Abstract
Iron is an essential but poorly bioavailable nutrient due to its low solubility, especially in alkaline soils. Here, we describe the discovery of a novel redox-active catecholic metabolite, termed sideretin, which derives from the coumarin fraxetin, and is the primary molecule exuded by Arabidopsis thaliana roots in response to iron deficiency. We have identified two enzymes that complete the biosynthetic pathway of fraxetin and sideretin. Chemical characterization of fraxetin and sideretin, and biological assays with pathway mutants, suggest that these coumarins are critical for iron nutrition in A. thaliana. Further, we show that sideretin production occurs in eudicot species only distantly related to A. thaliana. Untargeted metabolomics of the root exudates of various eudicots revealed production of structurally diverse redox-active molecules in response to iron deficiency. Our results indicate that secretion of small-molecule reductants by roots may be a widespread and previously underappreciated component of reduction-based iron uptake.
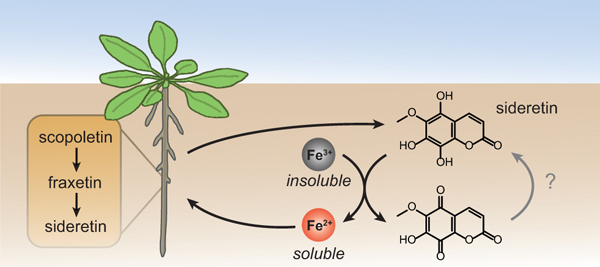
Graphical Abstract
Introduction
Iron (Fe) is a key micronutrient required for the growth of all organisms, fulfilling numerous biochemical roles. In plants, iron is essential for various cellular processes, such as respiration and chlorophyll biosynthesis, and serves as a cofactor for many enzymes involved in electron or oxygen transfer processes, such as cytochrome P450s (CYPs) and 2-oxoglutarate-dependent dioxygenases (2-ODDs)1,2.
While nominally abundant in the Earth’s crust, iron is only poorly bioavailable due to the low solubility product of the ferric form Fe(III) with several common soil anions, such as hydroxide and phosphate. Even though the ferrous form Fe(II) is more soluble, it is readily oxidized to Fe(III) in aerated soils. To cope with poor iron availability, land plants have evolved at least two distinct strategies for iron mobilization and uptake3,4. The chelation-based mechanism, also known as Strategy II, is confined to true grasses (family Poaceae) and relies on the exudation5 and re-uptake of mugineic acid-type phytosiderophores6,7, which solubilize and bind Fe(III)8. All other plant families are thought to employ a reduction-based mechanism (Strategy I), which involves acidification of the rhizosphere via proton secretion9, enzymatic reduction of iron chelates at the root surface10, and secondary active transport of Fe(II) across the plasma membrane11,12. Efficient root-mediated Fe(III) reduction in Strategy I plants depends on the presence of soluble Fe(III)-chelates derived, e.g., from the soil13. Several phenylpropanoid pathway enzymes are upregulated under iron deficiency in Arabidopsis14,15, suggesting that low molecular-weight compounds released by Strategy I plants are also involved in mobilization of iron from insoluble pools in the soil or root apoplast16,17; however, this observation has not been explained mechanistically.
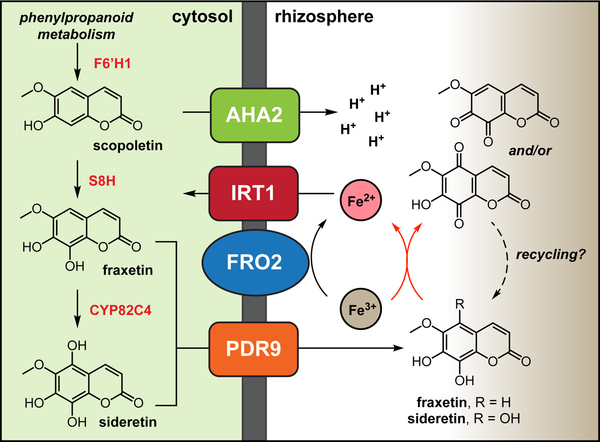
Recently, several groups have reported that root exudation of phenylpropanoid-derived coumarins are required for efficient iron uptake, especially in alkaline conditions15,18–20. The oxoglutarate-dependent oxidase F6’H1 (refs. 15,19–21) and the ABC-type transporter PDR9 (refs. 18,22) have been shown to be involved in coumarin biosynthesis and export, respectively. In line with a role for coumarins in Fe acquisition, both f6’h1 and pdr9 mutants showed severe symptoms of iron deficiency relative to wild-type plants when iron was supplied in an insoluble form15,18–20. Catecholic coumarins can potentially contribute to Fe acquisition by two mechanisms: solubilization of ferric Fe precipitates and reduction of Fe(III) to Fe(II) for direct import23. However, in reports to date, the two known natural catecholic coumarins found in the exudates of iron-deficient wild-type plants, esculetin and fraxetin, were produced at insufficient levels to explain the Fe deficiency of f6’h1 and pdr9 plants, while scopoletin, lacking a catechol motif, is incapable of iron chelate formation or reduction18,19. It has been suggested that scopoletin may be further oxidized to catecholic coumarins, perhaps in the rhizosphere18,19.
Here, we describe the identification of a novel catecholic coumarin, termed sideretin (5,7,8-trihydroxy-6-methoxycoumarin), produced by A. thaliana. We show that sideretin is the major coumarin exuded into the rhizosphere in response to iron deficiency under acidic conditions, whereas alkaline conditions stimulate exudation of less-oxidized coumarins such as fraxetin. Fraxetin and sideretin are biosynthesized from scopoletin by two hydroxylases: a 2-ODD (S8H), which generates fraxetin, and a CYP enzyme (CYP82C4), which can further oxidize fraxetin to generate sideretin. Both sideretin and fraxetin efficiently mobilize and reduce insoluble Fe(III), and rescue chlorotic phenotypes of susceptible mutants and wild-type plants grown under conditions of low iron availability. We also show that the sideretin biosynthetic pathway arose early during angiosperm evolution, but appears to have been lost in various lineages. Interestingly, several Strategy I plants that do not produce catecholic coumarins exude structurally distinct redox-active molecules under iron deficiency. Together, these data suggest that exudation of various redox-active and/or iron-binding small molecules into the rhizosphere is a key component of iron acquisition in plants.
Results
Fraxetin and sideretin are biosynthesized from scopoletin
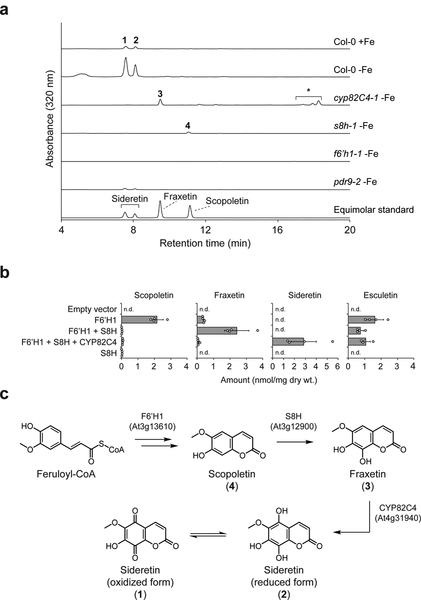
In order to obtain a comprehensive picture of the metabolic changes that occur in A. thaliana plants under iron deficiency, we carried out comparative metabolomic studies of Columbia-0 (Col-0) ecotype seedlings grown hydroponically under iron-abundant and iron-depleted conditions at pH 5.7. In our setup, seeds were germinated on a buoyant PTFE mesh floating on liquid medium. This cultivation method allowed for axenic growth, easy separation of root and aerial tissue, and collection of spent medium without injuring the roots, which could lead to spurious observation of naturally root-confined metabolites in the medium. Comparative analysis of spent medium methanolic extracts of 12 day-old Col-0 seedlings by HPLC-ESI-MS revealed two UV-active peaks (1 and 2) that strongly accumulated under iron-depleted conditions (Fig. 1a). The corresponding m/z [M+H]+ values of 223.0237 and 225.0394 are consistent with neutral compound molecular formulas of C10H6O6 (1) and C10H8O6 (2), respectively. Compound 2 differs from scopoletin, the primary coumarin present in A. thaliana roots, by two additional hydroxylations, while 1 is the dehydrogenated counterpart of 2. We observed only trace quantities of the known coumarins scopoletin, fraxetin, and esculetin in Col-0 exudates (Supplementary Fig. 1a). We note that m/z = 225.0394 has been detected as a highly-induced signal in root exudates under iron deficiency in previous MS-based metabolomic studies18–20, but neither the structure nor the importance of this metabolite has been elucidated. In root tissues grown under iron-depleted conditions, we primarily observed accumulation of mass signals corresponding to the glucosides of 1 (Supplementary Figs. 1b and 2), while no substantial metabolic differences were found between iron-sufficient and iron-deficient aerial tissues using our reverse-phase liquid chromatography analysis method.
Figure 1 |. Metabolomics of T-DNA insertion lines and heterologous gene expression reveal the sideretin biosynthetic pathway in A. thaliana.
(a) Representative UV-Vis absorbance traces for spent medium extracts of hydroponically grown A. thaliana lines under Fe deficiency at pH = 5.7; data are representative of three biological replicates. Numbered peaks correspond to compounds shown in c. *, non-specific cycloaddition products of fraxetin with certain alcohols (cleomiscosins). (b) Levels of oxidized coumarins in glucosidase-treated extracts of N. benthamiana leaves transiently expressing subsets of sideretin pathway enzymes. Data represent mean ± s.d. of four biological replicates; n.d.: not detected. Both experiments were repeated two times with similar results. (c) Schematic of sideretin biosynthesis in A. thaliana.
We postulated that compound 2 represents a doubly hydroxylated analog of scopoletin with a catechol moiety, and 1 is the corresponding quinone. Since we could not easily obtain enough of these compounds for structural characterization directly from spent medium, we chose instead to profile various T-DNA insertion lines potentially deficient in their biosynthesis in order to gain clues about their structures. First, we profiled knockout lines of feruloyl-CoA 6’-hydroxylase 1 (F6’H1), the enzyme responsible for scopoletin biosynthesis in A. thaliana21. A complete absence of both compounds as well as other prominent UV-active peaks in medium of f6’h1 lines grown under iron-depleted conditions demonstrates that they are indeed scopoletin-derived (Fig. 1a). Next, we sought to identify biosynthetic enzymes responsible for the oxidation of scopoletin. One strong candidate is CYP82C4 (At4g31940), which encodes a cytochrome P450 enzyme, whose expression is upregulated by iron deficiency and correlates to the Fe(II) root importer IRT1 under various abiotic stress conditions (Supplementary Fig. 3)24.
Furthermore, CYP82C4 has been shown to hydroxylate 8-methoxypsoralen, a furanocoumarin structurally similar to scopoletin25. We therefore obtained two independent cyp82C4 T-DNA insertion lines for profiling. Quantitative PCR (qPCR) analysis confirmed that CYP82C4 expression in roots is strongly induced under iron deficiency, but abolished in these T-DNA lines (Supplementary Fig. 4a). Metabolomic profiling under iron-depleted conditions further showed that both 1 and 2 are absent from root exudates of these lines. However, unlike for f6’h1, another prominent UV-active peak (3) was observed in cyp82C4 spent medium, with m/z [M+H]+ = 209.0444, corresponding to C10H8O5. When compared with an authentic standard, this compound was identified as fraxetin (8-hydroxyscopoletin, 3) (Fig. 1a). Therefore, we suspected that fraxetin is the direct precursor leading to 1 and 2. To further test this hypothesis, we expressed CYP82C4 in yeast and incubated the microsomal fraction with fraxetin and NADPH. As expected, we observed in vitro formation of compounds identical to Col-0-derived 1 and 2 (Supplementary Fig. 5a). Taken together, these data indicate that CYP82C4 is responsible for the final step in the biosynthetic pathway leading to 1 and 2 from scopoletin. Interestingly, cyp82C4 spent medium contains additional UV-active peaks that correspond to non-specific cycloaddition products of fraxetin with coniferyl and sinapyl alcohols (also referred to as “cleomiscosins”26; Supplementary Fig. 6).
Based on the known activity of CYP82C4 as an 8-methoxypsoralen 5-hydroxylase25, we inferred that it likely hydroxylates fraxetin at the structurally analogous position to yield a novel coumarin we have termed sideretin (5-hydroxyfraxetin). We synthesized sideretin from fraxetin using a phthaloyl peroxide-mediated hydroxylation strategy27 (Supplementary Fig. 7), and confirmed its identity to 2 (Supplementary Fig. 5b). Sideretin readily oxidizes (e.g. on exposure to air or during column chromatography) to yield 1, corresponding to the quinone analog (Supplementary Fig. 8), and can be re-reduced to the hydroquinone upon treatment with H2 over palladium.
To complete the biosynthetic pathway of sideretin, we sought an enzyme with scopoletin 8-hydroxylase (S8H) activity. By searching publicly available transcriptome data, we identified At3g12900, a gene encoding a 2-ODD, which is coexpressed with CYP82C4, upregulated under iron deficiency (Supplementary Fig. 3), and has 48% sequence identity to F6’H1 at the amino acid level. We therefore obtained two independent T-DNA insertion lines for functional analysis. Similarly to CYP82C4, qPCR analysis confirmed that At3g12900 expression is strongly induced in iron deficient-roots, but abolished in the two selected T-DNA lines (Supplementary Fig. 4b). Consistent with the proposed activity of this enzyme, we observed exudation of scopoletin (4) but no further oxidized coumarins in these lines (Fig. 1a). We attempted to express and purify At3g12900 protein in E. coli and N. benthamiana for in vitro biochemical characterization, but could not readily obtain active protein, evidently due to instability (Supplementary Fig. 9).
In lieu of complete in vitro biochemical characterization, we attempted to reconstitute the proposed pathway in vivo by transiently expressing subsets of pathway enzymes in N. benthamiana and analyzing metabolite extracts by LC-MS (Fig. 1b). None of the pathway coumarins are present at detectable levels in N. benthamiana leaves expressing only the empty vector. However, expression of F6’H1 results in accumulation of scopoletin (Fig. 1b). Notably, leaves expressing both F6’H1 and At3g12900 proteins accumulate fraxetin but not scopoletin, further confirming that At3g12900 functions as a scopoletin 8-hydroxylase (S8H). Finally, expression of all three pathway enzymes leads to accumulation of sideretin but no pathway intermediates, further confirming the activity of CYP82C4 (Fig. 1b). Together, these data outline the complete biosynthetic pathway of sideretin from feruloyl-CoA, a lignin precursor found ubiquitously in higher plants (Fig. 1c). Interestingly, expression of F6’H1 also leads to accumulation of esculetin (5), whose levels are not significantly affected by expression of downstream sideretin pathway enzymes (Fig. 1b). It is not clear whether esculetin arises here via demethylation of scopoletin or side activity of F6’H1 on caffeoyl-CoA, although this latter activity was not seen in vitro21.
Finally, we profiled the pdr9–2 mutant, which is known to be defective in export of various coumarins from the roots18,22. The level of sideretin in spent medium for this mutant is approximately 10-fold lower than for wild-type plants (Fig. 1a and Supplementary Fig. 1a), while levels of sideretin glucosides in the roots are significantly elevated (Supplementary Figs. 1b and 2), indicating that PDR9 is the principal sideretin root exporter.
Catecholic coumarins mobilize Fe(III) in vitro
The structure of sideretin is immediately suggestive of its function: Catecholic compounds bind to the Fe3+ ion, usually forming intensely colored complexes (Supplementary Fig. 10), can reduce Fe(III) to Fe(II), and are therefore well-suited for facilitating iron uptake in Strategy I plants. The possibility that catecholic coumarins are important for iron uptake in A. thaliana has been suggested previously, and it is known that f6’h1 knockouts incapable of producing scopoletin (and therefore, as we have now shown, fraxetin and sideretin) are notably impaired in their iron uptake capacity at elevated pH19,20. We therefore hypothesized that sideretin is the major catecholic coumarin facilitating iron uptake from soil in A. thaliana. To explore this possibility, we performed a variety of chemical and biological assays with relevant coumarins and pathway mutants.
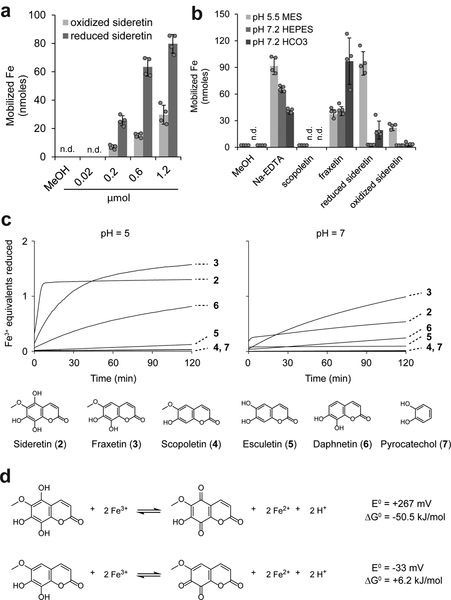
First, we compared the iron mobilization characteristics of coumarins in vitro (Fig. 2). Fraxetin and sideretin, but not scopoletin, can efficiently solubilize iron from hydroxide precipitates (Fig. 2a,b). A kinetic Fe(III) reduction assay further demonstrated that the configuration of the catechol moiety and presence of other substituents on the coumarin scaffold have substantial effects on Fe(III) reduction kinetics (Fig. 2c). In particular, fraxetin can reduce Fe(III) faster than esculetin and daphnetin (6), both of which are catecholic coumarins but lack a methoxy substituent. Sideretin, in turn, exhibits even faster initial kinetics, but reduces less than the expected two equivalents of Fe(III) under the conditions of our assay, possibly due to instability at higher pH (Fig. 2c). We also measured the redox potentials of fraxetin and sideretin by cyclic voltammetry (Fig. 2d and Supplementary Fig. 11): the additional hydroxylation on sideretin lowers the reduction potential by 300 mV with respect to fraxetin (Fig. 2d and Supplementary Fig. 11), consistent with the observation that sideretin is readily oxidized on exposure to air whereas fraxetin is not (Supplementary Fig. 8). In comparison to fraxetin and scopoletin, sideretin showed high sensitivity to light (Supplementary Fig. 12). Together, these data demonstrate that catecholic coumarins can in general solubilize and reduce Fe(III), and that specific structural variations have substantial impacts on kinetic and thermodynamic parameters that govern overall iron mobilization capacity.
Figure 2 |. Chemical characterization of the iron mobilization capacity of coumarins.
(a,b) Fe(III) mobilization from iron hydroxide precipitates (pH 7.2) by reduced or air-oxidized sideretin at various concentrations (a) and comparison to other catecholic coumarins at 0.6 micromoles (b). Data represent the mean ± s.d. of four independent in vitro assays. (c) Traces showing Fe(III) reduction kinetics by various compounds at pH 5 or 7, monitored spectrophotometrically by formation of Fe(II)–ferrozine complex. The experiment was repeated two times with similar results. (d) Standard free-energy changes and redox potentials for Fe(III) reduction by sideretin and fraxetin, calculated from cyclic voltammetry measurements (see Supplementary Fig. 11) and the literature value of +0.77 V (vs. SHE) for Fe(III) reduction (Fe3+ + e− → Fe2+).
Catecholic coumarins mediate iron uptake in vivo
To test the importance of the sideretin pathway for iron mobilization in vivo, we first investigated the localization of S8H and CYP82C4 expression. Similarly to F6’H1 (ref. 19), the Fe deficiency-dependent upregulation of S8H and CYP82C4 was mainly confined to roots, where the promoter activity of either gene was detected along the primary root and lateral roots, except for the apical root zones (Supplementary Figure 13a,c). At the cell type-specific level, both S8H and CYP82C4 were expressed most strongly in the root epidermal layer under iron-deficient conditions (Supplementary Fig. 13b,d), consistent with their function in biosynthesizing a compound secreted into the rhizosphere. Notably, F6’H1 is also expressed in the cortex19, suggesting that the conversion of F6’H1-generated scopoletin into fraxetin and sideretin takes place mainly in epidermal cells.
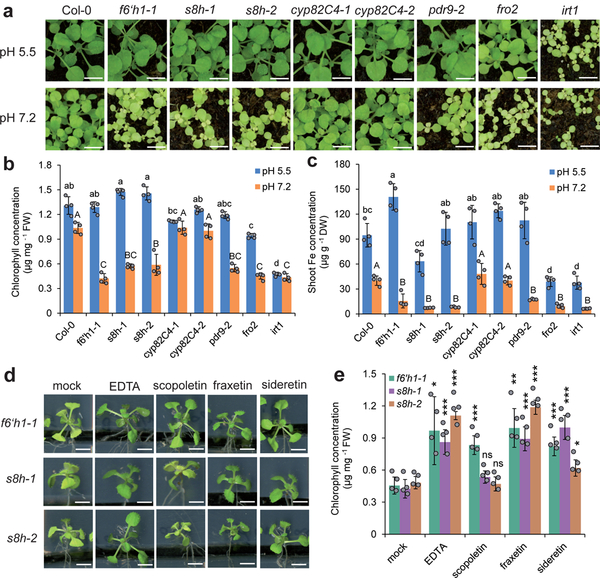
Next, we compared the growth of wild-type plants with that of various pathway mutants, as well as several other lines known to be impaired in aspects of iron uptake. Due to the photosensitivity of sideretin (Supplementary Fig. 12), we initially performed our phenotypic characterization in peat-based soil substrate. In unmodified soil (pH = 5.5), sideretin pathway mutants (f6’h1, s8h, cyp82C4) and the exporter mutant pdr9–2 showed no significant differences from wild-type seedlings in appearance (Fig. 3a), chlorophyll (Fig. 3b), or Fe concentrations (Fig. 3c). In contrast, when these plants were grown in soil in which iron availability was decreased by adjusting the pH to 7.2 with calcium carbonate and bicarbonate, f6’h1–1, pdr9–2 and s8h lines were visibly more chlorotic and stunted (Fig. 3a), with significantly lower chlorophyll (Fig. 3b) and Fe concentrations (Fig. 3c). Surprisingly, growth of cyp82C4 lines was not impaired under high pH conditions, and in fact appeared more robust than wild-type, at least in terms of biomass (Supplementary Fig. 14). Similar results were obtained on agar plate experiments with FeCl3 as the sole iron source, except that the f6’h1–1 line showed mild chlorosis even at pH = 5.5 (Supplementary Fig. 15). To rule out the possibility that exudation of scopoletin alone is capable of mediating iron uptake, we tested whether we could complement the phenotypes of f6’h1 and s8h seedlings grown on agar by exogenous addition of various coumarins. We found that fraxetin and sideretin could rescue chlorosis in both f6’h1–1 and s8h seedlings, while scopoletin could rescue f6’h1–1 but not s8h seedlings (Fig. 3d,e). Thus, whereas externally supplied scopoletin can be further processed to catecholic coumarins in f6’h1–1, this cannot occur in s8h lines, and scopoletin alone cannot reverse the phenotype, even when provided in abundance. Together, these results provide direct evidence for the importance of catecholic compound secretion into the rhizosphere in iron mobilization.
Figure 3 |. Phenotypic characterization and complementation assays for s8h, cyp82C4 and other mutants under conditions of low Fe availability.
(a–c) Appearance of shoots (a), leaf chlorophyll concentration (b), and Fe concentration (c) of wild-type (Col-0) and various mutant plants grown for 21 days on standard substrate at pH 5.5 or limed substrate at pH 7.2. Data represent mean ± s.d. of four independent replicates containing five shoots each. Different letters at pH 5.5 or pH 7.2 indicate significant differences according to one-way ANOVA with post hoc Tukey’s test (p < 0.05). (d,e) Appearance of shoots (d) and leaf chlorophyll concentration (e) of f6’h1–1 and s8h mutant lines grown for 7 days on low Fe availability and supplied with indicated compound (150 μM). Data represent mean ± s.d. of four replicates containing five shoots each. *p < 0.05, **p < 0.01, ***p < 0.001 and n.s., not significant (p > 0.05), two-tailed t-test (pairwise comparison with mock treatment for each line separately). Scale bars, 0.5 cm. Both experiments were repeated two times with similar results.
Interestingly, our data suggest that CYP82C4-mediated conversion of fraxetin to sideretin may actually be maladaptive for iron uptake at high pH: sideretin is apparently a poorer iron mobilization agent than fraxetin at higher pH (Fig. 2), and may not form stable complexes with Fe(III) (Supplementary Fig. 10). Moreover, cyp82C4 knockout lines slightly outperform wild-type plants in certain respects when grown under high pH conditions (Fig. 3c, Supplementary Figs. 14 and 15b,c). Given these observations, we decided to compare the exuded coumarin profile of iron-deficient plants grown at pH 5.7 and pH 7.3 in our hydroponic setup. We found that, at pH = 7.3, while sideretin is still the major exuded coumarin in Col-0 plants, substantial amounts of fraxetin, esculetin, and cleomiscosins also accumulate. This contrasts strongly with the exudates collected at pH = 5.7, in which only negligible amounts of coumarins besides sideretin could be detected (Supplementary Fig. 16). This observation suggests that the composition of root exudates of A. thaliana responds not only to iron nutritional status, but also to soil pH.
Various redox-active molecules are secreted by eudicots
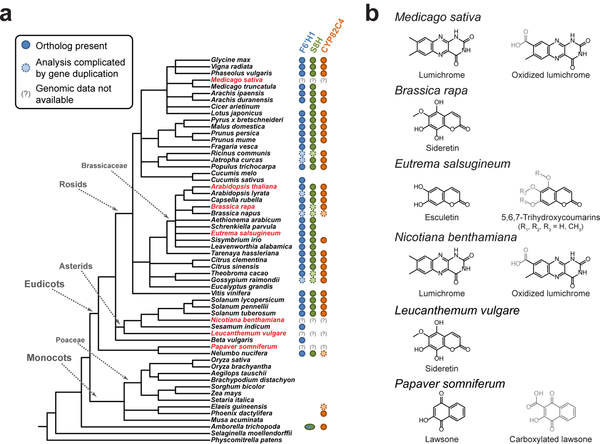
Finally, in order to explore the phylogenetic distribution of the sideretin pathway, we used two strategies to identify putative orthologs of the three A. thaliana sideretin biosynthetic genes in 54 sequenced plant species. For members of the Brassicaceae family, we performed shared synteny analysis using Genome Browser28 (Supplementary Fig. 17), whereas for more divergent species, we performed a reciprocal best BLAST hit search via the KEGG database29
(Supplementary Data Set 1). In general, sideretin pathway orthologs appear to be widespread in eudicots, with sporadic loss of all or part of the pathway in certain lineages (Fig. 4a). Interestingly, while F6’H1 and S8H are conserved in all Brassicaceae species analyzed, CYP82C4 has been independently lost in several members of this family. Furthermore, the basal angiosperm Amborella trichopoda contains a CYP82C4 ortholog, as well as a single F6’H1/S8H-like ortholog. However, all pathway orthologs are conspicuously absent from graminaceous (Strategy II) plants, suggesting that Fe acquisition mediated by coumarins and phytosiderophores likely do not co-exist in nature. Given these facts, coupled with the high sequence similarity of F6’H1 and S8H, we speculate that this pathway arose early in angiosperm evolution, partly via a gene duplication event that gave rise to the F6’H1/S8H paralog pair, but has been supplanted by alternative iron mobilization strategies in various lineages.
Figure 4 |. Phylogenetic distribution of sideretin pathway enzyme orthologs and Fe deficiency-induced small molecule exudation in various plants.
(a) Presence of sideretin pathway orthologs in sequenced plants, inferred from shared synteny or reciprocal best BLAST hit analysis. Amborella trichopoda, the basal angiosperm, contains a single f6’h1/s8h-like ortholog. Species highlighted in red were those profiled for small molecule exudation under iron deficiency in this study. (b) Structures of major redox-active and/or iron-binding compounds secreted in response to iron deficiency in profiled species.
To experimentally validate some of the conclusions drawn from the phylogenetic analysis, we profiled root exudates of several plant species amenable to axenic growth in modified versions of our hydroponic platform under iron-abundant and iron-depleted conditions at pH = 5.7. We first investigated whether loss of CYP82C4 in a member of the Brassicaceae family correlates with loss of sideretin production. Towards this end, we grew Brassica rapa and Eutrema salsugineum, which have, respectively, retained and lost CYP82C4. Notably, E. salsugineum is an extremophile that preferentially inhabits alkaline salt flats30, unlike A. thaliana or B. rapa. As expected, B. rapa exudes sideretin under iron-depleted conditions, whereas E. salsugineum exudes a mixture of oxidized coumarins, primarily esculetin, isoscopoletin (8), and various methylated isomers of 5,6,7-trihydroxycoumarin, but no quadruply oxidized coumarins like sideretin (Fig. 4b and Supplementary Fig. 18b). At present, how these coumarins are biosynthesized in E. salsugineum is unclear. However, this finding does suggest that different chemical strategies—in this case, exudation of oxidized coumarins with specific iron binding and/or redox properties—likely evolved to fit different species’ ecological requirements.
We further profiled four different eudicot species, Medicago sativa, Nicotiana benthamiana, Leucanthemum vulgare, and Papaver somniferum, for small molecule exudation in response to iron deficiency. The choice of these species was motivated chiefly by ease of obtaining seeds and cultivating seedlings in our hydroponic setup; while none of these species are presently included in the KEGG database, precluding ortholog analysis, they do sample widely divergent eudicot lineages (Fig. 4a). We found that L. vulgare exudes sideretin (Fig. 4b and Supplementary Fig. 19a), demonstrating that the complete biosynthetic pathway was in place before the divergence of the rosids and asterids approximately 125 million years ago31. In contrast, both M. sativa and N. benthamiana exude lumichrome (9), the redox-active moiety of the flavin cofactors, as well as an oxidized derivative (10) whose structure is not entirely clear (Fig. 4b and Supplementary Fig. 19b,c). The exudation of modified riboflavins in response to iron deficiency has previously been demonstrated in Beta vulgaris32 (which lacks S8H and CYP82C4 orthologs) and has recently been implied by transcriptomics analysis in Medicago truncatula15. Intriguingly, we also observed iron deficiency-induced exudation of acetosyringone (11) by N. benthamiana and various flavonoids (12, 13) by M. sativa (Supplementary Fig. 19b,c). These compounds do not possess obvious redox-active or iron-binding moieties, but are known to mediate plant-microbe interactions33. Finally, we detected lawsone (14) in P. somniferum exudates (Fig. 4b and Supplementary Fig. 19d), as well as a carboxylated derivative (15), which we suspect is produced via hydroxylation of 1,4-dihydroxy-2-naphthoic acid, a phylloquinone precursor that is ubiquitous in plants. Remarkably, lawsone possesses the same 1,2,4-trihydroxybenzene substitution pattern seen in sideretin, but on a naphthalene rather than a coumarin scaffold. Although we have not yet performed detailed biochemical characterization of these compounds, the fact that all eudicot species tested by us exude redox-active and/or iron-binding molecules under iron deficiency suggests this phenomenon may be a common and previously underappreciated feature of Strategy I iron uptake.
Discussion
While root exudation of compounds capable of binding and/or reducing iron in various Strategy I plants was first observed over four decades ago34,35, the relative importance of this phenomenon in iron mobilization and the underlying biochemical mechanisms have remained obscure. More recently, studies with A. thaliana have shown that synthesis19,20 and export18,36 of coumarins are crucial for efficient iron uptake under iron-limiting conditions, but still failed to explain the mechanisms underlying this effect. Here, we have identified a widely conserved pathway for the biosynthesis of catecholic coumarins in Strategy I plants and demonstrated the biochemical basis for the role of these compounds in iron mobilization from soil (Fig. 5).
Figure 5 |. Model for soil iron uptake in Arabidopsis thaliana.
Under Fe deficiency conditions, a branch of the phenylpropanoid pathway consisting of F6’H1, S8H and CYP82C4 is strongly induced, leading to the synthesis of scopoletin and its conversion into redox-active fraxetin and sideretin. These molecules are found as glycosides in the root (Supplementary Fig. 2) but are drawn as the aglycones for simplicity. Deglycosylation likely occurs as a step coupled to the export process. Secretion of oxygenated coumarins to the rhizosphere is mediated by the ATP-binding cassette protein PDR9. Sideretin is invariably the most abundant coumarin exuded by roots of Fe-deficient plants (and is shown as the sole secreted coumarin for simplicity), although at high pH considerable amounts of esculetin and fraxetin can be detected in the root exudates. In the rhizosphere, the redox-active coumarins act in concert with other components of the strategy I iron acquisition machinery (AHA2, FRO2 and IRT1) by solubilizing and reducing Fe from sparingly available sources. AHA2: Arabidopsis H+-ATPase 2 (At4g30190).
The secretion of redox-active metabolites from plant roots is reminiscent of the small-molecule electron shuttles used by certain bacterial species for mineral reduction. For instance, phenazines produced by Pseudomonas are thought to contribute to iron acquisition, while flavins serve as electron shuttles for anaerobic Shewanella, enhancing the rate of extracellular respiration of Fe(III)37,38. Although the reactivity of sideretin to light has made it difficult to probe whether a related mechanism of redox cycling occurs near plant roots, our findings are consistent with a role for sideretin in extracellular electron transport. It will be interesting to probe whether plant-derived catecholic coumarins act alone, or instead link root surface reductases to otherwise inaccessible pools of precipitated Fe(III).
While our work paves the way for a deeper mechanistic understanding of the role of small molecules in iron mobilization, several unresolved questions remain. Specifically, the importance of CYP82C4-mediated conversion of fraxetin to sideretin is unclear, and we have thus far been unable to find any beneficial phenotype for wild-type plants in comparison to cyp82C4 knockouts. We suspect that the unique redox capacity of sideretin and the marked preference for its production at low pH are relevant under certain conditions, which however may be difficult to capture experimentally. At high pH, production of less oxidized coumarins like fraxetin and esculetin could be preferred simply due to their enhanced stability and ability to stably chelate Fe(III), but the relative importance of chelation and reduction may be difficult to disentangle in vivo. It is also possible that secreted coumarins mediate interactions with rhizosphere microbes, and fraxetin and sideretin may play different roles in such interactions.
Further, despite the wide conservation of the sideretin pathway in eudicots, numerous additional chemical strategies for iron mobilization apparently exist, and a broader search than the one we have undertaken here will likely reveal others. Future efforts will entail uncovering the biosynthesis of these additional molecules and clarifying the link between specific chemical strategies and iron mobilization capacity under environmental conditions affecting iron bioavailability.
To date, most efforts in understanding soil iron uptake limitations have focused on the role of soil pH, ignoring other potentially relevant factors such as interactions with soil organic matter or other metals like Zn(II) or Mn(II), which are not redox-active in soil but are also imported by IRT1. With a detailed biochemical understanding of the pathways involved, we can now approach some of these problems. Ultimately, we hope that this knowledge can be applied towards engineering crop varieties with enhanced iron acquisition capacity, enabling growth in otherwise unsuitable environments.
Online Methods
Chemical synthesis
For detailed chemical synthesis procedures and compound characterization, please see the Supplementary Note.
Plant lines used
The following homozygous T-DNA insertion lines from the SALK or SM collections or previously described lines were used in this study (all mutants are in the Col-0 background unless otherwise noted, with locus of affected gene indicated): f6’h1–1 (At3g13610, SALK_132418C); f6’h1–2 (At3g13610, SALK_050137C); s8h-1 (At3g12900, SM_3_27151); s8h-2 (At3g12900, SM_3_23443); cyp82C4–1 (At4g31940, SALK_001585); cyp82C4–2 (At4g31940, SM_3_25388); pdr9–2 (At3g53280, SALK_050885); fro2 (At1g01580 (ref. 39)); irt1 (At4g19690, in the Wassilewskija (Ws) background40). Seeds for SM lines were obtained from the Nottingham Arabidopsis Stock Centre (NASC), irt1 seeds were a gift of Catherine Curie, while all other A. thaliana seeds were obtained from the Arabidopsis Biological Resource Center (ABRC). Zygosity was confirmed by standard PCR methods using primer sequences obtained from the SALK T-DNA primer tool (http://signal.salk.edu/tdnaprimers.2.html).
Seeds of Eutrema salsugineum (≡ Thellungiella salsuginea) Shandong ecotype (CS22504) were obtained from ABRC. Seeds of Rapid Cycling Brassica rapa (stock no. 1–33) were obtained from the Rapid Cycling Brassica Collection at the University of Wisconsin-Madison. Seeds of the following plant species were purchased from Horizon Herbs (Williams, OR): Papaver somniferum (Poppy, Turkish Red); Leucanthemum vulgare (≡ Chrysanthemum leucanthemum; Daisy, Oxeye); Medicago sativa (Alfalfa). Seeds of Nicotiana benthamiana were a gift of the Mary Beth Mudgett lab at Stanford University.
Preparation of MS liquid medium and agar plates
For plant metabolomics experiments, 1x Murashige-Skoog (MS) medium stock without Fe was prepared from individual components according to the following recipe (all amounts are per liter of medium):
| Sucrose | 5.0 g |
| Potassium nitrate | 1.9 g |
| Ammonium nitrate | 1.65 g |
| MES monohydrate | 0.5 g |
| Calcium chloride dihydrate | 0.44 g |
| Magnesium sulfate heptahydrate | 0.37 g |
| Monopotassium phosphate | 0.17 g |
| myo-Inositol | 0.1 g |
| Disodium EDTA | 29.2 mg |
| Manganese sulfate monohydrate | 16.9 mg |
| Zinc sulfate heptahydrate | 8.6 mg |
| Boric acid | 6.2 mg |
| Glycine | 2.0 mg |
| Potassium iodide | 0.83 mg |
| Nicotinic acid | 0.5 mg |
| Pyridoxine hydrochloride | 0.5 mg |
| Sodium molybdate dihydrate | 0.25 mg |
| Thiamine hydrochloride | 0.1 mg |
| Cobalt chloride hexahydrate | 25.0 μg |
| Copper chloride pentahydrate | 25.0 μg |
For hydroponics experiments at pH = 7.3, 0.5 g of HEPES was used instead of MES monohydrate in the above recipe; pH was adjusted to the desired value (5.7 or 7.3) using 2 M potassium hydroxide. Fe-free MS medium can be stored at room temperature in the dark for extended periods. Fe was added as sterile 100 mM FeSO4 solution immediately prior to use to avoid precipitation of iron phosphates.
Agar growth plates were prepared by adding agar (10 g/L) to 0.5x MS medium without sucrose or MES/HEPES buffer. Iron was added as iron(II) sulfate heptahydrate prior to autoclaving.
A. thaliana metabolomics growth conditions and sample harvest
For a typical seedling metabolomics experiment, A. thaliana seeds were surface sterilized for 5 minutes in a 20% Clorox bleach solution with 0.1% Tween-20, washed three times with water, and stratified at 4°C for 2 days before planting. Circular “rafts” 3 cm in diameter were cut out from a 0.025” x 0.005” opening size PTFE mesh (McMaster-Carr) and sterilized by autoclaving. 3 mL of standard MS medium (0.5% sucrose, 100 μM sodium EDTA, 5 mM MES, pH = 5.7) containing either 100 μM FeSO4 (Fe-sufficient condition) or 10 μM FeSO4 (Fe-deficient condition) was added to the wells of a 6-well plate (Corning), a PTFE mesh raft placed on the medium surface in each well, and 16 seeds individually placed in a square pattern on the surface of the raft with a pipette. The plates were sealed with microporous tape (EMD Millipore), and wrapped in aluminum foil to etiolate and thereby reduce wetting of aerial tissues. The plants were grown in a growth chamber under a 16 h light cycle (photon flux of 100 μmol m–2 s–1, 22°C, 50% relative humidity); the aluminum foil was removed 3 d after planting. 8 d after planting, medium was removed from all wells and replaced with 3 mL of fresh medium containing either 100 μM FeSO4 (iron-sufficient condition) or no added FeSO4 (iron-deficient condition). After a further 4 d of growth, the roots were washed in water, and the roots, aerial tissues, and medium harvested separately, transferred to pre-weighed (for tissue samples only) microfuge tubes and flash-frozen prior to storage at −80°C.
Growth conditions and tissue harvest for other plant species
The A. thaliana raft growth protocol described above was modified to accommodate growth of numerous other species under iron deficiency. Various aspects of the protocol – seed sterilization, stratification and etiolation times, growth container size and medium amount, raft mesh size, amount of iron initially supplied, and the total growth time – were adjusted to attain axenic growth resulting in observable differences in phenotype (specifically, chlorosis) between the +Fe and -Fe condition at the time of harvest. Specific details for all species are shown in Supplementary Table 1. In all cases, for the +Fe control condition, the medium contained 100 μM FeSO4, and was also exchanged for fresh medium containing 100 μM FeSO4 at the time indicated in Supplementary Table 1.
Tissue and medium extraction
Tissue and medium extraction were performed as described previously41, with the following modifications: 80:20 MeOH/H2O was used as the extraction solvent in all cases. For tissue extraction, 60–1,000 μL solvent per mg of dry tissue was used, the exact ratio depending on tissue mass to obtain sufficient extract for analysis. For medium extraction, 1 mL of spent medium was lyophilized and resuspended in 100 μL of solvent by brief vortexing, followed by 5 min of sonication.
LC-MS analysis
LC-MS analysis was performed as described previously41 except that 5 μL injection volumes were used in all cases.
Transcriptomics data mining
All microarray datasets for transcriptome mining and analysis were obtained from the AtGenExpress series (stress set data42) or Dinneny et al. (root iron deficiency timecourse data)43.
Bioinformatics analysis
Shared synteny analysis for Brassicaceae species was performed and visualized using the UCSC Genome Browser at http://mustang. biol.mcgill.ca:8885/cgi-bin/hgGateway.
To compile the list of putative sideretin pathway enzyme orthologs, a “Best-Best” (reciprocal best BLAST hit) ortholog search was initially performed using the KEGG database (http://www.genome.jp/kegg/) with each of the three A. thaliana sideretin pathway genes (At3g13610 (F6’H1), At3g12900 (S8H), At4g31940 (CYP82C4)) against all plant species (see e.g. ref. 44 for a discussion of this strategy). The “Best-Best” search was iterated with each of the candidates identified in the initial search, as well as any additional genes subsequently found, to generate a graph where vertices represent genes and edges represent “Best-Best” relationships. The (unique) highly connected subgraph containing the A. thaliana pathway gene defines the final set of putative orthologs (a subgraph with n vertices is considered “highly connected” if the number of edges incident to each vertex is at least[n/2]+1). For the purpose of this analysis allowance for recent duplication events (which can obfuscate “Best-Best” ortholog relationships) was made by considering as orthologs sets of paralogous genes in a given species that collectively satisfy the connectivity criterion, and moreover individually have at least three “Best-Best” hits in other species. KEGG identifiers for all putative orthologs, paralogs, and percent amino acid sequence identities for reciprocal best BLAST hit relationships are listed in Supplementary Data Set 1.
Cloning of sideretin pathway genes and enzyme purification
The CYP82C4 gene sequence was amplified from cDNA-bearing plasmid RAFL09–57-O03 (RIKEN resource number pda08472) using the following primers and cloned into pYeDP60 vector using standard methods (restriction sites in bold):
CYP82C4_F (BamHI):
5’-GTACTACTTAGGATCCATGGATACTTCCCTCTTTTCTTTG-3’
CYP82C4_R (EcoRI):
5’- GATATCGTAGAATTCACACAAAAAGTTCTTCCTTAATACGTGG-3’
The cDNA clone was found to contain an insertion within a stretch of 7 repeated T nucleotides (positions 726–732 within the coding sequence). This mutation was repaired via overlap extension PCR using the following pair of complementary primers:
CYP82C4_Repair_Fragment_2_F: 5’-CCGACACTAAGTTTTTTTGATTTGCAAGGACATGAGAAGGAG-3’
CYP82C4_Repair_Fragment_1_R:
5’-CTCCTTCTCATGTCCTTGCAAATCAAAAAAACTTAGTGTCGG-3’
Yeast (strain WAT11) transformation using the pYeDP60-CYP82C4 construct and microsome isolation were performed as described previously41.
The coding sequence of S8H (At3g12900) was amplified directly from A. thaliana Col-0 genomic DNA by PCR using the following primers and cloned into AgeI- and XmaI-digested pEAQ-HT vector45 using Gibson assembly46:
S8H_F:
5’-GTATATTCTGCCCAAATTCGCGACCGGTATGGGTATCAATTTCGAGGACCAAACCAC A-3’
S8H_+6xHis_R:
5’- GTGATGGTGATGGTGATGCCCGGGCTCGGCACGTGCGAAGTCGAGAGATTTTTT-3’
Transformation of A. tumefaciens GV3101, infiltration into N. benthamiana leaves, and attempted protein purification were performed as described previously41.
Enzyme assays with CYP82C4 microsomes
Enzymatic reactions were carried out at room temperature in 50 mM sodium phosphate buffer (pH = 6.5). The reaction mix contained 100 μM fraxetin, 1 mM NADPH, 2 mM tris(2-carboxyethyl)phosphine (TCEP), and 5 mg/mL total microsomal protein. After 1 h of incubation, reactions were quenched by adding one volume of MeOH, precipitates removed by centrifugation, and supernatants analyzed by LC-MS.
Heterologous pathway reconstitution in N. benthamiana
Sequences encoding sideretin pathway genes were amplified from previously generated plasmids or from a cDNA library with the following primers (primers for S8H are listed above):
F6’H1_pEAQ_F:
#x2019;-GTATATTCTGCCCAAATTCGCGACCGGTATGGCTCCAACACTCTTGACAACCC-3’
F6’H1_pEAQ_R:
5’-GTGATGGTGATGGTGATGCCCGGGGATCTTGGCGTAATCGACGGTTTTCTTTC-3’
CYP82C4_pEAQ_F:
5’-GTATATTCTGCCCAAATTCGCGACCGGTATGGATACTTCCCTCTTTTCTTTGTTTGTT-3’
CYP82C4_pEAQ_R:
5’-GTGATGGTGATGGTGATGCCCGCACAAAAAGTTCTTCCTTAATACGTGG-3’
Amplicons were inserted into AgeI- and XmaI-digested pEAQ-HT vector using Gibson assembly46. Transformation of A. tumefaciens GV3101, infiltration into N. benthamiana leaves, and harvest 3 days post-infiltration were carried out as described previously41.
N. benthamiana tissue was initially extracted and analyzed as described in the “Tissue and medium extraction” section above; 80:20 MeOH/H2O was used for extraction, without any treatment. In this case, sideretin pathway products were detected exclusively as glucosides.
To enable direct quantification of oxidized coumarin aglycones, 100 μL volumes of N. benthamiana tissue methanolic extracts were dried under argon and resuspended in 200 μL of a 1 mg/mL solution of almond β-glucosidase (Sigma-Aldrich) in 100 mM sodium acetate buffer (pH = 5.0) and incubated at 37°C for 1 h. The reactions were subsequently frozen in liquid nitrogen, lyophilized, and resuspended in 200 μL of 80:20 MeOH/H2O prior to LC-MS analysis. The tissue content of coumarin aglycones measured in this way was found to be in excellent agreement (±5%) with the corresponding glucoside content measured in the untreated methanolic tissue extracts for those coumarins where a glucoside standard was available (scopoletin, fraxetin, and esculetin), allowing us to estimate the ionizability of glucosylated sideretin.
UV-Vis scanning kinetics with sideretin
Hydrogen gas was bubbled through a 50 μM solution of sideretin in 10 mM ammonium formate buffer (pH = 3.0) in the presence of granular palladium with vigorous stirring to obtain the pure reduced form of sideretin. The solution was degassed, put under argon, and transferred to a sealed quartz cuvette, which was immediately placed in an Agilent Cary 60 Spectrophotometer. The cuvette was unsealed to expose the solution to air and absorbance spectra (250–700 nm) were acquired for 6 hours at 5 min intervals as the sideretin slowly oxidized.
Static in vitro iron mobilization
Iron mobilization capacity was determined by incubating different compounds with freshly-prepared 0.1 mM Fe hydroxide. The pH was adjusted to either 5.5 with MES or 7.2 with HEPES or NaHCO3. Coumarins, EDTA or methanol (mock) were added to Fe hydroxide solution and incubated at constant shaking for 1 hour. Solutions were then filtered through Chromafil CA-45/25 (0.45 μm pore) filters, and the Fe contained in the filtrates was detected by sector-field high-resolution inductively coupled plasma mass spectrometry (HR)-ICP-MS (ELEMENT 2, Thermo Fisher Scientific, Germany). Elemental standards were prepared from certified reference single standards from CPI International (USA).
Spectrophotometric iron reduction kinetics assays
Britton-Robinson (BR) buffer spanning the pH range 2–12 in increments of 1 pH unit was prepared as described previously47; briefly, BR buffer contains 40 mM each boric, acetic, and phosphoric acid titrated to the desired pH with 200 mM sodium hydroxide. Assays were run in a 96-well plate format: each well contained 100 μL of BR buffer, 50 μL of 4 mM FeCl3 in water (800 μM final concentration), 50 μL of 10 mM ferrozine in water (2 mM final concentration), and 50 μL of 400 μM reducing agent in 50:50 DMSO/H2O (80 μM final concentration), or 50 μL of 50:50 DMSO/H2O as control. Upon addition of reducing agent, formation of Fe2+-ferrozine complex was monitored by measuring absorbance at 562 nm for 4 h in 20 s intervals on an xMark Microplate Absorbance Reader (Bio-Rad). Standard absorbance curves were generated for Fe2+-ferrozine complex (iron(II) sulfate heptahydrate serving as the source of Fe2+) at each pH tested to enable determination of absolute Fe2+ concentrations.
Cyclic voltammetry measurements
1 mM solutions of fraxetin and sideretin were prepared in 50 mM phosphate buffer, pH = 6.5 and cyclic voltammograms acquired over a glassy carbon electrode at 25°C using a CHI 760D electrochemical workstation (CH Instruments) with the following parameters: Scan rate: 0.1 V/s; Sample interval: 0.001 V; Quiet time: 0; Sensitivity: 0.1 mA/V. Measurements were taken with an Ag/AgCl reference electrode (data shown in Supplementary Fig. 12), corrected (+0.200 V) by comparison to a normal hydrogen electrode reference measurement (1 M HClO4), and further corrected for pH (59 mV per pH unit; +0.384 V) to yield values vs. the standard hydrogen electrode (pH = 0). The average of the two potentials at maximal current for each voltammogram was used to estimate the standard redox potential for each compound.
Coumarin light sensitivity assay
30 μL of sterilized 1 mM coumarin (scopoletin, fraxetin, or sideretin) solution was added to 3 mL of MS medium, pH = 5.7 containing 10 μM FeSO4, and the medium samples transferred a 6-well plate. The plates were incubated under standard hydroponic growth conditions described above; for the “dark” condition, the plate was wrapped in aluminum foil for the duration of the experiment. Every 24 h, a 750 μL sample was withdrawn from each well and analyzed by LCMS as described above to determine remaining coumarin concentrations.
Phenotypic characterization in soil substrate
Seeds were germinated and cultivated on peat-based substrate. The pH of the substrate was either maintained at pH 5.6 or increased to pH 7.2 by supplementation with 5 g/kg CaCO3 and 15 g/kg NaHCO3. Each insertion mutant was grown on substrate pH 5.6 and pH 7.2 (n = four pots containing 15 plants per mutant and treatment) for 21 days. The pots were placed inside a conditioned growth chamber with a 22°C/18°C and 16-h/8-h light/dark regime at a light intensity of 120 μmol photons m−2 s−1.
Phenotypic characterization on agar plates
Seeds were surface sterilized with a solution containing 70% (v/v) ethanol and 0.05% (v/v) Triton X-100. Seeds were then sown on sterile plates containing half-strength Murashige and Skoog (MS) medium with 40 μM Fe-EDTA, supplemented with 0.5% sucrose, 2.5 mM MES (pH 5.6) and solidified with 1% (w/v) Difco agar (Becton Dickinson). The plates were incubated at 4°C for 2 days and afterwards placed in a vertical position inside growth cabinets under a 22°C/18°C and 10/14-h light/dark regime and light intensity adjusted to 120 μmol photons m−2 s−1. After 10 days, seedlings were transferred to new agar plates containing half-strength MS medium with variable Fe availability and cultivated for another 6 days. Depending on the experiment (see figure legends), plants were supplemented with 40 μM Fe-EDTA or no external Fe at pH 5.6 (2.5 mM MES buffer); or supplied with 20 μM freshly prepared FeCl3 at pH 5.6 (2.5 mM MES buffer) or pH 7.2 (2.5 mM HEPES buffer or 100 μM NaHCO3). In coumarin resupply experiments, 150 μM of various coumarins (dissolved in methanol) were supplied to solidified agar plates 6 hours before transferring plants. Equivalent amounts of methanol (mock) and 150 μM Fe-EDTA served, respectively, as negative and positive controls in these experiments. In order to avoid shoot contact with the exogenously supplied compounds, agar plates were split horizontally and treatments were supplied only in the lower, root-containing compartment.
Shoot chlorophyll and Fe quantification
Chlorophylls were extracted from whole shoots with N,N’-dimethyl formamide (Merck) at 4°C for 24 hours. Absorption at 647 nm and 664 nm was then determined in the extracts following the protocol described by Porra et al.48. For Fe measurements, whole shoot samples were dried for 48 h at 65°C and digested with HNO3 in polytetrafluoroethylene vials in a pressurized microwave digestion system (UltraCLAVE IV, MLS GmbH). Iron concentrations were analyzed by sector-field high-resolution inductively coupled plasma mass spectrometry (HR-ICP-MS; ELEMENT 2, Thermo Fisher Scientific, Germany). Element standards were prepared from certified reference standards from CPI-International (USA).
Generation of GUS reporter lines
The promoter sequences of S8H (2023 bp) and CYP82C4 (1151 bp) were amplified from genomic DNA of A. thaliana (accession Col-0) using the following primers:
ggS8Hpro-for: 5’-AACAGGTCTCAACCTGCAGAACCGAAATTAGTACCG-3’
ggS8Hpro-rev: 5’-AACAGGTCTCATGTTTCTCCACACTTCTGCTTGAAAA-3’
ggCYP82C4pro-for: 5’-AACAGGTCTCAACCTGCTCTTTGTGGGCTTTTTGGAT-3’
ggCYP82C4pro-rev: 5’-AACAGGTCTCATGTTGAGAGTGCAGAAGAGATGTGTGT-3’
The constructs proS8H:GUS, proCYP82C4:GUS, proS8H:3xGFP and proCYP82C4:3xGFP were assembled in a pGREEN-IIS based binary vector using the GreenGate method as described previously49. The resulting plasmids were then transformed into Agrobacterium tumefaciens strain GV3101 containing the helper plasmid pSOUP. Transformation into A. thaliana plants (Col-0) was performed by the floral dip method50. Positive transformants were selected on agar plates containing 10 μg ml−1 phosphinothricin (proS8H:GUS and proCYP82C4:GUS), 50 μg ml−1 hygromycin (proS8H:3xGFP) or 50 μg ml−1 kanamycin (proCYP82C4:3xGFP).
Staining and imaging of GUS reporter lines
Whole seedlings of proS8H:GUS or proCYP82C4:GUS were incubated in GUS staining solution (2 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide, 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6·3H2O, 10 mM Na2EDTA, 0.1% Triton X-100, and 50 mM sodium phosphate, pH 7.0) at 37°C for 6 hours. Before imaging, samples were cleared with an ethanol series (30%, 50%, 70% and 100%). The GUS-stained plants were examined under a microscope (Axio Imager 2, Zeiss, Germany), and digital images were captured using Zeiss AxioVision software (version LE 4.8.2.0). Ten seedlings per independent transgenic line (six independent lines per construct) and treatment were analyzed and representative images are shown.
Confocal microscopy imaging of GFP reporter lines
Confocal images of proS8H:3xGFP and proCYP82C4:3xGFP roots were acquired with a LSM 780 (Zeiss, Germany) laser scanning microscope equipped with a 10x/0.45 M27 objective. Prior to imaging, roots were stained with propidium iodide (10 μg ml−1) for 10 min. GFP-dependent fluorescence was detected by excitation at 488 nm with an argon laser and filtering the emitted light at 536 nm. The 561-nm excitation and 648-nm emission lines were used to image the propidium iodide-derived fluorescence. ZEN software (Zeiss, Germany) was used for adjustments and image recording.
RT-PCR gene expression analysis
Plants were grown axenically on agar plates, and total RNA was extracted from roots of either Fe-sufficient or -deficient plants using NucleoSpin RNA Plant kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions.
Total RNA was reverse-transcribed into cDNA with the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Germany), using oligo(dT)-primers and following the manufacturer’s instructions. The complementary DNA samples were then used to investigate gene expression by quantitative real-time PCR using the CFX384 Touch Real-Time PCR Detection System (Bio-Rad, Germany) and iQ SYBR Green Supermix (Bio-Rad, Germany) using the primers listed below. Relative expression was calculated according to Pfaffl51.
qPCR_F6H1_for: 5’-TGATATCTGCAGGAATGAAACG-3’
qPCR_F6H1_rev: 5’-GGGTAGTAGTTAAGGTTGACTC-3’
qPCR_CYP82C4_for: 5’-CCTTACATGGGCCATTTCTC-3’
qPCR_ CYP82C4_rev: 5’-TCCTCGACGTTCCTGTCTCT-3’
qPCR_S8H_for: 5’-TCAAGAGTCTCTGTGCCCATT-3’
qPCR_S8H_rev: 5’-TCCTTGTAACGAGCCACTCC-3’
qPCR_UBQ2_for: 5’-CCAAGATCCAGGACAAAGAAGGA-3’
qPCR_UBQ2_rev: 5’-TGGAGACGAGCATAACACTTGC-3’
Statistical analysis
Unless otherwise noted, statistical analysis was performed using Microsoft Excel 2016 for two-tailed Student’s t test comparisons, or SigmaPlot 11.0 for one-way ANOVA comparisons with post hoc Tukey’s test for statistical significance.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This work was supported by the US National Institutes of Health grant DP2 AT008321 and an HHMI-Simons Faculty Scholar Award to E.S.S. and by a grant of the Deutsche Forschungsgemeinschaft to N.v.W. (WI1728/21–1). J.R. was supported by an NIH biotechnology training grant (T32 GM008412–20). We thank Nicole Schmid, Jacqueline Fuge, Annett Bieber, Heike Nierig, and Mathias Voges for valuable discussions and help with experiments, Sean Elliott for advice on redox potential measurements, Scott Fendorf for helpful discussions on rhizosphere iron, M. Kevin Brown and Justin Du Bois for suggestions for chemical synthesis of sideretin, and Thomas Veltman for help with cyclic voltammetry measurements.
Footnotes
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Hänsch R & Mendel RR Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Current Opinion in Plant Biology 12, 259–266 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Balk J & Pilon M Ancient and essential: the assembly of iron–sulfur clusters in plants. Trends in Plant Science 16, 218–226 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Palmer CM & Guerinot ML Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat Chem Biol 5, 333–340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi T & Nishizawa NK Iron Uptake, Translocation, and Regulation in Higher Plants. Annu. Rev. Plant Biol. 63, 131–152 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Nozoye T et al. Phytosiderophore Efflux Transporters Are Crucial for Iron Acquisition in Graminaceous Plants. J. Biol. Chem 286, 5446–5454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curie C, Panaviene Z, Loulergue C & Dellaporta SL Maize yellow stripe1 encodes a membrane protein directly involved in Fe (III) uptake. Nature 409, 346–349 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Murata Y et al. A specific transporter for iron(III)-phytosiderophore in barley roots. The Plant Journal 46, 563–572 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Takagi S-I Naturally occurring iron-chelating compounds in oat- and rice-root washings. Soil Science and Plant Nutrition 22, 423–433 (1976). [Google Scholar]
- 9.Santi S & Schmidt W Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytologist 183, 1072–1084 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Robinson NJ, Procter CM, Connolly EL & Guerinot ML A ferric-chelate reductase for iron uptake from soils. Nature 397, (1999). [DOI] [PubMed] [Google Scholar]
- 11.Eide D, Broderius M, Fett J & Guerinot ML A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proceedings of the National Academy of Sciences 93, 5624–5628 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Connolly EL Expression of the IRT1 Metal Transporter Is Controlled by Metals at the Levels of Transcript and Protein Accumulation. THE PLANT CELL ONLINE 14, 1347–1357 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romheld V & Marschner H Evidence for a Specific Uptake System for Iron Phytosiderophores in Roots of Grasses. PLANT PHYSIOLOGY 80, 175–180 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lan P et al. iTRAQ Protein Profile Analysis of Arabidopsis Roots Reveals New Aspects Critical for Iron Homeostasis. PLANT PHYSIOLOGY 155, 821–834 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Celma J et al. Mutually Exclusive Alterations in Secondary Metabolism Are Critical for the Uptake of Insoluble Iron Compounds by Arabidopsis and Medicago truncatula. PLANT PHYSIOLOGY 162, 1473–1485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin CW et al. Iron Deficiency-Induced Secretion of Phenolics Facilitates the Reutilization of Root Apoplastic Iron in Red Clover. PLANT PHYSIOLOGY 144, 278–285 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sisó-Terraza P, Rios JJ, Abadía J, Abadía A & Álvarez-Fernández A Flavins secreted by roots of iron-deficient Beta vulgaris enable mining of ferric oxide via reductive mechanisms. New Phytol 209, 733–745 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Fourcroy P et al. Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol 201, 155–167 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Schmid NB et al. Feruloyl-CoA 6’-Hydroxylase1-Dependent Coumarins Mediate Iron Acquisition from Alkaline Substrates in Arabidopsis. PLANT PHYSIOLOGY 164, 160–172 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt H et al. Metabolome Analysis of Arabidopsis thaliana Roots Identifies a Key Metabolic Pathway for Iron Acquisition. PLoS ONE 9, e102444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kai K et al. Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. The Plant Journal 55, 989–999 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Ziegler J, Schmidt S, Strehmel N, Scheel D & Abel S Arabidopsis Transporter ABCG37/PDR9 contributes primarily highly oxygenated Coumarins to Root Exudation. Scientific Reports 2017 7:1 7, 3704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mladěnka P et al. In vitro interactions of coumarins with iron. Biochimie 92, 1108–1114 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Murgia I, Tarantino D, Soave C & Morandini P Arabidopsis CYP82C4 expression is dependent on Fe availability and circadian rhythm, and correlates with genes involved in the early Fe deficiency response. Journal of Plant Physiology 168, 894–902 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Kruse T et al. In Planta Biocatalysis Screen of P450s Identifies 8-Methoxypsoralen as a Substrate for the CYP82C Subfamily, Yielding Original Chemical Structures. Chemistry & Biology 15, 149–156 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Ray AB et al. Structures of cleomiscosins, coumarinolignoids of cleome viscosa seeds. Tetrahedron 41, 209–214 (1985). [Google Scholar]
- 27.Yuan C et al. Metal-free oxidation of aromatic carbon–hydrogen bonds through a reverse-rebound mechanism. Nature 499, 192–196 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Fujita PA et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Research 39, D876–D882 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amtmann A Abiotic Stress and Plant Genome Evolution. Search for New Models. PLANT PHYSIOLOGY 138, 127–130 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith SA, Beaulieu JM & Donoghue MJ An uncorrelated relaxed-clock analysis suggests an earlier origin for flowering plants. Proceedings of the National Academy of Sciences 107, 5897–5902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Susín S et al. Flavin excretion from roots of iron-deficient sugar beet (Beta vulgaris L.). Planta 193, 514–519 (1994). [Google Scholar]
- 33.Peters NK Current Review Phenolic Compounds as Regulators of Gene Expression in Plant-Microbe Interactions. MPMI 3, 4 (1990). [DOI] [PubMed] [Google Scholar]
- 34.BROWN JC, CHANEY RL & AMBLER JE A New Tomato Mutant Inefficient in the Transport of Iron. Physiol Plant 25, 48–53 (1971). [Google Scholar]
- 35.BROWN JC & AMBLER JE ‘Reductants’ Released by Roots of Fe-Deficient Soybeans. Agronomy Journal 65, 311 (1973). [Google Scholar]
- 36.Sisó-Terraza P et al. Accumulation and Secretion of Coumarinolignans and other Coumarins in Arabidopsis thaliana Roots in Response to Iron Deficiency at High pH. Front. Plant Sci. 7, 327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brutinel ED & Gralnick JA in Microbial Metal Respiration 83–105 (Springer; Berlin Heidelberg, 2012). doi: 10.1007/978-3-642-32867-1_4 [DOI] [Google Scholar]
- 38.Price-Whelan A, Dietrich LEP & Newman DK Rethinking ‘secondary’ metabolism: physiological roles for phenazine antibiotics. Nat Chem Biol 2, 71–78 (2006). [DOI] [PubMed] [Google Scholar]
References (Online Methods)
- 39.Yi Y & Guerinot ML Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. The Plant Journal 10, 835–844 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Vert G IRT1, an Arabidopsis Transporter Essential for Iron Uptake from the Soil and for Plant Growth. THE PLANT CELL ONLINE 14, 1223–1233 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajniak J, Barco B, Clay NK & Sattely ES A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature 525, 376–379 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kilian J et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal 50, 347–363 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Dinneny JR et al. Cell Identity Mediates the Response of Arabidopsis Roots to Abiotic Stress. Science 320, 942–945 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Fang G, Bhardwaj N, Robilotto R & Gerstein MB Getting Started in Gene Orthology and Functional Analysis. PLoS Comput Biol 6, e1000703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sainsbury F, Thuenemann EC & Lomonossoff GP pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnology Journal 7, 682–693 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Gibson D One-step enzymatic assembly of DNA molecules up to several hundred kilobases in size. Protocol Exchange (2009). doi: 10.1038/nprot.2009.77 [DOI] [PubMed] [Google Scholar]
- 47.Britton HTS & Robinson RA LXI.—The use of the antimony–antimonous oxide electrode in the determination of the concentration of hydrogen ions and in potenliometric titrations. The Prideaux–Ward universal buffer mixture. J. Chem. Soc 0, 458–473 (1931). [Google Scholar]
- 48.Porra RJ, Thompson WA & Kriedemann PE Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA) - Bioenergetics 975, 384–394 (1989). [Google Scholar]
- 49.Lampropoulos A et al. GreenGate - A Novel, Versatile, and Efficient Cloning System for Plant Transgenesis. PLoS ONE 8, e83043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clough SJ & Bent AF Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Pfaffl MW A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, 45e–45 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.