ye K, Gadgil MS, Carlson SE, O’Brien KO, Zhang JY, Park HG, Ojukwu K, Zou J, Hyon SS, et al. 2016. Positive selection on a regulatory insertion-deletion polymorphism in FADS2 influences apparent endogenous synthesis of arachidonic acid. Mol Biol Evol. 33(7): 1726–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, et al. 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 536(7617): 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513(7518): 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616): 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gu S, Han Y, Xu Z, Pakstis AJ, Jin L, Kidd JR, Kidd KK.. 2011. Diversification of the ADH1B gene during expansion of modern humans. Ann Hum Genet. 75(4): 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR.. 2011. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 21(6): 940–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ong RT, Pillai EN, Elzein AM, Small KS, Clark TG, Kwiatkowski DP, Teo YY.. 2013. Detecting and characterizing genomic signatures of positive selection in global populations. Am J Hum Genet. 92(6): 866–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Lipson M, Patterson N, Moorjani P, Pickrell JK, Reich D, Berger B.. 2013. Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193(4): 1233–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, et al. 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538(7624): 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM.. 2010. Robust relationship inference in genome-wide association studies. Bioinformatics 26(22): 2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus JH, Novembre J.. 2017. Visualizing the geography of genetic variants. Bioinformatics 33(4): 594–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528(7583): 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JD, Truong T, Gaborieau V, Chabrier A, Chuang SC, Byrnes G, Zaridze D, Shangina O, Szeszenia-Dabrowska N, Lissowska J, et al. 2011. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 7(3): e1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F.. 2016. The Ensembl Variant Effect Predictor. Genome Biol. 17(1): 122.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338(6104): 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Moran E, Dieguez-Lucena JL, Fernandez-Arcas N, Peran-Mesa S, Reyes-Engel A.. 1998. Genetic selection and folate intake during pregnancy. Lancet 352(9134): 1120–1121. [DOI] [PubMed] [Google Scholar]
- Naeem Z, Newton V.. 1996. Prevalence of sensorineural hearing loss in Asian children. Br J Audiol. 30(5): 332–339. [DOI] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, et al. 2008. Genes mirror geography within Europe. Nature 456(7218): 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oota H, Pakstis AJ, Bonne-Tamir B, Goldman D, Grigorenko E, Kajuna SL, Karoma NJ, Kungulilo S, Lu RB, Odunsi K, et al. 2004. The evolution and population genetics of the ALDH2 locus: random genetic drift, selection, and low levels of recombination. Ann Hum Genet. 68(Pt 2): 93–109. [DOI] [PubMed] [Google Scholar]
- Pare G, Chasman DI, Parker AN, Zee RR, Malarstig A, Seedorf U, Collins R, Watkins H, Hamsten A, Miletich JP, et al. 2009. Novel associations of CPS1, MUT, NOX4, and DPEP1 with plasma homocysteine in a healthy population: a genome-wide evaluation of 13 974 participants in the Women's Genome Health Study. Circ Cardiovasc Genet. 2(2): 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, Kang TC, Nam YW, Kim GB, Shin HD, et al. 2013. Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum Genet. 132(6): 657–668. [DOI] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D.. 2012. Ancient admixture in human history. Genetics 192(3): 1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Shi H, Qi XB, Xiao CJ, Zhong H, Ma RL, Su B.. 2010. The ADH1B Arg47His polymorphism in east Asian populations and expansion of rice domestication in history. BMC Evol Biol. 10:15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BM. 2016. Admixture, population structure, and F-statistics. Genetics 202(4): 1485–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, Ge D, Rotter JI, Torres E, Taylor KD, et al. 2008. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 83(1): 132–135; author reply 135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, de Filippo C, Grote S, Mafessoni F, Korlevic P, Hajdinjak M, Vernot B, Skov L, Hsieh P, Peyregne S, et al. 2017. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358(6363): 655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481): 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW Jr, Orlando L, Metspalu E, et al. 2014. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505(7481): 87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon-Anderson J. 2005. Reluctant pioneers: China's expansion northward, 1644–1937. Stanford (CA: ): Stanford University Press. [Google Scholar]
- Rosenberg N, Murata M, Ikeda Y, Opare-Sem O, Zivelin A, Geffen E, Seligsohn U.. 2002. The frequent 5, 10-methylenetetrahydrofolate reductase C677T polymorphism is associated with a common haplotype in whites, Japanese, and Africans. Am J Hum Genet. 70(3): 758–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, Prufer K, Kelso J, Paabo S, Patterson N, Reich D.. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507(7492): 354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Patterson N, Reich D.. 2016. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr Biol. 26(9): 1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidore C, Busonero F, Maschio A, Porcu E, Naitza S, Zoledziewska M, Mulas A, Pistis G, Steri M, Danjou F, et al. 2015. Genome sequencing elucidates Sardinian genetic architecture and augments association analyses for lipid and blood inflammatory markers. Nat Genet. 47:1272–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, et al. 2016. The phenotypic legacy of admixture between modern humans and Neandertals. Science 351(6274): 737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siska V, Jones ER, Jeon S, Bhak Y, Kim HM, Cho YS, Kim H, Lee K, Veselovskaya E, Balueva T, et al. 2017. Genome-wide data from two early Neolithic East Asian individuals dating to 7700 years ago. Sci Adv. 3(2): e1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo C, Xu H, Khor CC, Ong RT, Sim X, Chen J, Tay WT, Sim KS, Zeng YX, Zhang X, et al. 2012. Natural positive selection and north-south genetic diversity in East Asia. Eur J Hum Genet. 20(1): 102–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter K, Min JL, Huang J, Crooks L, Memari Y, McCarthy S, Perry JRB, Xu CJ, Futema M, Lawson D, et al. 2015. The UK10K project identifies rare variants in health and disease. Nature 526(7571): 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meurs JB, Pare G, Schwartz SM, Hazra A, Tanaka T, Vermeulen SH, Cotlarciuc I, Yuan X, Malarstig A, Bandinelli S, et al. 2013. Common genetic loci influencing plasma homocysteine concentrations and their effect on risk of coronary artery disease. Am J Clin Nutr. 98(3): 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Akey JM.. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343(6174): 1017–1021. [DOI] [PubMed] [Google Scholar]
- Vernot B, Akey JM.. 2015. Complex history of admixture between modern humans and Neandertals. Am J Hum Genet. 96(3): 448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Szpiech ZA, Degnan JH, Jakobsson M, Pemberton TJ, Hardy JA, Singleton AB, Rosenberg NA.. 2010. Comparing spatial maps of human population-genetic variation using Procrustes analysis. Stat Appl Genet Mol Biol. 9: doi:10.2202/1544-6115.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. 2014. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42(Database issue): D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen B, Li H, Lu D, Song X, Zhang F, He Y, Li F, Gao Y, Mao X, Zhang L, et al. 2004. Genetic evidence supports demic diffusion of Han culture. Nature 431(7006): 302–305. [DOI] [PubMed] [Google Scholar]
- Whiffin N, Minikel E, Walsh R, O'Donnell-Luria AH, Karczewski K, Ing AY, Barton PJR, Funke B, Cook SA, MacArthur D, et al. 2017. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet Med. 19(10):1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Yin X, Li S, Jin W, Lou H, Yang L, Gong X, Wang H, Shen Y, Pan X, et al. 2009. Genomic dissection of population substructure of Han Chinese and its implication in association studies. Am J Hum Genet. 85(6): 762–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yafei W, Lijun P, Jinfeng W, Xiaoying Z.. 2012. Is the prevalence of MTHFR C677T polymorphism associated with ultraviolet radiation in Eurasia? J Hum Genet. 57(12): 780–786. [DOI] [PubMed] [Google Scholar]
- Yang J, Jin ZB, Chen J, Huang XF, Li XM, Liang YB, Mao JY, Chen X, Zheng Z, Bakshi A, et al. 2017. Genetic signatures of high-altitude adaptation in Tibetans. Proc Natl Acad Sci U S A. 114(16): 4189–4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM.. 2011. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 88(1): 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang MA, Gao X, Theunert C, Tong H, Aximu-Petri A, Nickel B, Slatkin M, Meyer M, Paabo S, Kelso J, et al. 2017. 40,000-Year-old individual from Asia provides insight into early population structure in Eurasia. Curr Biol. 27(20): 3202–3208. e3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Gao F, Wang D, Bar-Yosef O, Keinan A.. 2017. Dietary adaptation of FADS genes in Europe varied across time and geography. Nat Ecol Evol 1:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You QS, Xu L, Wang YX, Liang QF, Cui TT, Yang XH, Wang S, Yang H, Jonas JB.. 2013. Prevalence of retinitis pigmentosa in North China: the Beijing Eye Public Health Care Project. Acta Ophthalmol. 91(6): e499–e500. [DOI] [PubMed] [Google Scholar]
- Zhao YB, Zhang Y, Zhang QC, Li HJ, Cui YQ, Xu Z, Jin L, Zhou H, Zhu H.. 2015. Ancient DNA reveals that the genetic structure of the northern Han Chinese was shaped prior to 3,000 years ago. PLoS One 10(5): e0125676.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z. 2011. New archaeobotanic data for the study of the origins of agriculture in China. Curr Anthropol. 52(S4): S295–S306. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.