Summary
Genetically engineered mouse models harboring large sequence insertions or modifications are critical for a wide range of applications including endogenous gene tagging, conditional knockout, site-specific transgene insertion, and gene replacement; however, existing methods to generate such animals remain laborious and costly. To address this, we developed an approach called CRISPR-READI (CRISPR RNP Electroporation and AAV Donor Infection), combining adeno-associated virus (AAV) mediated HDR donor delivery with Cas9/sgRNA RNP electroporation to engineer large site-specific modifications in the mouse genome with high efficiency and throughput. We successfully targeted a 774 bp fluorescent reporter, a 2.1 kb CreERT2 driver, and a 3.3 kb expression cassette into endogenous loci in both embryos and live mice. CRISPR-READI is applicable to most widely used knock-in schemes requiring donor lengths within the 4.9 kb AAV packaging capacity. Altogether, CRISPR-READI is an efficient, high-throughput, microinjection-free approach for sophisticated mouse genome engineering with potential applications in other mammalian species.
Keywords: CRISPR, genome editing, mouse models, knock-in, electroporation, AAV
Introduction
Genetically modified mice are invaluable assets for investigating mammalian gene function, as well as for modeling human development, physiology, and disease. In particular, knock-in mice harboring large sequence insertions or substitutions are essential for a variety of applications including endogenous gene tagging, conditional gene knockout, site-specific transgene insertion, and gene replacement. While embryonic stem cells (ESCs) engineered by homologous recombination were classically used to generate these mouse models, the rapid adoption of CRISPR/Cas9 technology has provided an attractive alternative—direct microinjection of CRISPR components with a donor template to trigger homology-directed repair (HDR) and produce edited animals in one generation (Aida et al., 2015; Cong et al., 2013; Fujii et al., 2013; Jinek et al., 2012; Li et al., 2013; Nakagawa et al., 2016; Wang et al., 2013; Yang et al., 2013). Multiple technical refinements have also been developed to increase the rate of HDR in mouse zygotes through the use of ssDNA, linearized dsDNA, or chemically modified dsDNA donor templates, co-injection of HDR-stimulating compounds, and timed microinjection in 2-cell stage embryos (Gu et al., 2018; Maruyama et al., 2015; Miura et al., 2017; Quadros et al., 2017; Yao et al., 2018; Yoshimi et al., 2016). In spite of these improvements, complex editing in mice remains challenging due to the need for microinjection, a costly procedure with a high technical barrier and low throughput (Brinster et al., 1985; Nagy et al., 2003).
Recently, we and others developed electroporation-based methods to deliver CRISPR reagents into zygotes for highly efficient genome engineering; these strategies have garnered popularity due to improved throughput, technical ease, and cost-effectiveness across multiple mammalian species (Chen et al., 2016b; Hashimoto et al., 2016; Hashimoto and Takemoto, 2015; Hur et al., 2016; Miyasaka et al., 2018; Ohtsuka et al., 2018; Remy et al., 2017; Takahashi et al., 2015; Tanihara et al., 2016; Wang et al., 2016). While electroporation-based approaches are highly effective in introducing indel mutations, large deletions, and small insertions or substitutions, the use of short (typically <200 nt) single-stranded oligodeoxynucleotides (ssODNs) as HDR donors renders these techniques unsuitable for editing schemes involving targeted insertion of multi-kilobase sequences. Alternatively, long single-stranded oligodeoxynucleotides (lssODNs) can be applied for more complex editing, and several lssODN synthesis methods, including reverse-transcription, plasmid nicking followed by gel extraction, and selective strand phosphorylation/degradation, have been reported (Chen et al., 2011a; Li et al., 2017; Miura et al., 2017; Miyasaka et al., 2018; Ohtsuka et al., 2018; Quadros et al., 2017; Remy et al., 2017; Takahashi et al., 2015; Yoshimi et al., 2016). However, these protocols carry several notable drawbacks, including limited length (typically ≤2000 nt), GC-content and other sequence constraints, suboptimal yield, and costly reagents (Chen et al., 2011b; Li et al., 2017; Miura et al., 2017; Miyasaka et al., 2018; Quadros et al., 2017). Furthermore, electroporation of lssODNs typically yields much lower knock-in efficiencies than that of short ssODNs, likely due to inefficient lssODN delivery into zygotes (Ohtsuka et al., 2018; Yoshimi et al., 2016). Owing to these restrictions, targeted knock-ins are predominantly performed using microinjection or ESC-based approaches with plasmids or linearized dsDNA as HDR donors.
Here, we developed a strategy for complex mouse genome engineering that leverages the simplicity and throughput of CRISPR electroporation while overcoming the current limitations of large HDR donor delivery. Recombinant adeno-associated virus (rAAV) has emerged as a safe and efficient gene delivery vector with the innate ability to transduce mammalian cells and stimulate gene targeting by promoting homologous recombination (Hiramoto et al., 2018; Hirata et al., 2002; Khan et al., 2011; Kotterman et al., 2015; Russell and Hirata, 1998). In particular, rAAV donors packaged with single-stranded DNA genomes have successfully served as repair templates for HDR upon Cas9-mediated cleavage in mammalian cell lines, enhancing the efficiency of site-specific gene integration by >10-fold relative to plasmid donor nucleofection (Gaj et al., 2017). More recently, three separate rAAV6 vectors were used to deliver Cas9, sgRNA, and donor components into mouse zygotes to mediate gene targeting, demonstrating targeted knock-in of up to ~700 bp (Yoon et al., 2018). Of note, AAV’s naturally occurring single-stranded DNA genomes can be engineered into a duplexed form, termed self-complementary AAV (scAAV), which can also facilitate efficient gene targeting(Hirsch et al., 2010).
In this study, we identified AAV1 as an optimal naturally occurring serotype for highly efficient transduction of mouse zygotes. We then developed CRISPR-READI (CRISPR RNP Electroporation and AAV Donor Infection), a highly efficient method that combines AAV-mediated HDR donor delivery with Cas9/sgRNA RNP electroporation to generate complex genome modifications in mice. Using CRISPR-READI, we successfully inserted a 774 bp fluorescent reporter or a 2.1 kb CreERT2 cassette into the endogenous Sox2 locus, as well as a 3.3 kb gene expression cassette into the Rosa26 locus, in both preimplantation stage embryos and in mice. CRISPR-READI permits the use of HDR donors up to 4.9 kb, the packaging capacity of recombinant AAVs (Dong et al., 1996), and thus enables a broad range of complex genome modifications, including site-specific integration of reporters, Cre-drivers, and expression cassettes. Altogether, CRISPR-READI produces genetically edited animals harboring multi-kilobase modifications with unparalleled efficiency and throughput.
Results
Specific natural AAV serotypes efficiently transduce mouse zygotes
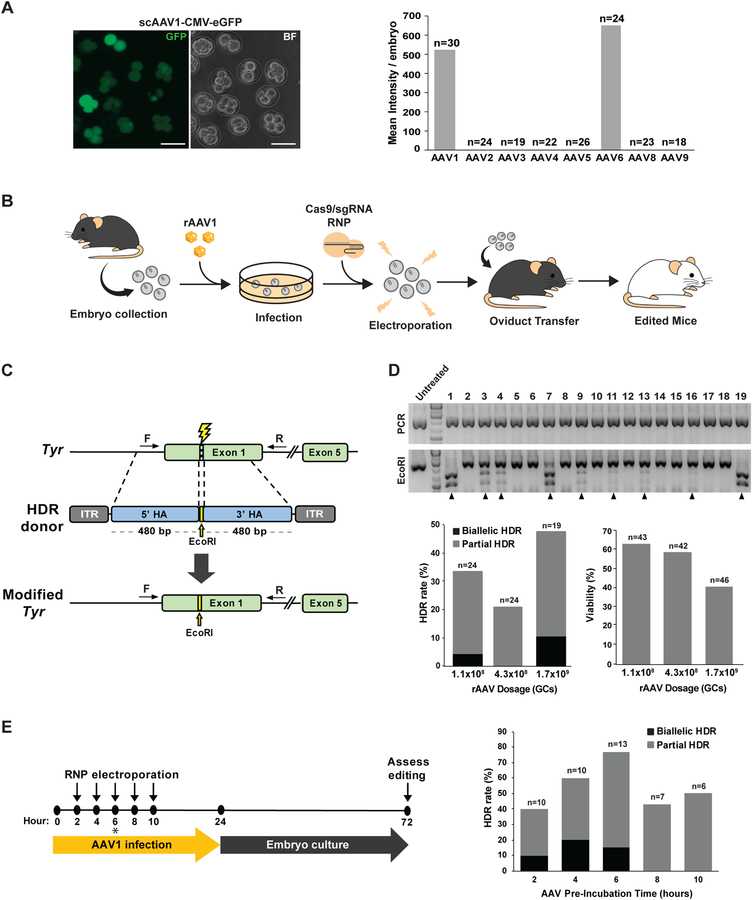
To characterize the native capacity of rAAV vectors to penetrate the zona pellucida and deliver the viral genome into mouse zygotes, we infected C57BL/6J mouse zygotes with a panel of eight natural AAV serotypes packaged with a self-complementary CMV-eGFP reporter (scAAV-CMV-eGFP) at a dose of 2×108 genome copies (GCs) per culture droplet (Fig. 1A; Fig. S1). Strong fluorescent signal was only detected for serotypes 1 and 6 (scAAV1 and scAAV6), which yielded comparable signal intensity by the 4 to 8-cell stage, suggesting that efficient rAAV-mediated DNA delivery can be achieved in mouse zygotes (Fig. 1A; Fig. S1). While ~20% of scAAV1- or scAAV6-transduced embryos exhibited bright eGFP fluorescence, all treated embryos displayed signal above background (Fig. S1). Although a previous study utilized rAAV6 for mouse editing (Yoon et al., 2018), we selected rAAV1 for knock-in experiments with CRISPRREADI due to the superior viral yield of rAAV1 compared to rAAV6 when produced in HEK293T cells (Vandenberghe et al., 2010).
Figure 1.
CRISPR-READI optimization for efficient HDR editing in mouse embryos. a Zygotes were transduced with a panel of AAV serotypes harboring a CMV-eGFP reporter and imaged by fluorescent microscopy 48 hours post-transduction. Representative embryos transduced with scAAV1-CMV-eGFP are shown (left), and mean fluorescence intensity per embryo was quantified for each serotype (right). Scale bars = 50 μm. b Cartoon depiction of CRISPR-READI workflow. Embryos are collected from superovulated female mice, transduced with rAAV1 harboring the donor template, electroporated with Cas9/sgRNA RNPs, and implanted into pseudopregnant females to generate edited mice. c Schematic of Tyr targeting strategy. The scAAV1-Tyr donor creates an EcoRI restriction site in exon 1 of the Tyr locus upon HDR editing. ITR: inverted terminal repeat, HA: homology arm, F/R: forward/reverse primers for RFLP analysis. d Optimization of rAAV1 dosage for HDR editing. Zygotes were transduced with scAAV1-Tyr at a dose of 1.1×108, 4.2×108, or 1.7×109 GCs, electroporated with RNPs 5 hours post-transduction, and then returned to rAAV1 incubation for another 19 hours. Treated embryos were cultured to the morula stage and genotyped by restriction fragment length polymorphism (RFLP) analysis (shown for dose of 1.7×109 GCs). Edited embryos yield 650 bp and 420 bp bands upon EcoRI digestion of the PCR amplicon (top, black arrows). HDR rate was quantified by RFLP analysis for each dose (bottom left), and embryo viability was scored as percentage of cultured embryos that reached the morula stage (bottom right). e Optimization of RNP electroporation timing relative to rAAV transduction. Zygotes were transduced with scAAV1-Tyr, electroporated at varying time points post-transduction (2, 4, 6, 8, or 10 hours), and returned to rAAV incubation for a total of 24 hours. Treated embryos were cultured to the morula stage, lysed, and assessed by RFLP analysis (right). 6 hours (*) was identified as the optimal time of RNP electroporation for maximal editing efficiency.
Optimization of CRISPR-READI enables efficient HDR editing in embryos
To generate edited animals with CRISPR-READI, we devised a workflow wherein mouse zygotes are harvested from superovulated females, infected with rAAV1 donors, electroporated with pre-assembled Cas9/sgRNA RNPs, and cultured to the 2-cell stage before transferring to the oviducts of pseudopregnant females (Fig. 1B). In a proof-of-principle CRISPR-READI experiment, we designed a strategy to insert an EcoRI restriction site into Tyrosinase (Tyr) exon 1 using a ~960 bp donor (including ~480 bp homology arms) that was packaged into an scAAV1 vector (scAAV1-Tyr) (Fig. 1C). As intracellular trafficking, nuclear localization, and capsid uncoating precede AAV-mediated HDR editing (Nonnenmacher and Weber, 2012), we pre-incubated the zygotes for 5 hours with three doses of scAAV1-Tyr (1.1×108, 4.3×108, and 1.7×109 GCs), introduced Cas9/sgRNA RNP by electroporation, and then continued viral incubation for a total of 24 hours (Fig. 1D). Treated embryos were grown for another 48 hours post-transduction until the morula / early blastocyst stage, and editing efficiency was assessed by screening for the engineered EcoRI site using restriction fragment length polymorphism (RFLP) analysis (Chen et al., 2016). As anticipated, HDR editing efficiency was dependent on viral titer. The highest AAV dose, 1.7×109 GCs, resulted in 48% (9 out of 19) embryos harboring the precise sequence substitution, with 11% (2 out of 19) showing bi-allelic HDR-mediated editing, while the lower doses of 1.1×108 and 4.3×108 GCs yielded HDR rates of 33% and 21%, respectively (Fig. 1D; Fig. S2A; Table S1). Although increased AAV dosage elicited a higher HDR rate, it was also correlated with a moderate reduction in embryo viability, as 41% (19 out of 46) embryos developed to the morula stage in culture when treated with 1.7×109 GCs, while 63% (25 out of 43) embryos did so with 1.1×108 GCs (Fig. 1D; Table S1). Altogether, these results suggest that the optimal AAV dose for CRISPR-READI ranges from 108 to 109 GCs, and a balance between HDR rate and embryo viability should be considered when determining AAV dosage for each CRISPR-READI experiment.
HDR requires nuclear co-localization of Cas9, sgRNA, and the donor template. As multiple intracellular trafficking steps must occur prior to nuclear import of the AAV genome (Nonnenmacher and Weber, 2012), we reasoned that optimizing the timing of RNP delivery relative to rAAV transduction would enhance HDR editing efficiency. Using the Tyr editing scheme described above (Fig. 1C), we pre-incubated zygotes with the rAAV donor vector for 0, 2, 4, 6, 8, or 10 hours prior to RNP electroporation (Fig. 1E). In all conditions, we achieved a minimum of 40% HDR-mediated editing (Fig. 1E; Fig. S2B; Table S1). We found that RNP electroporation at 6 hours after rAAV transduction resulted in maximal editing, with 77% (10 out of 13) assayed embryos harboring the precise sequence modification and 15% (2 out of 13) showing bi-allelic editing. Notably, this exceeds the HDR frequencies we previously achieved by CRISPR-EZ, using electroporation of short ssODNs (46%) (Chen et al., 2016b). Henceforth, we settled on 6 hours of rAAV pre-incubation prior to RNP electroporation for subsequent CRISPRREADI experiments.
As double-strand breaks induced by Cas9 can be repaired either by non-homologous end joining (NHEJ) or HDR, we next sought to characterize the extent of NHEJ in this system. To quantitatively measure the frequency of HDR versus NHEJ-mediated editing, we treated embryos using scAAV1-Tyr at a dosage of 1.7×109 GCs, amplified the edited region by PCR, and clonally analyzed editing events by Sanger sequencing (10 clones from each of 10 embryos). As expected, NHEJ occurs frequently, as 90% (9 out of 10) of embryos carried indels. In comparison, 70% (7 out of 10) harbored the desired HDR-mediated point mutation, most of which exhibited both HDR and NHEJ-mediated editing events (Fig. S3A). HDR frequencies of each edited embryo ranged from 10% to 67%, revealing a degree of editing mosaicism.
AAV vectors can be packaged in either single-stranded (ssAAV) or self-complementary (scAAV) forms, the latter of which bypasses the necessity of second-strand synthesis and in turn enhances transduction efficiency (McCarty et al., 2003). In a previous study, scAAV vectors were also shown to modestly enhance gene correction in mammalian cells in vitro relative to their single-stranded equivalents, possibly due to their increased stability, their two copies of the donor template in opposite polarities, or their ability to undergo double crossover events(Hirsch et al., 2010). Using the same Tyr editing scheme, we found that scAAV1-Tyr moderately outperformed ssAAV1-Tyr by ~17% in HDR efficiency, demonstrating that scAAV vectors promote HDR-mediated editing in mouse embryos (Fig. S3B; Table S1). However, it is important to note that scAAV vectors possess half the packaging capacity of ssAAV and thus limit the length of HDR donors to ~2.4 kb (McCarty et al., 2003).
CRISPR-READI enables efficient targeted integration of fluorescent reporters
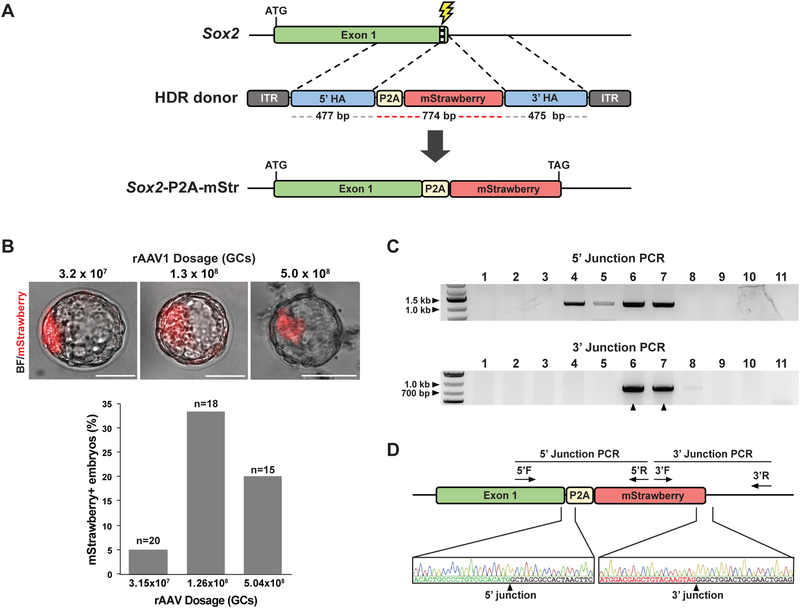
A common strategy to characterize the expression pattern of a gene is to insert a fluorescent reporter under the control of its endogenous promoter. We designed a 1.7 kb scAAV1 donor vector consisting of a 774 bp P2A-mStrawberry insert flanked by ~480 bp homology arms (scAAV1-Sox2-mStr) to knock in the P2A-mStrawberry cassette immediately downstream of the 3’ terminus of the Sox2 open reading frame (ORF) (Fig. 2A). Mouse zygotes were treated with CRISPRREADI using scAAV1-Sox2-mStr at three doses (3.2×107, 1.3×108, and 5.0×108 GCs) and cultured to the blastocyst stage. Robust red fluorescence was detected in the inner cell mass (ICM) of a subset of the resulting blastocysts, recapitulating endogenous Sox2 expression (Fig. 2B). scAAV1 donor at a dose of 1.3×108 GCs produced the highest number of correctly targeted embryos, with 33% (6 out of 18) of blastocysts showing ICM-specific fluorescent signal (Fig. 2B; Fig. S4A; Table S1). Since many widely used reporters, such as fluorescent proteins, luciferase, and HaloTag, can be targeted with an HDR donor under 2.4 kb in length, CRISPR-READI using scAAV1 donors can be applied to tag endogenous loci with many common reporter cassettes.
Figure 2.
CRISPR-READI enables efficient knock-in of fluorescent reporters in embryos and animals. a Schematic of strategy to engineer an mStrawberry fluorescent reporter downstream of the endogenous Sox2 locus. The scAAV1-Sox2-mStr vector contains a 774 bp P2A-mStrawberry cassette flanked by ~480 bp homology arms that mediate insertion at the 3’ terminus of the Sox2 ORF upon HDR editing. ITR: inverted terminal repeat, HA: homology arm. b CRISPR-READI efficiently generates embryos with an mStrawberry reporter driven by the endogenous Sox2 promoter. Embryos were treated with scAAV1-Sox2-mStr at a dose of 3.2×107, 1.3×108, or 5.0×108 GCs. Treated embryos were cultured to the late blastocyst stage and imaged by fluorescent microscopy. In edited blastocysts, merged brightfield and fluorescent images show localization of mStrawberry fluorescence to the inner cell mass, recapitulating the endogenous Sox2 expression pattern (top). HDR rate was quantified for each dose by the percentage of mStrawberry-positive embryos (bottom). Scale bars: 50 μm. c CRISPR-READI efficiently generates Sox2-P2A-mStrawberry knock-in mice. PCR genotyping confirmed correctly edited 5’ and 3’ junctions of the modified Sox2 locus in 2 out of 11 mice generated by CRISPR-READI, as indicated by black arrows. We also identified two animals harboring the 5’ but not the 3’ end of the donor sequence, which is likely due to incomplete HDR (lanes 4 and 5). Primers were designed such that one primer binds outside the homology arm and the other primer binds within the P2A-mStrawberry cassette. 5’ expected band size: 1,088 bp, 3’ expected band size: 836 bp. d Representative Sanger sequencing and chromatograms for correctly edited mice. 5’ F/R: forward/reverse primers for 5’ junction genotyping, 3’ F/R: forward/reverse primers for 3’ junction genotyping.
To assess the ability of our method to generate live knock-in mice, we treated zygotes with CRISPR-READI using scAAV1-Sox2-mStr at a dose of 1.3×108 GCs and transferred them to the oviducts of pseudopregnant females. Of the 11 live pups we obtained, 18% harbored the desired P2A-mStrawberry insertion, as validated by 5’ and 3’ integration of the targeted cassette using PCR analysis and sequencing confirmation (Fig. 2C-D; Table S1). Since Sox2 is an essential gene, we also assayed for the presence of indels in the Sox2 locus using RFLP analysis (Fig. S4B). 9 out of 11 pups (82%) carried indels in the Sox2 allele (Fig. S4C), confirming our previous observation that NHEJ is more prevalent than HDR (Fig. S3A). Notably, the high rate of NHEJ is not specific to CRISPR-READI, but is an inherent property of CRISPR/Cas9-based approaches for mouse zygotic editing (Chen et al., 2016a; Modzelewski et al., 2018), and thus may pose a challenge when editing essential genes. In fact, we encountered challenges with breeding our founder animals, most likely due to NHEJ-mediated Sox2 deficiency (Avilion et al., 2003; Ferri et al., 2004). Nevertheless, our results show that CRISPR-READI facilitates efficient knock-in of a fluorescent reporter in live animals, demonstrating its utility in generating reporter mouse models.
CRISPR-READI achieves site-specific knock-in of large gene cassettes
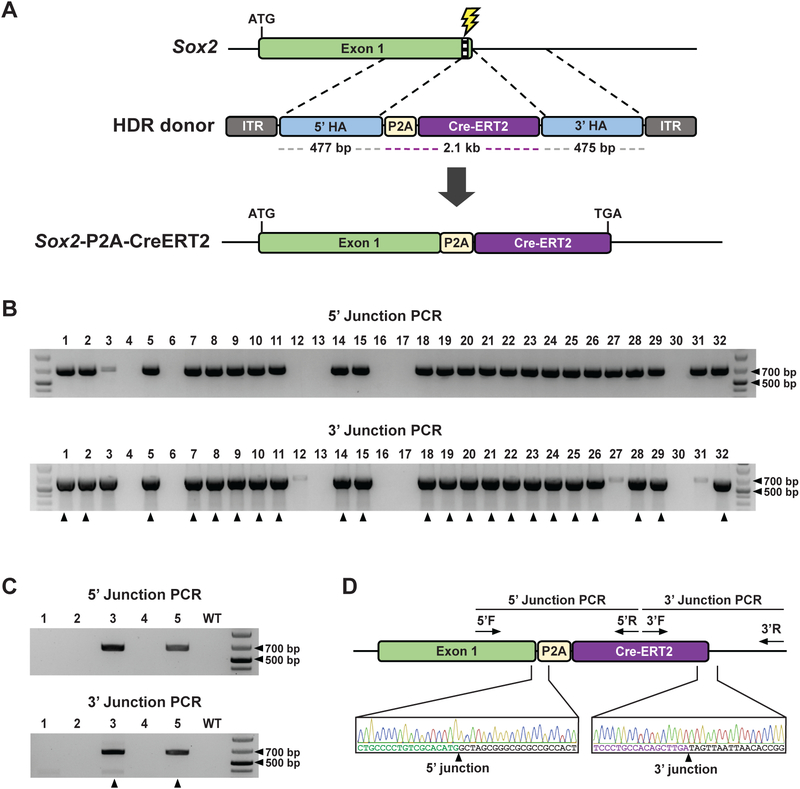
While CRISPR-READI using scAAV1 donors mediates efficient gene targeting, its 2.4 kb packaging capacity limits its usage for more complex editing schemes that require larger HDR donors. In contrast, ssAAV1 doubles the packaging capacity and can accommodate a donor construct of up to 4.9 kb (Dong et al., 1996). To demonstrate the ability of ssAAV1 to mediate knock-in of multi-kilobase cassettes, we applied CRISPR-READI to insert an inducible CreERT2 (Seibler et al., 2003) into the Sox2 locus. We designed a 3 kb ssAAV1 donor vector consisting of a 2112 bp Sox2-P2A-CreERT2 cassette flanked by two ~480 bp homology arms (ssAAV1-Sox2-CreERT2) (Fig. 3A). Treated embryos were cultured to the blastocyst stage for PCR genotyping analysis. Remarkably, 69% (22 out of 32) of treated blastocysts exhibited correct targeting of the P2A-CreERT2 cassette, highlighting the considerable HDR efficiency achieved by CRISPRREADI (Fig. 3B; Table S1). We then replicated this experiment and transferred the embryos to pseudopregnant females to generate live mice. 40% (2 out of 5) of the resulting pups were successfully edited, as confirmed by PCR genotyping and Sanger sequencing (Fig. 3C-D; Table S1). Similar to results obtained from the scAAV1-Sox2-mStr experiments, 60% (3 out of 5) of the mice carried indels in the Sox2 locus, with both HDR-positive animals also harboring indels (Fig. S4D). Since HDR donors containing recombinase platforms (Cre and Flp) and tetracyclineinducible systems can be easily accommodated by the multi-kilobase packaging capacities of scAAV1 (~2.4 kb) and ssAAV1 (~4.9 kb), CRISPR-READI is well-suited for engineering a wide range of complex mouse models with temporal and spatial gene regulation.
Figure 3.
CRISPR-READI efficiently engineers an inducible CreERT2 reporter driven by its endogenous promoter. a Schematic of strategy to create a Sox2-driven inducible CreERT2 reporter. The ssAAV1-Sox2-CreERT2 vector contains a 2112 bp P2A-CreERT2 cassette flanked by ~480 bp homology arms that mediate P2A-CreERT2 insertion at the 3’ terminus of the Sox2 ORF upon successful HDR editing. ITR: inverted terminal repeat, HA: homology arm. b CRISPR-READI efficiently produces embryos with an inducible CreERT2 reporter driven by the endogenous Sox2 promoter. PCR genotyping confirmed the correctly edited 5’ and 3’ junctions of the modified Sox2 locus in 22 out of 32 embryos, as indicated by black arrows. Primers were designed such that one primer binds outside the homology arm and the other primer binds within the CreERT2 cassette. 5’ expected band size: 658 bp, 3’ expected band size: 666 bp. c CRISPR-READI efficiently generates Sox2-P2A-CreERT2 mice. PCR genotyping confirmed correctly edited 5’ and 3’ junctions of the modified Sox2 locus in 2 out of 5 mice with previously described primers. d Representative Sanger sequencing and chromatograms for correctly edited mice. 5’ F/R: forward/reverse primers for 5’ junction genotyping, 3’ F/R: forward/reverse primers for 3’ junction genotyping.
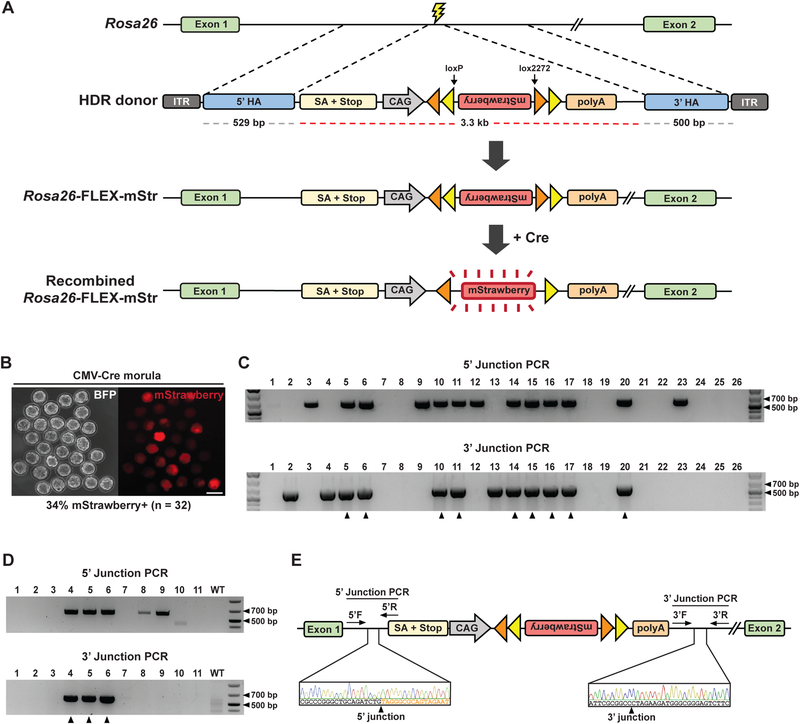
To evaluate the capacity of CRISPR-READI to engineer even larger knock-ins, we designed an editing scheme to insert a 3.3 kb FLEX-mStrawberry expression cassette into the Rosa26 safe harbor locus. The resulting 4.3 kb donor consists of a splice acceptor and a polyA element that terminate the endogenous Rosa26 transcript, a CAG promoter driving expression of a FLEX (flip-excision) mStrawberry switch, a chimeric SV40/bGH polyA signal, and two ~500 bp homology arms (ssAAV1-R26-FLEX-mStr) (Fig. 4A). We applied CRISPR-READI using ssAAV1-R26-FLEX-mStr to target the FLEX-mStrawberry cassette to the Rosa26 locus in CMV-Cre embryos (Schwenk et al., 1995). In successfully edited embryos, mStrawberry expression is irreversibly activated through consecutive Cre-mediated recombination events (Fig. 4A). We detected red fluorescent signal in 34% (11 out of 32) 8-cell stage embryos, with an increase in signal intensity over the following 24 hours, indicating functional recombination of the FLEX construct in developing embryos (Fig. 4B). In a separate experiment, we confirmed HDR-mediated editing in 34% (9 out of 26) of embryos by validating 5’ and 3’ integration of the targeted cassette using PCR (Fig. 4C; Table S1). Finally, we generated live animals using this editing scheme in a C57BL/6J background, obtaining 27% (3 out of 11) successfully edited pups, as confirmed by PCR and Sanger sequencing (Fig. 4D-E; Table S1). Our findings affirm the utility of CRISPRREADI for highly efficient, site-specific knock-in of large expression cassettes at endogenous loci.
Figure 4.
CRISPR-READI enables efficient site-specific insertion of a large expression cassette. a Schematic of strategy to knock a CAG-FLEX-mStrawberry cassette into the Rosa26 locus. The ssAAV1-R26-FLEX-mStr vector contains a splice acceptor + polyA element, a CAG promoter, an inverted mStrawberry ORF flanked by loxP and lox2272 sites, chimeric SV40/bGH polyA signal, and ~500 bp homology arms that direct targeting to the first Rosa26 intron. Upon Cre-mediated recombination, the mStrawberry ORF is irreversibly inverted, leading to robust mStrawberry expression. ITR: inverted terminal repeat, HA: homology arm, SA: splice acceptor. b Representative fluorescent image of CMV-Cre embryos treated with ssAAV1-R26-FLEX-mStr. Successful editing and Cre-mediated recombination lead to robust mStrawberry expression by the morula stage. Scale bar: 50 μm. c PCR genotyping confirmed the correctly edited 5’ and 3’ junctions of the modified Rosa26 locus in 9 out of 26 embryos, as indicated by black arrows. Primers were designed such that one primer binds outside the homology arm and one primer binds within the ssAAV1-R26-FLEX-mStr cassette. 5’ expected band size: 608 bp, 3’ expected band size: 626 bp. d CRISPR-READI efficiently generates Rosa-FLEX-mStr mice. PCR genotyping confirmed correctly edited 5’ and 3’ junctions of the modified Rosa26 locus in 3 out of 11 mice with previously described primers. Notably, we also detected two animals exhibiting partial sequence insertion, similar to our Sox2-P2A-mStr animals (lanes 8 and 9). e Representative Sanger sequencing and chromatograms for correctly edited mice. 5’ F/R: forward/reverse primers used for 5’ junction genotyping, 3’ F/R: forward/reverse primers used for 3’ junction genotyping.
Discussion
Electroporation-based CRISPR technologies offer significant advantages over microinjection in throughput, affordability, and ease of use. Previously, we developed CRISPR EZ, an approach that utilizes Cas9/sgRNA RNP and short ssODN (<200 nt) electroporation to achieve highly efficient mouse genome editing (Chen et al., 2016b; Modzelewski et al., 2018). Many groups have reported success using similar electroporation strategies, including both ex vivo and in utero approaches (Chen et al., 2016b; Hashimoto et al., 2016; Hashimoto and Takemoto, 2015; Hur et al., 2016; Ohtsuka et al., 2018; Takahashi et al., 2015; Wang et al., 2016). However, inefficient delivery of long HDR donors into pronuclear embryos by electroporation, possibly due to their highly anionic properties, limits its application for complex genome engineering. While lssODN electroporation has mediated large knock-ins in mice with some success, targeting efficiencies are typically suboptimal (Ohtsuka et al., 2018; Yoshimi et al., 2016). Furthermore, current lssODN synthesis protocols are hampered by low yield, costly reagents, and limited length (Chen et al., 2011b; Li et al., 2017; Miura et al., 2017; Miyasaka et al., 2018; Quadros et al., 2017). Hence, lssODNs remain unsuitable for many mouse engineering schemes involving large genomic modifications.
In contrast, CRISPR-READI harnesses rAAV vectors to deliver large donor constructs into mouse embryos for complex editing (Dong et al., 1996). With multi-kilobase packaging capacities, scAAVs (~2.4 kb) and ssAAVs (~4.9 kb) can accommodate many commonly used HDR donors for engineering large knock-ins in mice. The enhanced genome stability and transduction efficiency of scAAVs may further improve gene correction frequency in pronuclear embryos (Ferrari et al., 1996; Fisher et al., 1996; Hirsch et al., 2010; McCarty, 2008). With rAAV donors, CRISPR-READI enables a variety of sophisticated HDR editing schemes that are used for disease modeling and functional studies; these include precise sequence modifications (Fig. 1), as well as site-specific integration of reporters (Fig. 2), recombinases (Fig. 3), and large expression cassettes (Fig. 4). Our proof-of-concept studies demonstrate the potential of CRISPR-READI to efficiently produce a vast range of popular knock-in models, such as those harboring fluorescent proteins, luciferase reporters, HaloTag, Cre and Flp recombinase, Tet-on/off inducible systems, and many others. Moreover, we show that CRISPR-READI is an electroporation-based genome editing methodology capable of delivering a 4.3 kb donor template and inserting a 3.3 kb reporter cassette, which was previously only feasible with dsDNA microinjection or ESC-based approaches.
While our manuscript was in preparation, Yoon et al reported AAV6 as the most efficient natural AAV serotype for transduction of mouse zygotes (Yoon et al., 2018). In our studies, AAV1 and AAV6, which share 99.2% amino acid sequence identity (Huang et al., 2016), exhibited comparable infectivity, with AAV1 being superior in viral particle assembly. Yoon et al also described a triple AAV co-infection system to deliver CRISPR-Cas9 components (AAV-Cas9, AAV-sgRNA) and an HDR donor (AAV-donor). Using the Tyr editing scheme, we directly compared CRISPR-READI (scAAV1-Tyr and Cas9/sgRNA RNP) with a triple AAV1 co-infection strategy (scAAV1-Tyr, ssAAV1-SpCas9 and scAAV1-Tyr-gRNA) at the same AAV dosage (1×108 GCs). The two methods yielded similar HDR rates and embryo viability (Fig. S3C; Table S1); however, the triple AAV infection strategy resulted in multiple embryos harboring large deletions, while CRISPR-READI did not (Fig. S3C).
CRISPR-READI likely carries several notable advantages compared to the triple AAV co-infection method. First, CRISPR-READI harnesses pre-assembled Cas9/sgRNA complexes that are electroporated with 100% delivery efficiency (Chen et al., 2016b) and are active immediately upon delivery, whereas virally delivered Cas9 may exhibit delayed activity, as it must undergo transcription, translation, and RNP assembly prior to mediating cleavage. Second, it was recently shown that Cas9 can induce large-scale genomic deletions (Kosicki et al., 2018), which may be facilitated by the persistence of AAV episomes expressing Cas9 and sgRNA within the developing mouse embryo. In comparison, RNPs are less prone to these effects due to their rapid turnover (Kim et al., 2014). Finally, Yoon et al observed rare but detectable integration of both the AAVCas9 and AAV-sgRNA vectors at their target site in the Tyr locus (Yoon et al., 2018), leading to long-term expression of Cas9 and/or sgRNAs, whereas CRISPR-READI circumvents this undesirable knock-in through the use of RNPs.
We demonstrate that CRISPR-READI can be broadly applied to various gene targeting schemes to rapidly generate mouse lines for functional studies and disease modeling. The unprecedented efficiency of CRISPR-READI for knocking in gene cassettes up to 3.3 kb makes this approach highly appealing for creating multi-kilobase genome modifications. With such high editing efficiencies, CRISPR-READI could enable multiplexed gene targeting to simultaneously modify two different target loci. Furthermore, a dual rAAV donor system carrying a split gene has recently been shown to mediate insertion of large gene cassettes through sequential homologous recombination events in primary human cells (Bak and Porteus, 2017), and it would be interesting to explore this strategy in the context of mouse embryo editing. Taken together, CRISPR-READI provides a simple, efficient, and high-throughput alternative to microinjection and ESC-based methods for sophisticated mouse genome engineering.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lin He (lhe@berkeley.edu).
Experimental Model and Subject Details
Animal studies were approved by the Animal Care and Use Committee at the University of California, Berkeley (Protocol #AUP-2015-04-7485). 4-week old C57BL/6J female mice were produced on-site or purchased from the Jackson Laboratory, and housed in the Li Ka Shing animal facility (University of California, Berkeley.) 4-week old B6.C-Tg(CMV-cre)1Cgn/J female mice were purchased from the Jackson Laboratory and housed in the Li Ka Shing animal facility (University of California, Berkeley). 7–8 week old CD1 female mice were purchased from Charles River and housed in the Life Sciences Addition animal facility (University of California, Berkeley.)
Method Details
Plasmid construction
For construction of the scAAV-Tyr donor plasmid, a Tyr HDR donor fragment and a 1 kb stuffer sequence were assembled to optimize the genome size for vector packaging (Dong et al., 1996). Briefly, a 968 bp Tyr fragment with the engineered EcoRI site was amplified from a 1 kb megamer ssDNA fragment (IDT custom synthesis) using Tyr-XbaI-F and Tyr-XbaI-R primers (Table S2) and then digested with XbaI for subsequent cloning into rAAV destination vectors. To prepare the destination scAAV vector, a plasmid derivative of scAAV-CMV-GFP (Fu et al., 2003) containing a nanoluciferase cassette was digested with XbaI to remove the majority of the nanoluciferase ORF, leaving a 1 kb stuffer sequence. The digested Tyr insert was then ligated to the scAAV destination vector containing the stuffer sequence to generate the final scAAV-Tyr plasmid. To build the ssAAV-Tyr donor plasmid, an additional stuffer sequence was first amplified from pGEM-T easy vector (Promega, A1360) using pGEM-stuffer-HindIII-F and pGEM-stuffer-HindIII-R primers and digested with HindIII. A plasmid derivative of pX601 (Addgene #61591) containing a nanoluciferase cassette was digested with HindIII and ligated to the pGEM stuffer sequence to generate pX601-long. Subsequently, pX601-long was digested with XbaI and ligated to the previously described XbaI digested Tyr insert to create the ssAAV-Tyr plasmid.
For construction of the scAAV-Sox2-Str donor plasmid, a Sox2 targeting cassette was subcloned into the XbaI digested scAAV destination vector described above. Briefly, a 1726 bp fragment containing a P2A-mStrawberry cassette flanked by homology arms from the Sox2 locus was amplified from a Sox2-P2A-mStrawberry plasmid (gift from D Stafford) using Sox2-P2A-Straw-XbaI-F and Sox2-P2A-Straw-XbaI-R primers and then digested with XbaI. This insert was ligated to the XbaI digested scAAV vector described above to generate the scAAV-Sox2-Str plasmid. For the ssAAV-Sox2-Cre-ERT2 donor plasmid, the previously described Sox2 targeting cassette and a 981 bp stuffer sequence were assembled and subcloned into a ssAAV destination vector, and the P2A-mStrawberry cassette was then replaced with a P2A-Cre-ERT2 cassette. Briefly, the Sox2 targeting construct with an additional 981 bp stuffer sequence downstream of the donor was PCR amplified from an ssAAV-Sox2-Str plasmid (data not shown) using FseI-Sox2-donor fwd and NotI-Sox2-donor rev primers and subcloned into a plasmid derivative of pX601 using FseI and NotI to generate pX601-Sox2. The P2A-Cre-ERT2 donor was then PCR amplified from a plasmid derivative of WT1–2A-eGFP (Addgene #82333) containing a P2A-Cre-ERT2 cassette using NheI-AscI-CreERT2-fwd and SgrAI-PacI-CreERT2-rev primers, digested with NheI and SgrAI, and cloned into pX601-Sox2 to produce the final ssAAV-Sox2-Cre-ERT2 plasmid.
For construction of the ssAAV-R26-FLEX donor plasmid, a Rosa26 targeting construct was first subcloned into a ssAAV destination vector, and the original donor cassette was then replaced with a CAG-FLEX-mStrawberry cassette. Briefly, a Rosa26 donor construct containing ~500 bp homology arms was PCR amplified from pR26-CAG-AsiSI/MluI (Addgene #74286) with Rosa-FLEX-mStr-Fwd and Rosa-FLEX-mStr-Rev primers, digested with NotI and NsiI, and ligated into a plasmid derivative of pX601 (Addgene #61591) to produce ssAAV-Rosa. To generate the CAG-FLEX-mStrawberry cassette, mStrawberry was PCR amplified from the scAAV-Sox2-Str plasmid with mStr-Fwd and mStr-Rev primers, digested with KpnI and XhoI, and cloned into pAAV-FLEX-GFP (Addgene #28304), generating an intermediate pAAV-FLEX-mStr plasmid. The assembled CAG-FLEX-mStrawberry cassette was then PCR amplified from pAAV-FLEX-mStr with CAG-mStr-Fwd and CAG-mStr-Rev primers, digested with NheI and MluI, and ligated into ssAAV-Rosa to generate the final ssAAV-R26-FLEX plasmid.
For construction of the AAV1-SpCas9 vector, the pAAV-nEFCas9 plasmid (Addgene #87115) was digested with HindIII and AgeI to remove the truncated 5’LTR of the nEF promoter, and a 9 bp stuffer sequence was ligated in to generate a shortened EF1α (EFS) promoter. For construction of the AAV1-Tyr-gRNA vector, a plasmid derivative of scAAV-CMV-GFP(Fu et al., 2003) with a CAG promoter in place of the original CMV promoter and a short 48 bp polyA in place of the original SV40 polyA was digested with KpnI and NotI to produce scAAV-CAG-GFP. The sgTyr sequence was cloned into pX458 (Addgene #48138) with BbsI, and the U6-Tyr-gRNA expression cassette was PCR amplified using KpnI-U6-sgTyr and NotI-U6-sgTyr primers. The PCR amplicon was then digested with KpnI and NotI and ligated into scAAV-CAG-GFP to generate the final scAAV-U6-sgTyr-CAG-GFP plasmid.
Production and purification of recombinant AAV vectors
HEK293T cells were obtained from the American Type Culture Collection (Manassas, CRL-3216) and cultured in DMEM (GIBCO, 12800082) with 10% fetal bovine serum (Invitrogen, 10437028) and 1% Antibiotic-Antimycotic (GIBCO, 15240062) at 37°C and 5% CO2. All recombinant AAV vectors were packaged in HEK293T cells, as previously described (Gaj and Schaffer, 2016; Grieger et al., 2006). In summary, recombinant AAV vectors were produced by triple transient transfection of a helper plasmid encoding adenoviral helper genes, an AAV helper plasmid encoding AAV rep and cap genes, and a transfer plasmid containing donor constructs flanked by AAV ITRs, into HEK293T cells using polyethylenimine (PEI) (Polysciences, 23966–1). Culture media was changed 14–16 hours post-transfection to reduce PEI-associated toxicity. At 72 hours post-transfection, cells were dissociated from culture dishes using a cell scraper and pelleted by centrifugation at 1500 g for 2.5 min. Cells were then resuspended in AAV lysis buffer (50 mM Tris base, 150 mM NaCl, pH 8.2–8.5) and lysed with 3 freeze/thaw cycles using a dry ice / ethanol bath and a 37°C water bath. Supernatant was collected following centrifugation at 10,000 rpm for 10 min in a tabletop centrifuge. For additional purification of vector preparations (optional), clarified cell lysate and culture supernatant can be combined with a solution of 40% polyethylene glycol (PEG) 8000 (Sigma-Aldrich, P2139) and 2.5 M NaCl at a 4:1 ratio, incubated at 4°C overnight, centrifuged at 2500 g for 30 min to harvest precipitated AAV particles, and resuspended in AAV lysis buffer(Ayuso et al., 2010). Crude lysates were treated with 10 U benzonase (Sigma-Aldrich, E8263) per mL of lysate for 30 min at 37°C. For AAV purification by iodixanol density centrifugation, discontinuous gradients comprised of 15%, 25%, 40%, and 54% iodixanol layers were set up in Optiseal tubes (Beckman Coulter, 362185) using OptiPrep (Axis-Shield, AVS-1114542). Benzonase-treated lysates were then loaded onto iodixanol gradients and centrifuged at 174,000 g for 2 hr at 18°C. Tubes were punctured using 21-gauge needles attached to a 1 mL syringe at the 40% / 54% interface, and the 40% iodixanol fraction containing AAV particles was collected. Vector preps were then buffer exchanged and concentrated into PBS with 0.001% or 0.00025% Tween 20 (Sigma-Aldrich, 9416) using Amicon filtration (EMD Millipore, UFC910024) at 3000 g. DNase-resistant (Sigma-Aldrich, 04536282001) viral genomic titers were measured by real-time qPCR(Gaj and Schaffer, 2016) using SYBR Green I (Invitrogen, S7567) and the CFX96 Real-Time PCR cycler (Bio-Rad, 1855195).
Cas9/sgRNA RNP assembly
To synthesize sgRNAs in vitro, a DNA template that contained a T7 promoter, a 20-nt guide sequence, and a sgRNA scaffold was generated by overlapping PCR. Specifically, we performed PCRs using Phusion high fidelity DNA polymerase (New England Biolabs, catalog no. M0530), with the annealed product from a uniquely designed oligo (5′-GGA TCC TAA TAC GAC TCA CTA TAG–guide sequence–GTT TTA GAG CTA GAA-3′, 0.02 μM) and a common oligo T7RevLong (5′-AAA AAA GCA CCG ACT CGG TGC CAC TTT TTC AAG TTG ATA ACG GAC TAG CCT TAT TTT AAC TTG CTA TTT CTA GCT CTA AAA C-3′, 0.02 μM) as the template, and T7FwdAmp (5′-GGA TCC TAA TAC GAC TCA CTA TAG-3 ′, 1 μM) and T7RevAmp (5′-AAA AAA GCA CCG ACT CGG-3′, 1 μM) as two common primers. All oligo sequences are listed in Table S2. The thermocycler setting consisted of 30 cycles of 95 °C for 10 s, 57 °C for 10 s, and 72 °C for 10 s. A 20μl in vitro transcription reaction consisting of 25 ng/μl PCR-amplified DNA template, 10 mM NTPs, and 1 unit of T7 RNA polymerase (New England Biolabs, E2040S) was incubated at 37 °C for more than 18 h, followed by a brief treatment of RNase-Free DNase I (New England Biolabs, M0303S, 2 units) at room temperature for 20 min. The in vitro synthesized sgRNAs were purified by magnetic SeraMeg Speedbeads (GE Healthcare, 65152105050250). The in vitro transcription reaction was first brought to 100 μl in volume with 100% ethanol, followed by gentle mixing of 100 μl of SeraMeg Speedbeads 10 times before a 5-min room temperature incubation. The reaction was subsequently placed on a magnetic stand (Invitrogen, 12321D) for 5 min under room temperature to allow the formation of compact RNA/bead pellets. After the supernatant was carefully aspirated by pipette, we washed the pellets gently with 80% ethanol three times (2 min wash each time, without pipetting) and air-dried the pellets for 10 min. sgRNAs bound to the beads were eluted by incubating with 20 μl of RNase-free H2O (Ambion, AM9937) and stored at −80°C.
To assemble the Cas9/sgRNA RNPs, we incubated purified Cas9 protein (QB3 Macrolab, University of California at Berkeley) in a 1:1.5 molar ratio with sgRNAs to obtain a final concentration of 8 μM Cas9/sgRNA RNPs in a 10-μl solution containing 20 mM HEPES, pH 7.5 (Sigma, H3375), 150 mM KCl (Sigma, P9333), 1 mM MgCl2 (Sigma, M8266), 10% glycerol (Thermo Fisher, BP229), and 1 mM reducing agent tris(2-carboxyethyl)phosphine (TCEP, Sigma, C4706). This mixture was incubated at 37 °C for 10 min immediately before use.
CRISPR-READI
Three-to-five-week old female C57BL/6J mice (000664; Jackson Laboratory), or CMV-Cre mice (006054; Jackson Laboratory), were superovulated by intraperitoneal administration of 5 IU of pregnant mare serum gonadotropin (Calbiochem, Millipore, 367222), and 46–48 h later, 5 IU human chorion gonadotropin (Calbiochem, Millipore, 230734). Superovulated females were mated at a 1:1 ratio with 3–8-month-old C57BL/6J males to generate one-cell zygotes at 0.5 days post-coitum. Fertilized zygotes were harvested and washed, as previously described (Chen et al., 2016). Briefly, zygotes were harvested from the ampulla of euthanized females, washed in hyaluronidase/M2 solution (Millipore, MR-051-F) to remove cumulus cells, washed four times in M2 media (Zenith, ZFM2–100) supplemented with 4 mg/ml bovine serum albumin (BSA, Sigma, A3311), washed briefly in acid Tyrode’s solution (Sigma, T1788) to thin the zona pellucida, and washed again four times in M2 + BSA media. Embryos were then cultured in 20 μL droplets of KSOM + AA media (KCl-enriched simplex optimization medium with amino acid supplement, Zenith Biotech, ZEKS-050) containing the specified AAV vector dosage in 35 × 10 mm culture dishes (CellStar Greiner Bio-One, 627160) at 37°C with 95% humidity and 5% CO2 for 6 hours prior to RNP electroporation.
AAV-transduced embryos were electroporated with assembled RNPs as previously described (Chen et al., 2016b). In summary, embryos were transferred to 40 uL of Opti-MEM reduced serum media (Thermo Fisher, 31985062) to dilute the M2 + BSA media, and 10 uL of Opti-MEM containing the embryos were mixed with 10 uL assembled Cas9/sgRNA RNPs. The 20-μl embryo and RNP mixture was transferred to a 1-mm electroporation cuvette (Bio-Rad, 1652089) and electroporated (square wave, 6 pulses, 30 V, 3 ms duration, 100 ms interval) using a Gene Pulser XCell electroporator (Bio-Rad, 1652660). Zygotes were recovered from the cuvette by flushing three times with 100 μl of KSOM + AA media, then transferred into the culture droplets containing AAV for a total incubation length of 24 hours. The following day, embryos were transferred to fresh KSOM + AA media overlaid with mineral oil until analysis or oviduct transfer. For generating live mice, treated zygotes that successfully developed into two-cell embryos were surgically transferred into the oviducts of pseudopregnant CD1 females (Charles River, Strain 022), using 15 embryos per oviduct.
Fluorescence imaging and analysis of edited embryos
Embryos treated with rAAV CMV-GFP and scAAV1-Sox2-Str were imaged using a Zeiss Observer A1 fluorescent microscope 48 hours and 72 hours post-transduction, respectively. Images were processed using ZEN software (Zeiss), and fluorescence intensity was quantified for embryos transduced with rAAV-CMV-GFP using ImageJ software. Fluorescence intensity was calculated by the following equation: (total signal intensity – negative control background signal) / number of embryos.
RFLP and genotyping analyses of edited embryos or pups
Crude DNA extract was obtained from morula and blastocyst stage embryos by placing individual embryos in 10 uL of embryo lysis buffer (0.2 mg/ml Proteinase K, 50 mM KCl, 10 mM Tris-HCl pH 8.3, 2.5 mM MgCl2, 0.1 mg/ml gelatin, 0.45% NP40, 0.45% Tween 20), followed by heating at 55° for 4 hours to lyse the embryos, as previously described (Chen et al., 2016). Tail DNA was extracted by standard chloroform extraction. For all embryo RFLP and genotyping, two nested PCR reactions were performed using an external and internal pair of primers in order to obtain enough signal from crude embryo lysate, while only the internal set of the primers were used when amplifying from mouse tail samples. All PCRs were conducted using GoTaq (Promega, M712), except for Sox2-CreERT2 5’ junction and Rosa26-FLEX 5’ junction genotyping, which were conducted using Q5 polymerase with GC enhancer due to high GC content (New England Biolabs, M0491S). For Tyr editing experiments, successful editing replaces the endogenous HinfI restriction site with an EcoRI site, while for Sox2 editing experiments, the presence of indels over the sgRNA target sequence disrupts a PciI restriction site. RFLP was performed to screen for these editing events by digesting the PCR products with EcoRI (New England Biolabs, R0101S) or PciI (New England Biolabs, R0655S), respectively, for 4 hours at 37°. For Sox2-P2A-mStrawberry, Sox2-P2A-CreERT2 and Rosa26-FLEX-mStrawberry knock-in experiments, primer pairs used for PCR genotyping of the 5’ and 3’ junctions consisted of one primer designed from the genomic region flanking the homology arm and another from within the inserted sequence. All primers used for RFLP and genotyping analysis are listed in Table S2.
Quantification and Statistical Analysis
For all quantitative analyses, percentages were reported as the number of edited samples (i.e. embryos or animals) divided by the total number samples. For all data, n represents the total number of embryos or animals assayed per condition.
Data and Software Availability
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | |
|---|---|---|---|
| Antibodies | |||
| Bacterial and Viral Strains | |||
| Single-stranded AAV (Serotype 1) | This paper | N/A | |
| Self-complementary AAV (Serotypes 1, 2, 3, 4, 5, 6, 8, 9) | This paper | N/A | |
| Biological Samples | |||
| Chemicals, Peptides, and Recombinant Proteins | |||
| Cas9–3xNLS | QB3 MacroLab, University of California, Berkeley | N/A | |
| Pregnant mare serum gonadotropin | Millipore | 367222 | |
| Human chorionic gonadotropin | Millipore | 230734 | |
| Critical Commercial Assays | |||
| HiScribe T7 High Yield RNA Synthesis Kit | New England Biolabs | E2040S | |
| Deposited Data | |||
| Experimental Models: Cell Lines | |||
| Human: HEK293T Cells | Manassas | CRL-3216 | |
| Experimental Models: Organisms/Strains | |||
| Mouse: C57BL/6J | The Jackson Laboratory | 000664 | |
| Mouse: B6.C-Tg(CMV-cre)1Cgn/J | The Jackson Laboratory | 006054 | |
| Mouse: CD1 | Charles River | Strain 022 | |
| Oligonucleotides | |||
| See Table S2 of this paper | |||
| Recombinant DNA | |||
| scAAV-CMV-GFP | Fu et al., 2003. | N/A | |
| pGEM-T easy | Promega | A1360 | |
| pX601 | Ran et al., 2015. | Addgene #61591 | |
| Sox2-P2A-mStrawberry | Gift from Stafford, D., University of California, Berkeley | N/A | |
| WT1–2A-eGFP | Bao et al., 2016. | Addgene #82333 | |
| pR26-CAG-AsiSI/MluI | Chu et al., 2016. | Addgene #74286 | |
| pAAV-FLEX-GFP | Edward Boyden (unpublished) | Addgene #28304 | |
| pAAV-nEFCas9 | Suzuki et al., 2016. | Addgene #87115 | |
| pX458 | Ran et al., 2013. | Addgene #48138 | |
| Software and Algorithms | |||
| ImageJ | Schneider et al., 2012. | https://imagej.nih.gov/ij/ | |
| ZEN | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html | |
| Other | |||
Acknowledgements
We thank MA Dewitt, J Corn, AJ Modzelewski, S Jin, C Jeans and T Gaj, for stimulating discussion and input on experimental design. We also thank D Stafford for providing reagents. LH is a Thomas and Stacey Siebel Distinguished Chair Professor, supported by a Howard Hughes Medical Institute (HHMI) Faculty Scholar award, a Bakar Fellow award at UC Berkeley, and several grants from the National Institutes of Health (NIH; 1R01GM114414, 1R21HD088885, 2R01CA139067, GRANT12095758). SC was supported by the Cancer Research Coordinating Committee Fellowship. SS was supported by the American Heart Association Predoctoral Fellowship, NIH R21 EB021572–01, and NIH R01 EY022975.
Footnotes
Declaration of Interests
DVS is an inventor on patents involving AAV directed evolution and a co-founder and a shareholder of a company developing AAV vectors for clinical gene therapy. In addition, he is on the board of directors of and holds shares in a second company developing AAV vectors for clinical gene therapy. SC and LH are inventors on patents involving an electroporation-based CRISPR technology for mouse genome engineering and are founders of a company to further develop this technology for mammalian genome editing.
References
- Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF, Sakuma T, Yamamoto T, Tanaka K, 2015. Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol. 16, 1–11. 10.1186/s13059-015-0653-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R, 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 17, 126–40. 10.1101/gad.224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso E, Mingozzi F, Montane J, Leon X, Anguela XM, Haurigot V, Edmonson SA, Africa L, Zhou S, High KA, Bosch F, Wright JF, 2010. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 17, 503–510. 10.1038/gt.2009.157 [DOI] [PubMed] [Google Scholar]
- Bak RO, Porteus MH, 2017. CRISPR-Mediated Integration of Large Gene Cassettes Using AAV Donor Vectors. Cell Rep. 20, 750–756. 10.1016/j.celrep.2017.06.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao X, Lian X, Hacker TA, Schmuck EG, Qian T, Bhute VJ, Han T, Shi M, Drowley L, Plowright A, Wang Q-D, Goumans M-J, Palecek SP, 2016. Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat. Biomed. Eng 1 10.1038/s41551-016-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Chen HY, Trumbauer ME, Yagle MK, Palmiter RD, 1985. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc. Natl. Acad. Sci. U. S. A 82, 4438–42. 10.1073/PNAS.82.13.4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD, 2011a. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755. 10.1038/nmeth.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Pruett-Miller SM, Huang Y, Gjoka M, Duda K, Taunton J, Collingwood TN, Frodin M, Davis GD, 2011b. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat. Methods 8, 753–755. 10.1038/nmeth.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lee B, Lee AY-F, Modzelewski AJ, He L, 2016a. Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J. Biol. Chem 291, 14457–67. 10.1074/jbc.M116.733154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Lee B, Lee AYF, Modzelewski AJ, He L, 2016b. Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J. Biol. Chem 291, 14457–14467. 10.1074/jbc.M116.733154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu VT, Weber T, Graf R, Sommermann T, Petsch K, Sack U, Volchkov P, Rajewsky K, Kühn R, 2016. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 16, 4 10.1186/s12896-016-0234-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–23. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J-Y, Fan P-D, Frizzell RA, 1996. Quantitative Analysis of the Packaging Capacity of Recombinant Adeno-Associated Virus. Hum. Gene Ther. 7, 2101–2112. 10.1089/hum.1996.7.17-2101 [DOI] [PubMed] [Google Scholar]
- Ferrari FK, Samulski T, Shenk T, Samulski RJ, 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol 70, 3227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri ALM, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, Nicolis SK, 2004. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 131, 3805–19. 10.1242/dev.01204 [DOI] [PubMed] [Google Scholar]
- Fisher KJ, Gao GP, Weitzman MD, DeMatteo R, Burda JF, Wilson JM, 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol 70, 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Muenzer J, Samulski RJ, Breese G, Sifford J, Zeng X, McCarty DM, 2003. Self-complementary adeno-associated virus serotype 2 vector: Global distribution and broad dispersion of AAV-mediated transgene expression in mouse brain. Mol. Ther 10.1016/j.ymthe.2003.08.021 [DOI] [PubMed] [Google Scholar]
- Fujii W, Kawasaki K, Sugiura K, Naito K, 2013. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 41, e187–e187. 10.1093/nar/gkt772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Schaffer DV, 2016. Adeno-associated virus-mediated delivery of CRISPR-cas systems for genome engineering in mammalian cells. Cold Spring Harb. Protoc 2016, 941–952. 10.1101/pdb.prot086868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Staahl BT, Rodrigues GMC, Limsirichai P, Ekman FK, Doudna JA, Schaffer DV, 2017. Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery. Nucleic Acids Res. 1–11. 10.1093/nar/gkx154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, Choi VW, Samulski RJ, 2006. Production and characterization of adeno-associated viral vectors. Nat. Protoc 1, 1412–1428. 10.1038/nprot.2006.207 [DOI] [PubMed] [Google Scholar]
- Gu B, Posfai E, Rossant J, 2018. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol 36, 632–637. 10.1038/nbt.4166 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Takemoto T, 2015. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep 5, 1–3. 10.1038/srep11315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Yamashita Y, Takemoto T, 2016. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol 418, 1–9. 10.1016/j.ydbio.2016.07.017 [DOI] [PubMed] [Google Scholar]
- Hiramoto T, Li LB, Funk SE, Hirata RK, Russell DW, 2018. Nuclease-free Adeno-Associated Virus-Mediated Il2rg Gene Editing in X-SCID Mice. Mol. Ther 10.1016/j.ymthe.2018.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata R, Chamberlain J, Dong R, Russell DW, 2002. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol 20, 735–738. 10.1038/nbt0702-735 [DOI] [PubMed] [Google Scholar]
- Hirsch ML, Green L, Porteus MH, Samulski RJ, 2010. Self-complementary AAV mediates gene targeting and enhances endonuclease delivery for double-strand break repair. Gene Ther. 17, 1175–1180. 10.1038/gt.2010.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Patel A, Ng R, Miller EB, Halder S, Mckenna R, Asokan A, Agbandje-mckenna M, 2016. Characterization of the Adeno-Associated Virus 1 and 6 Sialic Acid Binding Site 90, 5219–5230. 10.1128/JVI.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur JK, Kim K, Been KW, Baek G, Ye S, Hur JW, Ryu S-M, Lee YS, Kim J-S, 2016. Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleoproteins. Nat. Biotechnol 34, 807–808. 10.1038/nbt.3596 [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–21. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IF, Hirata RK, Russell DW, 2011. AAV-mediated gene targeting methods for human cells. Nat. Protoc 6, 482–501. 10.1038/nprot.2011.301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim J-S, 2014. Highly Efficient RNA-guide genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 128, 1–32. 10.1101/gr.171322.113.Freely [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, Bradley A, 2018. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol 36, 765 10.1038/nbt.4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotterman MA, Chalberg TW, Schaffer DV, 2015. Viral Vectors for Gene Therapy: Translational and Clinical Outlook. Annu. Rev. Biomed. Eng 17, 63–89. 10.1146/annurev-bioeng-071813-104938 [DOI] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu Meizhen, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu Mingyao, 2013. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol 31, 681–683. 10.1038/nbt.2661 [DOI] [PubMed] [Google Scholar]
- Li H, Beckman KA, Pessino V, Huang B, Weissman JS, Leonetti MD, 2017. Design and specificity of long ssDNA donors for CRISPR-based knock-in. bioRxiv 178905. 10.1101/178905 [DOI] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL, 2015. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol 33, 538–542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM, 2008. Self-complementary AAV vectors; advances and applications. Mol. Ther 16, 1648–1656. 10.1038/mt.2008.171 [DOI] [PubMed] [Google Scholar]
- McCarty DM, Fu H, Monahan PE, Toulson CE, Naik P, Samulski RJ, 2003. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 10, 2112–2118. 10.1038/sj.gt.3302134 [DOI] [PubMed] [Google Scholar]
- Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M, 2017. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protoc 13, 195–215. 10.1038/nprot.2017.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka Y, Uno Y, Yoshimi K, Kunihiro Y, Yoshimura T, Tanaka Tomohiro, Ishikubo H, Hiraoka Y, Takemoto N, Tanaka Takao, Ooguchi Y, Skehel P, Aida T, Takeda J, Mashimo T, 2018. CLICK: one-step generation of conditional knockout mice. BMC Genomics 19, 318 10.1186/s12864-018-4713-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modzelewski AJ, Chen S, Willis BJ, Lloyd KCK, Wood JA, He L, 2018. Efficient mouse genome engineering by CRISPR-EZ technology. Nat. Protoc 13, 1253–1274. 10.1038/nprot.2018.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Gertenstein M, Vintersten K, Behringer R, 2003. Manipulating the Mouse Embryo: A Laboratory Manual Third Edition., Third. ed Cold Spring Harbor Laboratory Press; 10.1086/380032 [DOI] [Google Scholar]
- Nakagawa Y, Sakuma T, Nishimichi N, Yokosaki Y, Yanaka N, Takeo T, Nakagata N, Yamamoto T, 2016. Ultra-superovulation for the CRISPR-Cas9-mediated production of gene-knockout, single-amino-acid-substituted, and floxed mice. Biol. Open 5, 1142–8. 10.1242/bio.019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher M, Weber T, 2012. Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther 19, 649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka M, Sato M, Miura H, Takabayashi S, Matsuyama M, Koyano T, Arifin N, Nakamura S, Wada K, Gurumurthy CB, 2018. I-GONAD: A robust method for in situ germline genome engineering using CRISPR nucleases. Genome Biol. 19, 1–15. 10.1186/s13059-018-1400-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, Redder R, Richardson GP, Inagaki Y, Sakai D, Buckley SM, Seshacharyulu P, Batra SK, Behlke MA, Zeiner SA, Jacobi AM, Izu Y, Thoreson WB, Urness LD, Mansour SL, Ohtsuka M, Gurumurthy CB, 2017. Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol. 18, 92 10.1186/s13059-017-1220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F, 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. 10.1038/nature14299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F, 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy S, Chenouard V, Tesson L, Usal C, Ménoret S, Brusselle L, Heslan J-M, Nguyen TH, Bellien J, Merot J, De Cian A, Giovannangeli C, Concordet J-P, Anegon I, 2017. Generation of gene-edited rats by delivery of CRISPR/Cas9 protein and donor DNA into intact zygotes using electroporation. Sci. Rep 7, 16554 10.1038/s41598-017-16328-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DW, Hirata RK, 1998. Human gene targeting by viral vectors. Nat. Genet 18, 325–330. 10.1038/ng0498-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K, 1995. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23, 5080–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler J, Zevnik B, Küter-Luks B, Andreas S, Kern H, Hennek T, Rode A, Heimann C, Faust N, Kauselmann G, Schoor M, Jaenisch R, Rajewsky K, Kühn R, Schwenk F, 2003. Rapid generation of inducible mouse mutants. Nucleic Acids Res. 31, e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z, Kurita M, Hishida T, Li M, Aizawa E, Guo S, Chen S, Goebl A, Soligalla RD, Qu J, Jiang T, Fu X, Jafari M, Esteban R, Travis Berggren W, Lajara J, Nuñez-Delicado E, Guillen P, Campistol JM, Matsuzaki F, Liu G-H, Magistretti P, Zhang Kun, Callaway EM, Zhang Kang, Carlos Izpisua Belmonte J, 2016. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540 10.1038/nature20565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi G, Gurumurthy CB, Wada K, Miura H, Sato M, Ohtsuka M, 2015. GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: a novel microinjection independent genome engineering method in mice. Sci. Rep 5, 11406 10.1038/srep11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanihara F, Takemoto T, Kitagawa E, Rao S, Do LTK, Onishi A, Yamashita Y, Kosugi C, Suzuki H, Sembon S, Suzuki S, Nakai M, Hashimoto M, Yasue A, Matsuhisa M, Noji S, Fujimura T, Fuchimoto D.-i., Otoi T, 2016. Somatic cell reprogramming-free generation of genetically modified pigs. Sci. Adv 2, e1600803–e1600803. 10.1126/sciadv.1600803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberghe LH, Xiao R, Lock M, Lin J, Korn M, Wilson JM, 2010. Efficient Serotype-Dependent Release of Functional Vector into the Culture Medium During Adeno-Associated Virus Manufacturing. Hum. Gene Ther. 21, 1251–1257. 10.1089/hum.2010.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R, 2013. One-step generation of mice carrying mutations in multiple genes by CRISPR/cas-mediated genome engineering. Cell 153, 910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kutny PM, Byers SL, Longstaff CJ, DaCosta MJ, Pang C, Zhang Y, Taft RA, Buaas FW, Wang H, 2016. Delivery of Cas9 protein into mouse zygotes through a series of electroporation dramatically increased the efficiency of model creation. J. Genet. Genomics 10.1016/j.jgg.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R, 2013. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–9. 10.1016/j.cell.2013.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Zhang M, Wang X, Ying W, Hu X, Dai P, Meng F, Shi L, Sun Y, Yao N, Zhong W, Li Y, Wu K, Li W, Chen Z, Yang H, 2018. Tild-CRISPR Allows for Efficient and Precise Gene Knockin in Mouse and Human Cells. Dev. Cell 45, 526–536.e5. 10.1016/J.DEVCEL.2018.04.021 [DOI] [PubMed] [Google Scholar]
- Yoon Y, Wang D, Tai PWL, Riley J, Gao G, Rivera-Pérez JA, 2018. Streamlined ex vivo and in vivo genome editing in mouse embryos using recombinant adeno-associated viruses. Nat. Commun 9, 412 10.1038/s41467-017-02706-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K, Kunihiro Y, Kaneko T, Nagahora H, Voigt B, Mashimo T, 2016. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat. Commun 7, 10431 10.1038/ncomms10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.