ABSTRACT
Heterochromatic regions of the genome are epigenetically regulated to maintain a heritable ‘“silent state”’. In fission yeast and other organisms, epigenetic silencing is guided by nascent transcripts, which are targeted by the RNA interference pathway. The key effector complex of the RNA interference pathway consists of small interfering RNA molecules (siRNAs) associated with Argonaute, assembled into the RNA-induced transcriptional silencing (RITS) complex. This review focuses on our current understanding of how RITS promotes heterochromatin formation, and in particular on the role of Argonaute-containing complexes in many other functions such as quelling, release of RNA polymerases, cellular quiescence and genome defense.
KEYWORDS: RNAi, epigenetics, silencing, argonaute, RITS, RNA, quelling, dicer, genome defense, DNA repair, quiescence, cell cycle
Introduction
RNA interference was discovered two decades ago as a posttranscriptional sequence-specific gene silencing mechanism in the nematode Caenorhabditis elegans [1–3]. A few years later, transcriptional centromeric silencing in fission yeast (Schizosaccharomyces pombe) was also found to rely on small interfering RNA (siRNA) molecules (~21 to 24 nucleotides) and components of the RNA interference pathway [4]. RNA interference has since been shown to be well-conserved in the vast majority of the eukaryotes, and is mechanistically related to other RNA silencing processes previously described in fungi and plants, like quelling, co-suppression, transcript processing, polymerase elongation and release, and post-transcriptional gene silencing (PTGS) [5–10]. In addition, RNA interference has roles beyond cellular control of gene expression, such as in protection of the genome against mobile repetitive DNA sequences and more broadly on the epigenetic regulation of eukaryotic genomes [6,7,11–13]. RNA interference also functions as an antiviral mechanism in plants and insects [14]. Components of the RNA interference pathway have now been implicated in wide array of cellular functions including growth, development, apoptosis, but have also been implicated in non-physiological processes such as cancer [15].
Transcription-coupled RNA interference during the assembly of heterochromatin in fission yeast as a model system has provided considerable insight in understanding the molecular details of the process. The key factors involved in heterochromatin formation in higher eukaryotic species are conserved in fission yeast [16–18], and several studies suggest that the mechanisms of heterochromatin assembly might be conserved from fission yeast to humans [19–22]. Heterochromatin formation in fission yeast relies on the RNA interference pathway and in particular on a complex of proteins called RITS (RNA-induced initiation of transcriptional gene silencing complex) [18]. The RITS complex contains siRNAs and is the major effector of RNA interference. Heterochromatin assembly is associated with large-scale chromatin condensation to regulate a variety of chromosomal processes, such as centromere formation. Heterochromatin mediates proper segregation of chromosomes and facilitates long-range chromatin interactions between distant chromosomal regions [23–27]. Additionally, heterochromatin represses the transcription and recombination of repetitive DNA elements, which due to their ability to transpose or recombine with other elements are a major cause of genomic instability [28–30].
Argonaute is the catalytic component of the RITS complex and this review will largely focus on the role of Argonaute as an effector of RNA interference. It is also important to note that phylogenetically Argonautes can be divided into two subclades: the Ago subclade and the Piwi subclade. PIWI proteins bind a specific class of 24–32 nucleotide long small RNAs termed PIWI-interacting RNAs (piRNAs), which can also trigger heterochromatin formation via H3K9me and HP1 [31]. piRNAs are thought to constitute a defense against transposable elements in the germline, where PIWI proteins are predominantly expressed and play crucial roles in early development. Due to space limitations, we refer the reader interested in PIWI, piRNA biogenesis and its role in heterochromatin formation to extensive reviews already available on the subject [31–33].
Heterochromatin
Chromatin fibers, which make up chromosomes, are composed of nucleosome arrays, with each nucleosome consisting of an octamer of 4 core histones (H2A, H2B, H3, and H4), which are wrapped by 147 bp of double-stranded DNA [34]. Chromatin is classically considered to be organized in two main types: euchromatin, which is largely decondensed, gene-rich and transcriptionally active; and heterochromatin, which is highly condensed, gene-poor and transcriptionally inactive [35,36]. Core histone octamers bind DNA, but the amino-terminal tails interact with other proteins and are subject to various post-translational modifications [37]. Differences in heterochromatin and euchromatin can be attributed to the differences in post-translational modification of histone tails that can change the degree of chromatin compaction and provide docking sites for structural factors [37]. Although different types of post-translational modification can occur at multiple positions on the histone tails, in general, euchromatic regions of the genome are hyperacetylated whereas heterochromatic sites of the genome are hypoacetylated [38–40]. In heterochromatin acetylation is replaced by methylation on histone H3 lysine (K) 9, histone H3K27 and on histone H4K20; in contrast euchromatic regions are enriched for histone H3K4 and H3K36 methylation [37,39,41]. A hallmark of heterochromatin in most eukaryotes is the presence of H3K9 histone methylation, which serves as a specific binding site for the highly conserved HP1 family of chromodomain proteins [39].
RNA interference-mediated heterochromatin formation in fission yeast
Fission yeast chromosomes contain large blocks of heterochromatin at the centromeres, telomeres, mating-type region, and at the ribosomal DNA (rDNA) locus, in addition to small heterochromatic ‘islands’ associated with a handful of meiotic genes (Fig. 1) [30,42]. Assembly of these heterochromatic domains relies on a well-conserved set of histone-modifying enzymes, histone marks and histone-binding proteins [43,44]. Amongst these regions, pericentromeric heterochromatin is the most dependent on RNAi, while others depend only partially on RNAi (such as telomeric heterochromatin, mediated by Taz1) or are independent (such as heterochromatin islands, mediated by Mmi1, Red1 and Erh1) [45].
Figure 1.
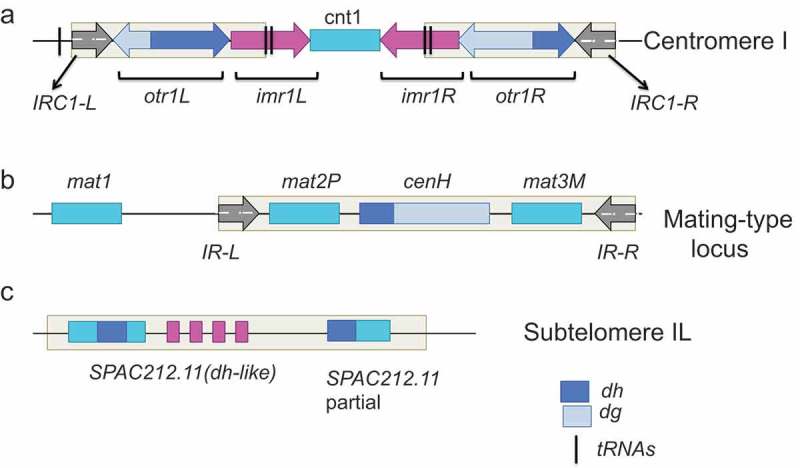
Sites of heterochromatin assembly in the fission yeast genome. This figure is adapted from [54] (a). Fission yeast centromeres contain a unique central domain consisting of central core region (cnt) and inner repeat sequences (imr), which is flanked by the outer repeat elements composed of one or more tandem array dg and dh repeats. Clusters of tRNA genes and/or IRC inverted repeats are present at the border of the pericentromeric heterochromatin. The number of dg/dh repeats differs between each chromosome arm, from 1 (cen1L) to 7–8 (cen3R) (b). At the mating type locus, mat2 and mat3 genes are located within a 20-kb heterochromatin domain. RNA interference regulates assembly of heterochromatin on the cenH element that shares strong homology with dg and dh centromeric repeats. Heterochromatin can also be nucleated in an RNA interference-independent manner within the mat2P/mat3M locus, including a 2.1 Kb region between cenH and mat3 by Atf1/Pcr1. The heterochromatic domain is restricted by boundary elements IR-L and IR- R. (c). cenH-like elements/dh-like sequences (SPAC212.11) within tlh1 and tlh2 genes are located at subtelomeric regions, which can nucleate heterochromatin both in an RNA interference-dependent and RNA interference-independent manner by the telomere specific factor Taz1.
Heterochromatin in fission yeast is distinguished by H3K9 methylation because unlike most eukaryotes that carry a fully functional RNA interference machinery, fission yeast lacks DNA methylation and H3K27 methylation [46]. Histone methylation provides a docking site for proteins that effectively silence RNA polymerase transcription, recruit chromatin modifiers and aid the process of establishment and maintenance of heterochromatin assembly [47]. In fission yeast H3K9 methylation is catalyzed by cryptic loci regulator 4 (Clr4), the homolog of Drosophila melanogaster and mammalian SU(VAR)3–9 [48]. The H3K9me mark in fission yeast is bound by the four chromodomain-containing proteins Swi6, Chp2 (which are both homologues of the vertebrates HP1), Chp1 and Clr4 itself [49–51]. Once bound to an H3K9me modified nucleosome, Swi6 recruits Clr4 and other proteins including histone deacetylases (HDACs) such as Clr3, Clr6 and Sir2, which initiates a new cycle of H3K9 methylation, histone deacetylation and histone-binding onto adjacent nucleosomes resulting in ‘spreading’ of heterochromatin [18,52,53].
In fission yeast, Drosophila and plants, RNA interference can influence gene expression at the level of chromatin in a mechanism referred to as transcriptional gene silencing (TGS) that occurs in the nucleus [54,55]. One such role of TGS is in RNA interference mediated heterochromatin formation [55]. Fission yeast was the first model demonstrating this, and has proven to be a powerful biological system to study the mechanism of RNA interference-mediated heterochromatin formation [56]. In particular, since fission yeast contains a fully functional ‘canonical’ RNA interference pathway with a single gene encoding each factor in the pathway, it makes this system particularly amenable to genetic and biochemical analyses of heterochromatin formation, assembly, maintenance and its various functions [45]. Key RNA interference proteins such as Dicer (Dcr1; the RNAse III enzyme that cleaves double-stranded RNA into siRNAs), Argonaute (Ago1; a member of the Argonaute family of proteins that contains the PAZ and PIWI-domains and can bind siRNAs) and RNA-directed RNA polymerase 1 (Rdp1, an enzyme that synthesizes double-stranded RNA from a RNA template) are essential for centromeric heterochromatin formation in fission yeast [45,57]. Mutations in any of these genes results in loss of small RNAs, loss of most H3K9 methylation and Swi6 recruitment; accumulation of forward and reverse transcripts of pericentromeric DNA repeats and strong defects in chromosome segregation [4,58]. Although RNA interference contributes to the formation of heterochromatin at other genomic regions such as at the mating type locus and the subtelomeric regions, deletion of dcr1∆, ago1∆ or rdp1∆ does not disrupt heterochromatin formation and transcriptional silencing at the mating-type interval and telomeres [59,60]. This is because RNA interference acts redundantly at these loci in parallel to RNA interference-independent heterochromatin formation pathways. At the mating-type locus, two stress-response transcription factors of the ATF/CREB family, Atf1 and Pcr1, work in parallel to RNA interference to initiate heterochromatin formation [61,62], whereas at the telomeres, the Taz1 protein nucleates heterochromatin formation in a RNA interference-independent manner [63,64]. Silencing re-initiation at the mating-type region has been shown to be very slow resulting in epigenetic inheritance in wild-type cells [59,65–67]. At centromeric regions, RNAi does initiate silencing more efficiently, where it is also required, at least in part, for maintenance [68].
The current model of RNAi-dependent centromeric heterochromatin assembly relies on transcription of the centromeric heterochromatin during early S-phase replication (Fig. 2) [4,66,69]. These transcripts derived from centromeric repeats can serve as template and are converted to double-stranded RNAs by the action of the RDRC complex, which contains Rdp1 (Fig. 2) [42,70]. The Rdp1 polymerases are well conserved in fission yeast, worms and plants and act in a primer-independent fashion to generate double-stranded RNA from single stranded centromeric transcripts to provide template for the RNase III enzyme Dcr1 (Fig. 2) [71,72]. Dcr1, which is associated with the RDRC complex, processes the long double-stranded RNA into siRNAs ranging in size from about 21 to 24 nucleotides (Fig. 2) [73]. These double-stranded siRNAs are first loaded onto the ARC complex (Argonaute siRNA chaperone complex), which contains Ago1 and two other chaperone proteins, Arb1 and Arb2 (argonaute binding) (Fig. 2) [74]. Like Dicer, Argonaute is a very well conserved protein and is characterized by the presence of PAZ and PIWI domains separated by the MID domain. The PAZ domain binds the 3ʹ end of the siRNA and its 5′ phosphate end is bound by the MID domain [75–77]. Like mammalian Ago2, the fission yeast Ago1 protein has endonucleolytic or ‘slicer’ activity, which is responsible for the passenger strand cleavage and release from duplex siRNA as well as cleavage of centromeric transcripts [78,79]. In the fission yeast ARC complex, Arb1 inhibits the slicer activity of Ago1 in this complex, thus ARC contains primarily double-stranded siRNAs [74]. These double-stranded siRNAs are passed onto another complex termed RITS, which in addition to Ago1 contains Chp1 and Tas3 (Figure 2) [42,74]. The slicer activity of Ago1 in the RITS complex promotes passenger strand release from duplex siRNA, enabling this effector complex to target homologous RNA through base pairing (Fig. 2) [72,80]. The mechanism(s) by which siRNAs induce chromatin modifications are not well understood. One possibility is that the RITS complex tethered to nascent transcripts via siRNAs and/or specific factors such as the LIM domain protein Stc1 (siRNA to chromatin) might mediate recruitment of chromatin-modifying factors such as Clr4 to heterochromatic repeats for H3K9 methylation (Fig. 2) [81–83]. Methylated H3K9 nucleosomes serve as binding sites for chromodomain proteins, including Swi6, Chp1, Chp2 and Clr4. Chp1 binding to methylated H3K9 can further strengthen the association between RITS and heterochromatin, creating a positive feedback loop (Fig. 2) [81,82]. A second positive feedback loop is created by the binding of Clr4 to methylated H3K9, which in turn promotes H3K9 methylation of the neighboring histones, thus allowing heterochromatin spreading in a sequence independent manner [82]. It is important to note that in Clr4 chromodomain mutants, there is a drastic reduction in H3K9me3 levels, but not H3K9me2, in the pericentromeric DNA repeats, demonstrating that Clr4 binding to the RNAi machinery was sufficient for its recruitment and to establish H3K9me2 [84]. In fission yeast, the formation of heterochromatin has an additional component of cell cycle regulation. The generation of centromeric siRNAs occurs along with centromeric transcription during S phase of the cell cycle and the siRNA-dependent recruitment of CLRC and assembly of H3K9me2 chromatin during late S-phase and G2-phase of the cell cycle [85]. All components in the RDRC, RITS and CLRC complexes are essential for robust siRNA biogenesis and efficient H3K9 methylation at pericentromeric heterochromatin. Consistent with this, deletion of any of these RNA interference proteins causes accumulation of transcripts from the outer repeats of the centromere, reduced centromeric H3K9me and Swi6 localization, increased H3/H4 acetylation and chromosome missegregation [58].
Figure 2.
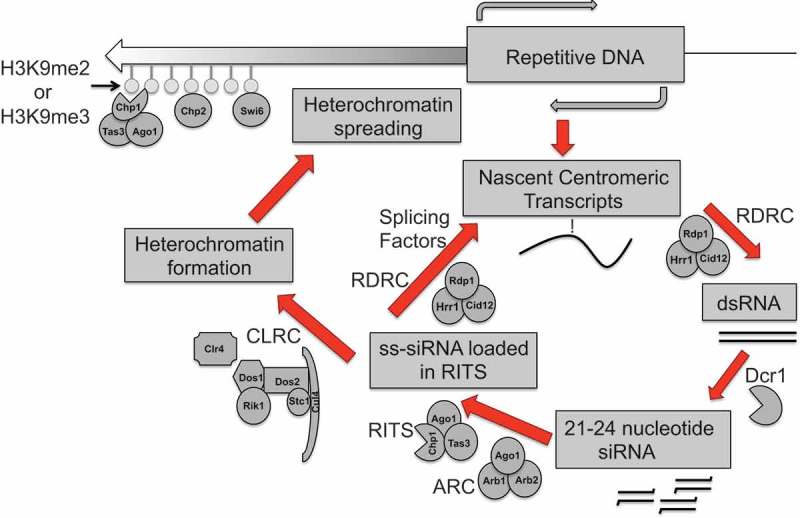
siRNA directed heterochromatin formation in fission yeast. This figure is adapted from [45,203,204] Rdp1, a member of the RDRC complex converts nascent transcripts from repetitive regions of the genome during S phase of the cell cycle into double-stranded RNA. The RNase III–like enzyme Dcr1, can process these double-stranded RNAs into siRNAs. These siRNAs then pass through the first Argonaute chaperone complex, ARC prior to loading into the RITS complex, where Ago1 endonuclease activity cleaves the passenger strand siRNA forming an effector complex that is recruited to chromatin by siRNA-nascent RNA base-pairing. This is followed by the recruitment of the CLRC histone-modifier complex to chromatin to reinstate transcriptional gene silencing by Clr4-mediated H3K9 methylation. The chromodomain protein Swi6 that along with Clr4 recruits other chromodomain proteins such as Chp1 and Chp2 then recognizes the H3K9 mark. This complex then methylates the adjacent nucleosome thus ‘spreading’ the H3K9 heterochromatin mark in an RNA interference-independent manner. The RITS effector complex can also target complementary nascent transcripts and recruit RDRC to promote the synthesis of double-stranded RNA.
The RITS complex: a RNA interference effector complex
The affinity purification of chromodomain protein Chp1 led to the identification and the subsequent biochemical characterization of the RITS complex [54,81]. Chp1 had previously been shown to be essential for centromeric heterochromatin assembly since deletion mutants of chp1Δ showed reduced CLRC activity at centromeres albeit these mutants retained heterochromatin at the mating-type locus and at telomeres [4,59,60,86,87]. The components of the RITS complex localize to all sites of heterochromatin and are essential for the assembly of centromeric heterochromatin. Deletion of chp1∆, tas3∆ or ago1∆ leads to disruption of heterochromatin formation and gene silencing at centromeres, and reduction of histone H3K9 methylation and recruitment of Swi6 to centromeres [51,68,81]. Unlike other organisms in which small RNAs are required for stability of Argonaute complexes, in fission yeast siRNAs are not critical for RITS formation or stability; however, in dcr1∆ mutants, where siRNAs are not produced, the RITS complex fails to localize to pericentromeric repeats, and centromeric heterochromatin does not form, indicating a role for siRNAs in targeting RITS to specific regions in the chromatin [81]. RITS recruitment to a transcript is enough to silence it, but only if Ago1 has catalytic activity [78,88]. Thus, the mechanism by which the RITS complex mediates the various stages of heterochromatin assembly has been an area of intense research.
It is important to note that while Ago1 is evolutionarily conserved, Chp1 and Tas3 are not; in fact, the related fission yeasts Schizosaccharomyces octosporus and S. cryophilus have a truncated Chp1 protein that does not localize to heterochromatin [89], and Tas3 is only found in S. pombe and S. japonicus [82]. However, Tas3 possesses a GW-rich domain, a feature that is conserved in Argonaute interactors, including GW182 proteins in mammals, Gawky in Drosophila, AIN-1 in C. elegans, RNA polymerase IV and RNA polymerase V in Arabidopsis [90,91], suggesting a case of functional conservation rather than conservation at the primary sequence level, with Argonaute as the catalytic component of the complex. Accordingly, a mutant in which, the GW-rich domain of RNA polymerase IV is replaced by the GW-rich domain of GW182 is functional [91]. RISC (RNA-induced silencing complex)–the ‘Argonaute effector’ PTGS ribonucleoproteic complex–is localized to GW/P-bodies along with GW182 [90], while in S. pombe, RITS, involved in TGS, is localized to chromatin [92], via the binding of H3K9me by the chromodomain of Chp1. Bioinformatic analyses indicate that many other GW-motif containing proteins exist, and may therefore potentially be interacting with Argonaute complexes [93].
Additional insights into the activities of the RITS complex were discovered by the purification of Rdp1, which led to the identification of a second RNA interference effector complex known as the RNA-Directed RNA polymerase Complex (RDRC) [72]. Rdp1 was found in complex with a putative RNA helicase (Hrr1) and poly-A polymerase (Cid12) [72]. Rdp1 plays the primary role in the generation of double-stranded RNA, which is subsequently processed to form siRNAs. Although transcription of sense and anti-sense RNA from both DNA strands can also generate double-stranded RNA, in rdp1Δ deletion mutants no siRNAs are detected in the RITS complex indicating that RDRC might act upstream of RITS in the siRNA-dependent heterochromatin formation pathway [72]. Like RITS, all subunits of RDRC are critical for siRNA production and formation of heterochromatin at centromeres [72,80]. Further biochemical analysis also discovered a physical interaction between RDRC and RITS that depends on Clr4 and Dcr1, suggesting RITS and RDRC interact in the context of specific chromatin regions i.e., in heterochromatin [72]. The physical interaction of Dcr1 and RDRC enhances the double-stranded RNA synthesis activity of Rdp1, thus increasing the production siRNAs [94]. Additionally, in clr4∆ cells, purified RITS contains little or no siRNAs, RITS and RDRC fail to interact, and Rdp1 fails to associate with centromeric DNA or RNA [72]. Put together, these observations suggest that the RNA interference machinery primarily acts in cis and is regulated by Clr4-dependent H3K9 methylation.
The structures of argonaute and the RITS complex
The first structural information on Argonaute (Fig. 3), the key effector of the RITS complex, was provided by the structure of the Drosophila Argonaute PAZ domain [75,76,95], followed by the complete structure of the bacterial Argonaute from Pyrococcus furiosus [79]. This structure showed that the PIWI domain is similar to ribonuclease H, including conserved catalytic amino acid residues and that Argonaute is indeed the Slicer protein [79,96]. Fission yeast Ago1 performs a similar catalytic role in fission yeast RITS [78]. More recently, the structure of human AGO2 was solved [97,98].
Figure 3.
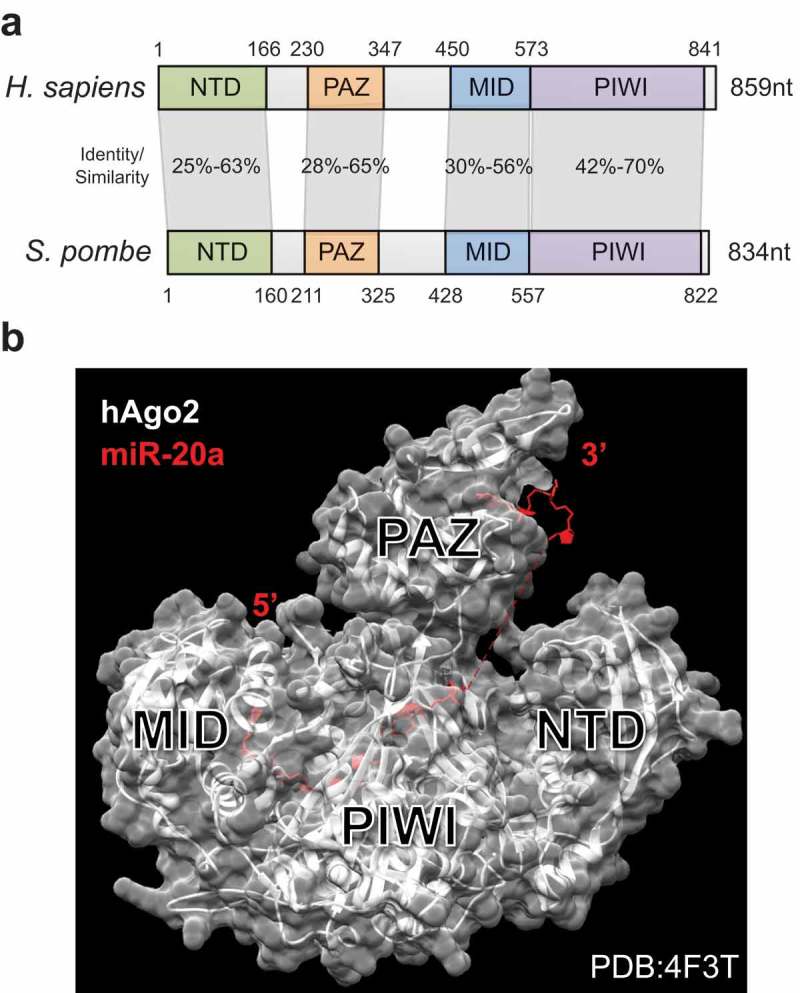
Structure of Argonaute. (a) Argonaute proteins are organized in 4 domains that are well-conserved from yeast to humans (>25% identity between S. pombe and H. sapiens). (b) Structure of human Argonaute (Ago2) in complex with miR-20a showing the binding of MID to the 5ʹ end of the small RNA, and of PAZ to the 3ʹ end [97].
The 5ʹ end of the small RNA binds to the MID domain and its 3ʹ end to the PAZ domain [97,99], the most mobile domain of the protein [100]. Tryptophan-binding pockets in the PIWI domain are the likely recognition sites for GW-motif proteins [98]. Argonautes bind small RNAs of different sizes, ranging from 21nt siRNAs to 40nt pre-piRNAs and the nucleic acid binding channel differs between PIWI-family and AGO-family Argonautes and accounts for this distinct size recognition [101]. Each Argonaute is still flexible, as seen in Mucor circinelloides where a single Argonaute protein is necessary for the production of three different types of small RNAs of distinct sizes [102]. In S. pombe, Argonaute can not only bind to 21-24nt small RNAs, but also to longer, Dicer-independent precursors known as priRNAs, which are subsequently trimmed in a 3ʹ-to-5ʹ fashion by the exonuclease Triman [103]. A similar mechanism was found in C. elegans and silkworm for piRNA trimming, by Trimmer/PARN-1 [104,105]
In S. pombe, further information on the structure of RITS was obtained from the structure of Argonaute-associated factors, and in particular of the core platform assembled by the Chp1/Tas3 association [106], between the C-terminal domain of Chp1 and N-terminal domain of Tas3. In addition, the cryo-EM structure of the Chp1 chromodomain (N-terminal), in association with one nucleosome, gave additional insights into its concurrent binding of H3K9me and nascent transcripts [107,108]. The C-terminal domain of Tas3 contains an alpha-helix, termed the TAM domain, which allows Tas3 polymerization, resulting in RITS spreading; mutations in this domain result in a strong loss of silencing and small RNA formation [109].
In most species, small RNAs show a 5ʹ-U/A nucleotide preference. Structural and genetic work has shown that the MID domain contributes to this bias, such as the 5ʹ-U bias of piRNAs [99] and the 5ʹ-U bias of AGO1-associated small RNAs in Arabidopsis [110]. However, it has also been argued that Argonaute is theoretically flexible enough to accommodate all four bases in 5ʹ [111]. One possibility is that the 5ʹ nucleotide is used to sort different classes of small RNAs to the appropriate Argonaute complex, as is the case in Arabidopsis [112]. Interestingly, human AGO2 is able to recognize chemically modified nucleotide analogs in the 5ʹ position [113], suggesting the possibility that some small RNAs could be 5ʹ modified in vivo. The 3ʹ end of the small RNA is commonly modified by 2ʹ-O-methylation, catalyzed by HEN1, as in plant miRNAs and siRNAs [114], Drosophila piRNAs [115,116] and Tetrahymena scnRNAs [117]. In addition to the 5ʹ and 3ʹ modifications, structural work has shown that Argonaute can tolerate a variety of nucleotide modifications within the siRNA guide [98].
The N-terminal domain of eukaryotic Argonaute proteins is the most variable and may account for the recognition and formation of distinct protein-protein complexes. However, it has been shown recently that it could also play a key role in Argonaute activity. Two motifs in the N-terminal domain along with PIWI regulate RNA cleavage activity of human Argonautes [118,119]. This discovery solved the puzzling observation that Ago3 is catalytically inactive despite having an active PIWI domain [118], and has allowed for re-engineering of slicer activity in inactive human Ago1 and Ago3 [119], and Ago4 [120].
Another important contribution of the N-terminal domain was found in relation to distant bacterial Argonaute homologs, which are unique in their capacity to recognize ssDNA as well as ssRNA in a RNA-guided manner, such as in Marinitoga piezophila [121]. Accordingly, the structure of M. piezophila Argonaute also revealed unique features of the N-terminal domain [121].
The role of the RITS complex in post-transcriptional gene silencing
Post-transcriptional gene silencing (PTGS) differs from transcriptional gene silencing (TGS) in that it does not rely on heterochromatin formation. PTGS reduces gene expression by transcript cleavage in the absence of chromatin modification [122]. PTGS thus also relies on the endonucleolytic activity of RITS [78]. In fission yeast, several protein-coding genes are regulated by antisense small RNAs [123]. In RNAi mutants, they are typically up-regulated at the protein level, but not at the mRNA level, indicating that RNAi can effect translational inhibition in fission yeast [123], although some genes (such as cum1) are regulated at the mRNA level [124]. However, while many loci can give rise to antisense transcription, such as during sexual differentiation, they are not targeted by RNAi [125], indicating that specific recruitment mechanisms must be present to allow RNAi to degrade the transcript or inhibit its translation. The mechanism of PTGS in fission yeast deserves further scrutiny.
Additionally, co-transcriptional gene silencing (CTGS) by Ago1 can occur independently of histone modification [55]. For instance, in the context of euchromatin, efficient RNA interference-dependent silencing can be achieved by tethering of Rik1, a single subunit of a complex to a euchromatic transcript [126]. Artificially tethered Rik1 can recruit RITS to mediate the cleavage of nascent transcripts via the siRNA directed endonucleolytic activity of Ago1 [126]. Here silencing occurs independently of any detectable H3K9 methylation, although the components of the Rik1 complex are required [126]. It is possible that Clr4 acts through additional non-H3K9 targets, such as Mlo3 [126]
Viral inhibitors of RITS
Most plant and animal viruses encode viral suppressors of RNA silencing proteins (VSRs) to counter-act host antiviral RNA interference. While most VSRs bind double-stranded RNA to preclude its cleavage, several directly target Argonaute complexes. For example, protein P1 from sweet potato mild mottle virus binds Argonaute by a GW-domain ‘decoy’ [14], which precludes target RNA recognition [127], protein 2b from cucumber mosaic virus binds Argonaute and inhibits its cleavage activity [128], and protein P0 from polerovirus binds and destabilizes Argonaute, leading to its degradation [129,130] by precluding RISC formation [131]. The HIV-1 accessory protein Nef also binds Ago2 via a GW-motif [132].
The role of argonaute in quelling
Quelling was one of the early RNA interference mechanisms to be described in eukaryotes [133]. Quelling is induced by repetitive transgenic sequences, resembling co-suppression in plants, giving rise to aberrant RNA (aRNA) in Neurospora crassa [133]. Silencing results when wild-type isolates of N. crassa are transformed with DNA from the carotenoid biosynthesis genes al-1 and al-3 (albino 1 and 3) (responsible for the orange pigment of N. crassa) [133]. The transformation resulted in pale yellow/white transformants with reduced al mRNA levels indicating the silencing of endogenous al genes by the transgenes [134]. The efficiency of the quelling correlated with the high copy number of the transgene suggesting the silencing to be a consequence of the repetitive transgene [134]. Additionally, a reduction in the copy number of the transgene resulted in the spontaneous reversion of quelled transformants to wild-type or intermediate phenotypes [134]. Mutations of the al genes are recessive, however the al quelled strains are dominant over wild-type strains in heterokaryon complementation experiments which indicates a diffusible and transacting cytoplasmic silencing molecule to be involved in quelling [135]. Furthermore, since quelling did not affect the levels of pre-mRNA, this observation led to the hypothesis that the production of aberrant RNA (aRNA) in the presence of multi-copy of transgenes causes PTGS [135].
Subsequently, quelling-deficient (qde) mutants were isolated using forward and reverse genetics approaches and divided into three complementation groups: qde-1, qde-2 (Argonaute), and qde-3 [136–138]. Also, Dicer protein genes dcl-1 and dcl-2, were identified [139].
The molecular mechanism of quelling
Quelling is initiated by the integration of multiple transgene copies, particularly tandem repeats in the genome of Neurospora crassa. During replication, aRNA structures are formed from these repetitive DNA sequences [140]. The model for quelling proposes that Replication Protein A (RPA-1) recruits QDE-3 to resolve the intermediates such as single-stranded DNA that are formed at the sites giving rise to aRNA [141]. QDE-3 is a RecQ helicase [138]. RecQ helicases are known to be involved in DNA replication, homologous recombination (HR) and DNA repair. This interaction of single-stranded DNA-RPA1-QDE-3 blocks the formation of DNA/RNA hybrids [141]. QDE-1, is then recruited to single-stranded DNA by RPA-1 and QDE-3 where it first uses its DNA-dependent RNA polymerase (DdRP) activity to convert single-stranded DNA to aRNA and then uses its RdRP activity to convert aRNA into double-stranded RNA [141]. QDE-1 was the first eukaryotic RNA interference component identified which encodes a cellular RNA-dependent RNA polymerase [142]. The requirement of a RdRP in quelling suggested that double-stranded RNA is a crucial intermediate of gene silencing in vivo. The production of double-stranded RNA activates the downstream RNA interference pathway [141], and could theoretically be promoted by tandem repeats [143]. This mechanism ensures that siRNAs are specifically produced from repetitive DNA loci but not from non-repetitive regions of the genome. These long double-stranded RNAs are then processed into 25 nt siRNA duplexes by Dicer protein [144]. Quelling in Neurospora relies on the two partially redundant Dicer proteins (DCL-1 and DCL-2), which process double-stranded RNA into 25 nt long siRNA in an ATP-dependent manner [139]. The deletion of both DCL genes completely abolishes quelling in Neurospora crassa [139]. The siRNA molecules are then incorporated into RISC as duplexes (inactive RISC) [145]. RISC contains QDE-2 (Argonaute) [137] and the exonuclease QIP (QDE-2 interacting protein) which act in a two-step process to form an active RISC [145]. First, the QDE-2-bound double-stranded siRNA duplex is nicked by QDE-2 using its slicer activity to release the passenger strand from the duplex to generate a QDE-2-bound single-stranded siRNA molecule also known as the active RISC [145]. Second, QIP degrades the nicked passenger strand of siRNA with its exonuclease activity [145]. The single-stranded siRNA acts as a guide molecule to detect homologous mRNA for degradation and silencing [137]. QDE-2 and QIP are essential components of the quelling pathway.
Quelling can also be triggered by DNA damage in vegetative tissue. A special class of small RNA named qiRNA for QDE-2-interacting small RNAs is induced after Neurospora crassa is treated with a DNA damaging agent [146]. qiRNAs are 21–23 nt in length, have a 5ʹ uridine and mostly originate from the rDNA locus, which is a highly repetitive sequence in the wild-type Neurospora genome [146]. It is important to note that despite the repetitive sequence of the rDNA it is normally protected from quelling, however in response to certain physiological stimuli quelling can be triggered at the rDNA. The biogenesis of qiRNAs requires QDE-1, QDE-3, and Dicers indicating that the RNA interference machinery upon DNA damage generates qiRNAs. Both transgene repeat silencing (quelling) and DNA damage-induced silencing (qiRNA) originate from aRNA precursors [146]. Also, both quelling and qiRNA pathways appear to share the same molecular machinery for silencing [146]. The repetitive nature of the rDNA and the transgenes integration loci is the most probable common trigger for quelling and qiRNA production [147]. It can be speculated that since aRNA are produced in response to DNA damage and replicative stress during damage-induced silencing it is likely quelling could also result from DNA damage or replication stress by transgene integration at fragile repetitive DNA sequences [147].
Role of RNA polymerase II in RNA interference-mediated heterochromatin formation
As the end-effect of TGS is silencing – inhibition of transcription –, a close association between RNA interference proteins and the RNA polymerase II transcription machinery has been suspected. Indeed, early genetic screens have unveiled that the structural integrity of RNA polymerase II is essential for siRNA-dependent heterochromatin formation in S. pombe [66,69]. Point mutations in Rpb2 (second largest subunit of RNA polymerase II) and Rpb7 (fourth largest subunit of RNA polymerase II) subunits of RNA polymerase II have been shown to disrupt siRNA generation and heterochromatin formation in the pericentromere [66,69]. Since the global transcription profile of protein-coding genes showed no major change in either of the two RNA polymerase II point mutants it suggested that RNA polymerase II might play a direct role in centromeric heterochromatin formation and structural maintenance, in conjunction with the small RNA processing machinery [69]. Experiments done in Drosophila suggest that RNA polymerase II transcription through centromeric heterochromatin generates a nascent transcript that acts as a substrate for Dcr2 generating endo-siRNAs, which in turn guides chromatin modifications at the heterochromatin [148]. Plants, such as Arabidopsis thaliana, possess two additional RNA polymerases specifically dedicated to TGS, RNA polymerase IV and V. RNA polymerase IV is required for small RNA biogenesis from repetitive DNA elements [149], while RNA polymerase V transcribes long non-coding RNAs (lncRNAs) that associate with AGO4 [150].
In accordance with a direct role for RNA polymerase II at heterochromatin, specific mutants in several co-transcriptional complexes are required for efficient silencing. Indeed, specific mutants in the Mediator complex – which bridges transcription factors and the RNA polymerase machinery to assemble the pre-initiation complex [151] – also lose pericentromeric silencing [152], and the Mediator head domain is involved in the efficiency of transcription and processing of heterochromatic transcripts [153]. Mutants in the Sgf73 subunit of the SAGA complex disrupt the integrity of the RITS complex and reduce binding of Ago1 at heterochromatin, effectively resulting in loss of silencing [154]. Mutants in the cyclophilin Rct1, which interacts with the RNA polymerase II C-terminal domain to coordinate transcription and pre-mRNA processing [155], also lose silencing [156].
The close association at the centromeres of the transcription machinery and the silencing machinery has been perceived as a ‘paradox’: why is a silent region transcribed? – It was subsequently shown that transcription occurs specifically during the S-phase of the cell cycle [157]. H3 is phosphorylated at Serine-10 during mitosis, which prevents Swi6/HP1 from binding H3K9 methylation, and allows an interval for pericentromeric transcription in S-phase [157]. RNA interference then triggers heterochromatin formation on both daughter strands, thereby allowing its epigenetic inheritance [158]. Accordingly, centromeric heterochromatin inheritance can be retained in mutants that only partially lose silencing, and is achieved by reducing centromeric transcription, such as in the deletion mutant of the MYST-domain histone acetyltransferase Mst2 [159]. Conversely, inhibiting histone deacetylases by trichostatin A results in loss of pericentromeric silencing [160]. More recently, an additional explanation has been proposed: while H3K9me3 is transcriptionally silent, H3K9me2 allows recruitment of RNA polymerase II and RNAi [84,161]
Because S-phase cells need to coordinate transcription with the concomitant replication of the genome, the activity of the RITS complex in silencing pericentromeric transcripts in S-phase must be tightly co-regulated with the replication fork. Indeed, our laboratory recently found that mutants in RNA interference components, including the RITS complex, have a defect in RNA polymerase II release, resulting in accumulation of stalled RNA polymerase II over the pericentromeres and at several euchromatic loci [10,162]. It has therefore been proposed that RNA interference resolves transcription-replication collisions at pericentromeric regions by releasing RNA polymerase II in parallel with recruiting CLRC to the replication fork [163], resulting in spreading of H3K9 methylation. In RNAi mutants, the stalled RNA polymerase II is unable to be released, and the fork then has to bypass this block via the activity of HR. Accordingly, a strong negative genetic interaction is found between RNA interference mutants and Rad51 [162]. A similar co-regulation of transcription and replication occurs at the rDNA, although in this case, only Dcr1 but not Ago1 is required [10]. Indeed, double-mutants between Dcr1 and the rDNA-specific replication fork terminator Reb1 [164] also display a very strong growth defect in cycling cells [165].
RNA interference is involved in RNA polymerase I release from rDNA in quiescent cells
We have recently identified a novel function of RNA interference in S. pombe, where RNA interference becomes essential during quiescence [165]. Cellular quiescence is classically defined as the state of a non-dividing (G0) cell that still has metabolic activity and is able to re-enter the cell cycle given the appropriate signal. This state is essential in the life-cycle of many micro-organisms, for which the nutritional environment is limited, but also in the context of multi-cellular organisms where distinct sets of quiescent cells may effect key functions; for example, in humans, stem cells are typically quiescent [166]. In S. pombe, a simple nutritional signal (nitrogen starvation) can be used to shift a culture of cells into quiescence, thus making it a good model organism for investigating the molecular mechanisms involved in the maintenance of G0 cells [167,168]. While wild-type cells retain near full viability after extended periods of quiescence [167], we observed that deletion mutants for the key RNA interference factors, such as Dcr1, Ago1 and Rdp1, display a dramatic loss of viability both at G0-entry and during quiescence maintenance [165]. By using a suppressor screen strategy specifically designed to identify suppressors for G0-defective strains, we were able to investigate the molecular mechanism underlying this inability to maintain quiescence.
We found that in G0, dcr1Δ mutants are defective in releasing RNA polymerase I from the rDNA repeats, resulting in accumulation of stalled RNA polymerase I and DNA damage (γ-H2AX S. pombe equivalent – H2AS128Phos) [165]. This accumulation subsequently results in over-silencing of the rDNA by H3K9 methylation, via recruitment of the methyltransferase Clr4 and the HP1 protein Swi6. Counter-intuitively, dcr1Δ deletion mutants therefore undergo a strong and specific over-heterochromatinization of the rDNA. The G0 defect can be suppressed either by reducing the accumulation of stalled RNA polymerase I at the rDNA, such as in a specific TBP (TATA-binding protein) mutant, by destabilizing RNA polymerase I itself, such as in a deletion mutant of the non-essential subunit A12, or downstream by preventing the heterochromatinization of rDNA repeats, such as in mutants in CLRC and HP1 [165]. Therefore, RNA interference plays a novel and essential role in control of RNA polymerase I regulation.
Interesting parallels can be drawn between the defect of dcr1Δ mutants in RNA polymerase II release from centromeric repeats during S-phase [10,162] and in RNA polymerase I release from rDNA repeats during G0 [165]. In particular, mutations in equivalent non-essential subunits are able to specifically suppress defects in pericentromeric silencing (Rpb9 and TFIIS in RNA polymerase II) [169] and in G0 defects (A12 in RNA polymerase I) [165,170], buttressing the model of an intimate association between RNA interference and RNA polymerases.
All RNA interference deletion mutants, including deletion mutants of the RITS complex proteins and a catalytic-dead Dicer allele, result in the loss of viability during quiescence maintenance. Therefore, it is likely that specific small RNAs are involved in this function in G0. However, the comparison of small RNA populations in wild-type and dcr1∆ mutants did not reveal novel classes of Dcr1-dependent small RNAs [165]. It is possible that G0 small RNAs are not abundant or too transient, and therefore below our detection limit, especially compared to the levels of rRNA fragments, which comprise the majority of small RNA reads from libraries in G0 cells.
RNA interference and genome stability
Recently, a novel role for RNA interference components, including the RITS complex, has emerged in genome stability. The maintenance of genome integrity is vital for cell and organism survival. However, DNA is constantly under threat of damage both by factors generated both within the cell and externally. DNA double strand breaks (DSBs) are a particularly cytotoxic lesion that can cause chromosomal aberrations and threaten cellular survival [171–175]. Exogenous sources of DSBs include ionizing radiation (IR) and chemical toxins [176]. DSBs can be formed as a consequence of normal cellular metabolism, in the form of reactive oxygen species that oxidize DNA bases, as result of DNA replication, meiotic recombination and DNA replication transcription collision [162,177,178]. Left unrepaired DSBs would lead to genomic rearrangements, aneuploidy and/or cell death. In order to meet the challenge in preserving genome integrity cells have evolved with conserved recombination mediated DNA repair pathways as a mean for repairing DSBs and restarting replication forks, thus allowing genome duplication to continue [179]. These DNA double strand break repair (DSBR) pathways identify and respond to the many lesions that afflict the genetic material [180].
Two major pathways repair DSBs: non-homologous end joining (NHEJ) and homologous recombination (HR). DSBR pathways are broadly classified based on whether a homologous sequence is used as a template to join the broken DNA ends [171]. NHEJ is an efficient repair pathway that functions throughout the cell cycle however, NHEJ is considered to be error-prone and an intrinsically mutagenic route to restart replication [181,182]. HR is a high fidelity, template-dependent, ubiquitous DNA repair pathway where the cell employs a homologous DNA or near homologous (homeologous) DNA sequences as template for the repair of the broken ends [183]. In recent years work from our laboratory and others have discovered a new role for the Argonaute and Dicer in homologous recombination mediated DSBR. In this section, we summarize the discovery and functions of these new factors and discuss the possible mechanisms by which they function in the DSBR pathway.
Depletion of Argonaute and Dicer in human cells significantly decreases the DNA repair efficiency indicating a role for Argonaute and Dicer in DSBR [184]. Additionally, the mammalian Ago2 physically interacts with Rad51, the central recombinase in catalyzing HR [185]. Studies in Arabidopsis and human cells have identified production of diRNA (DSB-induced small RNA) from sequences around the DSB site [184]. These diRNAs are produced from sense and antisense strands of DSB proximal sequence. Additionally, in both plants and human cells diRNAs are associated with Argonaute and are required for efficient HR [184,185]. As discussed in the section on quelling it is also notable that in Neurospora crassa, qiRNAs derived from rDNA repeats have been detected in cells treated with DNA damaging agents and small RNA in fission yeast accumulates after HU treatment [146]. These observations led to speculations that diRNAs may either function as guide molecules by base-pairing between diRNAs and the damaged DNA or scaffold transcripts made from the damage sites to recruit the DNA repair factors to the site of the DSB to facilitate repair. Since diRNAs are required for MDC1 and 53BP1 foci formation but not for γ-H2AX focus formation diRNAs may function downstream of or in parallel to γ-H2AX to recruit DNA repair factors onto DSB site to facilitate repair [184,186–191]. Alternatively, diRNAs may trigger a number of chromatin changes at the site of the DSB including DNA methylation, histone modifications and these modifications may facilitate DSB repair [192,193].
However, in fission yeast no diRNAs were found at the site of DSBs (Martienssen laboratory unpublished data). Additionally, RNAi mutants have not been recovered from DNA repair screens nor have they been found in a recent DSB-interacting proteome [194]. In both fission yeast and human cells, diRNAs are not necessary for the Argonaute-Rad51 interaction suggesting a possible function for Argonaute in DSBR that is independent of its catalytic activity [185]. It is important to note that HR proteins are required for quelling and meiotic silencing by unpaired DNA (MSUD) in Neurospora indicating additional evidence of crosstalk between RNAi and DNA repair [195,196]. Furthermore, a recent paper reports a role for Rad51 and Rad54 in promoting gene silencing in fission yeast centromeres, suggesting a role for HR in centromeric heterochromatin structural maintenance [197]. In conclusion, while RITS/RNAi and DNA repair proteins are associated, and affect each other’s function, the role of this association is still unclear.
Conclusion
After the identification of Argonaute over a decade ago as the ‘Slicer’ in PTGS and TGS, it is now clear that it plays many more roles along with other RNA interference factors. In particular, there is now evidence of a functional association of RNA interference proteins with RNA polymerase II [10,162], with RNA polymerase I [165], and with DNA polymerase ϵ [163,198], along with the silencing CLR complex. This reveals complex interactions between the transcription machinery and the silencing machinery in the control of gene expression and heterochromatin formation.
Another puzzling novel function of Argonaute and RNAi lies in the DNA damage response and DNA repair. While the precise molecular mechanism is not yet understood, this association raised the possibility that misregulation of RNAi may be tumorigenic. Indeed, a rare syndrome known as the ‘DICER1 syndrome’ occurs in patients harboring mutations in the Dicer1 gene, and is characterized by a high incidence of several cancers, in particular pleuropulmonary blastoma and Sertoli-Leydig ovarian cell tumors [199]. Notably, DICER1 mutations are frequently found in addition to biallelic TP53 mutations [200]. It has also appeared that RNAi plays a specific role at the rDNA locus, with distinct requirements in dividing and non-dividing cells [10,165,170]. In particular, its function becomes essential in non-dividing cells [165]. As misregulation of rDNA transcription is a recurring characteristic in cancer, ageing and disease [201], it will be interesting to study whether the RITS complex also contributes to these effects.
As Argonaute and Dicer proteins arose independently in evolution, with Argonaute deriving from an archaeal pAgo and Dicer from an ERCC4-like helicase [202], it is possible that their contribution will be distinct in DNA repair.
Overall, the accumulation of novel roles played by the RITS complex promises many avenues of research in the next decade (Fig. 4). In particular, a future question for the field will be to delineate precisely the molecular mechanisms by which the RITS complex – and RNA interference – mediates its multiple, novel, non-canonical cellular functions.
Figure 4.
Schematic describing the various cellular roles of Argonaute and its associated proteins.
Key wordsCLRC: This cullin-dependent E3 ubiquitin ligase complex consists of Clr4, Rik1, Dos1, Dos2, Stc1 and Cul4. The complex is responsible for placing the methyl marks on histone H3 lysine 9 and forming heterochromatin in the yeast Schizosaccharomyces pombe.CTGS: Co-transcriptional gene silencing is a mechanism of nuclear RNA degradation mediated by RNAiDSBR: Double strand break repair refers to the mechanism by which cells repair DNA double-strand breaks, which are one of the most toxic lesions to a cellHR: Homologous recombination is a template-dependent DNA repair pathway implicated in the repair and tolerance of DSBs.NHEJ: Non-homologous end joining is a template-independent DNA repair pathway implicated in the repair and tolerance of DSBs.PTGS: Post-transcriptional gene silencing is a gene silencing mechanism mediated by RNAi that reduces gene expression by transcript cleavage in the absence of chromatin modificationRDRC: RNA-Directed RNA polymerase Complex consists of the RNA polymerase Rdp1, a putative RNA helicase Hrr1 and polyA polymerase Cid12. This complex catalyzes the formation of double-stranded RNA from single-stranded heterochromatic transcripts by forming phosphodiester bonds between ribonucleotides in an RNA template-dependent fashion.RITS and RISC: RNA interference transcriptional silencing complex is the effector complex of RNAi and contains the Argonaute protein bound to small RNAs. It is also called RNA-induced silencing complex in mammalian cells.RNAi: RNA interference is a silencing mechanism widely employed in eukaryotes that can be characterized by small RNAs that are bound by Argonaute effector proteins and act as specificity factors to target homologous sequences for repressionTGS: Transcriptional gene silencing is a gene silencing mechanism mediated by RNAi that triggers DNA and/or chromatin modifications that leads to transcriptional silencing and heterochromatin formation.
Funding Statement
This work was supported by the National Institutes of Health [GM076396-08].
Acknowledgments
SB and BR contributed equally to this work. We would like to thank the fission yeast community, PomBase and the members of the Martienssen laboratory, for useful discussions and suggestions. We apologize to those colleagues whose work has not been cited due to space limitation. This work was funded by the NIH grant R01 GM076396-08 and by the Howard HughesMedical Institute and Gordon and Betty Moore Foundation Plant Biology Investigator Program (to RM).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- [1].Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. PMID i s 9486653 [DOI] [PubMed] [Google Scholar]
- [2].Mello CC, Conte D Jr.. Revealing the world of RNA interference. Nature. 2004;431(7006):p. 338–42. [DOI] [PubMed] [Google Scholar]
- [3].Zamore PD, Tuschl T, Sharp PA, et al. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. [DOI] [PubMed] [Google Scholar]
- [4].Volpe TA, Kidner C, Hall IM, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297(5588):1833–7. [DOI] [PubMed] [Google Scholar]
- [5].Baulcombe D. RNA silencing in plants. Nature. 2004;431(7006):356–63. [DOI] [PubMed] [Google Scholar]
- [6].Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299(5607):716–9. [DOI] [PubMed] [Google Scholar]
- [7].Mochizuki K, Fine NA, Fujisawa T, et al. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell. 2002;110(6):689–99. [DOI] [PubMed] [Google Scholar]
- [8].Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9(2):315–27. [DOI] [PubMed] [Google Scholar]
- [9].Fulci V, Macino G. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr Opin Microbiol. 2007;10(2):199–203. [DOI] [PubMed] [Google Scholar]
- [10].Castel SE, Ren J, Bhattacharjee S, et al. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell. 2014;159(3):572–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Buhler M, Moazed D. Transcription and RNAi in heterochromatic gene silencing. Nat Struct Mol Biol. 2007;14(11):1041–8. [DOI] [PubMed] [Google Scholar]
- [12].Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–4. [DOI] [PubMed] [Google Scholar]
- [13].Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–103. [DOI] [PubMed] [Google Scholar]
- [14].Giner A, Lakatos L, García-Chapa M, et al. Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 2010;6(7):e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. [DOI] [PubMed] [Google Scholar]
- [16].Cam H, Grewal SI. RNA interference and epigenetic control of heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2004;69:419–27. [DOI] [PubMed] [Google Scholar]
- [17].Grewal SI, Elgin SC. Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev. 2002;12(2):178–87. [DOI] [PubMed] [Google Scholar]
- [18].Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8(1):35–46. [DOI] [PubMed] [Google Scholar]
- [19].Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447(7143):399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hiragami K, Festenstein R. Heterochromatin protein 1: a pervasive controlling influence. Cell Mol Life Sci. 2005;62(23):2711–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ebert A, Lein S, Schotta G, et al. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14(4):377–92. [DOI] [PubMed] [Google Scholar]
- [22].Huisinga KL, Brower-Toland B, Elgin SC. The contradictory definitions of heterochromatin: transcription and silencing. Chromosoma. 2006;115(2):110–22. [DOI] [PubMed] [Google Scholar]
- [23].Bernard P, Maure JF., Partridge JF, et al. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294(5551):2539–42. [DOI] [PubMed] [Google Scholar]
- [24].Nonaka N, Kitajima T, Yokobayashi S, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4(1):89–93. [DOI] [PubMed] [Google Scholar]
- [25].Pidoux A, Mellone B, Allshire R. Analysis of chromatin in fission yeast. Methods. 2004;33(3):252–9. [DOI] [PubMed] [Google Scholar]
- [26].Blackwell C, Martin KA, Greenall A, et al. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol. 2004;24(10):4309–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Blackwell C, Martin KA, Greenall A, et al. The schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol. 2004;24(10):4309–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Birchler JA, Bhadra MP, Bhadra U. Making noise about silence: repression of repeated genes in animals. Curr Opin Genet Dev. 2000;10(2):211–6. [DOI] [PubMed] [Google Scholar]
- [29].Bhadra U, Pal-Bhadra M, Birchler JA. Histone acetylation and gene expression analysis of sex lethal mutants in Drosophila. Genetics. 2000;155(2):753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hall IM, Noma K, Grewal SI. RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc Natl Acad Sci U S A. 2003;100(1):193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ku HY, Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev. 2014;1(2):205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26(5):603–9. [DOI] [PubMed] [Google Scholar]
- [33].Ozata DM, Gainetdinov I, Zoch A, et al. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20(2):89–108. [DOI] [PubMed] [Google Scholar]
- [34].Luger K, Mäder AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. [DOI] [PubMed] [Google Scholar]
- [35].Dunleavy E, Pidoux A, Allshire R. Centromeric chromatin makes its mark. Trends Biochem Sci. 2005;30(4):172–5. [DOI] [PubMed] [Google Scholar]
- [36].Pidoux AL, Allshire RC. The role of heterochromatin in centromere function. Philos Trans R Soc Lond B Biol Sci. 2005;360(1455):569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–80. [DOI] [PubMed] [Google Scholar]
- [38].Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002;14(3):286–98. [DOI] [PubMed] [Google Scholar]
- [39].Lachner M, O’carroll D, Rea S, et al. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–20. [DOI] [PubMed] [Google Scholar]
- [40].Maison C, Bailly D, Peters AH, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30(3):329–34. [DOI] [PubMed] [Google Scholar]
- [41].Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–4. [DOI] [PubMed] [Google Scholar]
- [42].Cam HP, Sugiyama T, Chen ES, et al. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat Genet. 2005;37(8):809–19. [DOI] [PubMed] [Google Scholar]
- [43].Grewal SI, Klar AJ. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics. 1997;146(4):1221–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mandell JG, Bähler J, Volpe TA, et al. Global expression changes resulting from loss of telomeric DNA in fission yeast. Genome Biol. 2005;6(1):R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Creamer KM, Partridge JF. RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast. Wiley Interdiscip Rev RNA. 2011;2(5):632–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sanders SL, Portoso M, Mata J, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119(5):603–14. [DOI] [PubMed] [Google Scholar]
- [47].Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6(11):838–49. [DOI] [PubMed] [Google Scholar]
- [48].Rea S, Eisenhaber F, O’carroll D, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406(6796):593–9. [DOI] [PubMed] [Google Scholar]
- [49].van Wolfswinkel JC, Ketting RF. The role of small non-coding RNAs in genome stability and chromatin organization. J Cell Sci. 2010;123(Pt 11):1825–39. [DOI] [PubMed] [Google Scholar]
- [50].Zhang K, Mosch K, Fischle W, et al. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15(4):381–8. [DOI] [PubMed] [Google Scholar]
- [51].Partridge JF, Scott KS, Bannister AJ, et al. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002;12(19):1652–60. [DOI] [PubMed] [Google Scholar]
- [52].Grewal SI, Rice JC. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol. 2004;16(3):230–8. [DOI] [PubMed] [Google Scholar]
- [53].Shankaranarayana GD, Motamedi MR, Moazed D, et al. Sir2 regulates histone H3 lysine 9 methylation and heterochromatin assembly in fission yeast. Curr Biol. 2003;13(14):1240–6. [DOI] [PubMed] [Google Scholar]
- [54].Verdel A, Moazed D. RNAi-directed assembly of heterochromatin in fission yeast. FEBS Lett. 2005;579(26):5872–8. [DOI] [PubMed] [Google Scholar]
- [55].Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457(7228):413–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301(5634):798–802. [DOI] [PubMed] [Google Scholar]
- [57].Creamer KM, Partridge JF. Should I stay or should I go? Chromodomain proteins seal the fate of heterochromatic transcripts in fission yeast. Mol Cell. 2012;47(2):153–5. [DOI] [PubMed] [Google Scholar]
- [58].Volpe T, Schramke V, Hamilton GL, et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11(2):137–46. [DOI] [PubMed] [Google Scholar]
- [59].Hall IM, Shankaranarayana GD, Noma KI, et al. Establishment and maintenance of a heterochromatin domain. Science. 2002;297(5590):2232–7. [DOI] [PubMed] [Google Scholar]
- [60].Petrie VJ, Wuitschick JD, Givens CD, et al. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol. 2005;25(6):2331–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kim HS, Choi ES, Shin JA, et al. Regulation of Swi6/HP1-dependent heterochromatin assembly by cooperation of components of the mitogen-activated protein kinase pathway and a histone deacetylase Clr6. J Biol Chem. 2004;279(41):42850–9. [DOI] [PubMed] [Google Scholar]
- [62].Jia S, Noma K, Grewal SI. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004;304(5679):1971–6. [DOI] [PubMed] [Google Scholar]
- [63].Hansen KR, Ibarra PT, Thon G. Evolutionary-conserved telomere-linked helicase genes of fission yeast are repressed by silencing factors, RNAi components and the telomere-binding protein Taz1. Nucleic Acids Res. 2006;34(1):78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Kanoh J, Sadaie M, Urano T, et al. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol. 2005;15(20):1808–19. [DOI] [PubMed] [Google Scholar]
- [65].Obersriebnig MJ, Pallesen EM, Sneppen K, et al. Nucleation and spreading of a heterochromatic domain in fission yeast. Nat Commun. 2016;7:11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Djupedal I, Portoso M, Spåhr H, et al. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19(19):2301–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ekwall K, Ruusala T. Mutations in rik1, clr2, clr3 and clr4 genes asymmetrically derepress the silent mating-type loci in fission yeast. Genetics. 1994;136(1):53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sadaie M, Iida T, Urano T, et al. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. Embo J. 2004;23(19):3825–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kato H, Goto DB, Martienssen RA, et al. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309(5733):467–9. [DOI] [PubMed] [Google Scholar]
- [70].Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297(5588):1831. [DOI] [PubMed] [Google Scholar]
- [71].Verdel A, Vavasseur A, Le Gorrec M, et al. Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol. 2009;53(2–3):245–57. [DOI] [PubMed] [Google Scholar]
- [72].Motamedi MR, Verdel A, Colmenares SU, et al. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119(6):789–802. [DOI] [PubMed] [Google Scholar]
- [73].Paddison PJ, Caudy AA, Bernstein E, et al. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16(8):948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Buker SM, Iida T, Bühler M, et al. Two different Argonaute complexes are required for siRNA generation and heterochromatin assembly in fission yeast. Nat Struct Mol Biol. 2007;14(3):200–7. [DOI] [PubMed] [Google Scholar]
- [75].Yan KS, Yan S, Farooq A, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426(6965):468–74. [DOI] [PubMed] [Google Scholar]
- [76].Lingel A, Simon B, Izaurralde E, et al. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426(6965):465–9. [DOI] [PubMed] [Google Scholar]
- [77].Frank F, Sonenberg N, Nagar B. Structural basis for 5ʹ-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465(7299):818–22. [DOI] [PubMed] [Google Scholar]
- [78].Irvine DV, Zaratiegui M, Tolia NH, et al. Argonaute slicing is required for heterochromatic silencing and spreading. Science. 2006;313(5790):1134–7. [DOI] [PubMed] [Google Scholar]
- [79].Song JJ, Smith SK, Hannon GJ, et al. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305(5689):1434–7. [DOI] [PubMed] [Google Scholar]
- [80].Sugiyama T, Cam H, Verdel A, et al. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci U S A. 2005;102(1):152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Verdel A, Jia S, Gerber S, et al. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303(5658):672–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Jia S, Kobayashi R, Grewal SI. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005;7(10):1007–13. [DOI] [PubMed] [Google Scholar]
- [83].Bayne EH, White SA, Kagansky A, et al. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140(5):666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Jih G, Nahid Iglesias, Mark A. Currie, et al. Unique roles for histone H3K9me states in RNAi and heritable silencing of transcription. Nature. 2017;547(7664):p. 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Chen ES, Zhang K, Nicolas E, et al. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451(7179):p. 734–7. [DOI] [PubMed] [Google Scholar]
- [86].Partridge JF, Borgstrom B, Allshire RC. Distinct protein interaction domains and protein spreading in a complex centromere. Genes Dev. 2000;14(7):p. 783–91. [PMC free article] [PubMed] [Google Scholar]
- [87].Thon G, Verhein-Hansen J. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics. 2000;155(2):p. 551–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125(5):p. 873–86. [DOI] [PubMed] [Google Scholar]
- [89].Upadhyay U, Srivastava S, Khatri I, et al. Ablation of RNA interference and retrotransposons accompany acquisition and evolution of transposases to heterochromatin protein CENPB. Mol Biol Cell. 2017;28(8):p. 1132–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Pfaff J, Meister G. Argonaute and GW182 proteins: an effective alliance in gene silencing. Biochem Soc Trans. 2013;41(4):p. 855–60. [DOI] [PubMed] [Google Scholar]
- [91].El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007;21(20):p. 2539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Noma K, et al. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004;36(11):p. 1174–80. [DOI] [PubMed] [Google Scholar]
- [93].Karlowski WM, et al. Genome-wide computational identification of WG/GW Argonaute-binding proteins in Arabidopsis. Nucleic Acids Res. 2010;38(13):p. 4231–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Colmenares SU, et al. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007;27(3):p. 449–61. [DOI] [PubMed] [Google Scholar]
- [95].Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10(12):p. 1026–32. [DOI] [PubMed] [Google Scholar]
- [96].Joshua-Tor L. The Argonautes. Cold Spring Harb Symp Quant Biol. 2006;71:p. 67–72. [DOI] [PubMed] [Google Scholar]
- [97].Elkayam E, et al. The structure of human argonaute-2 in complex with miR-20a. Cell. 2012;150(1):p. 100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Schirle NT, MacRae IJ. The crystal structure of human Argonaute2. Science. 2012;336(6084):p. 1037–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Cora E, et al. The MID-PIWI module of Piwi proteins specifies nucleotide- and strand-biases of piRNAs. Rna. 2014;20(6):p. 773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kong R, et al. Exploring the RNA-bound and RNA-free human Argonaute-2 by molecular dynamics simulation method. Chem Biol Drug Des. 2017;90:753–763. [DOI] [PubMed] [Google Scholar]
- [101].Matsumoto N, Nishimasu H, Sakakibara K, et al. Crystal structure of silkworm PIWI-clade argonaute siwi bound to piRNA. Cell. 2016;167(2):p. 484–497 e9. [DOI] [PubMed] [Google Scholar]
- [102].Cervantes M, et al. A single argonaute gene participates in exogenous and endogenous RNAi and controls cellular functions in the basal fungus Mucor circinelloides. PLoS One. 2013;8(7):p. e69283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Marasovic M, Zocco M, Halic M. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol Cell. 2013;52(2):p. 173–83. [DOI] [PubMed] [Google Scholar]
- [104].Izumi N, et al. Identification and Functional Analysis of the Pre-piRNA 3ʹ Trimmer in Silkworms. Cell. 2016;164(5):p. 962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Tang W, et al. The RNase PARN-1 Trims piRNA 3ʹ Ends to Promote Transcriptome Surveillance in C. elegans. Cell. 2016;164(5):p. 974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Schalch T, et al. The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS. Nat Struct Mol Biol. 2011;18(12):p. 1351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Ishida M, et al. Intrinsic nucleic acid-binding activity of Chp1 chromodomain is required for heterochromatic gene silencing. Mol Cell. 2012;47(2):p. 228–41. [DOI] [PubMed] [Google Scholar]
- [108].Zocco M, et al. The Chp1 chromodomain binds the H3K9me tail and the nucleosome core to assemble heterochromatin. Cell Discov. 2016;2:p. 16004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Li H, et al. An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing. Mol Cell. 2009;34(2):p. 155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Zha XF, Xia QY, Yuan YA. Structural insights into small RNA sorting and mRNA target binding by Arabidopsis Argonaute Mid domains. FEBS Lett. 2012;586(19):p. 3200–3207. [DOI] [PubMed] [Google Scholar]
- [111].Kalia M, Willkomm S, Claussen J, et al. Novel Insights into Guide RNA 5ʹ-Nucleoside/Tide Binding by Human Argonaute 2. Int J Mol Sci. 2015;17:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Mi SJ, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5 ‘ terminal nucleotide. Cell. 2008;133(1):p. 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Deleavey GF, et al. The 5ʹ binding MID domain of human Argonaute2 tolerates chemically modified nucleotide analogues. Nucleic Acid Ther. 2013;23(1):p. 81–7. [DOI] [PubMed] [Google Scholar]
- [114].Li J, et al. Methylation protects miRNAs and siRNAs from a 3ʹ-end uridylation activity in Arabidopsis. Curr Biol. 2005;15(16):p. 1501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20(16):p. 2214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wang K, et al. Prediction of piRNAs using transposon interaction and a support vector machine. BMC Bioinformatics. 2014;15:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Kurth HM, Mochizuki K. 2ʹ-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. Rna. 2009;15(4):p. 675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Schurmann N, et al. Molecular dissection of human Argonaute proteins by DNA shuffling. Nat Struct Mol Biol. 2013;20(7):818–26. [DOI] [PubMed] [Google Scholar]
- [119].Hauptmann J, Dueck A, Harlander S, et al. Turning catalytically inactive human Argonaute proteins into active slicer enzymes. Nat Struct Mol Biol. 2013;20(7):814–7. [DOI] [PubMed] [Google Scholar]
- [120].Hauptmann J, et al. Generation of catalytic human Ago4 identifies structural elements important for RNA cleavage. Rna. 2014;20(10):1532–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Doxzen KW, Doudna JA. DNA recognition by an RNA-guided bacterial Argonaute. PLoS One. 2017;12(5). p. e0177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Sigova A, Rhind N, Zamore PD. A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004;18(19):2359–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Smialowska A, et al. RNAi mediates post-transcriptional repression of gene expression in fission yeast Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2014;444(2):254–259. [DOI] [PubMed] [Google Scholar]
- [124].Normant V, Beaudoin J, Labbe S. An antisense RNA-mediated mechanism eliminates a meiosis-specific copper-regulated transcript in mitotic cells. J Biol Chem. 2015;290(37):22622–22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Bitton DA, et al. Programmed fluctuations in sense/antisense transcript ratios drive sexual differentiation in S. pombe. Mol Syst Biol. 2011;7:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Gerace EL, Halic M, Moazed D. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell. 2010;39(3):p. 360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Kenesi E, et al. A viral suppressor of RNA silencing inhibits ARGONAUTE 1 function by precluding target RNA binding to pre-assembled RISC. Nucleic Acids Res. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Zhang X, et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006;20(23):3255–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Bortolamiol D, Pazhouhandeh M, Marrocco K, et al. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol. 2007;17(18):1615–21. [DOI] [PubMed] [Google Scholar]
- [130].Baumberger N, Tsai CH, Lie M, et al. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol. 2007;17(18):1609–14. [DOI] [PubMed] [Google Scholar]
- [131].Csorba T, et al. Polerovirus protein P0 prevents the assembly of small RNA-containing RISC complexes and leads to degradation of ARGONAUTE1. Plant J. 2010;62(3):463–72. [DOI] [PubMed] [Google Scholar]
- [132].Aqil M, Naqvi AR, Bano AS, et al. The HIV-1 Nef protein binds argonaute-2 and functions as a viral suppressor of RNA interference. PLoS One. 2013;8(9):e74472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Romano N, Macino G. Quelling - transient inactivation of gene-expression in neurospora-crassa by transformation with homologous sequences. Mol Microbiol. 1992;6(22):p. 3343–3353. [DOI] [PubMed] [Google Scholar]
- [134].Cogoni C, Macino G. Conservation of transgene-induced post-transcriptional gene silencing in plants and fungi. Trends Plant Sci. 1997;2(11):p. 438–443. [Google Scholar]
- [135].Cogoni C, et al. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. Embo J. 1996;15(12):3153–3163. [PMC free article] [PubMed] [Google Scholar]
- [136].Cogoni C, Macino G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc Natl Acad Sci U S A. 1997;94(19):10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Catalanotto C, Azzalin G, Macino G, et al. Gene silencing in worms and fungi. Nature. 2000;404(6775):245. [DOI] [PubMed] [Google Scholar]
- [138].Cogoni C, Macino G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999;286(5448):p. 2342–4. [DOI] [PubMed] [Google Scholar]
- [139].Catalanotto C, et al. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in neurospora crassa. Mol Cell Biol. 2004;24(6):2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Nolan T, et al. The RNA-dependent RNA polymerase essential for post-transcriptional gene silencing in Neurospora crassa interacts with replication protein A. Nucleic Acids Res. 2008;36(2):532–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Lee HC, Aalto AP, Yang Q, et al. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Cogoni C, Macino G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999;399(6732):p. 166–9. [DOI] [PubMed] [Google Scholar]
- [143].Martienssen RA. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat Genet. 2003;35(3):p. 213–4. [DOI] [PubMed] [Google Scholar]
- [144].Catalanotto C, et al. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16(7):790–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Maiti M, Lee HC, Liu Y. QIP, a putative exonuclease, interacts with the Neurospora Argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21(5):590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Lee HC, Chang SS, Choudhary S, et al. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459(7244):274–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Chang SS, Zhang Z, Liu Y. RNA interference pathways in fungi: mechanisms and functions. Annu Rev Microbiol. 2012;66:p. 305–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kavi HH, Birchler JA. Interaction of RNA polymerase II and the small RNA machinery affects heterochromatic silencing in Drosophila. Epigenetics Chromatin. 2009;2(1):p. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Onodera Y, et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120(5):p. 613–22. [DOI] [PubMed] [Google Scholar]
- [150].Bohmdorfer G, Sethuraman S, Rowley M J, et al. Long non-coding RNA produced by RNA polymerase V determines boundaries of heterochromatin. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Hantsche M, Cramer P. Conserved RNA polymerase II initiation complex structure. Curr Opin Struct Biol. 2017;47:17–22. [DOI] [PubMed] [Google Scholar]
- [152].Thorsen M, et al. Mediator regulates non-coding RNA transcription at fission yeast centromeres. Epigenetics Chromatin. 2012;5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Oya E, Kato H, Chikashige Y, et al. Mediator directs co-transcriptional heterochromatin assembly by RNA interference-dependent and -independent pathways. PLoS Genet. 2013;9:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Deng XL, Zhou H, Zhang G, et al. Sgf73, a subunit of SAGA complex, is required for the assembly of RITS complex in fission yeast. Sci Rep. 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Gullerova M, Barta A, Lorkovic ZJ. Rct1, a nuclear RNA recognition motif-containing cyclophilin, regulates phosphorylation of the RNA polymerase II C-terminal domain. Mol Cell Biol. 2007;27(10):p. 3601–3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Chang AY, et al. The conserved RNA binding cyclophilin, rct1, regulates small RNA biogenesis and splicing independent of heterochromatin assembly. Cell Rep. 2017;19(12):2477–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Kloc A, Zaratiegui M, Nora E, et al. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18(7):490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Kloc A, Martienssen R. RNAi, heterochromatin and the cell cycle. Trends Genet. 2008;24(10):511–517. [DOI] [PubMed] [Google Scholar]
- [159].Reddy BD, Wang Y, Niu L, et al. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 2011;25(3):214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Ekwall K, et al. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell. 1997;91(7):1021–1032. [DOI] [PubMed] [Google Scholar]
- [161].Yu R, Wang X, Moazed D. Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation. Nature. 2018;558(7711):p. 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Zaratiegui M, Castel SE, Irvine DV, et al. RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature. 2011;479(7371):135–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Li F, Martienssen R, Cande WZ. Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature. 2011;475(7355):244–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Singh SK, et al. Regulation of replication termination by Reb1 protein-mediated action at a distance. Cell. 2010;142(6):868–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Roche B, Arcangioli B, Martienssen RA. RNA interference is essential for cellular quiescence. Science. 2016;354:6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14(6):329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Su SS, Tanaka Y, Samejima I, et al. A nitrogen starvation-induced dormant G0 state in fission yeast: the establishment from uncommitted G1 state and its delay for return to proliferation. J Cell Sci. 1996;109(Pt 6):1347–57. [DOI] [PubMed] [Google Scholar]
- [168].Yanagida M. Cellular quiescence: are controlling genes conserved? Trends Cell Biol. 2009;19(12):705–715. [DOI] [PubMed] [Google Scholar]
- [169].Reyes-Turcu FE, Zhang K, Zofall M, et al. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol. 2011;18(10):1132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Roche B, Arcangioli B, Martienssen RA. Transcriptional reprogramming in cellular quiescence. RNA Biol. 2017;14:843–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Bhattacharjee S, Nandi S. Choices have consequences: the nexus between DNA repair pathways and genomic instability in cancer. Clin Transl Med. 2016;5(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Bhattacharjee S, Nandi S. Synthetic lethality in DNA repair network: A novel avenue in targeted cancer therapy and combination therapeutics. IUBMB Life. 2017;69(12):929–937. [DOI] [PubMed] [Google Scholar]
- [173].Bhattacharjee S, Nandi S. Rare Genetic Diseases with Defects in DNA Repair: opportunities and Challenges in Orphan Drug Development for Targeted Cancer Therapy. Cancers (Basel). 2018;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Bhattacharjee S, Nandi S. DNA damage response and cancer therapeutics through the lens of the Fanconi Anemia DNA repair pathway. Cell Commun Signal. 2017;15(1):p. 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Ghosh D, et al. CRISPR-Cas9 a boon or bane: the bumpy road ahead to cancer therapeutics. Cancer Cell Int. 2019;19:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Chu G. Double strand break repair. J Biol Chem. 1997;272(39):24097–24100. [DOI] [PubMed] [Google Scholar]
- [177].Hori A, Yoshida M, Shibata T, et al. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res. 2009;37(3):749–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Syeda AH, Hawkins M, McGlynn P. Recombination and replication. Cold Spring Harb Perspect Biol. 2014;6(11):p. a016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Branzei D, Szakal B. DNA damage tolerance by recombination: molecular pathways and DNA structures. DNA Repair (Amst). 2016;44:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Cahill D, Connor B, Carney JP. Mechanisms of eukaryotic DNA double strand break repair. Front Biosci. 2006;11:1958–76. [DOI] [PubMed] [Google Scholar]
- [181].Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5(5):a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Kass EM, Jasin M. Collaboration and competition between DNA double-strand break repair pathways. FEBS Lett. 2010;584(17):p. 3703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:p. 229–57. [DOI] [PubMed] [Google Scholar]
- [184].Wei W, et al. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149(1):101–12. [DOI] [PubMed] [Google Scholar]
- [185].Gao M, Wei W, Li M M, et al. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014;24(5):532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Stewart GS, Wang B, Bignell CR, et al. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421(6926):961–966. [DOI] [PubMed] [Google Scholar]
- [187].Stucki M, Clapperton JA, Mohammad D, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123(7):1213–26. [DOI] [PubMed] [Google Scholar]
- [188].Lou ZK, Minter-Dykhouse K, Franco S, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21(2):187–200. [DOI] [PubMed] [Google Scholar]
- [189].Stucki M, Jackson SP. gammaH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst). 2006;5(5):534–43. [DOI] [PubMed] [Google Scholar]
- [190].Bekker-Jensen S, Lukas C, Melander F, et al. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170(2):201–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25(5):p. 409–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25(5):p. 409–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Lukas J, Lukas C, Bartek J. More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol. 2011;13(10):p. 1161–9. [DOI] [PubMed] [Google Scholar]
- [194].Yu Y, et al. A proteome-wide visual screen identifies fission yeast proteins localizing to DNA double-strand breaks. DNA Repair (Amst). 2013;12(6):433–443. [DOI] [PubMed] [Google Scholar]
- [195].Samarajeewa DA, Sauls PA, Sharp KJ, et al. Efficient detection of unpaired DNA requires a member of the rad54-like family of homologous recombination proteins. Genetics. 2014;198(3):895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Zhang ZY, Chang SS, Zhang Z, et al. Homologous recombination as a mechanism to recognize repetitive DNA sequences in an RNAi pathway. Genes Dev. 2013;27(2):145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Onaka AT, Toyofuku N, Inoue T, et al. Rad51 and Rad54 promote noncrossover recombination between centromere repeats on the same chromatid to prevent isochromosome formation. Nucleic Acids Res. 2016;44(22):10744–10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].He HJ, Gonzalez M, Zhang F, et al. DNA replication components as regulators of epigenetic inheritance-lesson from fission yeast centromere. Protein Cell. 2014;5(6):411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Doros LA, Rossi CT, Yang J, et al. DICER1 mutations in childhood cystic nephroma and its relationship to DICER1-renal sarcoma. Mod Pathol. 2014;27(9):1267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Pugh TJ, Yu W, Yang J, et al. Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene. 2014;33(45):5295–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Diesch J, Hannan RD, Sanij E. Perturbations at the ribosomal genes loci are at the centre of cellular dysfunction and human disease. Cell Biosci. 2014;4:p. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Koonin EV. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: common ancestry vs convergence. Biol Direct. 2017;12(1):p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Martienssen R, Moazed D. RNAi and Heterochromatin Assembly. Cold Spring Harb Perspect Biol. 2015;7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Goto DB, Nakayama J. RNA and epigenetic silencing: insight from fission yeast. Dev Growth Differ. 2012;54(1):p. 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]


